Abstract
Protein kinase C epsilon (PKCε), a novel calcium-independent PKC isoform, has been shown to be a transforming oncogene. PKCε-mediated oncogenic activity is linked to its ability to promote cell survival. However, the mechanisms by which PKCε signals cell survival remain elusive. We found that signal transducers and activators of transcription 3 (Stat3), which is constitutively activated in a wide variety of human cancers, is a protein partner of PKCε. Stat3 has two conserved amino acid (Tyr705 and Ser727) residues, which are phosphorylated during Stat3 activation. PKCε interacts with Stat3α isoform which has Ser727 and not with Stat3β isoform which lacks Ser727. PKCε-Stat3 interaction and Stat3Ser727 phosphorylation was initially observed during induction of squamous cell carcinomas and in prostate cancer. Now we present that: 1) PKCε physically interacts with Stat3α isoform in various human cancer cells: skin melanomas (MeWo and WM266-4), gliomas (T98G and MO59K), bladder (RT-4 and UM-UC-3), colon (Caco-2), lung (H1650), pancreatic (PANC-1), and breast (MCF-7 and MDA:MB-231). 2) Inhibition of PKCε expression using specific siRNA inhibits Stat3Ser727 phosphorylation, Stat3-DNA binding, Stat3-regulated gene expression as well as cell invasion. 3) PKCε mediates Stat3Ser727 phosphorylation via integration with the MAPK cascade (RAF-1, MEK1/2, and ERK1/2). The results indicate that PKCε-mediated Stat3Ser727 phosphorylation is essential for constitutive activation of Stat3 and cell invasion in various human cancers.
Keywords: PKCε, Stat3, Human cancer
Introduction
Cancer ranks as the second leading cause of death, exceeded only by heart disease in the United States. Each year, about 1.7 million Americans are diagnosed with cancer, and more than a million Americans die of the disease. Cancer develops in almost any organ or tissue of the body, but certain types of cancers are more life threatening than others (Jemal et al., 2009). Defining the molecular mechanisms linked to the transition from normal to cancer cell is essential for planning strategies in the prevention and/or treatment of cancers. Protein Kinase Cε (PKCε) and signal transducers and activators of transcription 3 (Stat3) have been shown to play roles in the development of several cancers (Aziz et al., 2007a, b, c).
STATs are a family of six [Stat1 (α and β isoforms), Stat2, Stat3 (α and β isoforms), Stat4, Stat5 (α, β isoforms), and Stat6)] latent transcription factors which reside in the cytoplasm and are encoded by seven distinct genes (Klampfer, 2006). STAT activation is linked to cell proliferation, differentiation, apoptosis, embryogenesis, and immune responses (Nikitakis et al., 2004; Vinkemeier, 2004; Hodge et al., 2005; Kortylewski et al., 2005). STATs exhibit functional divergence in their roles in oncogenesis. Stat3 and Stat5 promote cell survival while Stat1 has been associated with growth inhibitory effects (Akira, 2000; Stephanou and Latchman, 2005). Constitutively activated STATs, in particular Stat3, are found in a number of human cancers (e.g., head and neck, squamous cell carcinoma (SCC), breast, ovary, prostate, and lung) (Chan et al., 2004; Alvarez et al., 2005; Aziz et al., 2007b, c; Rivat et al., 2005; Kobielak and Fuchs, 2006). Since naturally occurring mutations of Stat3 have not been observed, constitutive activation of Stat3 appears to be mediated by aberrant growth factor signaling (Hodge et al., 2005; Klampfer, 2006). Tyrosine phosphorylation of Stat3 (Tyrosine 705) is mediated by a wide variety of polypeptides and is essential for Stat3 dimerization and nuclear translocation. Stat3 also has a conserved serine727 residue, which is a target for phosphorylation (Decker and Kovarik, 2000). Evidence indicates that cooperation of both tyrosine and serine phosphorylation is necessary for full activation of Stat3 (Li and Shaw, 2004).
PKC is a major intracellular receptor for the mouse skin tumor promoter TPA (Kazanietz, 2007). PKC represents a large family of phosphatidylserine (PS)-dependent serine/threonine kinases (Mellor and Parker, 1998; Mochly-Rosen and Kauvar, 1998; Newton, 2001; Griner and Kazanietz, 2007). Based on structural similarities and co-factor dependency, eleven PKC isoforms have been classified into 3 subfamilies: classical (cPKC), novel (nPKC) and atypical (aPKC) isoforms. The cPKCs (α, βI, βII,γ) are dependent on phosphatidylserine (PS), diacylglycerol (DAG) and Ca2+. The nPKCs (δ, ε, η and θ) retain responsiveness to DAG and PS, but do not require Ca2+ for full activation. The aPKCs (λ and ς) only require PS for their activation (Mochly-Rosen and Kauvar, 1998). PKC isoforms exhibit functional specificity in their signals to oncogenesis (Griner and Kazanietz, 2007). PKCε participates in the regulation of diverse cellular functions including gene expression, neoplastic transformation, cell adhesion, mitogenicity, and cellular motility. Overexpression of PKCε in rodent fibroblasts leads to increase in growth rates, anchorage independence, and tumor formation in nude mice (reviewed in Basu and Sivaprasad, 2007). PKCε has been shown to be a transforming oncogene (Basu and Sivaprasad, 2007), and a predictive biomarker of various human cancers (Pan et al., 2005) including prostate cancer (PCa) (Wu et al., 2002). We found PKCε is linked to the development of SCC (Reddig et al., 2000; Jansen et al., 2001; Wheeler et al., 2003; 2004; 2005). PKCε is also overexpressed in human PCa (Cornford et al., 1999). Overexpression of PKCε transforms androgen dependent (AD) LNCaP tumor cells to an androgen independent (AI) variant (Wu et al., 2002). The transformation of AD to an AI variant was associated with increased cell proliferation and resistance to apoptosis (Wu et al., 2004).
We have shown that PKCε interacts with Stat3, phosphorylates Stat3Ser727, and increases both DNA-binding and transcriptional activity of Stat3 in skin (Aziz et al., 2007b, c) and prostate cancers (Aziz et al., 2007a). However, it is unknown whether association of PKCε with Stat3 is universal. We present novel results in this communication: 1) PKCε interacts with Stat3 and phosphorylates Stat3Ser727 in various human cancer cells: skin melanoma (MeWo and WM266-4), glioma (T98G and MO59K), bladder (RT-4 and UM-UC-3), colon (Caco-2), lung (H1650), pancreatic (PANC-1), and breast (MCF-7 and MDA:MB-231). 2) Inhibition of PKCε expression, using PKCε specific siRNA, inhibits Stat3Ser727 phosphorylation, Stat3-DNA binding, Stat3-regulated gene expression as well as cell invasion not only in PCa but also in several others cancer cell lines including melanoma, glioma, pancreas and lung. 3) PKCε may integrate with MAPK cascade (Raf-1, MEK1/2, and ERK1/2) to phosphorylate Stat3Ser727.
