Abstract
Flows of cells growing as a quasimonolayer in a confined space can exhibit streaming, with narrow streams of fast-moving cells flowing around clusters of slowly moving cells. We observed and analyzed this phenomenon experimentally for E. coli bacteria proliferating in a microfluidic cell trap using time-lapse microscopy. We also performed continuum modeling and discrete-element simulations to elucidate the mechanism behind the streaming instability. Our analysis demonstrates that streaming can be explained by the interplay between the slow adaptation of a cell to its local microenvironment and its mobility due to changes of cell-substrate contact forces.
Microorganisms employ a wide range of cooperative strategies for responding to adverse environmental conditions [1–5]. In many cases, these strategies lead to intricate patterns and complex shapes in bacterial colonies [6–8]. While such patterning is usually associated with long-range cell signaling and motility [9], microorganisms are often found in dense communities where direct cellular contact plays an important role in the dynamics of colony formation [10,11]. Moreover, bacteria often actively seek to aggregate in small cavities and crevices, which helps them to cope with environmental conditions [12,13]. In our recent work [14] we studied orientational ordering of bacteria caused by their growth and ensuing hydrodynamic flow. Here, we use microfluidic traps to characterize a general streaming instability occurring in a confined colony of nonmotile bacteria. In order to investigate the mechanism driving the streaming instability, we develop a continuum model and complementary discrete-element simulations with cells modeled as growing and dividing soft spherocylinders which adapt their size and mobility to local microenvironments.
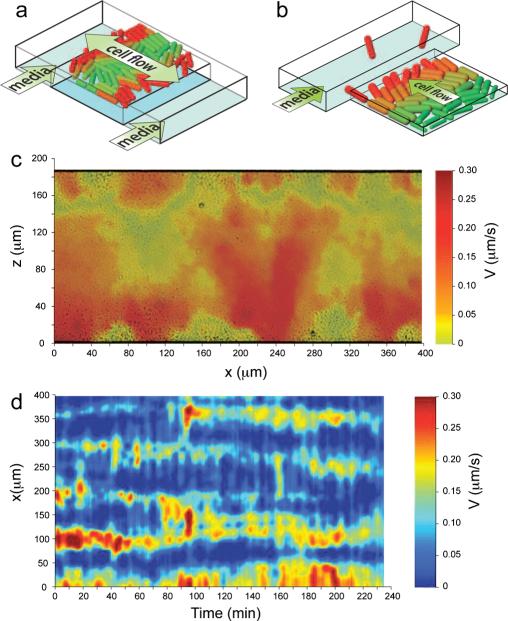
In order to study bacterial colony growth in a confined environment, we constructed microfluidic devices featuring two types of traps (open and side) capable of sustaining a two-dimensional colony of nonmotile bacteria E. coli over many generations. Open traps are ~1 μm-deep rectangular regions of different horizontal dimensions (up to 200 × 2000 μm2) embedded in the middle of the 6–10 μm-deep main channel [see Fig. 1(a)]. The external fluid flow through the main channel (~50 μm/ sec ) delivers nutrients to the open boundaries of the trap, allowing for their diffusion into the interior of the trap. The fluid flow in the channel also removes metabolic waste and cells ejected from the trap. Side traps have similar dimensions but are embedded in the side walls of the main channel and have only one open boundary [Fig. 1(b)].
FIG. 1.
(color online). (a),(b) Schematic views of the microfluidic devices with open and side traps. (c) Magnitude of the vertical component of cell velocity overlaid on a phase contrast image of the trap at time 210 min of the run shown in movie S1; (d) Space-time plot of the cells’ exit velocity component averaged over the lower 20 μm of the trap in (c).
In the beginning of each experiment we placed a few bacteria inside the trap and waited several hours (mean cell division time was about 20 min) until the colony grew and filled the trap region completely. The subsequent growth was balanced by a significant expansion flow towards the open boundaries of the trap. We found that the expansion flow of growing cells inside the traps was surprisingly nonuniform. This behavior was quantitatively analyzed by particle image velocimetry software MATPIV [15]. Figure 1(c) illustrates that cells escape from an open trap into the main channel in narrow rapidly moving streams that bypass regions of almost stagnant cells localized near the open boundaries (see also movie S1 [16]). The cell streams are dynamic [see Fig. 1(d)]; the number and positions of streams fluctuate over the duration of a typical experiment. Similar results were obtained in side-trap experiments; see [16]. More detailed inspection of the cells inside the traps revealed that stagnant cells are generally thicker than rapidly moving cells comprising the streams. Furthermore, the cell size is strongly dependent on its distance from the open boundary of the trap: in the trap interior the cell diameter is only half of that near the open boundaries (where the diameter is about 1 μm) [16]. There may be multiple factors which can cause this dependence, from nutrient depletion to waste accumulation and quorum signaling. Our estimate [16] gives the characteristic preferred carbon source depletion distance of the order of 25 μm from the open boundary, which is consistent with the observed transition to smaller cell sizes.
The instability in cellular streams can be understood in terms of the interplay between the cell size and its mechanical properties. Since the trap height is nearly equal to the cell diameter, the mechanical interaction between the cells and the top and bottom walls of the trap affects their mobility. Under the same pressure gradient, smaller cells experience less drag and move faster, while larger cells experience higher drag and move slower. As cells move towards the open boundary, they grow larger in diameter, which leads to their reduced mobility. The streaming instability occurs when the growth rate of the diameter is comparable to the time a cell spends moving from the back of the trap to the open boundary. Under this condition, slowly moving cells grow larger and can effectively stop moving and form obstacles that permit streaming patterns to emerge for smaller, fast-moving cells.
We developed a continuum model of the colony dynamics which generalizes equations of two-dimensional incompressible fluid dynamics to include the effects of cell growth and drag force, the latter arising from the interaction of the cells with the floor and ceiling of the trap. We did not include the effects of cell shape and position-dependent growth rate in the model since they do not appear to be essential for the basic streaming mechanism. The incompressible “cell fluid” of constant density (scaled to 1) is described by the following equations :
| (1) |
| (2) |
| (3) |
with the cell velocity field, p the pressure field, f a field characterizing cell diameter, g(f) an f-dependent coefficient of (top and bottom) drag for cells moving in the shallow trap, γ the rate of f adaptation to the local chemical environment, μ a coefficient of effective viscosity for cell flow, and α the volumetric growth rate of the “cell fluid.” According to Eq. (2), cell “diameter” f of a cell at a fixed position reaches an equilibrium value that is chosen to be highest near an open boundary of the cell trap. Thus, stagnant cells become largest near the trap opening. The somewhat unusual form of the incompressibility equation (3) is due to the presence of a distributed mass source due to exponential cell growth. This equation can be used to find the hydrodynamic pressure p. We impose the boundary condition of a constant pressure at the open sides of the trap. The drag coefficient g(f) is assumed to nonlinearly increase with f, due to the appearance of strong contact friction between the cell and the trap for large cells. In all results given below we have used g(f) = f2, although the specific form of the nonlinearity is not essential. We also neglect cell inertia and employ the over-damped limit for the momentum equation .
