Abstract
Sphingosine kinase 1 (SK1) is an enzyme that catalyses the phosphorylation of sphingosine to produce the bioactive lipid sphingosine 1-phosphate (S1P). We demonstrate here that FTY720 (Fingolimod™) and (S)-FTY720 vinylphosphonate are novel inhibitors of SK1 catalytic activity and induce the proteasomal degradation of this enzyme in human pulmonary artery smooth muscle cells, MCF-7 breast cancer cells and androgen-independent LNCaP-AI prostate cancer cells. Proteasomal degradation of SK1 in response to FTY720 and (S)-FTY720 vinylphosphonate is associated with the down-regulation of the androgen receptor in LNCaP-AI cells. (S)-FTY720 vinylphosphonate also induces the apoptosis of these cells. These findings indicate that SK1 is involved in protecting LNCaP-AI from apoptosis. This protection might be mediated by so-called ‘inside-out’ signalling by S1P, as LNCaP-AI cells exhibit increased expression of S1P2/3 receptors and reduced lipid phosphate phosphatase expression (compared with androgen-sensitive LNCaP cells) thereby potentially increasing the bioavailability of S1P at S1P2/3 receptors.
Keywords: Sphingosine kinase 1, Cancer, Proteasome, Fingolimod, Apoptosis, Sphingosine 1-phosphate signalling
1. Introduction
Sphingosine 1-phosphate (S1P) is a bioactive lipid that is produced by the enzyme sphingosine kinase (SK1 and SK2 isoforms), which catalyses the phosphorylation of sphingosine to produce S1P [1]. There are three N-terminal variants of SK1. SK1a (GenBank number: NM_001142601) is a 42 kDa protein, while SK1b (GenBank number: NM_182965) is a 51 kDa protein identical to SK1a, but with an 86 amino acid N-terminal extension. The third form has a molecular mass of 44 kDa and is identical to SK1a except for a 14 amino acid N-terminal extension (termed here SK1a + 14, GenBank number: NM_021972) and migrates with similar mobility as SK1a on SDS-PAGE. The annotation SK1a used herein therefore includes SK1a and SK1a + 14. S1P has an important role in regulating the growth, survival and migration of mammalian cells, mediating many of its effects by binding to a family of five G-protein coupled receptors (GPCR) termed S1Pn (where n = 1–5) that regulate various effectors, such as MAP kinase [1].
The immunosuppressant FTY720 (Fingolimod™) is a sphingosine analogue that is taken up by cells and is phosphorylated to FTY720-phosphate by SK2 [2], which is then released. FTY720-phosphate binds to four of the five S1P receptors [3] (S1P2 being the exception) and elicits polyubiquitination, endocytosis and degradation of S1P1 in T-lymphocytes [4]. S1P/S1P1 is essential for effective T-lymphocyte egress, and therefore the FTY720 phosphate-induced down-regulation of S1P1 on T-lymphocytes results in their retention in the lymph [5]. FTY720 is considered a T-lymphocyte specific immunosuppressant, and is currently in phase III trials for treatment of multiple sclerosis.
However, there are many studies that demonstrated that while FTY720 phosphate induces proliferation of cells, FTY720 induces apoptosis. For instance, treatment of human breast cancer cells (MCF-7, MDA-MB-231 and Sk-Br-3 cells) with FTY720 and the FTY720 analogue ISP-I-55 inhibits growth, activates JNK, and transiently inhibits ERK-1/2 activation with no effect on p38 MAPK in MCF-7 cells. In contrast, FTY720 phosphate induces growth of these cells [6]. FTY720 also inhibits growth, migration, colony formation and invasiveness of pancreatic cells (BxPC-3, AsPC-1 and PANC-1) linked with down-regulation of phosphorylated Akt and Bcl2 and increased caspase-3 activation [7]. FTY720 also suppresses liver tumour growth, associated with a reduction in phosphorylated Ser473 Akt and FAK levels and enhanced caspase-3 auto-cleavage [8]. FTY720 also induces apoptosis of androgen-insensitive DU145 prostate cancer cells via a caspase-3-dependent mechanism [9] and similarly in human renal cancer cells, resulting in reduced xenograft tumour growth [10]. FTY720 also induces apoptosis of hepatocellular carcinonoma cells through activation of PKCδ [11] and impairs chemical-induced lung carcinogenesis and early lung adenoma development by increasing caspase-3 activation [12]. Multiple myeloma cells [13] and several bladder cancer cell lines such as T24, UMUC3 and HT1197 also undergo apoptosis in response to FTY720 via a Bcl2-dependent and Fas-independent mechanism [14]. FTY720 is also cytotoxic to multiple myeloma cell lines and freshly isolated tumour cells from multiple myeloma patients. This cytotoxicity is mediated by activation of caspase-8, -9 and -2 and altered BAX cleavage and mitochondrial potential. There is also marked down-regulation of IL6-stimulated Akt phosphorylation, STAT3 and ERK-1/2 activation and IGF-I-stimulated Akt activation and TNFα-stimulated IKB and NFκB phosphorylation. FTY720 also induces apoptosis of B-cell malignancies and primary B cells from patients with chronic lymphocytic leukemia (CLL). However, this is mediated by down-regulation of Mcl-1 and not Bcl-2, and indeed survival cannot be rescued by enforced expression of Bcl-2. FTY720 also induces prolonged survival in a xenograft severe combined immunodeficiency (SCID) mouse model of disseminated B-cell lymphoma/leukemia [15]. Taking these findings together, it is clear that FTY720 functions as a bona-fide apoptotic agent.