Results
1. PKCε and Stat3 interaction in various human cancer cell lines
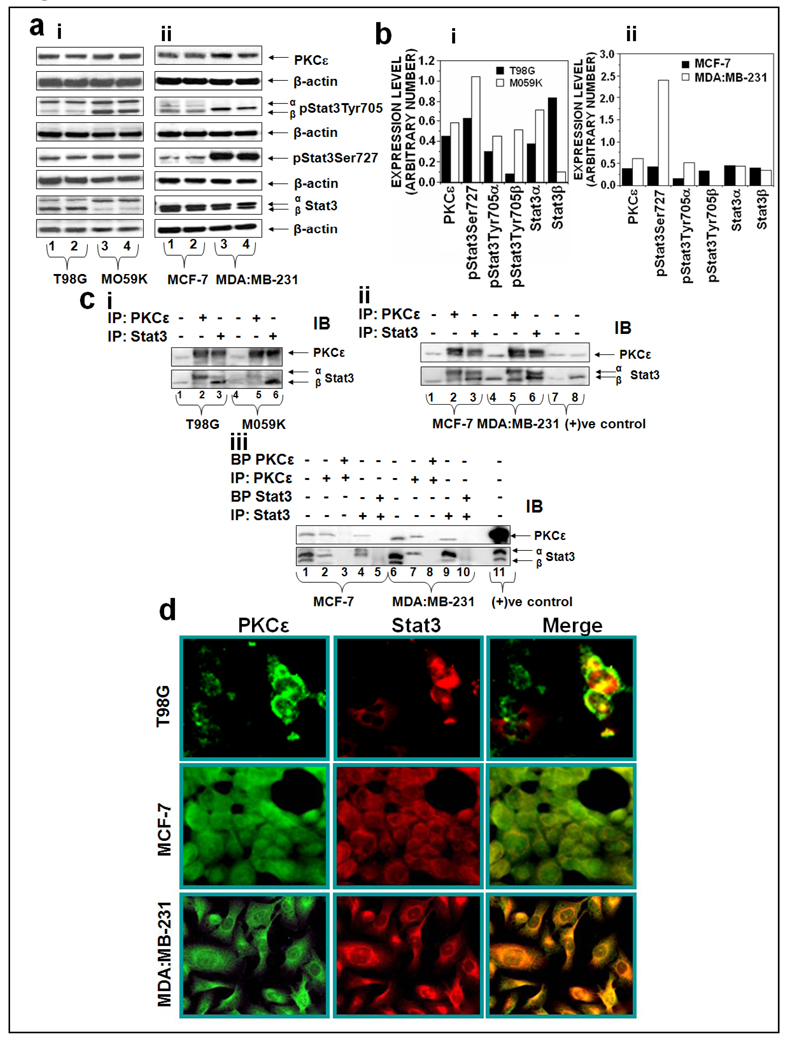
Various human cancer cell lines were used to test the hypothesis that the association of PKCε with Stat3 is universal. The interaction of PKCε with Stat3 in glioma (T98G and MO59K) and breast (MCF-7, MDA:MB-231) cancer cell lines is shown in Figure 1. T98G is a glioblastoma mutiformae fibroblast cell and not tumorigenic in nude mice whereas M059K is a malignant gliomblastoma fibroblast and forms tumors in SCID mice. Expression levels of PKCε and Stat3 in T98G and M059K are shown in Figure 1ai. The basal levels of PKCε expression were higher (32%) in M059K compared with T98G cells. Stat3 has two conserved amino acid (Tyr705 and Ser727) residues, which are phosphorylated during Stat3 activation. PKCε only interacts with Stat3α isoform which has Ser727 and not with Stat3β isoform which lacks Ser727. Also, MO59K elicited a high level of pStat3Tyr705α (53%), pStat3Tyr705β (537%), and pStat3Ser727 (64%) as compared to T98G cells (Figure 1 bi). PKCε and Stat3 interaction in estrogen-dependent MCF-7 human breast cancer cells and estrogen-independent MDA:MB-231 human breast cancer cells is also shown in Figure 1. MDA:MB-231 as compared to MCF-7, exhibited high expression levels of PKCε (63%) pStat3Tyr705 (225%) and pStat3Ser727 (255%) (Figure 1aii, 1bii). Reciprocal immunoprecipitation/blotting experiments indicate that PKCε physically interacts with Stat3 in glioblastoma and breast cell lines (Figure 1ci, ii). As shown in Figure 1ciii, the inclusion of blocking peptide (BP) in the immunoprecipitation experiments inhibited the interaction of PKCε with Stat3, providing straightforward evidence for the protein-protein interactions of PKCε and Stat3. Each immunoprecipitation experiment included a control that contained no primary antibody but pre-immune rabbit serum. Neither PKCε nor Stat3 was pulled down with the pre-immune serum. Furthermore, the immunoprecipitation experiments were repeated with both polyclonal and monoclonal antibodies and polyclonal antibody from different commercial suppliers. Irrespective of the source of the antibody, the results were identical. The co-localization of PKCε with Stat3 was confirmed by double immunofluorscence staining (Figure 1d). Merge images (yellow fluorescence) indicate localization of both PKCε and Stat3 in cytoplasm and nucleus.
Figure 1. PKCε associates with Stat3 in human glioblastoma and breast cancer cells. (a): PKCε and Stat3 expression in glioblastoma and breast cancer cells.
Cells at 70–80% confluency were homogenized in IP lysis buffer as described in Materials and Methods. 25 µg protein of whole cell lysates were fractionated by SDS-PAGE and immunoblotted (IB) with the indicated antibodies. β-actin was used as a control for gel loading variations. b: Protein quantification (normalized to β-actin) was performed as described in Materials and Methods. c: Association of PKCε with Stat3 in human (i) glioblastoma (T98G and M059K) and (ii) breast (MCF-7 and MDA:MB-231) cancer cell lines. Whole-cell lysates were used for immunoprecipitation (IP) with the indicated antibodies. The immunoprecipitated samples were analyzed by Western blot (IB) using the indicated antibodies. (iii): Whole cell lysate of the indicated cells were incubated with the indicated antibodies alone or in combination with their blocking peptide (1µg/mL) at 4°C for overnight before the immunoprecipitation. The immunoprecipitated samples were analyzed by Western blot (IB) using the indicated antibodies. d: double immunofluorescence indicates localization of PKCε and Stat3 in glioblastoma (T98G) and breast cancer cells (MCF-7 and MDA:MB-231). Localization of PKCε and Stat3 is shown by green and red fluorescence, respectively. Colocalization of PKCε with Stat3 is shown by yellow fluorescence. Images were captured at 20 × Magnification.