The analysis simplifies considerably in the case of narrow-channel flow (small x dimension), where Eqs. (1)–(3) in the overdamped limit can be reduced to the one-dimensional system
| (4) |
| (5) |
| (6) |
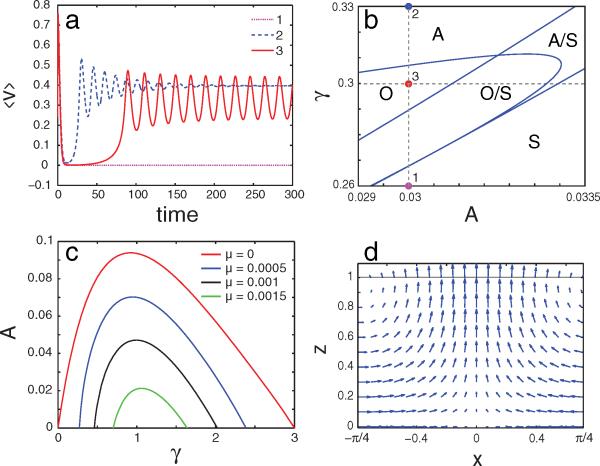
Equation (6) stipulates a linear velocity profile v(z, t) = αz + v0(t). In a side trap with the solid wall at z = 0 and the open boundary at z = Lz, v0(t) = 0, and the velocity, pressure, and f fields are asymptotically stationary and unique. The open-trap case (open boundaries at z = ±Lz) is more interesting, since v0(t) can be a function of time. Substituting the expression for v(z, t) in Eq. (4) and integrating the latter from —LZ to Lz with the boundary condition p(Lz) = p(—Lz) we obtain v0 (t) = —αG1[f]/G0[f], where . This formula defines the flow velocity for a known field f(z, t). The remaining Eq. (5) can be solved through a polynomial mode expansion , and truncation to a finite-dimensional set of nonlinear ODEs (see [16] for the straightforward derivation). The numerical bifurcation analysis of the system using MATCONT [17] reveals that the narrow-channel flow in an open trap exhibits a variety of dynamic regimes, including symmetric flow (v0 = 0), asymmetric flow (v0 = const ≠ 0), and oscillatory flow [v0(t) is periodic], depending on parameters; see Fig. 2(a) and 2(b). We indeed observed nonstationary asymmetric flow regimes in discrete-element simulations and the open-trap experiments [16]. It is also interesting to note the possibility of bistable regimes (e.g., bistability between oscillations and uniform flow).
FIG. 2.
(color online). Results of the continuum hydrodynamic model in the overdamped limit using c(z) = A + (z/Lz)2, Lz = 1, α = 1. (a) Three regimes of narrow-channel flows: symmetric (1), asymmetric (2), and periodic (3) flows; (b) Bifurcation diagram of the narrow-channel flows in the (A, γ) plane. Symbol S denotes symmetric flow, A, asymmetric, and O, oscillatory flow regimes, double symbols (A/S, O/S) indicate domains of bistability according to local bifurcation analysis. Points 1,2,3 correspond to time series shown in (a). (c) Streaming instability domain in the parameter plane (A, γ) for different values of viscosity coefficient μ. Streaming for a given μ occurs for A below the corresponding curve; (d) Velocity field for the eigenfunction near the onset of a streaming instability, with wave vector k = 2, γ = 1, A = 0.045, μ = 0.001. Since the flow is inverted for π/4 ≤ x ≤ 3π/4, only half the x period is displayed.
The onset of cell streaming in the full two-dimensional model can be determined by the linear stability analysis of the transversally uniform flow with respect to small periodic in x perturbations. We consider here the case of the stationary zero-order solution, v(0)(z), p(0)(z) and f(0)(z). In the first order, these solutions are perturbed by the functions {ṽz, ṽx; p̃, f̃} ={ V(z), iV′(z)/k, P(z), F(z)} × exp(ikx + λt) (the velocity components ṽx, ṽz satisfy incompressibility condition automatically). Substituting this ansatz in Eqs. (1)–(3) we arrive at a 1D eigenvalue problem for λ, V(z), P(z), and F(z) with corresponding boundary conditions for side or open-trap cases. In case of the side trap, the boundary conditions are V(0) = F(0) = P(Lz) = 0, and for a symmetric flow in an open trap, the b.c. are P(±Lz) = F(0) = 0. Additionally, we assume a continuous tangential stress condition (slip) at both the inner wall and the outer free boundary, k2V(z) + V′′(z) = 0, z = 0, L. This problem can be solved numerically by a shooting-matching method. The streaming instability first occurs near the wave number ; see Figs. 2(c) and 2(d). This instability requires the presence of a sufficiently steep gradient in the friction field g(f). Intermediate values for the relaxation rate (i.e. γ ≈ α) also are required for the instability, such that adaptation to the chemical microenvironment occurs on a time scale comparable to cell division time. Finally, the streaming instability was found to be sensitive to the value of the coefficient of viscosity μ [Fig. 2(c)].
The two-dimensional continuum analysis of streaming addressed only the linear stability of uniform flow. In addition, the continuum model does not include granular effects, including cell shape [18]. We therefore performed discrete-element simulations (DES) of growing and dividing rodlike cells in a two-dimensional monolayer using a generalization of the soft-particle algorithm described in Ref. [14]. We introduced an internal variable f carried with each “cell” and inherited by its offspring. The variable f for each cell obeys the equation analogous to Eq. (2). Cells grow at a rate proportional to their length and divide on average at the length . Escape of cells from the trap is treated by removing cells when their centers cross the open boundary of the trap.
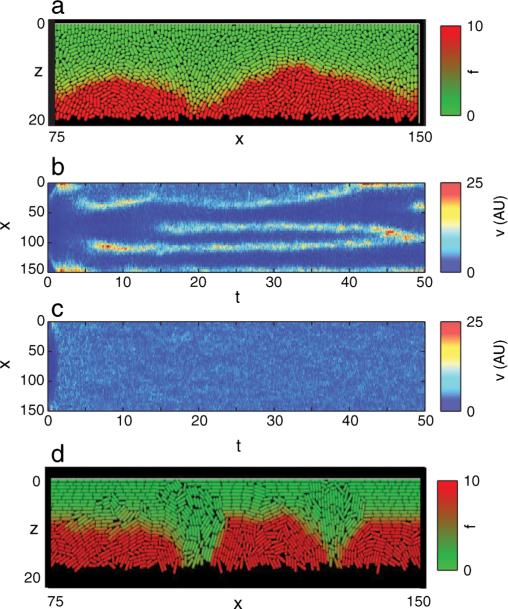
Simulations for cells in a side-trap geometry provide an extension of the linear stability analysis presented above. In the parameter region corresponding to the linear instability we have found significant streaming (see Fig. 3 and [16] movies S6 and S7). Similar to experiment, the structures in simulations remain dynamic, allowing drifting, merging, and spontaneous creation of streams. Since in most simulations traps of moderate horizontal dimensions (up to 150 cell diameters) were explored, as expected, the granular effects played a significant role in stabilizing streaming instability for cells with relatively small aspect ratios (e.g., ). In particular, we found that the system could be bistable between uniform flow and streaming pattern [see Fig. 3(b) and 3(c)]. These results demonstrate that the linear instability of uniform flow gives only a sufficient condition for streaming.
FIG. 3.
(color online). Streaming patterns in a wide side trap: (a) A snapshot of a cell population from a typical DES simulation of short rods at time t = 30 of the simulation shown in panel (b). Cells are colored according to their values of f. Panels (b) and (c) show (x, t) diagrams of the z component of cell velocity averaged over the z axis. Both simulations had the same parameters and were initiated at t = – 15) with a single cell, but for the simulation but in (b) f was allowed to evolve freely from the very beginning while in (c) f was fixed at c(z)/2 until time t = 5 and then relaxed. (d) Snapshot of the population of longer rods . Parameters are Lx = 150, Lz = 20, α = 0.5, γ = 0.5, c(z) = 1 + 200(z/Lz)4.
We also probed the effects of cell orientation on cell streaming by analyzing different cell aspect ratios. Longer cells within the streams tend to align their axis along the flow as expected [14]. This enhanced ordering locally reduces effective shear viscosity, since aligned long cells easily slide past each other (cell-cell friction is assumed small), and this further increases the intensity of streaming [see Fig. 3(d) and [16] movie S7 for ].
Our theoretical and numerical results indicate that the streaming instability arises due to the strong dependence of cell mobility on its size due to drag from top and bottom of the trap. This implies that the streaming instability should be sensitive to the depth of the trap. Additional experiments in deeper traps (1.65 μm) indeed demonstrated the loss of cell streaming [16].