In this study we have examined whether FTY720 and a novel analogue, (S)-FTY720 vinylphosphonate regulate SK1 expression in human pulmonary smooth muscle cells (hPASMC), MCF-7 breast cancer cells and androgen-independent LNCaP-AI prostate cancer cells. LNCaP-AI cells have been selected by culturing LNCaP cells in androgen-deprivation conditions for prolonged periods of time. This mimics androgen ablation therapy used for the treatment of prostate cancer [16]. LNCaP-AI cells are not dependent upon androgen for proliferation as they can grow in hormone-free medium. However, these cells do express the androgen receptor (AR) and respond to androgens, being able to express androgen-regulated genes such as the PSA (Prostatic Specific Antigen) gene [16]. Moreover, resistance to chemotherapeutic agents and irradiation of androgen-independent prostate cancer cells is purported to be mediated by SK1 [17–19]. In this article, we demonstrate for the first time that (S)-FTY720 vinylphosphonate inhibits SK1 activity and induces a novel proteasomal degradation of this enzyme. This is associated with the induction of apoptosis and a reduction in androgen receptor expression in androgen-independent prostate cancer cells.
2. Materials and methods
2.1. Materials
All general biochemicals and anti-actin antibody were from Sigma (Poole, UK). High glucose Dulbecco's modified Eagle's medium (DMEM), RPMI 1640 medium, minimum essential medium (MEM), European Fetal Calf Serum (EFCS) and penicillin-streptomycin (penicillin G sodium 10,000 units/ml-streptomycin sulphate 10,000 μg/ml) were from Invitrogen (Paisley, UK). Human pulmonary artery smooth muscle cells (hPASMC), human smooth muscle cell growth medium and passaging solutions were from TCS Cellworks (Buckingham, UK). Anti-phosphorylated ERK1/2 and anti-androgen receptor antibodies were from Santa Cruz (California, USA), anti-ERK2 antibody was from BD Transduction Biosciences (Oxford, UK), anti-SK1 antibodies were a gift from A. Huwiler (University of Bern, Switzerland [20]), anti-PARP and anti-cleaved caspase-3 antibodies were from Cell Signalling Technology (New England Biolabs (UK) Ltd., Hitchin, UK). MG132 was from Enzo Life Sciences (Exeter, UK). 2-(p-Hydroxyanilino)-4-(p-chlorophenyl)thiazole (SKi) was from Merck Biosciences (Nottingham, UK). N,N-Dimethylsphingosine (DMS) was from Avanti (Alabaster, Alabama, USA). FTY720 was from Cayman (Tallinn, Estonia). (S)-FTY720 vinylphosphonate and FTY720 phosphonates were synthesised according to Lu et al. [21]. (S)-FTY720 phosphate was synthesised according to Lu and Bittman [22].
2.2. Cell culture
hPASMC were cultured in human smooth muscle cell growth medium MCF-7 breast cancer cells were grown in DMEM supplemented with 10% EFCS and 1% penicillin-streptomycin, 0.4% geneticin and 15 μg/ml insulin. Human prostate cancer LNCaP and LNCaP-AI cell lines (derived from the parental LNCaP cells by prolonged androgen depletion) were maintained in RPMI 1640 medium supplemented with 10% EFCS or 10% delipidated serum, respectively, 1% penicillin-streptomycin and 1% L-Glutamine. All cells were maintained at 37 °C with 5% CO2. HEK 293 cells were maintained in MEM with 10% EFCS and 1% penicillin-streptomycin. For LNCaP-AI cells, FTY720, (S)-FTY720 vinylphosphonate, MG132 and SKi were replenished after 24 h. FTY720, FTY720 phosphate, FTY720 phosphonate, FTY720 (S)-vinylphosphonate, SKi and DMS were dissolved in DMSO and added to media at a final concentration of DMSO < 0.1% (v/v).
2.3. Transfection
HEK 293 cells were transfected with FLAG tagged hSK1 plasmid construct, using the Lipofectamine™2000 reagent according to the manufacturer's instruction.
2.4. Preparation of whole cell protein extracts
hPASMC protein lysates were prepared by washing treated cells (including floating cells, where appropriate, e.g. for analysis of apoptosis) with 5 ml of PBS, then resuspending the cell pellets in whole cell lysis buffer (137 mM NaCl, 2.7 mM KCl, 1 mM MgCl2, 1 mM CaCl2, 1% v/v NP40, 10% v/v glycerol, 20 mM Tris) (pH 8.0) containing 0.2 mM PMSF, 0.2 mM leupeptin, 0.2 mM aprotinin, 0.5 mM Na3VO4, 100 μM NaF, 10 mM β-glycerophosphate). Samples were passed through a 23-gauge needle and syringe (× 6) and rotated for 30 min at 4 °C to allow for efficient lysis. Cell debris was pelleted by centrifugation at 15,300 rpm for 10 min at 4 °C. The protein content of the supernatant (whole cell extract) was measured using the Pierce BCA assay kit (Fisher Scientific UK, Loughborough). LNCaP-AI cells were harvested in the same way. For each sample, 10–20 μg of protein, combined with sample buffer (0.5 M Tris, 2 mM Na4P2O7, 5 mM EDTA, 2% w/v SDS (pH 6.7) containing 5% v/v glycerol, 0.25% w/v bromophenol blue, 50 mM dithiothreitol) were used for SDS-PAGE and western blotting. MCF-7 cell lysates were prepared by adding sample buffer to adherent cells, and the samples were passed through a 23-gauge needle and syringe (× 6).
2.5. Western blotting
Analysis of proteins by SDS-PAGE and western blotting was performed as previously described [23] using anti-phosphorylated ERK1/2, anti-ERK2, anti-PARP, anti-cleaved caspase-3, anti-SK1, anti-actin and anti-AR antibodies.
2.6. Sphingosine kinase assay
Assays were performed according to Delon et al. [24]. SK1 activity was measured as the formation of [32P]S1P from sphingosine and [32P]-γ-ATP using purified recombinant SK1 (BioMol). Sphingosine (10 μM final concentration) was solubilised in 40 μl 5 mM Triton X-100 (0.313% w/v) and combined with 140 μl assay buffer (containing 20 mM Tris base (pH 7.4), 1 mM EDTA, 1 mM Na3VO4, 40 mM β-glycerophosphate, 1 mM NaF, 1 mM β-mercaptoethanol, 20% (v/v) glycerol, 10 μg/ml aprotinin, 10 μg/ml soyabean trypsin inhibitor, 1 mM PMSF, 0.5 mM 4-deoxypyridoxine) and 10 μl of diluted purified SK1 (15 ng) or HEK 293 cell lysates containing over-expressed recombinant SK1. Reactions were started by the addition of [32P]ATP (4.4 × 104 cpm/nmole, final concentration of ATP, 250 μM) and incubated at 30 °C for 15 min. Reactions were terminated by the addition of 500 μl 1-butanol and the samples mixed thoroughly before phase separation by the addition of 1 ml 2 M KCl. The lower aqueous phase was discarded, the upper phase washed twice with 1 ml 2 M KCl and [32P]S1P therein quantified by scintillation counting.
