Abstract
The use of enterococci as the primary fecal indicator bacteria (FIB) for the determination of recreational water safety has been questioned, particularly in sub/tropical marine waters without known point sources of sewage. Alternative FIB (such as the Bacteroidales group) and alternative measurement methods (such as rapid molecular testing) have been proposed to supplement or replace current marine water quality testing methods which require culturing enterococci. Moreover, environmental parameters have also been proposed to supplement current monitoring programs. The objective of this study was to evaluate the health risks to humans from exposure to subtropical recreational marine waters with no known point source. The study reported symptoms between one set of human subjects randomly assigned to marine water exposure with intensive environmental monitoring compared with other subjects who did not have exposure. In addition, illness outcomes among the exposed bathers were compared to levels of traditional and alternative FIB (as measured by culture-based and molecular-based methods), and compared to easily measured environmental parameters. Results demonstrated an increase in self-reported gastrointestinal, respiratory and skin illnesses among bathers vs. non-bathers. Among the bathers, a dose-response relationship by logistic regression modeling was observed for skin illness, where illness was positively related to enterococci enumeration by membrane filtration (odds ratio =1.46 [95% confidence interval=0.97-2.21] per increasing log10 unit of enterococci exposure) and positively related to 24 hour antecedent rain fall (1.04 [1.01 – 1.07] per increasing millimeters of rain). Acute febrile respiratory illness was inversely related to water temperature (0.74 [0.56-0.98] per increasing degree of water temperature). There were no significant dose response relationships between report of human illness and any of the other FIB or environmental measures. Therefore, for non-point source subtropical recreational marine waters, this study suggests that humans may be at increased risk of reported illness, and that the currently recommended and investigational FIB may not track gastrointestinal illness under these conditions; the relationship between other human illness and environmental measures is less clear.
Keywords: Recreational water quality, indicator organisms, enterococci, Bacteroidales, quantitative PCR, membrane filtration plate counts, chromogenic substrate, Gastrointestinal illness, respiratory illness, skin illness
1. INTRODUCTION
Currently, the levels of enterococci, a fecal indicator bacteria (FIB), as measured by membrane filtration plate counts or by chromogenic substrate method, is the accepted method for regulatory purposes to determine whether water, in particular marine water, is safe for recreational purposes. The current monitoring guidelines are based upon epidemiologic studies at sites impacted by point sources of sewage contamination which have found an increased risk for transmission of infectious diseases, including gastroenteritis, and acute febrile respiratory, skin, eye, and ear illnesses for bathers compared to non-bathers (Wade et al., 2003; Fleisher, 1991; Fleisher et al., 1993, 1996, 1998; Fujioka et al., 1994, Haile et al., 1999; Kay et al., 1994; Prieto et al., 2001; Pruss, 1998; Shuval, 2003).
Increasingly, the use of these fecal indicator bacteria to regulate recreational water bodies, particularly in sub/tropical marine waters without any known sources of sewage, has been questioned (US EPA, 2007; Boehm et al., 2009). In particular, the few epidemiologic studies conducted in sub/tropical environments characterized by non-point sources of fecal indicator microbes have not shown a statistically significant relationship between human health and the current recommended indicator microbes (Fujioka, 2001; Dwight et al., 2004; Colford et al., 2007; Prieto et al., 2001).
The US EPA is in the process of developing new Ambient Water Quality Critieria by 2012 for recreational waters (US EPA, 2007; Boehm et al., 2009; WERF, 2009). Evaluation of these new criteria includes both the use of alternative fecal indicators and rapid molecular methods that will give same-day results (within 4 hours or less). These alternatives include enterococci by qPCR and the Bacteroidales group by qPCR. Although not specifically included in the EPA studies, other researchers have also evaluated the relationship between Staphylococcus aureus, a common skin pathogen, and human health outcomes (Fujioka 2001); and, recently markers have been developed to delineate impacts from seabirds relative to mammals which frequent beach sites (Lu et al., 2008). The use of computer predictive modeling (i.e. “Now-Casting”) of indicator abundance from real-time measurement of physical environmental parameters (such as salinity, turbidity, water temperature, rainfall, etc.) is also being investigated by the US EPA and other groups (Boehm et al., 2009). However, there remains a number of critical information gaps, including the need for more epidemiologic studies (especially in subtropical environments characterized by non-point sources) to elucidate the relationships between: physical-chemical parameters; the abundance of traditional and alternative indicators as enumerated by both traditional culture-based and rapid molecular based methods; the detection and presence of pathogens; and the health outcomes of bathers exposed to recreational waters (US EPA, 2007; Boehm et al., 2009; WERF 2009).
This study was the first prospective randomized exposure study in the US, and the first randomized exposure study in sub/tropical non-point source recreational marine waters globally. The objectives of this study were to evaluate whether exposures at a recreational marine environment with no known point source of sewage result in an increased risk for illness, and to evaluate the use of culture-based and of rapid quantitative PCR methods plus readily measurable environmental parameters to assess their relation to human health outcomes. Although this study measured several various environmental parameters and different microbes including alternative indicators and source tracking markers, these measurements were only reported here in light of their potential predictive ability to reported health effects under the conditions of this particular study. The intent of the current report was to describe the associations between environmental parameters or abundance of traditional and alternative indicators and the reported health effects of water exposure by a randomized cohort of bathers at this particular non-point-source subtropical beach. The significance of source tracking marker abundance and those relationships to beach characteristics, population density, environmental parameters, and other non-health measures are beyond the scope of this particular report and are intended to be published in their own right as a separate paper.
2. MATERIALS AND METHODS
The study site has been utilized previously in a variety of other studies and has been well characterized by the investigators previously in terms of the indicator organisms in the water and sediments as being a non-point source sub/tropical beach with periodic use by people, dogs, and birds, as well as impacted by heavy seasonal rainfall (Fleming et al., 2004; Shibata et al., 2004; Elmir, 2006; Bonilla et al., 2006; Elmir et al., 2007; Fleming et al., 2008; Wright, 2008; Elmir et al., 2009, Wright et al., 2009; Abdelzaher et al., in press).
2.1. Study Design and Sample Collection
The investigators conducted a prospective randomized exposure study with participant exposure randomly assigned to either marine recreational waters or to beach-only exposure. The specific details regarding the epidemiologic study design and sample collection have been previously reported (Fleisher et al., in review) as well as detailed in the Supplement. In brief, the epidemiologic study data collection activities were conducted over 15 individual study days beginning December 15, 2007 and ending June 21, 2008. Local adult residents (≥ 18 years of age) who reported regularly bathing in recreational waters were recruited to participate. Participants were screened for current illness and symptoms, possible alternative risk factors for the study illnesses, and a health history. On the day of their beach exposure, participants were again interviewed about any current illnesses, symptoms, food consumption, and beach exposure since the baseline recruitment interview. Study subjects were then randomly assigned to either the (exposed) bather group or the (unexposed) non-bather group.