In summary, we have shown that flows of bacteria growing in confined spaces are prone to a streaming instability. The mechanism of the streaming instability is related to the coupling between the cell growth and mobility: larger cells experience greater drag force when moving within a confined space. Cell size, in turn, depends on local chemical environment. In our experiments, cells grow larger near the edge of the trap where the nutrient concentration is higher and waste concentration is lower. The longer the cell remains near the edge of the trap, the larger it becomes and the more difficult it becomes for it to leave the trap. Smaller cells, which are growing in the bulk of the colony, are forced to bypass the larger static cells and form narrow streams. These streams are reminiscent of Saffman-Taylor viscous fingers at an interface between two fluids with different viscosities [19,20]. While we observed the streaming instability in laboratory strains of bacteria grown in microfluidic environments, we believe that the phenomenon is fairly generic and is likely to occur in dense colonies in natural habitats, since bacteria often are found in dense populations in small cavities and crevices where they from biofilms. Future investigations of streaming in biofilms may benefit from the inclusion of effects not included in the present investigation, such as cell-cell and cell-trap adhesion due to an extracellular polymeric matrix. More generally, the interplay between physical properties of cells and their mobility may play an important role in other examples of morphogenesis, such as invasive tumor growth [21].
Supplementary Material
Streaming instability in growing cell populations: Supplementary Information
William Mather, Octavio Mondragón-Palomino, Tal Danino, Jeff Hasty, Lev S. Tsimring
I. EXPERIMENTAL METHODS
Microbial strain and growth conditions
Before each experiment we cultured non-motile strain of bacteria E. coli [1] in 50mL LB (10g/L NaCl) with antibiotics (100 μg/ml ampicillin(Amp) and 50 μg/ml kanamycin(Kan)) for approximately 2 hours from an overnight culture. Cells reached an OD600 of 0.05-0.1 and were spun down and concentrated in 5mL of fresh media with surfactant concentration of 0.075% Tween20 [Sigma-Aldrich, St.Louis,MO] before loading in a device. During the run cells received the same media(w/ 0.075% Tween20) via diffusion and advection, and grew exponentially filling the trapping regions in a monolayer.
Microscopy and image analysis
Images were acquired using an epifluorescent inverted microscope (TE2000-U, Nikon Instruments Inc.). A plexiglass incubation chamber encompassing the entire microscope was used to maintain the constant ambient temperature 37°C. Phase contrast images were taken at 20x or 60x every 1-2 minutes. Stitching of images and autofocusing were performed by Nikon Elements software. Each image was processed using grayscale morphology techiques in ImageJ [2] and particle-image velocimetry (MatPIV [3]) was used to measure coarse-grained velocity profiles.
II. SUPPLEMENTARY EXPERIMENTAL RESULTS
In addition to Fig. 1d of the main text that showed a space-time diagram for the average escape velocity of cells at the bottom edge of the open trap, here we present a similar plot for the the top part of the open trap, Fig. S1. It demonstrates that the dynamics on both open ends of the traps are qualitatively similar: cells organize in fast streams and slow clusters, which shift laterally as the clusters of large stagnant cells change in size and position.
In order to assess the importance of friction of cells with the wall chambers, we compared cell flows in trap with heights 1.0μm and 1.65μm. These traps are schematized in Fig. S2 a,b where the semi-closed geometry prevents the flow of media from sweeping cells away, allowing trap height to be larger than 1.0μm. The spatial distribution of vertical velocities in the 1.0μm case is shown in Fig. S2c and the corresponding space-time diagram of exit velocities is shown in Fig. S2 d. One stream is clearly identified at the middle of the trap with transient shifts in location and magnitude (Supplementary Movie 2). A snapshot of the cell flow in the ~1.65μm trap is shown in Fig. S2e and the space-time diagram shown in Fig. S2f. Here, cells are pushed out in a nearly uniform flow across the entire trap, and no streaming pattern is observed.
To study the effect of cell size distribution on streaming, we grew a colony of E. coli in a 300×90×0.95 μm3 open trap, which allows for better distribution of nutrients and more homogenous cell sizes throughout the trap (Fig. S3, Supplementary Movie 4). Because the trap is smaller, cells do not have time to grow large enough and form clusters near the periphery. Thus, cells leave the colony uniformly on both open boundaries of the trap. Interestingly, the PIV analysis of the experiment shows that the magnitude of the exit velocity on either side of the trap is anti-correlated (see the space-time plots in Figs. S3 b and c), which corresponds to the asymmetric regime of cell flow predicted by the continuum theory (see Sec. IV).
Fig. S4 illustrates the cell size dependence on the position within the side trap. In Panel a we show a snapshot of the side trap where the cells in the interior are significantly smaller than ones near the exit. Panel b shows the average cell area as a function of distance from the open side of the trap. As can be observed in this plot, cell area changes by a factor of 2 from the nutrient-rich open edge of the trap to the back wall. The cross section area of cells was measured by dividing the trap in five horizontal sections centered at z = 9, 27, 45, 63, and 81 μm and segmenting images of cells with ImageJ.
III. DEPTH OF NUTRIENT DEPLETION IN MICROFLUIDIC TRAPS
Our experimental data on the cell size dependence on the distance from the open boundaries (previous Section) suggest that there is variability in the environmental conditions across the trap. The most obvious candidate for such variability is the media which diffuses into the trap from the open boundary and is consumed by growing cells. Here we estimate the characteristic depth zd of the region near the open boundaries to which the nutrient can penetrate before it is being completely consumed by bacteria, and show that it is in a good agreement with the observed depth of the region in the microfluidic traps where large, healthy cells can be found. Derivation of this estimate uses a connection between the distribution of cells in a microfluidic trap and the growth of cells in a batch culture which has been previously studied in the literature [4].
There are several assumptions used in the analysis, but we do not believe these assumptions strongly affect our estimate. We assume that there exists a single preferred nutrient source (at concentration c which in general is a function of space and time) in the media (Luria-Bertani broth), and that fresh media contains this nutrient at a concentration c0. Cells are labeled “healthy” when c > 0 locally, while cells are “stressed” when c ≈ 0 locally. The temporal transition between healthy and stressed cells in batch culture appears as a sudden reduction in the apparent mass per cell [4]. We further assume that the consumption rate μ of the nutrient and the doubling time τ do not depend on the local concentration of the nutrient, although this assumption can be relaxed for more accurate estimates.
A. Depletion of nutrient in a well-stirred batch culture
We now show that the nutrient consumption rate μ can be estimated by analyzing the growth of cells in batch culture. The resulting expression for μ will be used in the next subsection.
Define n(t) as the concentration of cells at time t, and define c(t) as the limiting nutrient concentration at time t. Growth of the cells is exponential
| (1) |
The nutrient concentration is depleted by the cells at a rate μ
| (2) |
Eq. 2 can be integrated to find to the solution for c(t)
| (3) |
where c0 ≡ c(0). Depletion of the nutrient occurs at time td, such that c(td) = 0. By Eq. 3,
| (4) |
We assume that we work in the limit of a small inoculum, i.e. n0 ≈ 0. Then
| (5) |
Define nd ≡ n(td). Then, we can express μ in terms of nd, c0, and τ:
| (6) |
Experimentally, nd is indicated by a sudden change in the apparent mass per cell [4].
B. Distribution of nutrient in a trap
We now model distribution of the nutrient in a microfluidic trap in contact with the fresh media at fixed concentration c0 along the open boundary at z = 0. We assume that cells are present at a constant density n within the trap. We assume a reaction-diffusion model for the nutrient
| (7) |
where z is a depth coordinate for the trap, and D is the effective diffusion constant for the nutrient. Steady state of this system implies
| (8) |
which has the solution
| (9) |
with C1 a constant to be determined. For a sufficiently deep trap, c(z) will become zero at some critical value z = zd. At this point, both c(zd) and the diffusive flux are zero, i.e.
| (10) |
| (11) |
Eq. 10 can be used to find C1, such that
| (12) |
Furthermore, Eq. 11 implies
| (13) |
which leads to the expression for the depth of healthy cells
| (14) |
Using the batch result Eq. 6 for μ, Eq. 14 can be rewritten as
| (15) |
which is independent of c0.