2.7. Polymerase Chain Reaction (PCR)
PCR was performed to identify mRNA transcripts. The primers used for the reactions were as follows:
- S1P2 FWD
CACTCGGCAATGTACCTGTTTC
- RV
GACGCCTAGCACGATGGTGAC
- S1P3 FWD
GACTGCTCTACCATCCTGCCC
- RV
GTAGATGACCGGGTTCATGGC
- LPP1 FWD
CTTCAAGGCATACCCCCTTCCAAC
- REV
GCCCAGTCTCCCTTCATCCTG
- LPP2 FWD
CTGCTGTATACAAGGTGCTGGGG
- REV
CGTGCCCACTTCCAACAGAGTC
- LPP3 FWD
CTCGACCTCTTCTGCCTCTTCATG
- REV
GCTTCCTGGACTTTGCTGTCATCAC
- GAPDH FWD
TGAAGGTCGGTGTCAACGGATTTGGC
- RV
CATGTAGGCCATGAGGTCCACCAC
The PCR reaction conditions were: 1 cycle initial denaturation at 94 °C for 2 min, 30 cycles amplification at 94 °C for 1 min 30 s, 52 °C (LPP1, LPP3, GAPDH) or 54 °C (LPP2) or 56 °C (S1P2 and S1P3 for 30 s and 72 °C for 1 min 40 s, followed by a final extension at 72 °C for 5 min.
3. Results and discussion
3.1. FTY720 and (R and S)-FTY720 vinylphosphonate inhibit SK1 activity
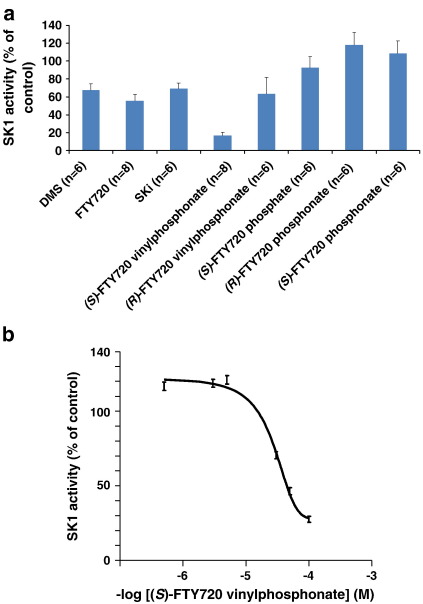
Using sphingosine (10 μM) and [32P]ATP as substrates, we found that 50 μM FTY720 (see Fig. 1) inhibited the activity of purified SK1 by ∼ 40% (Fig. 2a). The % inhibition by FTY720 was equivalent to that observed with the same concentration of DMS or SKi, which are established SK1 inhibitors [25–27] (Fig. 2a). Vessey and colleagues also demonstrated that FTY720 inhibited SK1 activity in a chromatographic fraction containing an anti-SK1 immunoreactive protein [28]. However, our findings are unequivocal in that they have been obtained using purified SK1. Others have demonstrated that FTY720 is a substrate for SK2 [2–4] but not SK1, which we confirmed using FTY720 and [32P]ATP as substrates (data not shown). These findings indicate that FTY720 binds to SK1, but is not phosphorylated. We also tested a number of FTY720 analogues (see Fig. 1). We found that both (S)-FTY720 vinylphosphonate and (R)-FTY720 vinylphosphonate inhibited SK1 activity (Fig. 2a). The S enantiomer was significantly more effective than the R enantiomer at 50 μM. At this concentration, (S)-FTY720 vinylphosphonate inhibited purified SK1 by more than 80%, whereas the R enantiomer inhibited the enzyme by 40% (Fig. 2a). On a molar basis, (S)-FTY720 vinylphosphonate is much more effective in inhibiting SK1 compared with FTY720, SKi or DMS (Fig. 2a). Inhibition of SK1 activity by FTY720 analogues is reversible (data not shown). The R and S enantiomers of FTY720 phosphonate and (S)-FTY720 phosphate do not significantly inhibit the enzyme (Fig. 2a). (S)-FTY720 vinylphosphonate also inhibited SK1 activity that had been ectopically over-expressed in HEK 293 cells and this inhibition was concentration-dependent (Fig. 2b) with an IC50 = 24 ± 5.7 μM (n = 3).
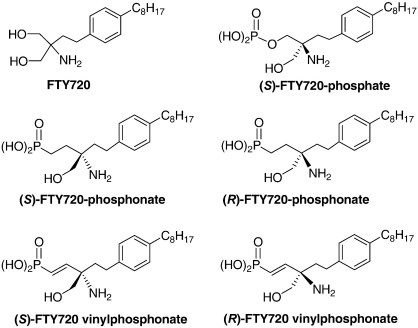
Fig. 1.
Structures of FTY720, (S)-FTY720 vinylphosphonate, (R)-FTY720 vinylphosphonate, (S)-FTY720 phosphonate, (R)-FTY720 phosphonate and (S)-FTY720 phosphate.
Fig. 2.
Assessment of FTY720 analogues on SK1 activity. (a) Effect of DMS, SKi and various FTY720 analogues on purified SK1 activity (assayed using 10 μM sphingosine and [32P]ATP (250 μM) as substrates). Results, expressed as percentage of vehicle control are means and standard deviations of 3–4 independent experiments performed in duplicate samples. All inhibitors were screened at 50 μM; (b) Concentration response of the inhibitory effect of (S)-FTY720 vinylphosphonate on SK1 activity (using 10 μM sphingosine and 250 μM [32P]ATP as the substrates) in lysates of HEK 293 cells in which SK1 has been ectopically over-expressed.