Bathers were required to spend 15 minutes in knee-deep water and to immerse their head three times completely under water. Each subject was given a sterile 5-liter personal sampling container and instructed to sample their own personal water space at 5 minute intervals during their water exposure (additional details are described within the Supplement). Staff members instructed all participants in the aseptic sample collection of their water space before their exposure. When subjects left the water, they gave their individual water samples to the environmental research study staff for microbial analysis and processing. Participants randomized to be in the non-bather group were restricted to sitting on chairs on plastic sheeting in a covered roped-off area distant from water and sand exposure for 15 minutes. Out of 1341 adult regular bathers randomly assigned to bather or non-bather groups, 1303 (i.e. 97% follow up) individuals completed the subsequent follow-up telephone interviews for reported illness after 7 days following the beach exposure.
2.2. Disease Endpoint Criteria
The following definitions were used to derive disease endpoint categories prior to data analysis based on the results of the 7 day follow-up questionnaire. “Gastrointestinal illness” was defined as the report of the following symptoms: all cases of vomiting or diarrhea, or all reported cases of indigestion or nausea accompanied by a fever; “Diarrhea” as having three or more runny stools within a 24-hour period; “Acute febrile respiratory illness” as report of at least one of each of the symptoms listed in each of the following 3 categories: (1) fever; (2) headache and/or body-aches and/or unusual fatigue and/or loss of appetite; and, (3) sore throat and/or runny nose and/or dry or productive cough; “Skin illness” as report of at least one of the following symptoms: (1) skin rash; (2) skin ulcer/sore; or (3) itching/irritation on the follow-up questionnaire. These illness definitions were compatible with past studies by the Investigators in Europe, and are roughly compatible with the “highly creditable illness” definitions used in the majority of the US recreational water cohort studies (Wade et al., 2003; Fleisher, 1991; Fleisher et al., 1993, 1996, 1996; Kay, 1994; Wiedenmann, 2006,Fleisher et al., in review).
2.3. Water Sample Processing
Immediately after collection by the participants, 5-L water samples were vigorously shaken and then aliquoted as follows: a) 500 mL poured into a Whirl-pak® bag for enterococci and S. aureus analyses by the membrane filtration (MF) method; b) 1 L poured into a Whirl-pak® bag for physical–chemical parameter analysis; and c) 2 L of beach water remaining in the container for additional enterococci analyses (by the chromogenic substrate (CS) and two quantitative polymerase chain reaction (qPCR) methods), for Bacteroidales analyses (2 human markers and 1 dog marker by qPCR), and for 1 seagull marker analysis (Catellicoccus marimammalium by qPCR). The physico-chemical parameters measured included: pH, salinity, temperature (YSI Model 650, Yellow Springs, OH) and turbidity (Model VWR Model 66120-200, Newark, DE). From the recorded time of sample collection, each sample was also associated with additional hydrometeorological conditions which included: tidal height, rainfall (24hrs), wind direction and speed, and solar radiation from the closest weather stations which were operated by either the Rosenstiel School of Marine and Atmospheric Science at the University of Miami (Miami, FL) or the National Oceanic and Atmospheric Administration (NOAA).
Within two hours of collection, all samples were processed for culture assays and filtered (1 liter filtration volume) for community genetic archive (frozen at -80°C for subsequent DNA extraction and qPCR analysis). The culture methods included enumeration of enterococci by MF (Method 1600, US EPA 2000) and the CS method using Enterolert® (IDEXX Laboratories, Inc., Westbrook, Maine, ASTM 2005); S. aureus was analyzed by MF method using Baird Parker agar plates (Charoenca and Fujioka, 1993). Large (2 to 3 mm) black circular colonies were counted and reported as presumptive counts in units of CFU/100 mL after incubation at 37 ± 0.5°C for 24 hours. Confirmation tests and methicillin resistance for S. aureus were performed based on a coagulation test, followed by PCR analysis of DNA extracts from coagulase positive colonies.
The nucleic acid was extracted from the qPCR filters using quantitative extraction and inhibition controls within a maximum 3 weeks of original sample filtration. Quantitative PCR analysis of the samples included two enterococci assays (Haugland et al. 2005; Siefring et al. 2008), bringing the total number of methods for enterococci analysis to 4 (MF, CS, and two qPCR). To simplify the description of the enterococci data by qPCR, the results corresponding the Haugland et al. (2005) and Siefring et al. (2008) methods are referenced as qPCR-a and qPCR-b, respectively. Quantitative PCR measurements also included three Bacteroidales markers, which included 2 human markers (HF8 as described by Bernhard and Field 2000a, 2000b, and UCD as described by Kildare et al., 2007) and one dog marker (based on Dick et al., 2005). In addition, one recently developed gull marker (C. marimammalium) was also included in the qPCR analyses (based on Lu et al., 2008). The choice of the Bacteroidales markers and gull marker was based upon extensive information obtained from beach surveys and camera image analyses which showed considerable numbers of humans, dogs, and birds, in particular seagulls (Larus atricilla and delawarensis), at the beach (Wright et al., 2009).
Complete details of sample DNA extraction, purification, and qPCR analysis are described in the Supplement. In brief, for the qPCR assays, the filters were spiked just before extraction with a control cell suspension of PBS-washed Lactococcus lactis whole cells (1 × 105 cells per filter) to measure and correct for any inter-sample variations in extraction efficiency and inhibition. Several different studies, including this one, have shown no Lactococcus in background environmental samples of this particular beach. In addition the relationship between Lactococcus and Enterococcus relative extraction efficiency and the prior use of Lactococcus as a calibrator for both Enterococcus and Bacteroides, as well as its use as an environmental inhibition control, has previously been characterized (Siefring et al, 2008), thereby making Lactococcus a suitable candidate to control for extraction efficiency and sample inhibition in the study reported here (More details about the use of the Lactococcus control is available in the supplemental text).
DNA from each filter was extracted by bead beating in lysis buffer, followed by purification, and the purified total community genomic DNA (gDNA) was eluted in a final volume of 100 μl (representing the total community DNA harvested from 1 liter of original water sample). Each qPCR reaction was run in singleplex, requiring at least 7 different 1 μl aliquots from the 100 μl DNA extract (two for enterococci - one for each method, three for Bacteroidales - HF8, UCD, dog, one for the Catellicoccus gull marker, and one for the Lactococcus control). Quantitation was determined from a serial dilution standard curve of target DNA concentrations prepared from genomic controls. Calculated sample quantitations were then corrected for recovery efficiency and inhibition using the measured recovery efficiencies of the Lactococcus controls for each sample. Potential inhibition was also independently assessed by the amplification efficiency of the other multiple targets assayed for each sample. Any samples demonstrating inhibition were diluted and reanalyzed.
An Enterococcus faecalis culture from the American Type Culture Collection (ATCC) was used as genomic control standards for enterococci, and a Bacteroides dorei culture from the DSMZ (Deutsche Sammlung von Mikroorganismen und Zellkulturen) culture collection was used as genomic controls for human-host Bacteroidales HF8 and UCD measurements, while an ATCC culture of Lactococcus lactis was used as the genomic control standard for the Lactococcus extraction control qPCR assay. The Bacteroidales dog-host assay and the Catellicoccus gull-host assay did not have genomic controls at the time of analyses, so plasmid controls containing a single copy number of the target sequence were utilized instead. More details about the analysis methods are included within the Supplement.
The concentrations of enterococci based on the MF and CS were expressed in colony forming units (CFU) and most probable number (MPN) per 100 mL, respectively. The concentrations of enterococci by qPCR and Bacteroidales human markers were expressed in genomic equivalent units (GEU) per 100 mL, whereas the concentrations of Bacteroidales dog marker and Catellicoccus gull marker were expressed in units of target sequence copy (TSC) per 100 ml.