In order to estimate for the value of the depletion depth, zd, we obtained parameter values from the literature:
nd = 8.3 × 10–14 M, corresponding to 5 × 107 cells per mL when OD600=0.3 [4].
n = 5.5 × 10–10 M, approximating the close packing of cells with cell volume ~ 3 μm3.
D = 880 μm2/s, the diffusion constant reported for serine, a representative amino acid, in water at 25°C [5]. This is consistent with the suggestion that the limiting nutrient is an amino acid [4].
τ = 20 min = 1200 s, often reported as the doubling time for E. coli [4].
Using the above quantities, we find zd ≈ 21 μm. However the estimate can be revised by noticing that diffusion should be faster in our experiments, which occur at a higher temperature (37°C). Supposing that D scales with respect to temperature and viscosity according to the Stokes formula for the diffusion constant of a sphere, we revise our estimate to be zd ≈ 25 μm.
IV. NARROW CHANNEL FLOW - POLYNOMIAL EXPANSION
The equations of motion for the narrow channel flow (4)-(6) from the Main text can be further reduced in the case that f(z, t) and c(z) are polynomials in z. Suppose
| (16) |
where many cn may be zero (e.g. cn = 0 for n > 4). Then Eqs. (4)-(6) of the Main text lead to
| (17) |
By identifying corresponding coefficients, we finally arrive at the set of ODEs
| (18) |
| (19) |
Equations (18)-(19) provide a closed set of ODEs for narrow channel flow which was used for the numerical bifurcation analysis shown in the Main Text.
Notice that high order coefficients of f remain zero if initially zero. That is, if cn = 0 and fn(t = 0) = 0 for M ≤ n ≤ N, then fn(t) = 0 for M ≤ n ≤ N.
For the side trap geometry (v0 = 0), all fn decouple from one another. Equations (18) and (19) can then be used to show that symmetric flow is globally stable.
V. LINEARIZED EQUATIONS FOR SMALL PERTURBATIONS ABOUT ZEROTH-ORDER SOLUTION IN A SIDE TRAP
Streaming is simplest to analyze in the case where narrow channel-like asymmetric instabilities are forbidden by geometric constraints. This can be done by analyzing a side trap, where instead of two open walls at z = ±Lz, there is an open wall at z = Lz and a solid wall at z = 0. In the following, we present a brief derivation of the equations governing eigenfunctions in a side trap geometry. These equations can be investigated with a mathematical analysis package capable of solving boundary value problems. We do this using the program Maple (version 11). Solutions are first extended from z = 0 to z = ∈ by a high order, but approximate, polynomial solution, in order to avoid singular behavior of the solution near the solid wall. Maple solves the boundary value problem with this polynomial-extended boundary condition.
A. μ = 0 linearized equations
First consider the case of negligible granular viscosity, i.e. μ = 0. We assume that the zeroth-order solutions, v0(z) and f0(z), are perturbed by the functions
| (20) |
| (21) |
| (22) |
| (23) |
Note that ∂ṽx/∂x + ∂ṽz/∂z = 0, such that the full divergence . The linearized equations that govern the growth of perturbations are straightforward to derive, with the result
| (24) |
| (25) |
| (26) |
where g′(f) = dg(f)/df. Equations 24-26 must satisfy the boundary conditions
| (27) |
| (28) |
| (29) |
The boundary condition in Eq. (28) follows from Eq. (26) if λ ≠ —γ, since it can be expected that v0(∂f/∂z) + v(∂f0/∂z) = 0 at z = 0. f(0) may be nonzero if λ = —γ exactly, but because these eigenfunctions are always stable, they are not relevant to cell streaming.
Eigenfunctions for the side trap geometry can be associated with the eigenfunctions for the open trap geometry with symmetric uniform flow as the zeroth order approximation. The boundary conditions are then p(±Lz) = 0 and f(0) = 0 (when λ ≠ —γ). Symmetry of the open trap eigenfunctions satisfying Eqs. (24)-(26) is chosen such that v0 and f0 are odd and even, respectively, with respect to z. This choice ensures that v(0) = 0, as is necessary for a side trap.
1. μ = 0 lowest order solutions for k → 0
Instead of v(z), consider the scaled function w(z) = v(z)/k2. The boundary condition for w(z) is w(0) = 0. Then Eqs. (24)-(26) can be rewritten
| (30) |
| (31) |
| (32) |
In lowest order in k2 (assuming w is order 1) these equations are
| (33) |
| (34) |
| (35) |
Analytic solutions for the k = 0 case can be indexed by a nonnegative integer m, such that
| (36) |
| (37) |
| (38) |
| (39) |
B. Linearized equations for non-zero viscosity
The condition μ = 0 is somewhat unrealistic for a granular flow. While μ ≠ 0 effects do not appear for narrow channel flow, we find that stability of uniform flow is significantly increased by a small value for μ.
Linearization of the dynamics for μ ≠ 0 can be done as in the μ = 0 case. The equations analogous to Eqs. (30)-(31) are now
| (40) |
| (41) |
The boundary conditions in Eqs. (27)-(29) continue to apply. Additionally, we assume a continuous tangential stress condition (a slip condition) at both the inner wall and the outer free boundary, which can be written in this case as
| (42) |
For the velocity function v(z), this condition leads to boundary conditions
| (43) |
| (44) |
Calculations reported in the main text solve these equations using solutions that are odd in v.
VI. DISCRETE ELEMENT SIMULATIONS
A. Details of simulation algorithm
Our basic algorithm for soft-particle simulations of growing and dividing spherocylinders has been described previously [6]. It calculates normal and tangential forces between cells based on the overlap of virtual soft spheres centered at the nearest points on the axes of interacting spherocylinders. The motion of the cylinders is obtained by the integrating the Newton's equations using 4th order predictor-corrector scheme, and each cell's length ℓ and f-factor are governed by the first-order ODEs associated with each cell. After the cell length exceeds a certain prescribed value , the cell is replaced by two collinear cells with half its length at the same location.
Cells experience negligible sliding friction from motion against the solid side walls of the trap (defining the x and z boundaries) or against other cells. The time step is ~ 0.25 × 10–5 (AU). The average length of division is typically short, i.e. , but is used for the simulations mentioned in Section VI B. The actual division length is chosen randomly from a Gaussian distribution with mean and coefficient of variation 0.2. Drag force on the cells is proportional to velocity of the cell times the factor g(f) = 2(f/d)2M, with the dimensionless mass of the cell, and ℓ the current length of the cell. Most of the other parameters in the simulation are the same as in Ref. [6].
In Fig. 6 we show an example of DES simulation of a periodic cell flow in a narrow open channel, and compare it with analytical theory discussed in the main text.
B. The role of cell shape in streaming
Though cell streaming can be theoretically investigated without including the effects of cell shape, simulations suggest that colonies of longer cells are more prone to destabilize into streams. Additionally, streams of long cells tend to be more highly focused. Fig. S7a presents space-time diagrams demonstrating streaming of long cells, with a snapshot of the cell configuration appearing in Fig. S8a. Figures S7b, c and S8b, c show the dynamics of short cells for comparison. The full time-lapse movie of the corresponding simulation runs are shown in Supplementary Movies 6 and 7. We conjecture that long rods enhance streaming by (i) reducing granular viscosity by local ordering of cells into flowing layers, and (ii) orienting the stress tensor along the direction of streams.
[1] J. Stricker, S. Cookson, M. R. Bennett, W. H. Mather, L. S. Tsimring, and J. Hasty, Nature 456, 516 (2008).
[2] http://rsbweb.nih.gov/ij/.
[3] J. K. Sveen, An introduction to MatPIV v.1.6.1, Eprint no. 2, ISSN 0809-4403, Dept. of Mathematics, University of Oslo (2004), http://www.math.uio.no/~jks/matpiv.