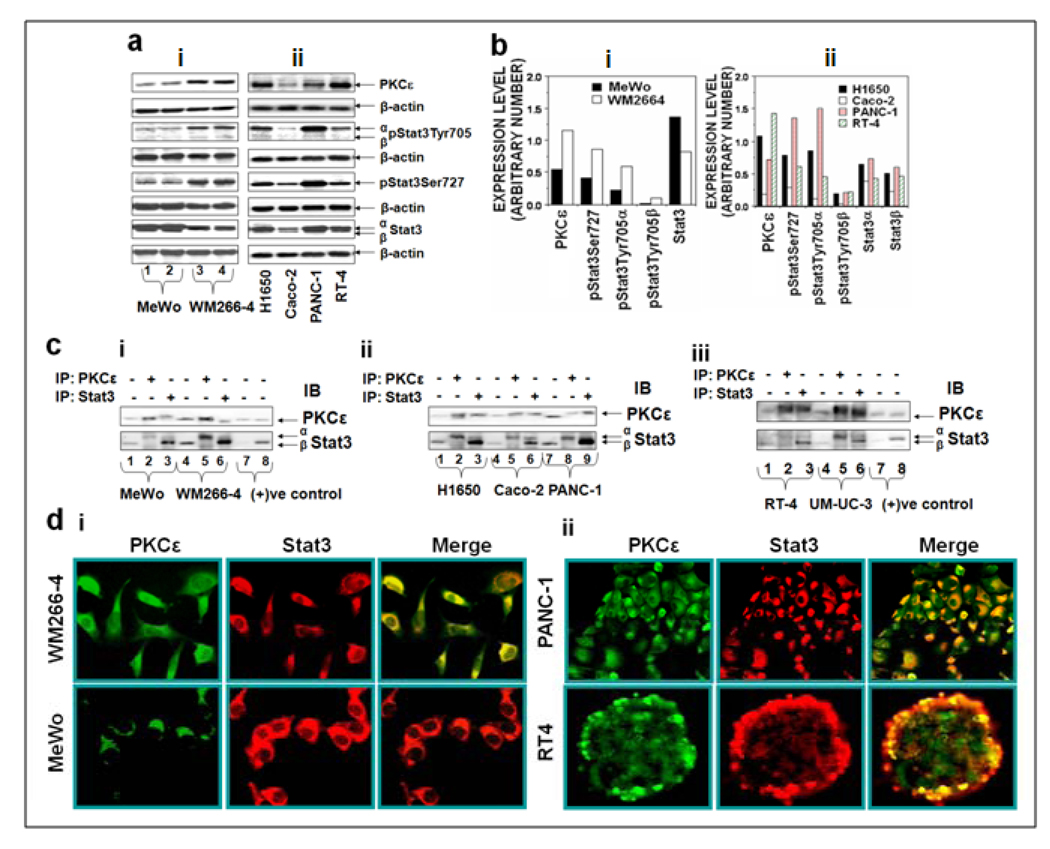
The interaction of PKCε with Stat3 in human melanoma (MeWo and WM266-4), lung adenocarcinoma cell lines (H1650), colon adenocarcinoma Caco2 cells, pancreatic carcinoma (PANC-1) and bladder cancer cells (RT-4, UM-UC-3) is illustrated in Figure 2. WM266-4 is an aggressive metastatic melanoma cell line, while MeWo is a nonmetastatic melanoma cell line. As compared to MeWo, WM266-4 cells elicited higher expression levels of PKCε (161%), pStat3Tyr705α (195%), pStat3Tyr705β (133%) and pStat3Ser727 (167%) (Figure 2ai, 2bi). Similarly, merge images depict significant localization of PKCε and Stat3 in metastatic melanoma cell lines WM266-4 (Fig. 2di). Reciprocal immunoprecipitation/blotting experiments (Figure 2c) reveal that PKCε physically interacts with Stat3. PKCε and Stat3 expression levels in lung (H1650), pancreatic (PANC-1) and bladder cancer (RT-4, UM-UC-3) were high, as compared to human colon adenocarcinoma (Caco-2) (Figure 2aii). It is noteworthy that shift in the mobility of PKCε-immunoprecipitated Stat3 in the western blots (Figure 2c) is perhaps due to the fact that only phosphorylated Stat3 interacts with PKCε. In accord with our results with other human cancer cell lines (Figure 1), PKCε and Stat3 colocalize (Figure 2d) and PKCε̣ interacts with Stat3 (Figure 2c) in lung (H1650), pancreatic (PANC-1) and bladder cancer (RT-4 and UM-UC-3) cell lines. . PKCε-Stat3 interaction was not cell-type specific
Figure 2. PKCε associates with Stat3 in human melanoma, lung, pancreatic, bladder and colon cancer cells. (a): Basal PKCε and Stat3 expression in human melanoma, lung, pancreatic, bladder and colon cancer cells.
Cells at 70–80% confluency were homogenized in IP lysis buffer as described in Materials and Methods. 25 µg of whole cell lysates were fractionated by SDS-PAGE and immunoblotted (IB) for individual antibodies. β-actin was used as a control for gel loading variations. b: Protein quantification (normalized to β-actin) was performed as described in Materials and Methods. c: Association of PKCε with Stat3 in human (i) melanoma (MeWo and WM266-4), (ii) lung (H1650), colon (Caco-2), pancreatic (PANC-1) and (iii) bladder (RT-4 and UM-UC-3), cancer cell lines. Whole-cell lysates were used for IP with the indicated antibodies. The immunoprecipitated samples were analyzed by Western blot (IB) using the indicated antibodies. d: Double immunofluorescence localization of PKCε and Stat3 in (i) melanoma (MeWo and WM266-4), (ii) bladder (RT-4) and pancreatic (PANC-1) cancer cells. Localization of PKCε and Stat3 is shown by green and red fluorescence, respectively. Colocalization of PKCε with Stat3 is shown by yellow fluorescence. Images were captured at 20 × Magnification
2. Functional consequence of PKCε interaction with Stat3
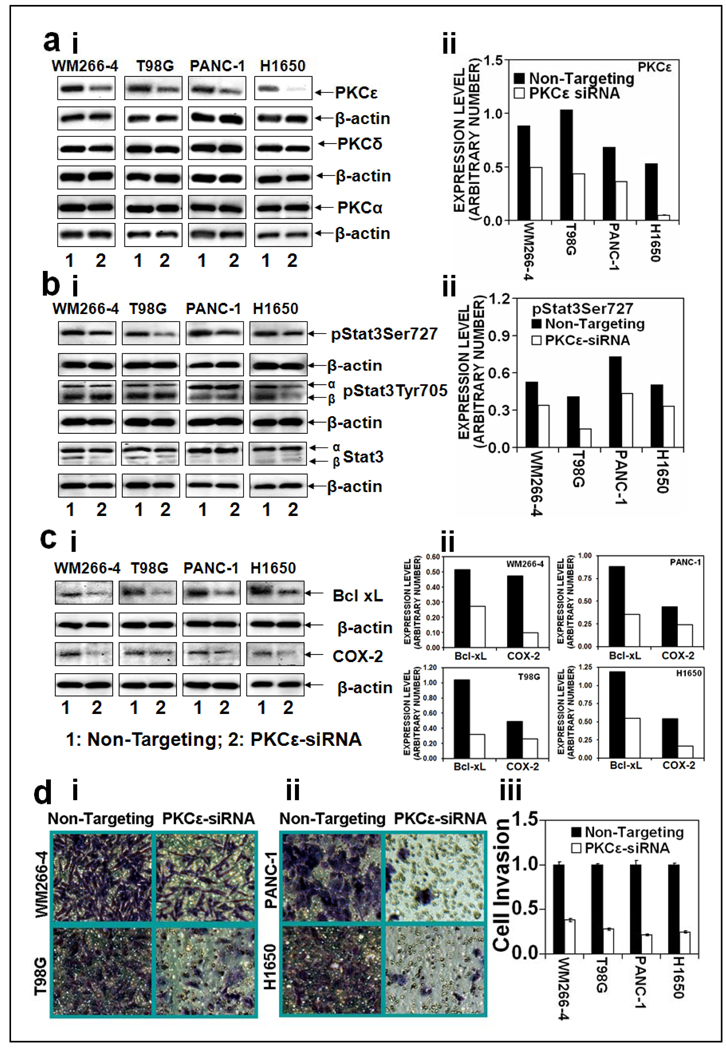
To determine that PKCε-mediated Stat3Ser727 phosphorylation is essential for Stat3 DNA-binding, Stat3 regulated gene expression and cell invasion, we used PKCε specific siRNA to silence PKCε in melonoma (WM266-4), glioma (T98G), pancreatic (PANC-1), and lung (H1650) cancer cells. In this experiment (Figure 3), PKCε specific siRNA was transfected in all the cancer cells to inhibit PKCε expression. Silencing of PKCε using PKCε siRNA resulted in significant inhibition of PKCε without inhibition of expression of other Protein Kinase C isoforms such as PKCδ and PKCα (Figure 3a).
Figure 3. PKCε mediates phosphorylation of Stat3Ser727, Stat3-regulated genes expression and cell invasion in human cancer cells.
Melanoma (WM266-4), glioma (T98G), pancreatic (PANC-1), and lung (H1650) cancer cells were transfected with 15 µg of non-targeting siRNA plasmid (lane 1) or PKCε specific siRNA plasmid (lane 2) (Ambion, Austin, TX) for 48hr and whole cell lysates were prepared as described in Materials and Methods. The whole cell lysates (25 µg protein) were immunoblotted and indicated protein expression levels were detected with appropriate antibodies. β-actin was used as a control for gel loading variations. Protein quantification (normalized to β-actin) was done as described in Materials and Methods (right side). Expression levels of: a (i and ii), PKC isoforms (PKCε, PKCδ and PKCα), b (i and ii), pStat3Ser727, pStat3Tyr705, Stat3, and c (i and ii), Stat3 regulated genes (Bcl-xL, cdc25A and COX-2). d: Human cancer cell invasion. Cells were transfected with non-targeting siRNA plasmid or PKCε specific siRNA plasmid (Ambion, Austin, TX) and cell invasion was determined as described in Materials and Methods. (i): Photographs of invading cells. The migrant cells were stained with crystal violet and photographed the invading cells (40× magnification), (ii): Number of invading cells was estimated by colorimetric measurements at 560 nm according to assay instructions (Chemicon International, Temecula, CA). Each value in the graph is the mean ± S.E. from three separate wells.