2.4. Statistical Analysis of Health Effect Associations
The statistical software packages SAS, and where necessary StatXact, were used to perform the statistical analyses. Participants reporting a particular illness on the study day were excluded from the statistical analysis of that particular illness. Basic analyses were focused on evaluating whether 1) bathers were at increased risk for illness relative to non-bathers, and 2) any relationships between indicator organism density or environmental parameters and the incidence of illnesses among bathers (i.e. a dose response relationship). The indicator organisms evaluated included enterococci by MF, enterococci by CS, enterococci by two qPCR methods (qPCR-a and qPCR-b), Bacteroidales UCD human marker, Bacteroidales HF-8 human marker, Bacteroidales dog marker, Catellicoccus gull marker, and S. aureus; the environmental parameters evaluated included pH, salinity, water temperature, turbidity, tidal height, time since last high tide, wind speed, wind direction, solar radiation, and antecedent rainfall (24 hour). Each outcome illness was modeled separately.
Simple bivariate logistic regression was used to screen each indicator organism or environmental parameter for signs of a dose-response relationship with each of the illnesses. Modeling was conducted by entering each indicator organism or environmental parameter into a separate logistic regression model as a continuous variable. The odds ratio, 95% confidence interval, and p value for that indicator organism are reported where the associated p value was statistically significant or nearly statistically significant (i.e. near a value of 0.05). In addition, a multiple logistic regression model that included all 9 environmental variables was calculated; backward selection procedure was used in this model. To assess the discrimination power of each of these models, the area under the receiver operating characteristic (ROC) curve was computed for each of the models that showed a statistically significant or nearly statistically significant dose-response with an outcome illness (Metz, 1998).
3. RESULTS
The water sample data sets used for each illness outcome were slightly different due to exclusion criteria based upon pre-existing illness on the day of exposure of each participant. The number of positive data points for gastrointestinal, acute febrile respiratory, and skin illnesses among the bathers corresponded to 31 (4.75%), 12 (1.94%), and 47 (7.47%), respectively, out of a total of 668 water samples. Results from the water sample measurements showed that the mean values for enterococci by membrane filtration exceeded regulatory guidelines levels (35 CFU/100ml) in 35% of the individual samples collected, with a mean= 71 CFU/100ml (standard deviation=244). Enterococci by CS averaged 63 MPN/100ml (192), while enterococci by qPCR-a averaged 471 GEU/100ml (2250) and by qPCR-b averaged 444 GEU/100ml (2250); Bacteroidales human markers UCD averaged 66 GEU/100ml (474); HF-8 averaged 1.6 GEU/100ml(15); Bacteroidales dog marker averaged 1100 TSC/100ml (5856); and Catellicoccus gull marker averaged 925 TSC/100 ml (2530). S. aureus was confirmed positive for 37% of the total samples evaluated, while 1.1% of samples tested positive for MRSA.
Environmental parameters were within the ranges typically observed for subtropical marine waters: average pH = 7.96 (0.30), salinity=37.73 (1.18) ppt, temperature = 26.02 (2.31)°C, turbidity =11.80 (13.51) ntu; similarly, hydro-meterological conditions were typical of subtropical environments over a 7 month period: average tidal height = 0.36 (0.20) m, wind speed = 5.84 (2.79) m/s, wind direction = 159.63 (84.49) degrees clockwise from north, solar radiation = 338.20 (206.12) W/m2, and 24 hour antecedent rainfall, range from 0.00 to 28.19 mm. With the exception of the positive association between enterococci MF and the report of skin illness (odds ratio 1.43 [95% confidence interval=0.96-2.14; p=0.08] per increasing log10 unit of enterococci exposure), there were no significant dose response relationships between any other indicator organisms and the report of human illness (Table 1). In addition to the enterococci MF and skin illness, plots of the probability of an association between an indicator organism and report of human illness and the area under the ROC curves were examined based on the p values that were statistically significant or nearly statistically significant (i.e. human Bacteroidales UCD and human Bacteroidales HF8 with gastrointestinal Illness; enterococci qPCR-b with skin illnesses; and enterococci qPCR-a with acute febrile respiratory ilIness). Only the logistic regression model for enterococci MF and skin illness (Figure 1) showed some level of positive discrimination (area under the ROC curve=0.60), and thus the most acceptable dose response relationship (Metz 1998).
Table 1.
Bivariate logistic regression analyses: illness vs indicator organism
| Illness vs. Indicator Organism Abundance | |||
|---|---|---|---|
| Measured parameter | GastroIntestinal Odds Ratio (95% Confidence Interval) P value | Skin Odds Ratio (95% Confidence Interval) P value | Acute Febrile Respiratory Odds Ratio (95% Confidence Interval) P value |
| Enterococci MF | 1.00 (1.00-1.01)* | 1.43 (0.96-2.14) | 1.00 (0.99-1.00)* |
| p=0.80 | p=0.08 | p=0.84 | |
| Enterococci IDEXX | 1.00 (0.99-1.00)* | 1.00 (0.99-1.00)* | 1.00 (0.99-1.00)* |
| p=0.71 | p=0.76 | p=0.89 | |
| Enterococci qPCR-a | 1.00 (0.99-1.00)* | 1.00 (1.00-1.00)* | 1.00 (1.00-1.00)* |
| p=0.62 | p=0.70 | p=0.009 | |
| Enterococci qPCR-b | 0.99 (0.95–1.03) | 1.01(0.99-1.02) | 1.01 (0.99-1.02) |
| p=0.59 | p=0.07 | p=0.19 | |
| Human Bacteroidales UCD | 1.00 (1.00-1.00)* | 1.00 (0.99-1.00)* | 1.00 (0.99-1.00)* |
| p=0.051 | p=0.99 | p=0.90 | |
| Human Bacteroidales HF8 | 1.05 (0.99-1.02) | 1.01 (1.00-1.02) | ** |
| p=0.09 | p=0.99 | ||
| Dog Bacteroidales | 1.00 (1.00-1.00)* | 1.00 (1.00-1.00)* | 1.00 (0.99-1.00)* |
| p=0.84 | p=0.60 | p=0.37 | |
| Gull Catellicoccus marker | 1.00 (1.00–1.00) | 1.00 (1.00–1.00)* | 1.00 (1.00–1.00)* |
| P=0.57 | P=0.12 | P=0.86 | |
| S. aureus | 1.00 (0.99 – 1.00) | 1.00 (0.99-1.01) | 0.98 (0.93 – 1.03) |
| P=0.87 | P=0.44 | P=0.48 | |
both odds ratio and confidence intervals rounded to one;
model does not converge
Figure 1.