3.2. FTY720 and (S)-FTY720 vinylphosphonate induce proteasomal degradation of SK1 in hPASMC and MCF-7 cells
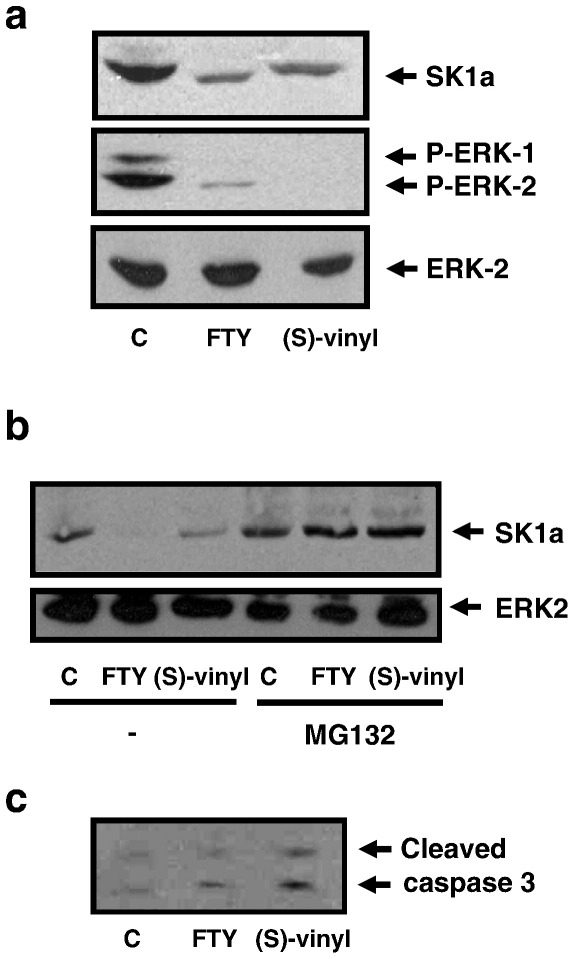
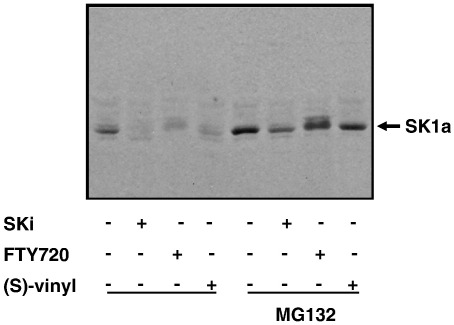
We have demonstrated that DMS and SKi induce degradation of SK1 via the ubiquitin-proteasomal pathway to promote apoptosis of normal and cancer cells unpublished. Therefore, since FTY720 and (S)-FTY720 vinylphosphonate are novel SK1 inhibitors, we explored the possibility that these compounds may also induce the proteasomal degradation of SK1. Both hPASMC and MCF-7 cells express SK1a (Mr = 42 kDa) (Figs. 3a, b and 4). Chronic treatment (24 h) of hPASMC (Fig. 3a, b) or MCF-7 breast cancer cells (Fig. 4) with FTY720 (10 μM) or (S)-FTY720 vinylphosphonate (10 μM) induced the down-regulation of SK1a. Moreover, a 24-h treatment of hPASMC or MCF-7 cells with the proteasomal inhibitor MG132 (10 μM) inhibited the down-regulation of SK1a induced by either compound (Figs. 3b and 4). SKi (10 μM) was used as a reference compound and was also shown to induce the MG132-sensitive proteasomal degradation of SK1a in MCF-7 cells (Fig. 4). It is well established that in order for proteins to be targeted for proteasomal degradation, the protein must first be modified by ubiquitination. Therefore, our findings suggest that SK1a might be regulated by the ubiquitin-proteasomal pathway and that the flux of SK1a through this pathway is increased by FTY720 and (S)-FTY720 vinylphosphonate. Indeed, Kihara and colleagues [29] reported that SK1a and ‘SK1b’ (a smaller 34 kDa species) are regulated by the ubiquitin-proteasomal degradation pathway with ‘SK1b’ being more susceptible to polyubiquitination. We did not detect the 34 kDa ‘SK1b’ in MCF-7 cells or hPASMC.
Fig. 3.
Effect of FTY720 and (S)-FTY720 vinylphosphonate on proteasomal degradation of SK1a in hPASMC. Western blots showing the effect of: (a) FTY720 and (S)-FTY720 vinylphosphonate (both 10 μM, 24 h) on SK1a, ERK-2 and phosphorylated ERK-1/2 levels in hPASMC; (b) MG132 (10 μM, 24 h) on the FTY720- and (S)-FTY720 vinylphosphonate-induced down-regulation of SK1a; (c) FTY720 and (S)-FTY720 vinylphosphonate (both 10 μM, 24 h) on caspase-3 activation in hPASMC. C = control. In each case, results are representative of 2–4 separate experiments.
Fig. 4.
Effect of SKi, FTY720 and (S)-FTY720 vinylphosphonate on proteasomal degradation of SK1a in MCF-7 cells. Western blot showing the effect of MG132 (10 μM, 24 h) on SKi-, FTY720- and (S)-FTY720 vinylphosphonate-induced degradation of SK1a in MCF-7 cells. The cells were treated for 24 h with the inhibitors at 10 μM. Results are representative of 3 separate experiments.
We next investigated the functional significance of the FTY720- and (S)-FTY720 vinylphosphonate-induced proteasomal degradation of SK1 in terms of cellular response. In this regard, treatment of hPASMC with either compound induced the onset of apoptosis as evidenced by increased caspase-3 activation (p17/p19 formation, Fig. 3c). Moreover, FTY720 and (S)-FTY720 vinylphosphonate reduced the phosphorylation state of ERK-1/2 in hPASMC (Fig. 3a), an enzyme required for maintenance of cell survival. These findings demonstrate that SK1a is critical for hPASMC survival.