Inhibition of PKCε attenuated Stat3Ser727 phosphorylation but not Stat3Tyr705 phosphorylation (Figure 3b). Inhibition of PKCε-mediated Stat3Ser727 phosphorylation (Figure 3b) accompanied inhibition of Stat3-DNA binding (data not shown), Stat3-regulated gene expression (Figure 3c). The results indicate that inhibition of PKC epsilon results in suppression of both Stat3Ser727 phosphorylation and Stat3-regulated gene expression. However, the effects are indeed cell-type specific. Silencing of PKCε significantly (p<0.01) inhibited cell invasion in melonoma (WM266-4), glioma (T98G), pancreatic (PANC-1), and lung (H1650) cancer cells.
3. PKCε-mediated Stat3Ser727 phosphorylation involves integration with MAPK cascade (RAF-1, MEK1/2, and ERK1/2)
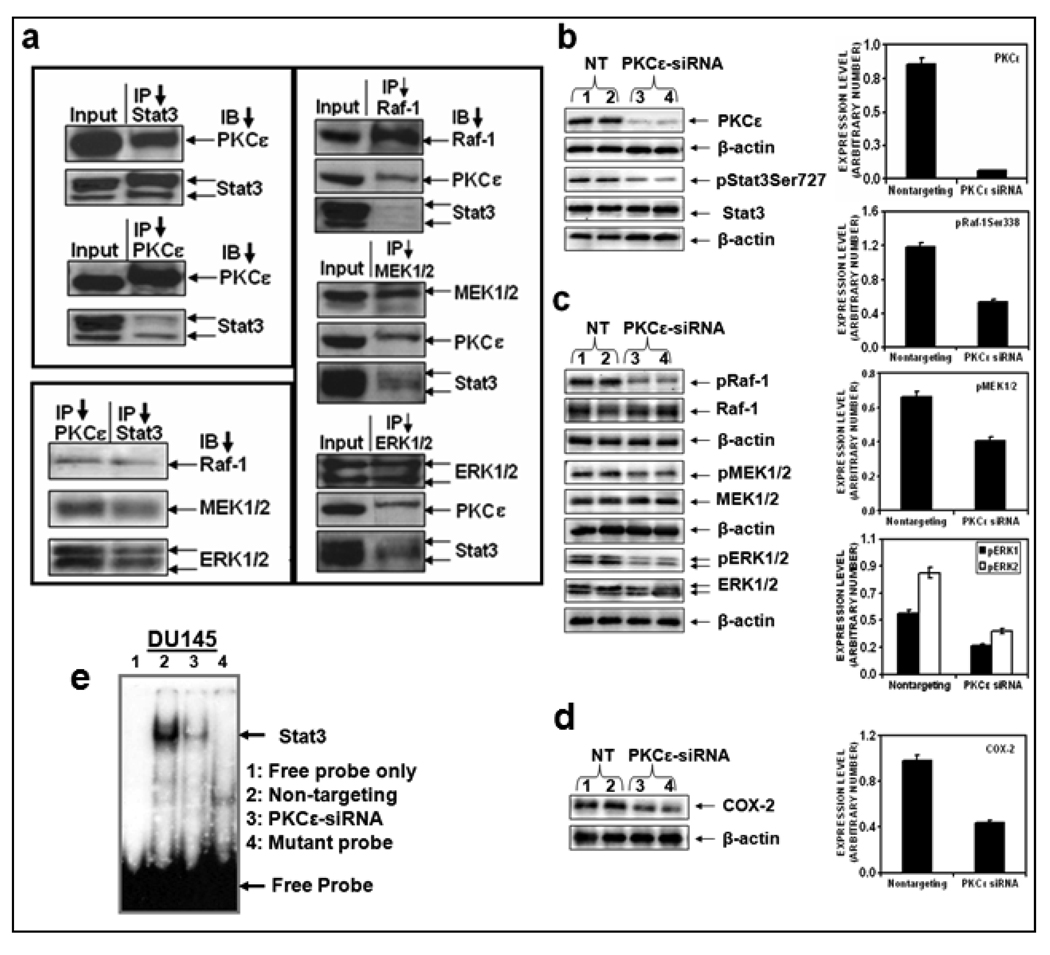
The results (Figure 1–Figure 3) presented clearly indicate that PKCε interacts with Stat3. However, it is unknown whether PKCε interacts with other protein kinase cascade to phosphorylate Stat3Ser727. To explore the possibility that PKCε-Stat3 interaction is mediated through other protein kinases, we used prostate cancer from TRAMP (Transgenic Adenocarcinoma of Mouse Prostate) mice and DU145 cells. In reciprocal immunoprecipitation/ blotting experiments, employing PCa from TRAMP mice, Raf-1, MEK-1/2, and ERK1/2 co-immunoprecipitated with PKCε and Stat3 (Figure 4a).
Figure 4. PKCε integrates with MAPK cascade to phosphorylate Stat3Ser727.
a: Tissue extracts of prostate cancer from TRAMP (Transgenic Adenocarcinoma of Mouse Prostate) mice (50 µg protein) were used for reciprocal IP experiments with antibodies specific to PKCε, Stat3, Raf-1, MEK1/2, and ERK1/2. The immunoprecipitates were subjected to western blot analysis using the indicated antibodies. b, c and d: DU145 cells were transfected with non-targeting siRNA (Lanes 1 and 2) or PKCε specific siRNA (Lanes 3 and 4) (from Dharmacon Inc., Lafayette, CO), and whole cell lysates were prepared as described before (3). The lysates (25 µg protein) were immunoblotted and indicated protein expression levels were detected with the appropriate antibodies. β-actin was used as a control for gel loading variations. Protein quantification (normalized to β-actin) was performed as described in Materials and Methods. Each value is the mean ± S.E. of three independent experiments. e: EMSA. DU145 total cells were suspended in buffer A [10 mmol/L HEPES (pH 7.9), 1.5 mmol/L MagCl2, 10 mmol/L KCl, 0.5 mmol/L DTT, 0.2 mmol/L PMSF]. After 15 min of incubation on ice, the cells were pelleted and resuspended in buffer B [20 mmol/L HEPES (pH 7.9), 20 mmol/L NaF, 1.5 mmol/L MgCl2, 1 mmol/L Na3VO4, 1 mmol/L EDTA, 1 mmol/L EGTA, 1 mmol/L DTT, 0.5 mmol/L PMSF, 420 mmol/L NaCl, 20% glycerol, 1 µg/mL leupeptin, 1 µg/mL aprotinin]. The samples were then centrifuged and the clear supernatant was used for EMSA as described in Materials and Methods. Lane 1, free probe only, Lane 2, nontargeting; lane 3, PKCε siRNA and Lane 4, mutant probe.