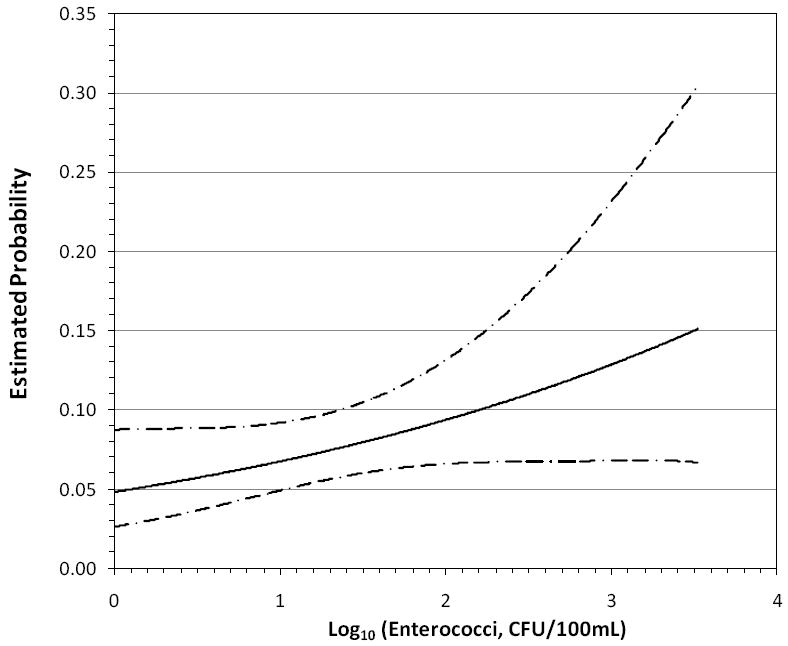
Dose response of reported skin illness vs. log 10 enterococci via MF method, with 95% confidence intervals
The results of the multiple logistic regression analysis focusing on the relationship between the environmental variables and individual health outcomes showed that skin illness was positively related to the 24 hour antecedent rain fall (1.04 [1.01 – 1.07] per increasing millimeter of rain in the antecedent 24 hours), while temperature was inversely related to acute febrile respiratory illness (0.74 [0.56-0.98]). The areas under the ROC curve equaled 0.57 and 0.69, respectively, which indicated an acceptable degree of discrimination (Figures 2 & 3).
Figure 2.
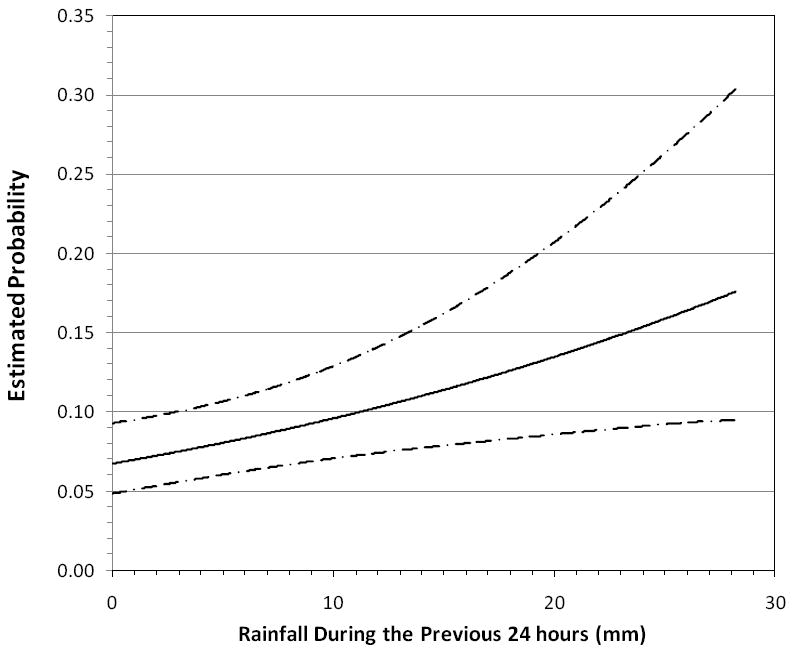
Dose response of reported Skin Illness vs. 24 hour antecedent rain, with 95% confidence intervals
Figure 3.
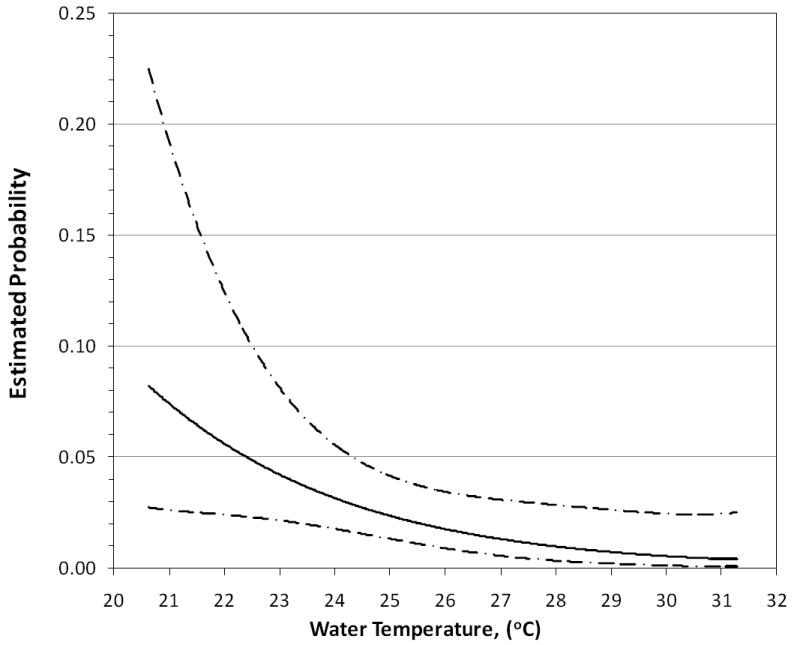
Dose response of reported acute febrile respiratory illness vs. water temperature, with 95% confidence intervals (note the inverse association).
4. DISCUSSION
Historically, fecal indicator bacteria (including total and fecal coliforms and enterococci) have been used as indicators for the presence of bacterial, viral and protozoan pathogens (Savichtchevaa et al., 2005). These microorganisms are of fecal origin from mammals and birds, and their presence in water may indicate fecal pollution and possible association with enteric pathogens. However, there are major problems with these bacterial indicators, including: short survival in water bodies (McFeters et al., 1974; McFeters, 1990); non-fecal source (Scott et al., 2002; Simpson et al., 2002); ability to multiply after release into the water column (Desmarais et al., 2002; Solo-Gabriele et al., 2000); susceptibility to disinfection processes (Hurst et al., 2002); an inability to be used to identify the source of fecal contamination (Field et al., 2003); low levels of correlation with the presence of pathogens (Deetz et al. 1984; Gerba and Rose 1990; Jiang et al., 2001; Griffin et al., 2003; Jiang and Chu 2004; Noble and Fuhrman 2005); and a low sensitivity of detection methods (Horman et al., 2004; Winfield and Groisman, 2003). As a result, none of the bacterial indicators currently used meet all the criteria for an ideal indicator. Furthermore, the only detection methodology currently accepted for regulatory purposes (i.e. enterococci) depends upon culture-based growth and enrichment of the target organisms for an incubation period of at least 18 to 24 hours, so current regulatory methods lack the ability to assess the same-day water quality status of tested water bodies.