3.3. FTY720 and (S)-FTY720 vinylphosphonate induce proteasomal degradation of SK1 in LNCaP-AI prostate cancer cells
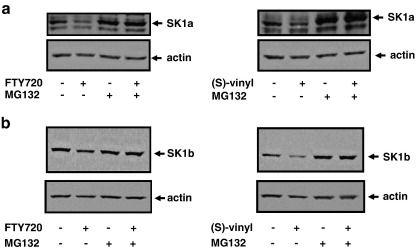
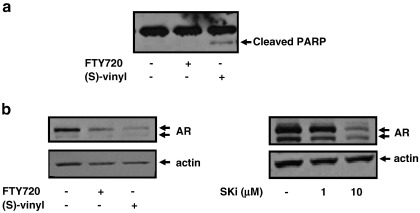
We also investigated whether the proteasomal degradation of SK1 induced by FTY720 and (S)-FTY720 vinylphosphonate is more widespread and is evident in prostate cancer cells. LNCaP-AI cells express both SK1a (Mr = 42 kDa) and SK1b (Mr = 51 kDa) (Fig. 5a, b). The treatment of LNCaP-AI cells with FTY720 or (S)-FTY720 vinylphosphonate reduced SK1a expression via the MG132-sensitive proteasomal degradation pathway (Fig. 5a). Similar results were also obtained for SK1b, although (S)-FTY720 vinylphosphonate was more effective than FTY720 in promoting the proteasomal degradation of SK1b (Fig. 5b). This can be correlated with the extent to which these compounds inhibit purified SK1 catalytic activity (Fig. 2a). In terms of the effect of these agents on apoptosis, we found that at equimolar concentrations, (S)-FTY720 vinylphosphonate induced PARP cleavage, while FTY720 was not effective (Fig. 6a). This might suggest that there is a threshold level of SK1b, below which LNCaP-AI cells are forced to undergo apoptosis. Thus, (S)-FTY720 vinylphosphonate might reduce SK1b expression to below this threshold, while FTY720 fails to do so.
Fig. 5.
Effect of FTY720 and (S)-FTY720 vinylphosphonate on proteasomal degradation of SK1a and SK1b in LNCaP-AI cells. Western blots showing the effect of MG132 (10 μM, 48 h) on the FTY720- and (S)-FTY720 vinylphosphonate-induced degradation of (a) SK1a and (b) SK1b in LNCaP-AI cells. The cells were treated for 48 h with 10 μM of each inhibitor. Western blots were reprobed with anti-actin antibody to ensure comparable protein loading. In each case, results are representative of 3 separate experiments.
Fig. 6.
Effect of FTY720 and (S)-FTY720 vinylphosphonate on PARP cleavage and AR expression in LNCaP-AI cells. Western blots showing (a) the effect of FTY720 and (S)-FTY720 vinylphosphonate (both 10 μM, 48 h) on PARP cleavage in LNCaP-AI cells; (b) the effect of SKi (1 and 10 μM, 48 h), FTY720 (10 μM, 48 h) or (S)-FTY720 vinylphosphonate (10 μM, 48 h) on AR expression in LNCaP-AI cells. Western blots were re-probed with anti-actin antibody to ensure comparable protein loading. In each case, results are representative of 3 separate experiments.
Treatment of LNCaP-AI cells with 10 μM FTY720 or (S)-FTY720 vinylphosphonate also induced down-regulation of AR expression (Fig. 6b). Moreover, a similar effect was observed in cells treated with SKi (Fig. 6b), suggesting that this might be a common feature of SK1 inhibitors. The elimination of constitutively active AR by FTY720 and (S)-FTY720 vinylphosphonate suggests that these compounds may be effective inhibitors of the growth of androgen-independent prostate cancer cells.
There are at least two possible models that describe the interaction between FTY720/(S)-FTY720 vinylphosphonate and SK1 that results in its proteasomal degradation. First, FTY720 or (S)-FTY720 vinylphosphonate may bind to SK1, thereby inducing a conformational change (leading to protein unfolding) that functions as a signal for its ubiquitin-proteasomal degradation. Second, the binding of inhibitor to SK1 induces a reduction in intracellular S1P formation and the accumulation of sphingosine and ceramide (i.e. an increase in the ceramide:S1P ratio). Ceramide, in turn, may activate the proteasome to remove SK1 that is already polyubiquitinated. Indeed, our preliminary results indicate that SK1 is polyubiquitinated under basal conditions (data not shown), and a short-chain analogue of ceramide, C2-ceramide has been shown to activate ubiquitin-mediated proteolytic pathway in astrocytes [30]. The two models described here are not necessarily mutually exclusive and may operate together.
Our findings concerning SK1, extend the number of intracellular targets of FTY720, which has been reported to also inhibit cytosolic phospholipase A2 [31], S1P lyase [32] and ceramide synthase [33,34] and to activate PP2A and PP2A-like phosphatases [35,36]. The effects on these intracellular targets may explain the anti-cancer activity of FTY720, of which there are numerous examples. For instance, FTY720 inhibits the invasive ability of androgen-independent prostate cancer cells (DU145 and PC-3) via a mechanism that involves down-regulation of RhoA-GTP [37]. FTY720, via functional antagonism of S1P1 receptors, also reduces VEGF- and S1P-induced angiogenesis and vascular permeability in vivo in growth factor implant and corneal models [38]. FTY720 also reduces tumour metastasis in a mouse B16/BL6 melanoma model, accompanied by a decrease in tumour cell proliferation and an increase in apoptosis [38]. FTY720 also dramatically decreases metastasis in a mouse model of breast cancer created by inoculating JYgMC(A) cells into the flank of BALB/c nu/nu mice [39].