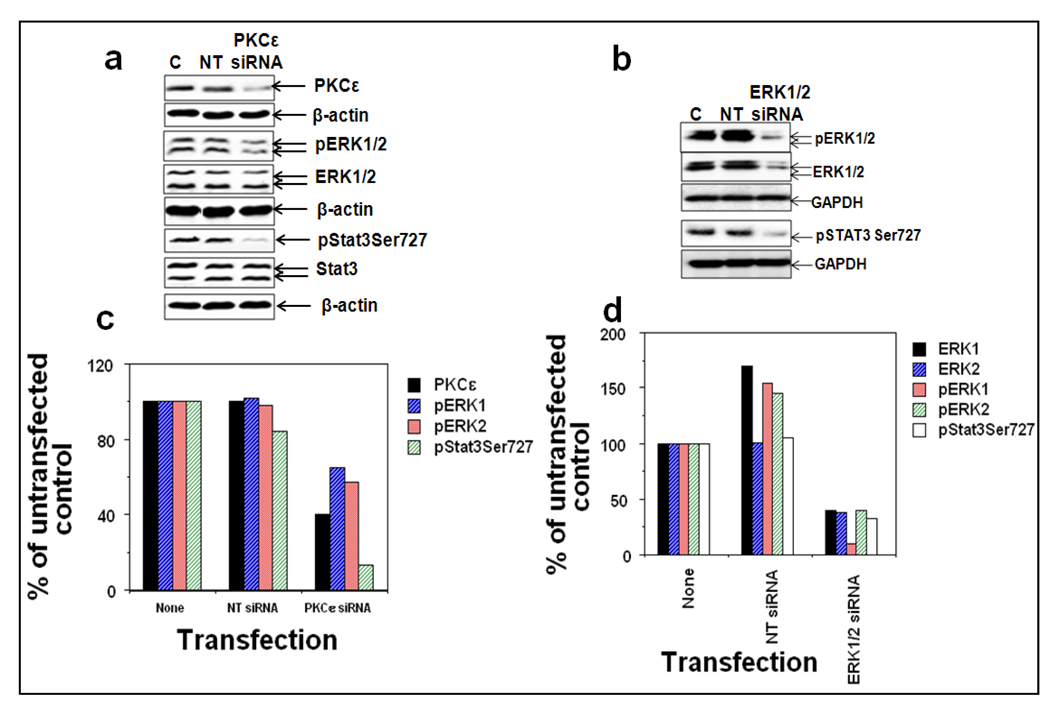
To further determine whether PKCε mediates Stat3Ser727 phosphorylation via activation of MAPK cascade (Raf-1, MEK1/2, and ERK1/2), we used siRNAs to silence PKCε in DU145 cells (Figure 4b). The transfection was done as per the manufacturer’s instructions (Dharmacon, Inc., Lafayette, CO). A pool of four specific siRNA oligonucleotides directed against PKCε was transfected into DU-145 cells to inhibit PKCε synthesis and non-targeting siRNA was used as a control (Figure 4b). Inhibition of PKCε (93%) in DU145 cells using PKCε specific siRNA inhibited the phosphorylation of Raf-1Ser338 (55%), pMEK1/2 (38%), pERK1 (53%), pERK2 (58%) and COX-2 (59%) without affecting total Raf-1, MEK1/2 and ERK1/2 levels (Figure 4c and e). In a separate experiment (Figure 5), silencing of ERK1/2 using ERK1/2 specific siRNA inhibited PKCε-mediated Stat3Ser727 phosphorylation. Taken together, the results indicate that PKCε may integrate with MAPK cascade to phosphorylate Stat3Ser727.
Figure 5. PKCε integrates with ERK1/2 to phosphorylate Stat3Ser727.
DU145 cells were untransfected (lane 1), or transfected with nontargeting ERK1/2 siRNA (lane 2), or ERK1/2 specific siRNA (lane 3), or nontargeting PKCε siRNA (lane 4), or PKCε-specific siRNA (lane 5) (ERK1/2 specific siRNA from Santa Cruz Biotechnology, Santa Cruz, CA and PKCε-specific siRNA from Dharmacon Inc., Lafayette, CO), for 48hr and whole-cell lysates were prepared as described in Materials and Methods. a: The protein extracts (25 µg protein) were immunoblotted and indicated protein expression levels were detected with the appropriate antibodies. β-actin was used as a control for gel loading variations. b: The quantification of proteins (normalized to β-actin) was done as described in Materials and Methods. i: % of control not treated with PKCε-siRNA, ii: % of control not treated with ERK1/2-siRNA.
Discussion
PKCε, a Ca2+-independent-phospholipid–dependent PKC, is linked to cancer induction, progression, and metastasis (Basu and Sivaprasad, 2007; Griner and Kazanietz, 2007). PKCε is constitutively activated in various human cancers and PKCε levels correlates with aggressiveness of human cancers including breast (Pan et al., 2005), HNSCC (Pan et al., 2006), prostate (Aziz et al., 2007a), lung (Bae et al., 2007), brain (Okhrimenko et al., 2005), and SCC (Aziz et al., 2007b, c). Also, when PKCε is over expressed, it transforms rat fibroblasts, colonic epithelial cells, and androgen-dependent LNCaP cells to an androgen-independent variant (Wu et al., 2002). Constitutively activated PKCε regulates the activation of signaling networks linked to cell survival (Basu and Sivaprasad, 2007). We have previously reported both in skin (Aziz et al., 2007b, c) and prostate (Aziz et al., 2007a) cancer that PKCε may mediate inhibition of apoptosis and promotion of survival of neoplastic cells via its association with Stat3, a transcription factor that is constitutively activated in various human cancers (Kobielak and Fuchs, 2000; Chan et al., 2004; Alvarez et al., 2005; Aziz et al., 2007c; Rivat et al., 2005). PKCε interacts with Stat3, phosphorylates Stat3Ser727, and regulates both Stat3-DNA binding and transcriptional activity (Aziz et al., 2007a, c). Now, we present that PKCε regulates Stat3Ser727 phosphorylation, not only in skin and prostate, but also in several cancer cell lines: skin melanomas (MeWo and WM266-4), glioma (T98G and MO59K), bladder (RT-4 and UM-UC-3), colon (Caco-2), lung (H1650), pancreatic (PANC-1), and breast (MCF-7 and MDA:MB-231). PKCε integrates with MAPK cascade to phosphorylate Stat3Ser727 (Figure 4 and Figure 5). PKCε-mediated Stat3Ser727 phosphorylation is essential for cancer cell invasion (Figure 1–Figure 5).
PKCε is overexpressed and constitutively activated in various human cancers (e.g., glioma, melanoma, breast, prostate) (Wu et al., 2002, 2004; Okhrimenko et al., 2005; Pan et al., 2005; Aziz et al., 2007a, c). The present results with various human cancer cell lines further support the evidence for increased expression levels of PKCε in cancer (Figure 1–Figure 3). The mechanism linked to increased PKCε protein stability in cancer is not defined. The signaling lifetime of PKC is under the control of multiple mechanisms (Chen et al., 2007). PKC is synthesized in the cytoplasm as an inactive precursor. A series of ordered phosphorylation converts PKC into a mature stable species. Binding to lipid second messenger controls the propagation of PKC signals. Termination of PKC signaling is achieved by mechanisms which include: dephosphorylation and proteolytic degradation and degradation of fully phosphorylated PKC by the ubiquitin-proteosome pathway (Chen et al., 2007). Several studies indicate that PKC isozymes become ubiquitinated following activation. PKCα, δ, and ε have been reported to become ubiquitinated following treatment of cells with phorbol esters or another potent PKC agonist, bryostatin. Both proteasome-sensitive and –insensitive pathways have been proposed to regulate PKC degradation. The specific machinery controlling the degradation of unprocessed or activated PKC remains to be elucidated. Chen et al., 2007 from Dr. Alexandra C. Newton’s laboratory discovered the role of novel E3 ubiquitin ligase in ubiquitination and degradation of PKC isozymes. Increased PKCε expression level in aggressive cancers may be result of increased PKCε synthesis and/or decreased degradation. Decreased degradation may be the result of either decreased expression of E3 ligase (RINCK1) or/and lack of recognition of PKCε by E3 ligase. Many RING domain proteins such as MDM2, C-Chol, a BRCA1, have intrinsic E3 ligase activities and regulate cellular proteins. It is also notable that breast cancer-associated gene-2 (BCA2), a novel RING domain protein, has E3 ubiquitin ligase activity and correlates with the outcome in invasive breast cancer (Burger et al., 2005; 2006).