In the epidemiologic portion of this study (Fleisher et al., in review), we have shown that bathers had a significantly greater risk of reporting gastrointestinal, respiratory and skin illnesses when exposed to non-point source subtropical recreational marine waters than non-bathers. In the environmental monitoring portion of this study, despite performing a range of FIB and environmental parameter monitoring, we only found a dose response relationship between: a) enterococci measured by membrane filtration vs. report of skin illness; b) 24 hour antecedent rain vs. report of skin illness; and c) water temperature vs. report of acute respiratory febrile illness among the bathers (inverse relationship). Nevertheless, these data suggest that even in the absence of a known point source of sewage contamination, exposure to recreational waters may have public health impacts at subtropical marine beaches.
Enterococci is traditionally used to track GI illness in recreational marine waters (Wade et al., 2003; Pruss, 1998; Shuval, 2003); however, for the current study, enterococci tracked only with skin illnesses. In addition, there was a positive association between skin illness and 24 hour antecedent rain in this study. Enterococci are not typically associated with skin disease; therefore, the correlation with skin illnesses may suggest that environmental conditions that impact enterococci at this recreational beach may also affect an unknown skin pathogen in a similar fashion. Measurements at this study site showed a greater number of enterococci in the water column immediately after storm events (within 6 hours); the hydrology of the site should be evaluated further to understand the sources that could contribute possible pathogens, in particular skin pathogens, a period of 24 hours after a storm event. Also of interest is that the skin pathogen measured as part of the study, S. aureus, did not show a statistically significant relationship with skin or any other illnesses. Part of the lack of relationship between skin illness and S. aureus may be due to the lack of virulence of the environmentally isolated strains, patchiness of the detection (only 37% of the water samples were positive), and relatively small sample sizes to evaluate dose response relationships.
The inverse relationship between water temperature and acute respiratory febrile illness is of particular interest. One hypothesis that could potentially explain this inverse relationship is that viruses or bacteria which cause acute respiratory febrile illness in the participants may be sensitive to water temperature. In particular, viruses can be vulnerable to higher water temperatures (Wetz et al., 2004), therefore perhaps the increase in water temperature in this study resulted in the decreased virulence of viruses which cause respiratory-related illnesses.
The unique feature of the environmental monitoring program implemented during this study was the availability of a wide range of bacteria and environmental parameters data on a per bather basis. This differs from all prior epidemiologic studies which required clustering illness reports to a set of environmental measurements. In this study, the basic unit of measurement was the individual bather rather than the average rates of Illness among many bathers on different study days. This use of an aggregated measure of individual exposures (i.e. rates or rate differences) as the basic unit of measurement in all previous prospective cohort designs would lead to significant misclassification of exposure bias (Fleisher et al., 1991). Thus, the design of the study reported herein would minimize this source of misclassification of exposure bias. Moreover, the variability of fecal indicator measurements has been well documented (EPA, 2007; Boehm et al., 2009), and the sampling strategy implemented in the current study minimizes the impacts from temporal and spatial variations in fecal indicator organisms through collection of a water sample at the time and location of exposure.
4.1. Limitations
Although the health data were based on self-reported symptoms, it should be noted that the bathers and the researchers were inherently blinded to the levels of bacteria in the water and to the other environmental measures, since they were not aware of the results from the monitoring data at the time of exposure. The randomization of individual study participants who reported bathing regularly in recreational marine waters into exposed and unexposed groups solely for the purposes of the study avoided a bias possibly inherent in the prospective cohort designs used in previous recreational bather studies: the hypothesized phenomena that persons who report regular non-bathing might constitute a less healthy group relative to regular bathers, leading to a self-selection bias (Kay et al., 2004; Fleisher et al., 2006). While this particular study had sufficient power to resolve the main question of relative health risk between bathers and non-bathers, the relative rareness of the individual diseases required a much larger sample size than that used here to elucidate clear health associations between the alternative markers or methods and reported illnesses. Investigations based on other study designs using the same methods and markers utilized in this study have suggested relationships that were not seen here. Finally, the bather cohort used in this study consisted of regular adult bathers; thus care must be used when interpreting these findings, especially with respect to susceptible subpopulations (such as small children or anyone with a compromised immune system) (EPA, 2007).
The disadvantage of the environmental monitoring program used in this study was the fact that the water samples collected included not only the ambient water quality, but also potentially the individual bather’s impacts from their individual shedding. It is not clear whether shedding by an individual can cause illness in that same individual. For example in the case of S. aureus, a person colonized with this bacteria could potentially become self-infected in another area of their body, perhaps in an open wound, through the release of their own bacteria into water in which they swim. So, it is not clear whether exposure to one’s own bacterial flora in recreational marine waters can impact health outcomes.
In addition, practical considerations of the study design (i.e. cost) limited the total number of participants that could be included, and thus limited the power of the study to elucidate finer-scale associations, such as associations between specific alternative assays and specific health outcomes. Such associations have been seen previously for some other epidemiologic studies of different design (Wade et al.,2006, 2008). However, it should also be noted that for these other studies the health associations with alternative assays were observed for beaches with exposure to known point-sources of human fecal contamination. To date there has been relatively little similar investigation of non-point-source beaches that utilize these same alternative assays, but at least one other such study at a non.point-source beach has observed no associations between any of the traditional or alternative FIB assay methods and reported gastrointestinal illness (Colford et al., 2007). Those observations are supportive of the similar findings the authors report here in this study. Overall, the authors of this study believe that the advantages gained from the epidemiologic study design used in this study outweigh the disadvantages. At a minimum, this study can be used as a comparison against the more traditional study designs used to evaluate recreational waters in the U.S. Similarities in the results between the current epidemiologic design and the traditional study design provide support to the conclusions reached from both.
4.2. Conclusions
In conclusion, it is apparent that exposure to even non-point source beaches in subtropical marine environments may have associated human health effects. Furthermore, in these non-point source subtropical marine waters, currently recommended and possibly future recommended fecal indicator bacteria may not be sufficient for the prevention of human illness. Further epidemiologic studies are needed to confirm these results, as well as explore in greater depth possible dose-response relationships between human illness with monitoring organisms and environmental parameters.
Supplementary Material
Table 2.
Multivariate logistic regression analyses: illness vs all environmental parameters
| Illness vs Environmental Parameters | |||
|---|---|---|---|
| Measured parameter | GastroIntestinal Odds Ratio (95% Confidence Interval) P value | Skin Odds Ratio (95% Confidence Interval) P value | Acute Febrile Respiratory Odds Ratio (95% Confidence Interval) P value |
| Turbidity | 1.03 (0.95 – 1.11) | 1.00 (0.93–1.07) | 1.06 (0.91 – 1.23) |
| p=0.56 | p=0.99 | p=0.45 | |
| Salinity | 1.08 (0.74 – 1.58) | 0.95 (0.69 – 1.30) | 0.99 (0.49 – 2.00) |
| p=0.69 | p=0.75 | p=0.98 | |
| Temperature | 0.91 (0.73 – 1.14) | 1.19 (0.96 – 1.47) | 0.74 (0.56-0.98) |
| p=0.21 | p=0.12 | p=0.03 | |
| Tidal height | 3.52 (0.46 – 26.60) | 1.41 (0.25 – 7.82) | 0.38 (0.01 – 14.91) |
| P=0.22 | p=0.69 | p=0.60 | |
| Time since last high tide | 1.04 (0.91–1.18) | 0.97(0.87 – 1.07) | 0.93 (0.78 – 1.11) |
| P=0.56 | P=0.54 | P=0.40 | |
| Wind direction | 1.00 (0.99 – 1.01) | 1.00 (0.99 – 1.01) | 0.99 (0.99 – 1.00) |
| p=0.99 | p=0.27 | p=0.27 | |
| Wind speed | 0.96 (0.79 – 1.16) | 0.94 (0.82 – 1.08) | 1.06 (0.75 – 1.48) |
| p=0.68 | p=0.44 | p=0.75 | |
| Solar radiation | 1.00 (0.99 – 1.01) | 1.00 (0.99 – 1.00) | 1.00 (0.99 – 1.00) |
| p=0.48 | p=0.71 | p=0.76 | |
| 24 hour antecedent rainfall | 1.04 (0.96–1.12) | 1.04 (1.01–1.07) | 1.03 (0.90–1.18) |
| p=0.32 | p=0.009 | p=0.65 | |
P values represent significance level at which the individual predictive variable was withdrawn from the multivariate logistic regression using backwards stepwise procedure
Acknowledgments
The researchers would like to dedicate this research to the memory of Ms. Seana Campbell, a very talented, hardworking and creative young researcher who enriched all people whose lives she touched, and who died too young.