[4] G. Sezonov, D. Joseleau-Petit, and R. D'Ari, J. Bacteriol. 189, 8746 (2007).
[5] P. S. Stewart, Journal Of Bacteriology 185, 1485 (2003).
[6] D. Volfson, S. Cookson, J. Hasty, and L. S. Tsimring, Proc. Natl. Acad. Sci. USA 105, 15346 (2008).
[7] A. Dhooge, W. Govaerts, and Y. A. Kuznetsov, ACM T. Math. Software 29, 141 (2003).
[8] J. Guckenheimer and P. Holmes, Nonlinear Oscillations, Dynamical Systems, and Bifurcations of Vector Fields (Springer-Verlag, New York, 1983).
List of Supplementary Movies
Movie S1. Time-lapse microscopy of 400 μm-long segment from the 2 mm-long open trap. Images were taken every minute with a 60x objective using phase contrast and stitched together.
Movie S2. Time-lapse microscopy of 90×100×1 μm3 side trap. Images were taken every minute with a 60x objective using phase contrast.
Movie S3. Time-lapse microscopy of 90×100×1.65 μm3 side trap. Images were taken every minute with a 60x objective using phase contrast. No streaming is observed in this trap because of the higher depth.
Movie S4. Time-lapse microscopy of 125 μm-long segment from a 90×300×1.65 μm3 open trap. Images were taken every 2.5 min with a 60x objective using phase contrast.
Movie S5. Numerical simulation of the oscillating flow in a narrow open channel (see Fig. S6 for simulation details)
Movie S6. Numerical simulation of the streaming instability of short cells in a wide side channel (see Fig. S7 for simulation details)
Movie S7. Numerical simulation of the streaming instability of long cells in a wide side channel (see Fig. S7 for simulation details)
Supplementary Figure S1: Space-time plot of the average exit velocity calculated over ~20 μm near the top edge of the monolayer segment shown in Fig. 1c of the main text.
Supplementary Figure S2: a. Schematic diagram of the microfluidic device with side traps (light blue rectangles) on either side of the main channel (black). The traps have been magnified 300% for visualization. Traps are seeded with cells from the cell loading port. Cells are supplied with nutrients from the media port, and as they escape from the trap, they are transported by the flow to one of the waste ports. b. Sketch of one cell trap. Color indicates the cell “size” c. Snapshot of the z-component of velocity overlaid with a phase contrast image of a cell monolayer confined in a 1μm-high side trap. A single “red” stream is flanked by two clusters of large slow moving cells. Deeper in the trap, cells are smaller and almost immobile. This snapshot corresponds to the frame at time 149 minutes in the Supplementary Movie 2. d. Space-time plot of the exit velocity calculated over ~20 μm strip at the bottom edge of the monolayer shown in Panel c. In this plot, a stream of cells shows up as a horizontal band along the middle. The blue areas around this band represent the flanking slow cells. e, f. Plots analogous to c, d, but for a 1.65μm-high side trap (see Supplementary Movie 3). In this case, the friction of cells along the wall is reduced to a minimum, and streaming is not pronounced.
Supplementary Figure S3: a) Magnitude of the vertical component of velocity overlaid with a phase contrast image of a colony in an open trap that is half as wide (~90μm) as the chamber shown in Fig. 1 of the main text. This snapshot corresponds to time 122.5 min in Supplementary Movie 4. Unlike in other one micron high traps, here cell size seems to be uniform and less affected by any chemical gradients that may exist within the colony, and cells seem to be pushed out without streaming. Space time plots for the average exit velocity at the bottom (b) and top(c) of the trap respectively. The average of velocity was calculated over 20 μm from each edge. These plots show that the escape velocity on both sides of the trap is anticorrelated.
Supplementary Figure S4: a. Snapshot of a colony growing in the ~1μm-high chamber described in Fig. S2. b. Plot of the average cross sectional area of cells as a function of the distance from the open edge of the trap.
Supplementary Figure S5: A local bifurcation analysis of the narrow channel flow (global bifurcations exist, but are not treated in detail). a. One parameter local bifurcation diagram (in coordinates A ≡ c(0) and f1, the first-order term of the f polynomial). Oscillations appear between Hopf bifurcations H1 and H2, while fixed points corresponding to asymmetric solutions exist left of H2 and between H1 and the saddle-node point N. Unstable fixed points exist left of C and between C and N. Stable symmetric fixed points (f1 = 0) are right of C. Parameters are α = 1, γ = 0.3, c(z) = A + (z/Lz)4, g(f) = f2, Lz = 1. b. A two-parameter local bifurcation diagram for the system in a in parameters A and γ. Symbols S, A, and O indicate regions with symmetric fixed points, asymmetric fixed points, or oscillations, respectively. Bistable attractors are listed together, e.g. O/S. BT represents a Bogdanov-Takens bifurcation. The dashed line indicates the value of γ used in panel a. c. A wider view of the bifurcation diagram b. The majority of space not belonging to symmetric flow is associated with a pair of asymmetric fixed points. These bifurcation diagrams are derived from local bifurcation analysis in Matcont [7]. Consistent with the appearance of a Bogdanov-Takens bifurcation [8], a global bifurcation analysis is necessary to fully understand the behavior of this system. Numerical investigation confirms the existence of infinite-period homoclinic bifurcations that lead to large-amplitude limit cycles (data not shown).
Supplementary Figure S6: Space-time diagrams of the narrow-channel flow. DES simulations were started from a single cell placed in the middle of the trap (narrow width Lx = 10, length 2Lz = 80). Color characterizes the cell “diameter” f averaged within a strip of width 2 along z dimension. a. Stationary asymmetric regime is seen for parameters γ = 0.5, c(z) = 1 + 20 (z/Lz)4; b. Oscillatory behavior is seen for parameters γ = 0.1, c(z) = 1 + 100 (z/Lz)4. Both simulations have parameters , α = 0.5; c. Space-time diagram of f for the continuum description of the narrow-channel flow with the same parameter values.
Supplementary Figure S7: Space-time diagrams for simulations of cell flows in wide side traps (Lx = 150, Lz = 20) with different cell aspect ratios. a. Streaming flow of long cells (average length 5 at division), b. Uniform flow of short cells (average length 3 at division), c. Short cells with streaming flow. Panels b and c correspond to the simulations in Fig. 4 of the main text. Other than differing average cell size and elongation rate (the two are balanced to keep the division rate of long cells the same as short cells), the parameters for the simulation in a are the same as in b and c.
Supplementary Figure S8: Snapshots at time t = 30 of the three simulations in Fig. S7. Green and red represent low (f = 0) and high (f ≥ 10) values of f, respectively, for each cell.
Acknowledgments
This work was supported by the NIH and UC-MEXUS. We are grateful to Dmitri Volfson for writing the original numerical code adapted for our discrete-element simulations, and to Denis Boyer for useful discussions.