3.4. Molecular basis for overcoming chemotherapeutic resistance
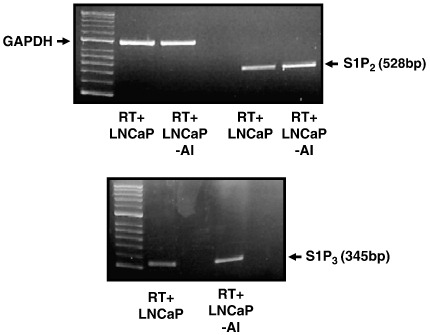
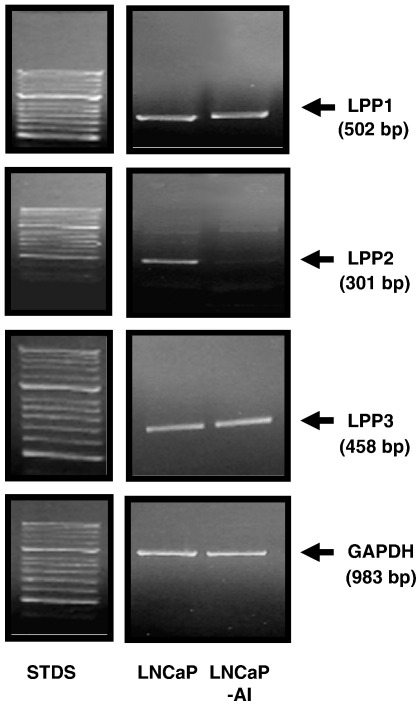
The ability of (S)-FTY720 vinylphosphonate to induce proteasomal degradation of SK1 and to promote apoptosis demonstrate that SK1 is critical for cell survival. How is this achieved at the molecular level? The most obvious candidate is S1P, which has both extracellular actions at S1P receptors and intracellular actions at undefined targets [1]. Indeed, there is substantial evidence that S1P formed by SK1 is released from cells or partitions into a lipid microenvironment in close proximity to S1P receptors, whereupon binding to the S1P receptor(s) induces ‘inside-out’ signalling [40] that, in this case, might protect LNCaP-AI cell from apoptosis. Indeed, S1P2 and S1P3 mRNA transcripts are substantially increased in LNCaP-AI cells compared with LNCaP cells (Fig. 7), suggesting that S1P formed by SK1 is accommodated for by increased expression of S1P2/3 in LNCaP-AI cells. In addition, we have investigated possible changes in the enzymes that regulate S1P degradation termed lipid phosphate phosphatases (LPP1-3) [41]. These enzymes catalyse dephosphorylation of lysophospholipids such as S1P, ceramide 1-phosphate and lysophosphatidic acid [41]. In this regard, we demonstrate here that LPP2, but not LPP1 or LPP3 mRNA transcript is markedly reduced in LNCaP-AI cells compared with LNCaP cells (Fig. 8). These findings suggest that the bioavailability of S1P at S1P2/3 receptors might be increased in LNCaP-AI cells. Therefore, the ‘inside out’ signalling by S1P might be the critical element in the acquisition of resistance to chemotherapeutic agents in androgen-independent prostate cancer. Notably, this resistance can be overcome by (S)-FTY720 vinylphosphonate. This might be a consequence of reducing SK1a and SK1b levels to below a threshold. Moreover, in the accompanying paper, Valentine and colleagues [42] have demonstrated that (S)-FTY720 vinylphosphonate is a pan S1P receptor antagonist, and this activity might contribute to its apoptotic action in LNCaP-AI cells by preventing ‘inside-out’ signalling by SK1.
Fig. 7.
Comparison of mRNA transcript levels of S1P2 and S1P3 in LNCaP and LNCaP-AI cells. RT-PCR was conducted using S1P2 or S1P3 specific primers using 0.15 μg of RNA isolated from either LNCaP or LNCaP-AI cells. An RT-PCR using GAPDH primers was performed to confirm that similar amounts of RNA were used between cell types. In each case, results are representative of 3 separate experiments.
Fig. 8.
Comparison of mRNA transcript levels of LPP1-3 in LNCaP and LNCaP-AI cells. RT-PCR was conducted using LPP1 or LPP2 or LPP3 specific primers using 0.15 μg of RNA isolated from either LNCaP or LNCaP-AI cells. An RT-PCR using GAPDH primers was performed to confirm that similar amounts of RNA were used between cell types. In each case, results are representative of 3 separate experiments.
Valentine and colleagues [42] have also demonstrated that low nM concentrations of (S)-FTY720 vinylphosphonate elicits an anti-apoptotic effect in IEC-6 intestinal epithelial cells challenged with camptothecin (CPT). The concentration-response is bell shaped such that the anti-apoptotic effect is lost at higher concentrations of (S)-FTY720 vinylphosphonate. This bell shaped response is therefore consistent with our observation that higher concentrations of (S)-FTY720 vinylphosphonate promote apoptosis. CPT also induces an increase in S1P receptor and SK1 expression in androgen-independent prostate cancer cells and this has been proposed to underlie the resistance to CPT-stimulated apoptosis in androgen-independent prostate cancer cells [43]. If this is a generic mechanism, then the increase in S1P receptor/SK1 expression with CPT might reduce sensitivity of IEC-6 intestinal epithelial cells to (S)-FTY720 vinylphosphonate in terms of the induction of apoptosis, thereby accounting for the predominance of its anti-apoptotic action at lower concentrations.
3.5. Summary
Our findings demonstrate that (S)-FTY720 vinylphosphonate is a more effective inhibitor of SK1 catalytic activity than DMS and SKi. (S)-FTY720 vinylphosphonate also promotes the novel proteasomal degradation of SK1 in hPASMC, MCF-7 and LNCaP-AI cells. The ability of (S)-FTY720 vinylphosphonate to overcome the chemotherapeutic resistance of LNCaP-AI prostate cancer cells suggests that this compound may be a useful agent for treating androgen-independent prostate cancer, for which there are currently limited therapeutic options. In this regard, combination therapies with γ-irradiation might prove highly efficacious as γ-irradiation-induced formation of ceramide, which induces apoptosis [44], should theoretically be potentiated by the (S)-FTY720 vinylphosphonate-induced proteasomal degradation of SK1.