The researchers would like to thank the following collaborators at the following Institutions: NOAA Atlantic Oceanographic and Meteorological Laboratory - Miami; Miami-Dade County Dept. of Health; Miami Seaquarium; University of Florida; BCS Laboratories; University of South Florida; NOAA Hollings Marine Laboratory - Charleston; Woods Hole Oceanographic Institute; and the University of Missouri. The researchers would also like to thank the many University of Miami and Florida International University students, other student researchers, and other researchers for their assistance in the performance of this study.
6.1. This Epidemiology Proposal was funded in part from the following sources: the National Center for Environmental Health (NCEH), Centers for Disease Control and Prevention (CDC); Florida Dept of Health (FL DOH) through monies from the Florida Dept of Environmental Protection (FL DEP); the Environmental Protection Agency (EPA) Internship Program; the National Science Foundation (NSF) and the National Institute of Environmental Health Sciences (NIEHS) Oceans and Human Health Center at the University of Miami Rosenstiel School [NSF 0CE0432368/0911373] and [NIEHS P50 ES12736] and NSF REU in Oceans and Human Health, and the NSF SGER [NSF SGER 0743987] in Oceans and Human Health. Development of the dog-host-specific Bacteroides qPCR assay was funded in part by the Northern Gulf Institute, a NOAA Cooperative Institute (NOAA’s Office of Ocean and Atmospheric Research, U.S. Department of Commerce award NA06OAR4320264). In addition, we would like to thank Source Molecular Corporation (Miami, FL) for supplies and logistic support for the development of the gull-host-specific Catellicoccus qPCR assay. We would also like to thank IDEXX Corporation for their support of our project through the provision of suppli es needed for the chromogenic substrate analysis of enterococci.
Abbreviations
- BEACHES
Beach Environmental Assessment and Characterization Human Exposure Study
- CFU
colony forming units
- CS
Chromogenic Substrate
- FIB
Fecal Indicator Bacteria
- EPA
US Environmental Protection Agency
- GEU
genome equivalent units
- L
Liter
- MF
membrane filtration
- mL
milliliter
- mm
millimeter
- MRSA
Methicillin resistant Staphylococcus aureus
- MPN
most probable number
- PBS
sterile 1X Phosphate Buffered Saline solution
- qPCR
quantitative real-time Polymerase Chain Reaction
- ROC
receiver operating characteristic
- TSC
Target Sequence Copies
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abdelzaher A, Wright M, Ortega C, Solo-Gabriele H, Miller G, Elmir S, Newman X, Shih P, Bonilla JA, Bonilla TD, Palmer CJ, Scott T, Lukasik J, Harwood VJ, McQuaig S, Sinigalliano C, Gidley M, Plano L, Zhu X, Wang JD, Fleming L. Presence of pathogens and indicator microbes at a non-point source subtropical recreational marine beach. Applied and Environmental Microbiology. 200X doi: 10.1128/AEM.02127-09. accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ASTM. Book of Standards Volume: 11.02. D6503-99 Standard Test Method for Enterococci in Water Using Enterolert. ASTM International; West Conshohocken, PA: 2005. [Google Scholar]
- Bernhard AE, Field KG. A PCR assay to discriminate human and ruminant feces on the basis of host differences in Bacteroides-Prevotella genes encoding 16S rRNA. Applied and Environmental Microbiology. 2000a;66:4571–4574. doi: 10.1128/aem.66.10.4571-4574.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhard AE, Field KG. Identification of nonpoint sources of fecal pollution in coastal waters by using host-specific 16S ribosomal DNA genetic markers from fecal anaerobes. Applied and Environmental Microbiology. 2000b;66:1587–1594. doi: 10.1128/aem.66.4.1587-1594.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm AB, Ashbolt NJ, Colford JM, Dunbar LE, Fleming LE, Gold MA, Hansel J, Hunter PR, Ichida AM, McGee CD, Soller JA, Weisberg SB. A sea change ahead for recreational water quality criteria. Journal of Water and Health. 2009;7(1):9–20. doi: 10.2166/wh.2009.122. [DOI] [PubMed] [Google Scholar]
- Bonilla TD, Nowosielski K, Esiobu N, McCorquodale DS, Rogerson A. Species assemblages of Enterococci indicate potential sources of fecal bacteria at a south Florida recreational beach. Mar Poll Bull. 2006;52:800–815. doi: 10.1016/j.marpolbul.2006.03.004. [DOI] [PubMed] [Google Scholar]
- Charoenca N, Fujioka R. Assessment of staphylococcus bacteria in Hawaii marine recreational waters. Wat Sci Tech. 1993;27(3-4):283–289. [Google Scholar]
- Colford JM, Jr, Wade TJ, Schiff KC. Water quality indicators and the risk of illness at beaches with nonpoint sources of fecal contamination. Epidemiology. 2007;18(1):27–35. doi: 10.1097/01.ede.0000249425.32990.b9. [DOI] [PubMed] [Google Scholar]
- Deetz TR, Smith EM, Goyal SM, Gerba CP, Vollet JJ, Tsai YL, Du Pont HL, Keswick BH. Occurrence of rota- and enterovirus in drinking water and environmental water in a developing nation. Water Res. 1984;18:567–571. [Google Scholar]
- Desmarais TR, Solo-Gabriele HM, Palmer CJ. Influence of soil on fecal indicator organisms in a tidally influenced subtropical environment. Applied and Environmental Microbiology. 2002;68:1165–1172. doi: 10.1128/AEM.68.3.1165-1172.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick LK, Bernhard AE, Brodeur TJ, Santo Domingo JW, Simpson JM, Walters SP, Field KG. Host distributions of uncultivated fecal Bacteroidales bacteria reveal genetic markers for fecal source identification. Appl Environ Microbiol. 2005;71:3184–3191. doi: 10.1128/AEM.71.6.3184-3191.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwight RH, Baker DB, Semenza JC, Olson BH. Health effects associated with recreational coastal water use: urban versus rural California. Am J Public Health. 2004;94(4):565–7. doi: 10.2105/ajph.94.4.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmir SM. PhD Thesis, University of Miami. Coral Gables, FL: 2006. Development of A Water Quality Model Which Incorporates Non-Point Microbial Sources. [Google Scholar]