References
- 1.Shapiro JA. Annu. Rev. Microbiol. 1998;52:81. doi: 10.1146/annurev.micro.52.1.81. [DOI] [PubMed] [Google Scholar]
- 2.Bassler BL. Cell. 2002;109:421. doi: 10.1016/s0092-8674(02)00749-3. [DOI] [PubMed] [Google Scholar]
- 3.Stoodley P, Sauer K, Davies DG, Costerton JW. Annu. Rev. Microbiol. 2002;56:187. doi: 10.1146/annurev.micro.56.012302.160705. [DOI] [PubMed] [Google Scholar]
- 4.Donlan RM. Emerging Infectious Diseases. 2002;8:881. doi: 10.3201/eid0809.020063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stewart PS, Franklin MJ. Nat. Rev. Microbiol. 2008;6:199. doi: 10.1038/nrmicro1838. [DOI] [PubMed] [Google Scholar]
- 6.Ben-Jacob E, Cohen I, Shochet O, Aranson I, Levine H, Tsimring LS. Nature (London) 1995;373:566. doi: 10.1038/373566a0. [DOI] [PubMed] [Google Scholar]
- 7.Levine H, Aranson I, Tsimring L, Truong TV. Proc. Natl. Acad. Sci. U.S.A. 1996;93:6382. doi: 10.1073/pnas.93.13.6382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dockery J, Klapper I. SIAM J. Appl. Math. 2002;62:853. [Google Scholar]
- 9.Park S, Wolanin PM, Yuzbashyan EA, Lin H, Darnton NC, Stock JB, Silberzan P, Austin R. Proc. Natl. Acad. Sci. U.S.A. 2003;100:13 910. doi: 10.1073/pnas.1935975100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tolker-Nielsen T, Brinch UC, Ragas PC, Andersen JB, Jacobsen CS, Molin S. J. Bacteriol. 2000;182:6482. doi: 10.1128/jb.182.22.6482-6489.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drasdo D. Phys. Rev. Lett. 2000;84:4244. doi: 10.1103/PhysRevLett.84.4244. [DOI] [PubMed] [Google Scholar]
- 12.Shimp R, Pfaender F. Appl. Environ. Microbiol. 1982;44:471. doi: 10.1128/aem.44.2.471-477.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Monier J, Lindow S. Proc. Natl. Acad. Sci. U.S.A. 2003;100:15 977. doi: 10.1073/pnas.2436560100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Volfson D, Cookson S, Hasty J, Tsimring LS. Proc. Natl. Acad. Sci. U.S.A. 2008;105:15 346. doi: 10.1073/pnas.0706805105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sveen JK. An Introduction to MatPIV v.1.6.1. Dept. of Mathematics, University of Oslo; Oslo: 2004. http://www.math.uio.no/~jks/matpiv. [Google Scholar]
- 16.See supplementary material at http://link.aps.org/supplemental/10.1103/PhysRevLett.104.208101.
- 17.Dhooge A, Govaerts W, Kuznetsov YA. ACM Trans. Math. Softw. 2003;29:141. [Google Scholar]
- 18.Jaeger HM, Nagel SR, Behringer RP. Rev. Mod. Phys. 1996;68:1259. [Google Scholar]
- 19.Casademunt J, Magdaleno FX. Phys. Rep. 2000;337:1. [Google Scholar]
- 20.Bensimon D, Kadanoff LP, Liang SD, Shraiman BI, Tang C. Rev. Mod. Phys. 1986;58:977. [Google Scholar]
- 21.Anderson ARA, Quaranta V. Nat. Rev. Cancer. 2008;8:227. doi: 10.1038/nrc2329. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Streaming instability in growing cell populations: Supplementary Information
William Mather, Octavio Mondragón-Palomino, Tal Danino, Jeff Hasty, Lev S. Tsimring
I. EXPERIMENTAL METHODS
Microbial strain and growth conditions
Before each experiment we cultured non-motile strain of bacteria E. coli [1] in 50mL LB (10g/L NaCl) with antibiotics (100 μg/ml ampicillin(Amp) and 50 μg/ml kanamycin(Kan)) for approximately 2 hours from an overnight culture. Cells reached an OD600 of 0.05-0.1 and were spun down and concentrated in 5mL of fresh media with surfactant concentration of 0.075% Tween20 [Sigma-Aldrich, St.Louis,MO] before loading in a device. During the run cells received the same media(w/ 0.075% Tween20) via diffusion and advection, and grew exponentially filling the trapping regions in a monolayer.
Microscopy and image analysis
Images were acquired using an epifluorescent inverted microscope (TE2000-U, Nikon Instruments Inc.). A plexiglass incubation chamber encompassing the entire microscope was used to maintain the constant ambient temperature 37°C. Phase contrast images were taken at 20x or 60x every 1-2 minutes. Stitching of images and autofocusing were performed by Nikon Elements software. Each image was processed using grayscale morphology techiques in ImageJ [2] and particle-image velocimetry (MatPIV [3]) was used to measure coarse-grained velocity profiles.
II. SUPPLEMENTARY EXPERIMENTAL RESULTS
In addition to Fig. 1d of the main text that showed a space-time diagram for the average escape velocity of cells at the bottom edge of the open trap, here we present a similar plot for the the top part of the open trap, Fig. S1. It demonstrates that the dynamics on both open ends of the traps are qualitatively similar: cells organize in fast streams and slow clusters, which shift laterally as the clusters of large stagnant cells change in size and position.
In order to assess the importance of friction of cells with the wall chambers, we compared cell flows in trap with heights 1.0μm and 1.65μm. These traps are schematized in Fig. S2 a,b where the semi-closed geometry prevents the flow of media from sweeping cells away, allowing trap height to be larger than 1.0μm. The spatial distribution of vertical velocities in the 1.0μm case is shown in Fig. S2c and the corresponding space-time diagram of exit velocities is shown in Fig. S2 d. One stream is clearly identified at the middle of the trap with transient shifts in location and magnitude (Supplementary Movie 2). A snapshot of the cell flow in the ~1.65μm trap is shown in Fig. S2e and the space-time diagram shown in Fig. S2f. Here, cells are pushed out in a nearly uniform flow across the entire trap, and no streaming pattern is observed.
To study the effect of cell size distribution on streaming, we grew a colony of E. coli in a 300×90×0.95 μm3 open trap, which allows for better distribution of nutrients and more homogenous cell sizes throughout the trap (Fig. S3, Supplementary Movie 4). Because the trap is smaller, cells do not have time to grow large enough and form clusters near the periphery. Thus, cells leave the colony uniformly on both open boundaries of the trap. Interestingly, the PIV analysis of the experiment shows that the magnitude of the exit velocity on either side of the trap is anti-correlated (see the space-time plots in Figs. S3 b and c), which corresponds to the asymmetric regime of cell flow predicted by the continuum theory (see Sec. IV).
Fig. S4 illustrates the cell size dependence on the position within the side trap. In Panel a we show a snapshot of the side trap where the cells in the interior are significantly smaller than ones near the exit. Panel b shows the average cell area as a function of distance from the open side of the trap. As can be observed in this plot, cell area changes by a factor of 2 from the nutrient-rich open edge of the trap to the back wall. The cross section area of cells was measured by dividing the trap in five horizontal sections centered at z = 9, 27, 45, 63, and 81 μm and segmenting images of cells with ImageJ.
III. DEPTH OF NUTRIENT DEPLETION IN MICROFLUIDIC TRAPS
Our experimental data on the cell size dependence on the distance from the open boundaries (previous Section) suggest that there is variability in the environmental conditions across the trap. The most obvious candidate for such variability is the media which diffuses into the trap from the open boundary and is consumed by growing cells. Here we estimate the characteristic depth zd of the region near the open boundaries to which the nutrient can penetrate before it is being completely consumed by bacteria, and show that it is in a good agreement with the observed depth of the region in the microfluidic traps where large, healthy cells can be found. Derivation of this estimate uses a connection between the distribution of cells in a microfluidic trap and the growth of cells in a batch culture which has been previously studied in the literature [4].
There are several assumptions used in the analysis, but we do not believe these assumptions strongly affect our estimate. We assume that there exists a single preferred nutrient source (at concentration c which in general is a function of space and time) in the media (Luria-Bertani broth), and that fresh media contains this nutrient at a concentration c0. Cells are labeled “healthy” when c > 0 locally, while cells are “stressed” when c ≈ 0 locally. The temporal transition between healthy and stressed cells in batch culture appears as a sudden reduction in the apparent mass per cell [4]. We further assume that the consumption rate μ of the nutrient and the doubling time τ do not depend on the local concentration of the nutrient, although this assumption can be relaxed for more accurate estimates.
A. Depletion of nutrient in a well-stirred batch culture
We now show that the nutrient consumption rate μ can be estimated by analyzing the growth of cells in batch culture. The resulting expression for μ will be used in the next subsection.