Acknowledgements
This work was supported by grants from CRUK (23158/A7536) and British Heart Foundation (PG/06/137/21825) to SP and NJP, USPHS (CA 92160) to GT and (HL 083187) to RB, and the Fay Fuller Foundation to SMP. SMP is a recipient of a Senior Research Fellowship from the National Health and Medical Research Council of Australia.
References
- 1.Pyne S., Pyne N.J. Biochem. J. 2000;349:385. doi: 10.1042/0264-6021:3490385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sanchez T., Estrada-Hernandez T., Paik J.H., Wu M.T., Venkataraman K., Brinkmann V., Claffey K., Hla T. J. Biol. Chem. 2003;278:47281. doi: 10.1074/jbc.M306896200. [DOI] [PubMed] [Google Scholar]
- 3.Brinkmann V., Davis M.D., Heise C.E., Albert R., Cottens S., Hof R., Bruns C., Prieschl E., Baumruker T., Hiestand P., Foster C.A., Zollinger M., Lynch K.R. J. Biol. Chem. 2002;277:21453. doi: 10.1074/jbc.C200176200. [DOI] [PubMed] [Google Scholar]
- 4.Gräler M.H., Goetzl E.J. FASEB J. 2004;18:551. doi: 10.1096/fj.03-0910fje. [DOI] [PubMed] [Google Scholar]
- 5.Mandala S., Hajdu R., Bergstrom J., Quackenbush E., Xie J., Milligan J., Thornton R., Shei G.J., Card D., Koehane C., Rosenbach M., Hale J., Lynch C.L., Rupprecht K., Parsons W., Rosen H. Science. 2002;296:346. doi: 10.1126/science.1070238. [DOI] [PubMed] [Google Scholar]
- 6.Nagaoka Y., Otsuki K., Fujita T., Uesato S. Bio. Pharm. Bull. 2008;6:1177. doi: 10.1248/bpb.31.1177. [DOI] [PubMed] [Google Scholar]
- 7.Shen Y., Cai M., Xia W., Liu J., Zhang Q., Xie H., Wang C., Wang X., Zheng S. Cancer Lett. 2007;254:288. doi: 10.1016/j.canlet.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 8.Ng K.T., Man K., Ho J.W., Sun C.K., Lee T.K., Zhao Y., Lo C.M., Poon R.T., Fan S.T. Int. J. Oncol. 2007;30:375. [PubMed] [Google Scholar]
- 9.Wang J.D., Takahara S., Nonomura N., Ichimaru N., Toki K., Azuma H., Matsumiya K., Okuyama A., Suzuki S. Prostate. 1999;40:50. doi: 10.1002/(sici)1097-0045(19990615)40:1<50::aid-pros6>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 10.Ubai T., Azuma H., Kotake Y., Inamoto T., Takahara K., Ito Y., Kiyama S., Sakamoto T., Horie S., Muto S., Takahara S., Otsuki Y., Katsuoka Y. Anticancer Res. 2007;27:75. [PubMed] [Google Scholar]
- 11.Hung J.H., Lu Y.S., Wang Y.C., Ma Y.H., Wang D.S., Kulp S.K., Muthusamy N., Byrd J.C., Cheng A.L., Chen C.S. Cancer Res. 2008;68:1204. doi: 10.1158/0008-5472.CAN-07-2621. [DOI] [PubMed] [Google Scholar]
- 12.Salinas N.R., Lopes C.T., Palma P.V., Oshima C.T., Bueno V. Pathol. Oncol. Res. 2009;15:549. doi: 10.1007/s12253-009-9152-2. [DOI] [PubMed] [Google Scholar]
- 13.Yasui H., Hideshima T., Raje N., Roccaro A.M., Shiraishi N., Kumar S., Hamasaki M., Ishitsuka K., Tai Y.T., Podar K., Catley L., Mitsiades C.S., Richardson P.G., Albert R., Brinkmann V., Chauhan D., Anderson K.C. Cancer Res. 2005;65:7478. doi: 10.1158/0008-5472.CAN-05-0850. [DOI] [PubMed] [Google Scholar]
- 14.Azuma H., Takahara S., Horie S., Muto S., Otsuki Y., Katsuoka Y. J. Urol. 2003;169:2372. doi: 10.1097/01.ju.0000064938.32318.91. [DOI] [PubMed] [Google Scholar]
- 15.Liu Q., Zhao X., Frissora F., Ma Y., Santhanam R., Jarjoura D., Lehman A., Perrotti D., Chen C.S., Dalton J.T., Muthusamy N., Byrd J.C. Blood. 2008;111:275. doi: 10.1182/blood-2006-10-053884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Halkidou K., Gnanapragasam V.J., Mehta P.B., Logan I.R., Brady M.E., Cook S., Leung H.Y., Neal D.E., Robson C.N. Oncogene. 2003;22:2466. doi: 10.1038/sj.onc.1206342. [DOI] [PubMed] [Google Scholar]
- 17.Pchejetski D., Golzio M., Bonhoure E., Calvet C., Doumerc N., Garcia V., Mazerolles C., Rischmann P., Teissié J., Malavaud B., Cuvillier O. Cancer Res. 2005;65:11667. doi: 10.1158/0008-5472.CAN-05-2702. [DOI] [PubMed] [Google Scholar]
- 18.Pchejetski D., Doumerc N., Golzio M., Naymark M., Teissié J., Kohama T., Waxman J., Malavaud B., Cuvillier O. Mol. Cancer Ther. 2008;7:1836. doi: 10.1158/1535-7163.MCT-07-2322. [DOI] [PubMed] [Google Scholar]
- 19.Nava V.E., Cuvillier O., Edsall L.C., Kimura K., Milstien S., Gelmann E.P., Spiegel S. Cancer Res. 2000;60:4468. [PubMed] [Google Scholar]
- 20.Huwiler A., Döll F., Ren S., Klawitter S., Greening A., Römer I., Bubnova S., Reinsberg L., Pfeilschifter J. Biochim. Biophys. Acta. 2006;1761:367. doi: 10.1016/j.bbalip.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 21.Lu X., Sun S., Valentine W.J., Shuyu E., Liu J., Tigyi G., Bittman R.L. J. Org. Chem. 2009;74:3192. doi: 10.1021/jo900023u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu X., Bittman R. Tetrahedron Lett. 2006;47:825. [Google Scholar]