The novel finding is the observation that PKCε is an initial signal that regulates human cancer cell invasion (Figure 3d). This is accomplished via phosphorylation of Stat3Ser727. The experiments involving use of PKCε specific siRNA provide unequivocal evidence that PKCε-mediated Stat3Ser727 phosphorylation is the key event in the constitutive activation of Stat3, Stat3-DNA binding, and Stat3-regulated gene expression.
PKCε is linked to the induction and progression of human cancers (Aziz et al., 2007a, c; Griner and Kazanietz, 2007). PKCε accomplishes its oncogenic role via mediation of anti-apoptotic and pro-survival signals (Basu and Sivaprasad, 2007; Griner and Kazanietz, 2007). We were the first to discover that PKCε signals oncogenic activity through activation of Stat3 (Aziz et al., 2007a, b). Stat3 is a protein partner of PKCε. Stat3 is constitutively activated in human cancers. PKCε interacts with Stat3 to phosphorylate Stat3Ser727 which is essential for Stat3 transcription activity and cell invasion. The results of reciprocal/blotting experiments (Figure 1 and Figure 2) clearly illustrate that PKCε physically interacts with Stat3. PKCε-Stat3 interaction was observed in various human cancer cell lines, implying that PKCε activation may be an initial signal in the constitutive activation of Stat3 in wide-variety of human cancers.
Depending upon the cellular context, Stat3 has been shown to be a substrate for several protein kinases (Jain et al., 1999). Stat3 has been shown to be an in vitro substrate for MAP kinase and/or ERKs, which raises the possibility that Stat3 integrates signals from MAP kinases. Members of the MAPK and JNK families of serine kinases may mediate serine 727 phosphorylation (Xuan et al., 2005). Pioneering research from Dr. Zigang Dong’s laboratory shows that UV-induced Ser727 phosphorylation in both Stat1 and Stat3 involves the integration of multiple kinase pathways (Zhang et al., 2001, 2003; Zykova et al., 2005). Our results indicate, using PKCε RNA interference experiments, that PKCε-mediated Stat3Ser727 phosphorylation involves integration of the MAPK cascade (Raf-1, MEK1/2, and ERK1/2) (Figures 4a–c).
In summary, PKCε activation is an initial signal in the constitutive activation of Stat3 that is observed in a wide variety of human cancer cells (Figure 1–Figure 3). PKCε integrates with MAPK cascade to phosphorylate Stat3Ser727 (Figure 4 and Figure 5). PKCε-mediated Stat3Ser727 phosphorylation is essential for constitutive activation of Stat3 (Figure 3). PKCε is constitutively activated in human cancers (Figure 1 and Figure 2) and is linked to cancer invasion (Figure 3). We conclude that PKCε is an initial signal which directs its partner Stat3 to maintain the invasive cancer. PKCε and Stat3 are potential targets for human cancer prevention and treatment.
Materials and Methods
Materials
The antibodies and their sources used in this study were: PKCε, Stat3, pStat3Tyr705, Bcl-xL, COX-2, β-actin, donkey anti-goat immunoglobulin (IgG)-FITC for PKCε and donkey anti-rabbit IgG-rhodamine for Stat3 (Santa Cruz Biotechnology, Santa Cruz, CA); and pStat3Ser727 (BD Biosciences, San Jose, CA). Double-stranded Stat3 consensus DNA binding motif 5'-GATCCTTCTGGGAATTCCTAGATC-3' was obtained from Santa Cruz Biotechnology, Santa Cruz, CA. PKCε-siRNA and siRNA transfection reagents were purchased from Dharmacon, Inc., Lafayette, CO. PKCε-siRNA plasmid was purchased from Ambion, Austin, TX. ERK1/2 siRNA, non-targeting siRNA and transfection reagent were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Collagen-Based Cell Invasion Assay kit was from Millipore, Temecula, CA.
Cell lines
Various human cancer cell lines: skin melanoma (MeWo and WM266-4), glioma (T98G and MO59K), bladder (RT-4), colon (Caco-2), lung (H1650), pancreatic (PANC-1), and breast (MCF-7 and MDA:MB-231) were obtained from ATCC (Manassas, VA). Cancer cell lines were cultured as follows: Skin melanomas (MeWo and WM266-4) were grown in MEM containing 10% fetal bovine serum and 1% penicillin-streptomycin), gliomas T98G was grown in MEM containing 10% fetal bovine serum and 1% penicillin-streptomycin and MO59K was grown in a 50:50 solution of DMEM and Ham’s F12 containing 15 mmol/L HEPES, 10% fetal bovine serum and 1% penicillin-streptomycin, bladder RT4 cell line was grown in McCoy’s 5a Medium containing 10% fetal bovine serum and 1% penicillin-streptomycin and UM-UC-3 was grown in MEM containing 10% fetal bovine serum and 1% penicillin-streptomycin. Colon Caco-2 cell line was grown in MEM containing 20% fetal bovine serum and 1% penicillin-streptomycin, lungs H1650 cell line was grown in RPMI-1640 medium containing 10% fetal bovine serum and 1% penicillin-streptomycin), pancreatic PANC-1 cell line was grown in DMEM containing 10% fetal bovine serum and 1% penicillin-streptomycin, and breast MCF-7 cells were grown in MEM containing 0,01% bovine insulin, 10% fetal bovine serum and 1% penicillin-streptomycin and MDA:MB-231 cells were grown in Leibovitz’s L-15 medium containing 10% fetal bovine serum and 1% penicillin-streptomycin. All the cells were incubated at 5% Carbon dioxide (CO2), 37°C except MDA:MB-231, which was incubated at 37°C without CO2.
Western Blot Analysis
Indicated human cancer cells were lysed in immunoprecipitation (IP) lysis buffer (50 mM N-2-hydroxyethylpiperazine-N’-2-ethanesulfonic acid [HEPES, pH 7.5], 150 mM NaCl, 10% glycerol, 1% Triton X-100, 1.5 mM MgCl2, 10 µg/ml aprotinin, 10 µg/ml leupeptin, 1 mM phenylmethylsulfonyl fluoride [PMSF], 200 mM Na3VO4, 200 mM NaF and 1 mM EGTA). The homogenate was centrifuged at 14 000×g for 30 min at 4°C. Twenty five µg of whole cell lysate was fractionated on on 10–15% Tris-glycine SDS-polyacrylamide gels for western blot analysis as described before (Aziz et al., 2007a, c).
Determination of PKCε and Stat3 localization by double immunofluorescence staining
Formalin-fixed human cancer cells were used to determine the localization of PKCε and Stat3 as described before (Aziz et al., 2007a, c).
Transfection
Cells were transiently transcfected with PKCε-siRNA (2µmol/L/100-mm Petri dish), PKCε-siRNA plasmid (15 µg/dish) as per the manufacturer’s instruction (Dharmacon Inc., Lafayette). For ERK1/2 siRNA transfection, DU-145 cells were serum starved with for 24 hrs prior to transfection. Cells were transiently transfected with (120 nmoles of ERK1 siRNA+ 120 nmoles ERK2 siRNA) or 240 nmoles of non-targeting siRNA and were harvested 48 hrs post-transfection. Whole cell lysates were made and 25 µg of protein was subjected for Western blot analysis.