Define n(t) as the concentration of cells at time t, and define c(t) as the limiting nutrient concentration at time t. Growth of the cells is exponential
| (1) |
The nutrient concentration is depleted by the cells at a rate μ
| (2) |
Eq. 2 can be integrated to find to the solution for c(t)
| (3) |
where c0 ≡ c(0). Depletion of the nutrient occurs at time td, such that c(td) = 0. By Eq. 3,
| (4) |
We assume that we work in the limit of a small inoculum, i.e. n0 ≈ 0. Then
| (5) |
Define nd ≡ n(td). Then, we can express μ in terms of nd, c0, and τ:
| (6) |
Experimentally, nd is indicated by a sudden change in the apparent mass per cell [4].
B. Distribution of nutrient in a trap
We now model distribution of the nutrient in a microfluidic trap in contact with the fresh media at fixed concentration c0 along the open boundary at z = 0. We assume that cells are present at a constant density n within the trap. We assume a reaction-diffusion model for the nutrient
| (7) |
where z is a depth coordinate for the trap, and D is the effective diffusion constant for the nutrient. Steady state of this system implies
| (8) |
which has the solution
| (9) |
with C1 a constant to be determined. For a sufficiently deep trap, c(z) will become zero at some critical value z = zd. At this point, both c(zd) and the diffusive flux are zero, i.e.
| (10) |
| (11) |
Eq. 10 can be used to find C1, such that
| (12) |
Furthermore, Eq. 11 implies
| (13) |
which leads to the expression for the depth of healthy cells
| (14) |
Using the batch result Eq. 6 for μ, Eq. 14 can be rewritten as
| (15) |
which is independent of c0.
In order to estimate for the value of the depletion depth, zd, we obtained parameter values from the literature:
nd = 8.3 × 10–14 M, corresponding to 5 × 107 cells per mL when OD600=0.3 [4].
n = 5.5 × 10–10 M, approximating the close packing of cells with cell volume ~ 3 μm3.
D = 880 μm2/s, the diffusion constant reported for serine, a representative amino acid, in water at 25°C [5]. This is consistent with the suggestion that the limiting nutrient is an amino acid [4].
τ = 20 min = 1200 s, often reported as the doubling time for E. coli [4].
Using the above quantities, we find zd ≈ 21 μm. However the estimate can be revised by noticing that diffusion should be faster in our experiments, which occur at a higher temperature (37°C). Supposing that D scales with respect to temperature and viscosity according to the Stokes formula for the diffusion constant of a sphere, we revise our estimate to be zd ≈ 25 μm.
IV. NARROW CHANNEL FLOW - POLYNOMIAL EXPANSION
The equations of motion for the narrow channel flow (4)-(6) from the Main text can be further reduced in the case that f(z, t) and c(z) are polynomials in z. Suppose
| (16) |
where many cn may be zero (e.g. cn = 0 for n > 4). Then Eqs. (4)-(6) of the Main text lead to
| (17) |
By identifying corresponding coefficients, we finally arrive at the set of ODEs
| (18) |
| (19) |
Equations (18)-(19) provide a closed set of ODEs for narrow channel flow which was used for the numerical bifurcation analysis shown in the Main Text.
Notice that high order coefficients of f remain zero if initially zero. That is, if cn = 0 and fn(t = 0) = 0 for M ≤ n ≤ N, then fn(t) = 0 for M ≤ n ≤ N.
For the side trap geometry (v0 = 0), all fn decouple from one another. Equations (18) and (19) can then be used to show that symmetric flow is globally stable.
V. LINEARIZED EQUATIONS FOR SMALL PERTURBATIONS ABOUT ZEROTH-ORDER SOLUTION IN A SIDE TRAP
Streaming is simplest to analyze in the case where narrow channel-like asymmetric instabilities are forbidden by geometric constraints. This can be done by analyzing a side trap, where instead of two open walls at z = ±Lz, there is an open wall at z = Lz and a solid wall at z = 0. In the following, we present a brief derivation of the equations governing eigenfunctions in a side trap geometry. These equations can be investigated with a mathematical analysis package capable of solving boundary value problems. We do this using the program Maple (version 11). Solutions are first extended from z = 0 to z = ∈ by a high order, but approximate, polynomial solution, in order to avoid singular behavior of the solution near the solid wall. Maple solves the boundary value problem with this polynomial-extended boundary condition.
A. μ = 0 linearized equations
First consider the case of negligible granular viscosity, i.e. μ = 0. We assume that the zeroth-order solutions, v0(z) and f0(z), are perturbed by the functions
| (20) |
| (21) |
| (22) |
| (23) |
Note that ∂ṽx/∂x + ∂ṽz/∂z = 0, such that the full divergence . The linearized equations that govern the growth of perturbations are straightforward to derive, with the result
| (24) |
| (25) |
| (26) |
where g′(f) = dg(f)/df. Equations 24-26 must satisfy the boundary conditions
| (27) |
| (28) |
| (29) |
The boundary condition in Eq. (28) follows from Eq. (26) if λ ≠ —γ, since it can be expected that v0(∂f/∂z) + v(∂f0/∂z) = 0 at z = 0. f(0) may be nonzero if λ = —γ exactly, but because these eigenfunctions are always stable, they are not relevant to cell streaming.
Eigenfunctions for the side trap geometry can be associated with the eigenfunctions for the open trap geometry with symmetric uniform flow as the zeroth order approximation. The boundary conditions are then p(±Lz) = 0 and f(0) = 0 (when λ ≠ —γ). Symmetry of the open trap eigenfunctions satisfying Eqs. (24)-(26) is chosen such that v0 and f0 are odd and even, respectively, with respect to z. This choice ensures that v(0) = 0, as is necessary for a side trap.
1. μ = 0 lowest order solutions for k → 0
Instead of v(z), consider the scaled function w(z) = v(z)/k2. The boundary condition for w(z) is w(0) = 0. Then Eqs. (24)-(26) can be rewritten
| (30) |
| (31) |
| (32) |
In lowest order in k2 (assuming w is order 1) these equations are
| (33) |
| (34) |
| (35) |
Analytic solutions for the k = 0 case can be indexed by a nonnegative integer m, such that
| (36) |
| (37) |
| (38) |
| (39) |
B. Linearized equations for non-zero viscosity
The condition μ = 0 is somewhat unrealistic for a granular flow. While μ ≠ 0 effects do not appear for narrow channel flow, we find that stability of uniform flow is significantly increased by a small value for μ.
Linearization of the dynamics for μ ≠ 0 can be done as in the μ = 0 case. The equations analogous to Eqs. (30)-(31) are now
| (40) |
| (41) |
The boundary conditions in Eqs. (27)-(29) continue to apply. Additionally, we assume a continuous tangential stress condition (a slip condition) at both the inner wall and the outer free boundary, which can be written in this case as
| (42) |
For the velocity function v(z), this condition leads to boundary conditions
| (43) |
| (44) |
Calculations reported in the main text solve these equations using solutions that are odd in v.
VI. DISCRETE ELEMENT SIMULATIONS
A. Details of simulation algorithm
Our basic algorithm for soft-particle simulations of growing and dividing spherocylinders has been described previously [6]. It calculates normal and tangential forces between cells based on the overlap of virtual soft spheres centered at the nearest points on the axes of interacting spherocylinders. The motion of the cylinders is obtained by the integrating the Newton's equations using 4th order predictor-corrector scheme, and each cell's length ℓ and f-factor are governed by the first-order ODEs associated with each cell. After the cell length exceeds a certain prescribed value , the cell is replaced by two collinear cells with half its length at the same location.
Cells experience negligible sliding friction from motion against the solid side walls of the trap (defining the x and z boundaries) or against other cells. The time step is ~ 0.25 × 10–5 (AU). The average length of division is typically short, i.e. , but is used for the simulations mentioned in Section VI B. The actual division length is chosen randomly from a Gaussian distribution with mean and coefficient of variation 0.2. Drag force on the cells is proportional to velocity of the cell times the factor g(f) = 2(f/d)2M, with the dimensionless mass of the cell, and ℓ the current length of the cell. Most of the other parameters in the simulation are the same as in Ref. [6].