- 23.Alderton F., Rakhit S., Kong K.C., Palmer T., Sambi B., Pyne S., Pyne N.J. J. Biol. Chem. 2001;276:28578. doi: 10.1074/jbc.M102771200. [DOI] [PubMed] [Google Scholar]
- 24.Delon C., Manifava M., Wood E., Thompson D., Krugmann S., Pyne S., Ktistakis K.T. J. Biol. Chem. 2004;279:44763. doi: 10.1074/jbc.M405771200. [DOI] [PubMed] [Google Scholar]
- 25.Kohama T., Olivera A., Edsall L., Nagiec M.M., Dickson R., Spiegel S. J. Biol. Chem. 1998;273:23722. doi: 10.1074/jbc.273.37.23722. [DOI] [PubMed] [Google Scholar]
- 26.Liu H., Sugiura M., Nava V.E., Edsall L.C., Kono K., Poulton S., Milstien S., Kohama T., Spiegel S. J. Biol. Chem. 2000;275:19513. doi: 10.1074/jbc.M002759200. [DOI] [PubMed] [Google Scholar]
- 27.French K.J., Schrecengost R.S., Lee B.D., Zhuang Y., Smith S.N., Eberly J.L.M., Yun J.K., Smith C.D. Cancer Res. 2003;63:5962. [PubMed] [Google Scholar]
- 28.Vessey D.A., Kelley M., Zhang J., Li L., Tao R., Karliner J.S. J. Biochem. Mol. Toxicol. 2007;21:273. doi: 10.1002/jbt.20193. [DOI] [PubMed] [Google Scholar]
- 29.Kihara A., Anada Y., Igarashi Y. J. Biol. Chem. 2005;281:4532. doi: 10.1074/jbc.M510308200. [DOI] [PubMed] [Google Scholar]
- 30.Calatayud C.A., Pasquini L.A., Pasquini J.M., Soto E.F. Dev. Neurosci. 2005;27:397. doi: 10.1159/000088454. [DOI] [PubMed] [Google Scholar]
- 31.Payne S.G., Oskeritzian C.A., Griffiths R., Subramanian P., Barbour S.E., Chalfant C.E., Milstien S., Spiegel S. Blood. 2007;109:1077. doi: 10.1182/blood-2006-03-011437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bandhuvula P., Tam Y.Y., Oskouian B., Saba J.D. J. Biol. Chem. 2005;280:33697. doi: 10.1074/jbc.C500294200. [DOI] [PubMed] [Google Scholar]
- 33.Lahiri S., Park H., Laviad E.L., Bittman R., Futerman A.H. J. Biol. Chem. 2009;284:16090. doi: 10.1074/jbc.M807438200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Berdyshev E.V., Gorshkova I., Skobeleva A., Bittman R., Lu X., Dudek S.M., Mirzapoiazova T., Garcia J.G., Natarajan V. J. Biol. Chem. 2009;284:5467. doi: 10.1074/jbc.M805186200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neviani P., Santhanam R., Oaks J.J., Eiring A.M., Notari M., Blaser B.W., Liu S., Trotta R., Muthusamy N., Gambacorti-Passerini C., Druker B.J., Cortes J., Marcucci G., Chen C.S., Verrills N.M., Roy D.C., Caligiuri M.A., Bloomfield C.D., Byrd J.C., Perrotti D. J. Clin. Invest. 2007;117:2408. doi: 10.1172/JCI31095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matsuoka Y., Nagahara Y., Ikekita M., Shinomiya T. Br. J. Pharmacol. 2003;138:1303. doi: 10.1038/sj.bjp.0705182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou C., Ling M.T., Kin-Wah Lee T., Man K., Wang X., Wong Y.C. Cancer Lett. 2006;233:36. doi: 10.1016/j.canlet.2005.02.039. [DOI] [PubMed] [Google Scholar]
- 38.LaMontagne K., Littlewood-Evans A., Schnell C., O'Reilly T., Wyder L., Sanchez T., Probst B., Butler J., Wood A., Liau G., Billy E., Theuer A., Hla T., Wood J. Cancer Res. 2006;66:221. doi: 10.1158/0008-5472.CAN-05-2001. [DOI] [PubMed] [Google Scholar]
- 39.Azuma H., Horie S., Muto S., Otsuki Y., Matsumoto K., Morimoto J., Gotoh R., Okuyama A., Suzuki S., Katsuoka Y., Takahara S. Anticancer Res. 2003;23:3183. [PubMed] [Google Scholar]
- 40.Takabe K., Paugh S.W., Milstien S., Spiegel S. Pharmacol. Rev. 2008;60:181. doi: 10.1124/pr.107.07113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pyne S., Kong K.C., Darroch P.I. Cell. Dev. Biol. 2004;15:492. doi: 10.1016/j.semcdb.2004.05.007. [DOI] [PubMed] [Google Scholar]
- 42.W.J. Valentine, G.N. Kiss, J. Liu, E. Shuyu, M. Gotoh, K. Murokami-Murofushi, T.C. Pham, D. Baker, A.B. Parrill, X. Lu, C. Sun, R. Bittman, N.J. Pyne, & G. Tigyi, Cell. Signal. (submitted for publication).
- 43.Akao Y., Banno Y., Nakagawa Y., Hasegawa N., Kim T., Murate T., Igarashi Y., Nozawa Y. Biochem. Biophys. Res. Commun. 2006;342:1284. doi: 10.1016/j.bbrc.2006.02.070. [DOI] [PubMed] [Google Scholar]
- 44.Haimovitz-Friedman A., Kan C.C., Ehleiter D., Persaud R.S., McLoughlin M., Fuks Z., Kolesnick R.N. J. Exp. Med. 1994;180:525. doi: 10.1084/jem.180.2.525. [DOI] [PMC free article] [PubMed] [Google Scholar]