Cell invasion assay
Cell invasion was assayed using a Collagen-Based Cell Invasion Assay kit as per the manufacturer's instructions (Millipore, Temecula, CA). Briefly, WM266-4 (melanoma), PANC-1 (pancreatic), Caco-2 (colon) and H1650 (lung) cancer cells at 80% confluency were serum starved 18 to 24 h before the assay. The cells were harvested and the pellets was gently resuspended in serum-free medium. In the upper chamber, 0.5 × 106 cells per well were plated in triplicates and incubated for 2 h at 37°C in a humidified incubator with 5% CO2 before PKCε specific siRNA plasmid transfection. Both the insert and the holding well were subjected to the same medium composition with the exception of serum. The insert contained no serum, whereas the lower well contained 10% fetal bovine serum that served as a chemoattractant. 15 µg of PKCε specific siRNA plasmid was then transfected into the cells. An equal amount of non-targeting siRNA plasmid was used as a control (Ambion, Austin, TX). Forty-eight hours after siRNA transfection, the cell invasion assay was done as per the manufacturer's instructions. The cells in the insert were removed by wiping gently with a cotton swab. Migrated cells sticking to the bottom side of the insert were stained with Cell Stain. Invading cells on the bottom side of the membrane were photographed using a light inverted microscopy (Nikon Eclipse TS 100) at 40× magnification. In addition, the number of cells migrated to the bottom side was estimated by colorimetric measurements at 560 nm. Mean ± SE was calculated from three independent experiments.
Electrophoretic mobility shift assay (EMSA)
Nuclear protein extracts from the indicated cells were prepared by lysing cells in a hypotonic solution (10 mM HEPES, pH 7.5; 10 mM KCl; 0.1 mM EDTA, pH 8.0; 0.1 mM EGTA pH 8.0; 1 mM DTT; 0.5 mM PMSF; 0.5 mg/ml benzamide; 2 µg/ml aprotinin; 2 µg/ml leupeptin), with detergent (NP-40 at 6.25% (v/v)) followed by low speed (1500 × g for 30 secs) to collect nuclei. Nuclear proteins were extracted in a high-salt buffer (20 mM HEPES, pH 7.5; .4 M NaCl; 1 mM EDTA, pH 8.0; 1 mM EGTA pH 8.0; 1 mM DTT; 1 mM PMSF; 0.5 mg/ml benzamide; 2 µg/ml aprotinin; 2 µg/ml leupeptin) and incubated on ice for 15 mins; nuclear membranes and genomic DNA removed by high-speed (16,000×g) centrifugation at 4°C for 5 minutes. The nuclear protein extract was incubated in a final volume of 20 µl of 10 mM HEPES (pH 7.9), 80 mM NaCl, 10% glycerol, 1 mM DTT, 1 mM EDTA, and 100 µg/ml poly(deoxyinosinic-deoxycytidylic acid) for 15 minutes. g-p32-radiolabeled double-stranded oligonucleotides of the consensus binding sequences of Stat3, were then added and the complexes were incubated for 20 minutes at room temperature. The protein-DNA complexes were resolved on a 4.5% acrylamide gel containing, 2.5% glycerol and 0.5X Tris-borate EDTA at room temperature. Gels were dried and autoradiographed to determine binding activity (Aziz et al., 2007a)
Acknowledgements
This work was supported in parts by DOD Grant W81XWH and NIH Grant CA35368 to AKV.
Abbreviations
- PCa
Prostate cancer
- STAT
signal transducers and activators of transcription
- TRAMP
Transgenic Adenocarcinoma of Mouse Prostate
- TPA
12-O-tetradecanoylphorbol-13-acetate
- PKC
Protein Kinase C
- PS
Phosphatidylserine
- DAG
diacylglycerol
- EGFR
epidermal growth factor receptor
- SCC
squamous cell carcinoma
Footnotes
Conflict of Interest
No conflict of Interest.
References
- Akira S. Roles of STAT3 defined by tissue-specific gene targeting. Oncogene. 2000;19:2607–2611. doi: 10.1038/sj.onc.1203478. [DOI] [PubMed] [Google Scholar]
- Alvarez JV, Febbo PG, Ramaswamy S, Loda M, Richardson A, Frank DA. Identification of a genetic signature of activated signal transducer and activator of transcription 3 in human tumors. Cancer Res. 2005;65:5054–5062. doi: 10.1158/0008-5472.CAN-04-4281. [DOI] [PubMed] [Google Scholar]
- Aziz MH, Manoharan HT, Church DR, Dreckschmidt NE, Zhong W, Oberley TD, et al. Protein kinase Cepsilon interacts with signal transducers and activators of transcription 3 (Stat3), phosphorylates Stat3Ser727, and regulates its constitutive activation in prostate cancer. Cancer Res. 2007a;67:8828–8838. doi: 10.1158/0008-5472.CAN-07-1604. [DOI] [PubMed] [Google Scholar]
- Aziz MH, Manoharan HT, Sand JM, Verma AK. Protein kinase Cepsilon interacts with Stat3 and regulates its activation that is essential for the development of skin cancer. Mol Carcinog. 2007b;46:646–653. doi: 10.1002/mc.20356. [DOI] [PubMed] [Google Scholar]
- Aziz MH, Manoharan HT, Verma AK. Protein kinase C epsilon, which sensitizes skin to sun's UV radiation-induced cutaneous damage and development of squamous cell carcinomas, associates with Stat3. Cancer Res. 2007c;67:1385–1394. doi: 10.1158/0008-5472.CAN-06-3350. [DOI] [PubMed] [Google Scholar]
- Bae KM, Wang H, Jiang G, Chen MG, Lu L, Xiao L. Protein kinase C epsilon is overexpressed in primary human non-small cell lung cancers and functionally required for proliferation of non-small cell lung cancer cells in a p21/Cip1-dependent manner. Cancer Res. 2007;67:6053–6063. doi: 10.1158/0008-5472.CAN-06-4037. [DOI] [PubMed] [Google Scholar]
- Basu A, Sivaprasad U. Protein kinase Cepsilon makes the life and death decision. Cell Signal. 2007;19:1633–1642. doi: 10.1016/j.cellsig.2007.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger A, Amemiya Y, Kitching R, Seth AK. Novel RING E3 ubiquitin ligases in breast cancer. Neoplasia. 2006;8:689–695. doi: 10.1593/neo.06469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger AM, Gao Y, Amemiya Y, Kahn HJ, Kitching R, Yang Y, et al. A novel RING-type ubiquitin ligase breast cancer-associated gene 2 correlates with outcome in invasive breast cancer. Cancer Res. 2005;65:10401–10412. doi: 10.1158/0008-5472.CAN-05-2103. [DOI] [PubMed] [Google Scholar]
- Chan KS, Sano S, Kiguchi K, Anders J, Komazawa N, Takeda J, et al. Disruption of Stat3 reveals a critical role in both the initiation and the promotion stages of epithelial carcinogenesis. J Clin Invest. 2004;114:720–728. doi: 10.1172/JCI21032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Gould C, Garza R, Gao T, Hampton RY, Newton AC. Amplitude control of protein kinase C by RINCK, a novel E3 ubiquitin ligase. J Biol Chem. 2007;282:33776–33787. doi: 10.1074/jbc.M703320200. [DOI] [PubMed] [Google Scholar]
- Cornford P, Evans J, Dodson A, Parsons K, Woolfenden A, Neoptolemos J, et al. Protein kinase C isoenzyme patterns characteristically modulated in early prostate cancer. Am J Pathol. 1999;154:137–144. doi: 10.1016/S0002-9440(10)65260-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker T, Kovarik P. Serine phosphorylation of STATs. Oncogene. 2000;19:2628–2637. doi: 10.1038/sj.onc.1203481. [DOI] [PubMed] [Google Scholar]
- Griner EM, Kazanietz MG. Protein kinase C and other diacylglycerol effectors in cancer. Nat Rev Cancer. 2007;7:281–294. doi: 10.1038/nrc2110. [DOI] [PubMed] [Google Scholar]
- Hodge DR, Hurt EM, Farrar WL. The role of IL-6 and STAT3 in inflammation and cancer. Eur J Cancer. 2005;41:2502–2512. doi: 10.1016/j.ejca.2005.08.016. [DOI] [PubMed] [Google Scholar]