In Fig. 6 we show an example of DES simulation of a periodic cell flow in a narrow open channel, and compare it with analytical theory discussed in the main text.
B. The role of cell shape in streaming
Though cell streaming can be theoretically investigated without including the effects of cell shape, simulations suggest that colonies of longer cells are more prone to destabilize into streams. Additionally, streams of long cells tend to be more highly focused. Fig. S7a presents space-time diagrams demonstrating streaming of long cells, with a snapshot of the cell configuration appearing in Fig. S8a. Figures S7b, c and S8b, c show the dynamics of short cells for comparison. The full time-lapse movie of the corresponding simulation runs are shown in Supplementary Movies 6 and 7. We conjecture that long rods enhance streaming by (i) reducing granular viscosity by local ordering of cells into flowing layers, and (ii) orienting the stress tensor along the direction of streams.
[1] J. Stricker, S. Cookson, M. R. Bennett, W. H. Mather, L. S. Tsimring, and J. Hasty, Nature 456, 516 (2008).
[2] http://rsbweb.nih.gov/ij/.
[3] J. K. Sveen, An introduction to MatPIV v.1.6.1, Eprint no. 2, ISSN 0809-4403, Dept. of Mathematics, University of Oslo (2004), http://www.math.uio.no/~jks/matpiv.
[4] G. Sezonov, D. Joseleau-Petit, and R. D'Ari, J. Bacteriol. 189, 8746 (2007).
[5] P. S. Stewart, Journal Of Bacteriology 185, 1485 (2003).
[6] D. Volfson, S. Cookson, J. Hasty, and L. S. Tsimring, Proc. Natl. Acad. Sci. USA 105, 15346 (2008).
[7] A. Dhooge, W. Govaerts, and Y. A. Kuznetsov, ACM T. Math. Software 29, 141 (2003).
[8] J. Guckenheimer and P. Holmes, Nonlinear Oscillations, Dynamical Systems, and Bifurcations of Vector Fields (Springer-Verlag, New York, 1983).
List of Supplementary Movies
Movie S1. Time-lapse microscopy of 400 μm-long segment from the 2 mm-long open trap. Images were taken every minute with a 60x objective using phase contrast and stitched together.
Movie S2. Time-lapse microscopy of 90×100×1 μm3 side trap. Images were taken every minute with a 60x objective using phase contrast.
Movie S3. Time-lapse microscopy of 90×100×1.65 μm3 side trap. Images were taken every minute with a 60x objective using phase contrast. No streaming is observed in this trap because of the higher depth.
Movie S4. Time-lapse microscopy of 125 μm-long segment from a 90×300×1.65 μm3 open trap. Images were taken every 2.5 min with a 60x objective using phase contrast.
Movie S5. Numerical simulation of the oscillating flow in a narrow open channel (see Fig. S6 for simulation details)
Movie S6. Numerical simulation of the streaming instability of short cells in a wide side channel (see Fig. S7 for simulation details)
Movie S7. Numerical simulation of the streaming instability of long cells in a wide side channel (see Fig. S7 for simulation details)
Supplementary Figure S1: Space-time plot of the average exit velocity calculated over ~20 μm near the top edge of the monolayer segment shown in Fig. 1c of the main text.
Supplementary Figure S2: a. Schematic diagram of the microfluidic device with side traps (light blue rectangles) on either side of the main channel (black). The traps have been magnified 300% for visualization. Traps are seeded with cells from the cell loading port. Cells are supplied with nutrients from the media port, and as they escape from the trap, they are transported by the flow to one of the waste ports. b. Sketch of one cell trap. Color indicates the cell “size” c. Snapshot of the z-component of velocity overlaid with a phase contrast image of a cell monolayer confined in a 1μm-high side trap. A single “red” stream is flanked by two clusters of large slow moving cells. Deeper in the trap, cells are smaller and almost immobile. This snapshot corresponds to the frame at time 149 minutes in the Supplementary Movie 2. d. Space-time plot of the exit velocity calculated over ~20 μm strip at the bottom edge of the monolayer shown in Panel c. In this plot, a stream of cells shows up as a horizontal band along the middle. The blue areas around this band represent the flanking slow cells. e, f. Plots analogous to c, d, but for a 1.65μm-high side trap (see Supplementary Movie 3). In this case, the friction of cells along the wall is reduced to a minimum, and streaming is not pronounced.
Supplementary Figure S3: a) Magnitude of the vertical component of velocity overlaid with a phase contrast image of a colony in an open trap that is half as wide (~90μm) as the chamber shown in Fig. 1 of the main text. This snapshot corresponds to time 122.5 min in Supplementary Movie 4. Unlike in other one micron high traps, here cell size seems to be uniform and less affected by any chemical gradients that may exist within the colony, and cells seem to be pushed out without streaming. Space time plots for the average exit velocity at the bottom (b) and top(c) of the trap respectively. The average of velocity was calculated over 20 μm from each edge. These plots show that the escape velocity on both sides of the trap is anticorrelated.
Supplementary Figure S4: a. Snapshot of a colony growing in the ~1μm-high chamber described in Fig. S2. b. Plot of the average cross sectional area of cells as a function of the distance from the open edge of the trap.
Supplementary Figure S5: A local bifurcation analysis of the narrow channel flow (global bifurcations exist, but are not treated in detail). a. One parameter local bifurcation diagram (in coordinates A ≡ c(0) and f1, the first-order term of the f polynomial). Oscillations appear between Hopf bifurcations H1 and H2, while fixed points corresponding to asymmetric solutions exist left of H2 and between H1 and the saddle-node point N. Unstable fixed points exist left of C and between C and N. Stable symmetric fixed points (f1 = 0) are right of C. Parameters are α = 1, γ = 0.3, c(z) = A + (z/Lz)4, g(f) = f2, Lz = 1. b. A two-parameter local bifurcation diagram for the system in a in parameters A and γ. Symbols S, A, and O indicate regions with symmetric fixed points, asymmetric fixed points, or oscillations, respectively. Bistable attractors are listed together, e.g. O/S. BT represents a Bogdanov-Takens bifurcation. The dashed line indicates the value of γ used in panel a. c. A wider view of the bifurcation diagram b. The majority of space not belonging to symmetric flow is associated with a pair of asymmetric fixed points. These bifurcation diagrams are derived from local bifurcation analysis in Matcont [7]. Consistent with the appearance of a Bogdanov-Takens bifurcation [8], a global bifurcation analysis is necessary to fully understand the behavior of this system. Numerical investigation confirms the existence of infinite-period homoclinic bifurcations that lead to large-amplitude limit cycles (data not shown).
Supplementary Figure S6: Space-time diagrams of the narrow-channel flow. DES simulations were started from a single cell placed in the middle of the trap (narrow width Lx = 10, length 2Lz = 80). Color characterizes the cell “diameter” f averaged within a strip of width 2 along z dimension. a. Stationary asymmetric regime is seen for parameters γ = 0.5, c(z) = 1 + 20 (z/Lz)4; b. Oscillatory behavior is seen for parameters γ = 0.1, c(z) = 1 + 100 (z/Lz)4. Both simulations have parameters , α = 0.5; c. Space-time diagram of f for the continuum description of the narrow-channel flow with the same parameter values.
Supplementary Figure S7: Space-time diagrams for simulations of cell flows in wide side traps (Lx = 150, Lz = 20) with different cell aspect ratios. a. Streaming flow of long cells (average length 5 at division), b. Uniform flow of short cells (average length 3 at division), c. Short cells with streaming flow. Panels b and c correspond to the simulations in Fig. 4 of the main text. Other than differing average cell size and elongation rate (the two are balanced to keep the division rate of long cells the same as short cells), the parameters for the simulation in a are the same as in b and c.
Supplementary Figure S8: Snapshots at time t = 30 of the three simulations in Fig. S7. Green and red represent low (f = 0) and high (f ≥ 10) values of f, respectively, for each cell.