- Elmir SM, Wright ME, Solo-Gabriele HM, Abdelzaher A, Fleming LE, Miller G, Rybolowik M, Shih M-TP, Pillai SP, Cooper JA, Quaye EA. Quantitative Evaluation of Bacteria Released by Bathers in Marine Water. Water Research. 2007;41:3–10. doi: 10.1016/j.watres.2006.10.005. with Supplement available on line. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmir SM, Shibata T, Solo Gabriele HM, Sinigalliano CD, Gidley ML, Miller G, Plano L, Kish J, Withum K, Fleming LE. Quantitative Evaluation of Enterococci and Bacteroidales Released by Adults and Toddlers in Marine Water. Water Research. 2009;43:4610–4616. doi: 10.1016/j.watres.2009.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleisher JM. A Re-Analysis of the Data Supporting U.S. Federal Bacteriological Water Quality Criteria Governing Marine Recreational Waters. Water Pollution Control Federation Journal. 1991;63:259–265. [Google Scholar]
- Fleisher JM, Jones F, Kay D, Stanwell-Smith R, Wyer M, Morano R. Water and non-water risk factors for gastroenteritis among bathers exposed to sewage-contaminated marine waters. International Journal of Epidemiology. 1993;22:698–708. doi: 10.1093/ije/22.4.698. [DOI] [PubMed] [Google Scholar]
- Fleisher JM, Kay D, Salmon RL, Jones F, Wyer MD, Godfree AF. Marine Waters Contaminated with Domestic Sewage: Nonenteric Illness Associated with Bather Exposure in the United Kingdom. American Journal of Public Health. 1996;86(9):1228–1234. doi: 10.2105/ajph.86.9.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleisher JM, Kay D, Wyer MD, Godfree AF. Estimates of the severity of illnesses associated with bathing in marine recreational waters contaminated with domestic sewage. International Journal of Epidemiology. 1998;27:722–726. doi: 10.1093/ije/27.4.722. [DOI] [PubMed] [Google Scholar]
- Fleisher JM, Fleming LE, Solo Gabriele HM, Kish JK, Sinigalliano CD, Plano L, Elmir SM, Wang JD, Withum K, Shibata T, Gidley ML, Abdelzaher AM, He G, Ortega C, Zhu X, Wright M, Hollenbeck J, Backer LC. The BEACHES Study: Health Effects and Exposures from Non-point Source Microbial Contaminants in Subtropical Recreational Marine Waters. doi: 10.1093/ije/dyq084. Submitted – In Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleisher JM, Kay D. Risk perception bias, self-reporting of illness, and the validity of reported results in an epidemiologic study of recreational water associated illnesses. Mar Pollut Bull. 2006;52(3):264–8. doi: 10.1016/j.marpolbul.2005.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field KG, Bernhard AE, Brodeur TJ. Molecular approaches to microbiological monitoring: fecal source detection. Environmental Monitoring and Assessment. 2003;81:313–326. [PubMed] [Google Scholar]
- Fleming LE, Solo Gabriele H, Elmir S, Shibata T, Squicciarini D, Quirino W, Arguello M, Van De Bogart G. A Pilot Study of Microbial Contamination of Subtropical Recreational Waters. Fl J Env Health. 2004:29–33. [PMC free article] [PubMed] [Google Scholar]
- Fleming LE, Solo Gabriele HM, Fleisher JM, Elmir S, Sinigalliano C, Plano L, Kish J, Wang J, Backer L, Shibata T, Abdelzaher A, Garza A, He G, Ortega C, Wright M, Scott T, Bonilla F, Withum K, Gidley MB, Harwood V, Zhu X, Hollenbeck J. The NSF NIEHS Oceans and Human Health Center, Rosenstiel School of Marine and Atmospheric Sciences. University of Miami; Miami, FL: 2008. Final Report on the Pilot Epidemiologic Assessment of Microbial Indicators for Monitoring Recreational Water Quality in Marine Sub/Tropical Environments. [Google Scholar]
- Fujioka RS, Roll K, Morens D. Hawaii Water Resources Research Center. 1999. A Pilot Epidemiological Study of Health Risks Associated with Swimming at Kuhio Beach. [Google Scholar]
- Fujioka RS, Byappanahalli MN. Draft of Final Report, Tropical Indicator Workshop. USEPA Office of Water; Washington, DC: 2001. [Google Scholar]
- Gerba CP, Rose JB. Viruses in source and drinking water. In: McFeters GA, editor. Drinking water microbiology. Springer-Verlag; New York: 1990. pp. 380–395. [Google Scholar]
- Griffin DW, Donaldson KA, Paul JH, Rose JB. Pathogenic human viruses in coastal waters. Clin Miicrobiol Rev. 2003;16:129–143. doi: 10.1128/CMR.16.1.129-143.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haile RW, Witte JS, Gold M. The health effects of swimming in ocean water contaminated by storm drain runoff. Epidemiology. 1999;10(4):355–63. [PubMed] [Google Scholar]
- Haugland RA, Siefring SC, Wymer LJ, Brenner KP, Dufour AP. Comparison of Enterococci measurements in freshwater at two recreational beaches by quantitative polymerase chain reaction and membrane filtration culture analysis. Water Research. 2005;39(4):559–568. doi: 10.1016/j.watres.2004.11.011. [DOI] [PubMed] [Google Scholar]
- Horman A, Rimhanen-Finne R, Maunula L, von Bonsdorff C-H, Torvela N, Heikinheimo A, Hanninen M-L. Campylobacter spp., Giardia spp., Cryptosporidium spp., Noroviruses, and indicator organisms in surface water in southwestern Finland, 2000–2001. Appl Environ Microbiology. 2004;70:87–95. doi: 10.1128/AEM.70.1.87-95.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst CJ, Crawford RL, Knudsen GR, McInerney MJ, Stetzenbach LD. Manual of Environmental Microbiology. second. ASM Press; Washington, DC: 2002. [Google Scholar]
- Jiang SC, Chu W. PCR detection of pathogenic viruses in southern California urban rivers. J Appl Microbiol. 2004;97:17–28. doi: 10.1111/j.1365-2672.2004.02269.x. [DOI] [PubMed] [Google Scholar]
- Jiang S, Noble R, Chui WP. Human adenoviruses and coliphages in urban runoff-impacted coastal waters of Southern California. Appl Environ Microbiology. 2001;67:179–184. doi: 10.1128/AEM.67.1.179-184.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay D, Bartram J, Prüss A. Derivation of numerical values for the World Health Organization guidelines for recreational waters. Water Res. 2004;38(5):1296–304. doi: 10.1016/j.watres.2003.11.032. [DOI] [PubMed] [Google Scholar]
- Kay D, Fleisher JM, Salmon RL. Predicting likelihood of gastroenteritis from sea bathing: results from randomised exposure. Lancet 1. 1994;344(8927):905–909. doi: 10.1016/s0140-6736(94)92267-5. [DOI] [PubMed] [Google Scholar]
- Kildare BJ, Leutenegger CM, McSwain BS, Bambic DG, Rajal VB, Wuertz S. 16S rRNA-based assays for quantitative detection of universal, human-, cow-, and dog-specific fecal Bacteroidales: A Bayesian approach. Water Research. 2007;41:3701–3715. doi: 10.1016/j.watres.2007.06.037. [DOI] [PubMed] [Google Scholar]
- Lu J, Santo Domingo JW, Lamendella R, Edge T, Hill S. Phylogenetic Diversity and Molecular Detection of Bacteria in Gull Feces. Applied and Environmental Microbiology. 2008;74(13):3969–3976. doi: 10.1128/AEM.00019-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFeters GA, Bissonnette GK, Jezeski JJ, Thomson CA, Stuart DG. Comparative survival of indicator bacteria and enteric pathogens in well water. Applied and Environmental Microbioliology. 1974;27:823–829. doi: 10.1128/am.27.5.823-829.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFeters GA. Drinking Water Microbiology: Progress and Recent Developments. Springer; New York: 1990. [Google Scholar]