- Jain N, Zhang T, Kee WH, Li W, Cao X. Protein kinase C delta associates with and phosphorylates Stat3 in an interleukin-6-dependent manner. J Biol Chem. 1999;274:24392–24400. doi: 10.1074/jbc.274.34.24392. [DOI] [PubMed] [Google Scholar]
- Jansen AP, Verwiebe EG, Dreckschmidt NE, Wheeler DL, Oberley TD, Verma AK. Protein kinase C-epsilon transgenic mice: a unique model for metastatic squamous cell carcinoma. Cancer Res. 2001;61:808–812. [PubMed] [Google Scholar]
- Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- Kazanietz MG. Eyes wide shut: protein kinase C isozymes are not the only receptors for the phorbol ester tumor promoters. Mol Carcinog. 2000;28:5–11. doi: 10.1002/(sici)1098-2744(200005)28:1<5::aid-mc2>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- Klampfer L. Signal transducers and activators of transcription (STATs): Novel targets of chemopreventive and chemotherapeutic drugs. Curr Cancer Drug Targets. 2006;6:107–121. doi: 10.2174/156800906776056491. [DOI] [PubMed] [Google Scholar]
- Kobielak A, Fuchs E. Links between alpha-catenin, NF-kappaB, and squamous cell carcinoma in skin. Proc Natl Acad Sci U S A. 2006;103:2322–2327. doi: 10.1073/pnas.0510422103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortylewski M, Jove R, Yu H. Targeting STAT3 affects melanoma on multiple fronts. Cancer Metastasis Rev. 2005;24:315–327. doi: 10.1007/s10555-005-1580-1. [DOI] [PubMed] [Google Scholar]
- Li L, Shaw PE. A STAT3 dimer formed by inter-chain disulphide bridging during oxidative stress. Biochem Biophys Res Commun. 2004;322:1005–1011. doi: 10.1016/j.bbrc.2004.08.014. [DOI] [PubMed] [Google Scholar]
- Mellor H, Parker PJ. The extended protein kinase C superfamily. Biochem J. 1998;332(Pt 2):281–292. doi: 10.1042/bj3320281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochly-Rosen D, Kauvar LM. Modulating protein kinase C signal transduction. Adv Pharmacol. 1998;44:91–145. doi: 10.1016/s1054-3589(08)60126-x. [DOI] [PubMed] [Google Scholar]
- Newton AC. Protein kinase C: structural and spatial regulation by phosphorylation, cofactors, and macromolecular interactions. Chem Rev. 2001;101:2353–2364. doi: 10.1021/cr0002801. [DOI] [PubMed] [Google Scholar]
- Nikitakis NG, Siavash H, Sauk JJ. Targeting the STAT pathway in head and neck cancer: recent advances and future prospects. Curr Cancer Drug Targets. 2004;4:637–651. doi: 10.2174/1568009043332736. [DOI] [PubMed] [Google Scholar]
- Okhrimenko H, Lu W, Xiang C, Hamburger N, Kazimirsky G, Brodie C. Protein kinase C-epsilon regulates the apoptosis and survival of glioma cells. Cancer Res. 2005;65:7301–7309. doi: 10.1158/0008-5472.CAN-05-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q, Bao LW, Kleer CG, Sabel MS, Griffith KA, Teknos TN, et al. Protein kinase C epsilon is a predictive biomarker of aggressive breast cancer and a validated target for RNA interference anticancer therapy. Cancer Res. 2005;65:8366–8371. doi: 10.1158/0008-5472.CAN-05-0553. [DOI] [PubMed] [Google Scholar]
- Pan Q, Bao LW, Teknos TN, Merajver SD. Targeted disruption of protein kinase C epsilon reduces cell invasion and motility through inactivation of RhoA and RhoC GTPases in head and neck squamous cell carcinoma. Cancer Res. 2006;66:9379–9384. doi: 10.1158/0008-5472.CAN-06-2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddig PJ, Dreckschmidt NE, Zou J, Bourguignon SE, Oberley TD, Verma AK. Transgenic mice overexpressing protein kinase C epsilon in their epidermis exhibit reduced papilloma burden but enhanced carcinoma formation after tumor promotion. Cancer Res. 2000;60:595–602. [PubMed] [Google Scholar]
- Rivat C, Rodrigues S, Bruyneel E, Pietu G, Robert A, Redeuilh G, et al. Implication of STAT3 signaling in human colonic cancer cells during intestinal trefoil factor 3 (TFF3) -- and vascular endothelial growth factor-mediated cellular invasion and tumor growth. Cancer Res. 2005;65:195–202. [PubMed] [Google Scholar]
- Stephanou A, Latchman DS. Opposing actions of STAT-1 and STAT-3. Growth Factors. 2005;23:177–182. doi: 10.1080/08977190500178745. [DOI] [PubMed] [Google Scholar]
- Vinkemeier U. Getting the message across, STAT! Design principles of a molecular signaling circuit. J Cell Biol. 2004;167:197–201. doi: 10.1083/jcb.200407163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler DL, Li Y, Verma AK. Protein kinase C epsilon signals ultraviolet light-induced cutaneous damage and development of squamous cell carcinoma possibly through Induction of specific cytokines in a paracrine mechanism. Photochem Photobiol. 2005;81:9–18. doi: 10.1562/2004-08-12-RA-271. [DOI] [PubMed] [Google Scholar]
- Wheeler DL, Martin KE, Ness KJ, Li Y, Dreckschmidt NE, Wartman M, et al. Protein kinase C epsilon is an endogenous photosensitizer that enhances ultraviolet radiation-induced cutaneous damage and development of squamous cell carcinomas. Cancer Res. 2004;64:7756–7765. doi: 10.1158/0008-5472.CAN-04-1881. [DOI] [PubMed] [Google Scholar]
- Wheeler DL, Ness KJ, Oberley TD, Verma AK. Protein kinase Cepsilon is linked to 12-O-tetradecanoylphorbol-13-acetate-induced tumor necrosis factor-alpha ectodomain shedding and the development of metastatic squamous cell carcinoma in protein kinase Cepsilon transgenic mice. Cancer Res. 2003;63:6547–6555. [PubMed] [Google Scholar]
- Wu D, Foreman TL, Gregory CW, McJilton MA, Wescott GG, Ford OH, et al. Protein kinase cepsilon has the potential to advance the recurrence of human prostate cancer. Cancer Res. 2002;62:2423–2429. [PubMed] [Google Scholar]
- Wu D, Thakore CU, Wescott GG, McCubrey JA, Terrian DM. Integrin signaling links protein kinase Cepsilon to the protein kinase B/Akt survival pathway in recurrent prostate cancer cells. Oncogene. 2004;23:8659–8672. doi: 10.1038/sj.onc.1207900. [DOI] [PubMed] [Google Scholar]
- Xuan YT, Guo Y, Zhu Y, Wang OL, Rokosh G, Messing RO, et al. Role of the protein kinase C-epsilon-Raf-1-MEK-1/2-p44/42 MAPK signaling cascade in the activation of signal transducers and activators of transcription 1 and 3 and induction of cyclooxygenase-2 after ischemic preconditioning. Circulation. 2005;112:1971–1978. doi: 10.1161/CIRCULATIONAHA.105.561522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Cho YY, Petersen BL, Bode AM, Zhu F, Dong Z. Ataxia telangiectasia mutated proteins, MAPKs, and RSK2 are involved in the phosphorylation of STAT3. J Biol Chem. 2003;278:12650–12659. doi: 10.1074/jbc.M210368200. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu G, Dong Z. MSK1 and JNKs mediate phosphorylation of STAT3 in UVA-irradiated mouse epidermal JB6 cells. J Biol Chem. 2001;276:42534–42542. doi: 10.1074/jbc.M106044200. [DOI] [PubMed] [Google Scholar]
- Zykova TA, Zhang Y, Zhu F, Bode AM, Dong Z. The signal transduction networks required for phosphorylation of STAT1 at Ser727 in mouse epidermal JB6 cells in the UVB response and inhibitory mechanisms of tea polyphenols. Carcinogenesis. 2005;26:331–342. doi: 10.1093/carcin/bgh334. [DOI] [PubMed] [Google Scholar]