- Metz CE. Basic principles of ROC analysis. Seminars in Nuclear Medicine. 1998;8:283–298. doi: 10.1016/s0001-2998(78)80014-2. [DOI] [PubMed] [Google Scholar]
- Noble RT, Fuhrman JA. Enteroviruses detected by reverse transcriptase polymerase chain reaction from the coastal waters of Santa Monica Bay, California: low correlation to bacterial indicator levels. Hydrobiologia. 2005;460:175–184. [Google Scholar]
- Prieto MD, Lopez B, Juanes JA, Revilla JA, Llorca J, Delgado Rodriguez M. Recreation in coastal waters: health risks associated with bathing in sea water. J Epidemiology Community Health. 2001;55:442–447. doi: 10.1136/jech.55.6.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruss A. Review of epidemiological studies on health effects from exposure to recreational water. International Journal of Epidemiology. 1998;27(1):1–9. doi: 10.1093/ije/27.1.1. [DOI] [PubMed] [Google Scholar]
- Savichtcheva O, Okayama N, Ito T, Okabe S. Application of a direct fluorescent-based live/dead staining combined with fluorescent in situ hybridization for assessment of survival rate of Bacteroides spp. in oxygenated water. Biotechnology and Bioengineering. 2005;92:356–363. doi: 10.1002/bit.20608. [DOI] [PubMed] [Google Scholar]
- Scott T, Rose JB, Jenkins TM, Farrah SR, Lukasik J. Microbial source tracking: current methodology and future directions. Applied and Environmental Microbiology. 2002;68:5796–5803. doi: 10.1128/AEM.68.12.5796-5803.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata T, Solo Gabriele HM, Fleming LE, Elmir S. Monitoring Marine Recreational Water Quality Using Multiple Microbial Indicators in an Urban Tropical Environment. Water Research. 2004;38:3119–3131. doi: 10.1016/j.watres.2004.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuval H. Estimating the global burden of thalassogenic diseases: human infectious diseases caused by wastewater pollution of the marine environment. J Water Health. 2003;1(2):53–64. [PubMed] [Google Scholar]
- Siefring S, Varma M, Atikovic E, Wymer L, Haugland RA. Improved real-time PCR assays for the detection of fecal indicator bacteria in surface waters with different instrument and reagent systems. Journal of Water and Health. 2008;6(2):225–37. doi: 10.2166/wh.2008.022. [DOI] [PubMed] [Google Scholar]
- Simpson JM, Santo Domingo JW, Reasoner DJ. Microbial source tracking: state of the science. Environmental Science and Technology. 2002;24:5279–5288. doi: 10.1021/es026000b. [DOI] [PubMed] [Google Scholar]
- Solo-Gabriele HM, Wolfert MA, Desmarais TR, Palmer CJ. Sources of E. coli in a coastal subtropical environment. Applied and Environmental Microbiology. 2000;66:230–237. doi: 10.1128/aem.66.1.230-237.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solo Gabriele HM, Abdelzaher A, Wright M. Pathogen Monitoring of a South Florida Beach Which Has Been Characterized by Elevated Microbe Levels. Final Report to the Florida Department of Health; Tallahassee, FL: 2008. [Google Scholar]
- U.S. Environmental Protection Agency (US EPA) Improved Enumeration Methods for the Recreational Water Quality Indicators: Enterococci and Escherichia coli. U.S Environmental Protection Agency, Office of Science and Technology; Washington DC: 2000. [Google Scholar]
- U.S. Environmental Protection Agency (US EPA) Report of the Experts Scientific Workshop on Critical Research Needs for the Development of New or Revised Recreational Water Quality Criteria. Washington, DC: 2007. EPA 823-R-07-006, http://www.epa.gov/waterscience/criteria/recreation/ [Google Scholar]
- Wade TJ, Pai N, Eisenberg JN, Colford JM., Jr Do U.S. Environmental Protection Agency water quality guidelines for recreational waters prevent gastrointestinal illness? A systematic review and meta-analysis. Environmental Health Perspectives. 2003;111(8):1102–9. doi: 10.1289/ehp.6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade TJ, Calderon RL, Sams E, Beach M, Brenner KP, Williams AH, Dufour AP. Rapidly measured indicators of recreational water quality are predictive of swimming-associated gastrointestinal illness. Environ Health Perspect. 2006;114(1):24–28. doi: 10.1289/ehp.8273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade TJ, Calderon RL, Brenner KP, Sames M, Beach M, Haugland R, Wymer L, Dufour AP. High sensitivity of children to swimming-associated gastrointestinal illness: Results using a rapid assay of recreation water quality. Epidemiology. 2008;19(3):375–383. doi: 10.1097/EDE.0b013e318169cc87. [DOI] [PubMed] [Google Scholar]
- WERF (Water Environment Research Foundation) Report on the Expert Scientific Workshop on Critical Research and Science Needs for the Development of Recreational Water Quality Criteria for Inland Waters. Water Environment Research Foundation; Alexandria, VA: 2009. [Google Scholar]
- Wetz JJ, Lipp EK, Griffin DW, Lukasik J, Wait D, Sobsey MD, Scott TM, Rose JB. Presence, infectivity, and stability of enteric viruses in seawater: relationship to marine water quality in the Florida Keys. Marine Poll Bull. 2004;48:698–704. doi: 10.1016/j.marpolbul.2003.09.008. [DOI] [PubMed] [Google Scholar]
- Wiedenmann A, Krüger P, Dietz K, López-Pila JM, Szewzyk R, Botzenhart K. A randomized controlled trial assessing infectious disease risks from bathing in fresh recreational waters in relation to the concentration of Escherichia coli, intestinal enterococci, Clostridium perfringens, and somatic coliphages. Environ Health Perspect. 2006;114(2):228–36. doi: 10.1289/ehp.8115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winfield MD, Groisman EA. Role of nonhost environment in the lifestyle of Salmonella and E. coli. Appl Environ Microbiol. 2003;69:3687–3694. doi: 10.1128/AEM.69.7.3687-3694.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright ME. Masters Thesis in Engineering. Coral Gables (FL); University of Miami: 2008. Evaluation of Enterococci, an Indicator Microbe, and the Sources that Impact the Water Quality at a Subtropical Non-Point Source Recreational Beach. [Google Scholar]
- Wright ME, Solo Gabriele HM, Elmir S, Fleming LE. Microbial Load from Animal Feces at a Recreational Beach. Marine Pollution Bulletin. 2009;58:1649–1656. doi: 10.1016/j.marpolbul.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


