Abstract
In order for the cell's genome to be passed intact from one generation to the next, the events of the cell cycle (DNA replication, mitosis, cell division) must be executed in the correct order, despite the considerable molecular noise inherent in any protein-based regulatory system residing in the small confines of a eukaryotic cell. To assess the effects of molecular fluctuations on cell-cycle progression in budding yeast cells, we have constructed a new model of the regulation of Cln- and Clb-dependent kinases, based on multisite phosphorylation of their target proteins and on positive and negative feedback loops involving the kinases themselves. To account for the significant role of noise in the transcription and translation steps of gene expression, the model includes mRNAs as well as proteins. The model equations are simulated deterministically and stochastically to reveal the bistable switching behavior on which proper cell-cycle progression depends and to show that this behavior is robust to the level of molecular noise expected in yeast-sized cells (∼50 fL volume). The model gives a quantitatively accurate account of the variability observed in the G1-S transition in budding yeast, which is governed by an underlying sizer+timer control system.
Keywords: bistability, cell-cycle variability, size control, stochastic model, transcription–translation coupling
Introduction
The cell cycle is the sequence of events by which a growing cell replicates all its components and divides them more or less evenly between two daughter cells, so that each daughter cell receives the information and machinery necessary to repeat the process (Murray and Hunt, 1993; Morgan, 2007). The most important component to be replicated and partitioned to the daughter cells is the genome. In eukaryotes, the replication and partitioning of DNA molecules occurs in distinct temporal phases. During S phase, each double-stranded DNA molecule (chromosome) is replicated to form a pair of identical sister chromatids, held together by cohesin proteins. During M phase, the cell forms a bipolar mitotic spindle, which captures the replicated chromosomes and aligns them on the metaphase plate with one chromatid attached to one pole of the spindle and its sister attached to the other pole, with the cohesin proteins maintaining tension between the two. When the replicated chromosomes are all properly aligned on the mitotic spindle, the cell activates a protease (called ‘separase’) that cleaves cohesin, thereby promoting separation of the sister chromatids to opposite poles of the spindle (a process called anaphase). Subsequently, in telophase, the cell divides to form two daughter cells, each containing precisely one copy of every chromosome.
Three fundamental properties characterize mitotic cell cycles in eukaryotes. First, the processes of DNA synthesis and mitosis must alternate with each other for a cell lineage to maintain its ploidy generation after generation. (That there exist exceptional polyploid tissues does not detract from the general applicability of this rule.) In a typical eukaryotic cell cycle, S and M phases are separated by two gap phases: G1-S-G2-M. Second, the alternating cycle of S and M must be balanced with other biosynthetic processes that achieve two-fold increases of all other cell components. To maintain a stable and favorable DNA-to-size ratio, a cell lineage must ensure that, on average, the period of the DNA replication-division cycle is exactly balanced by the mass-doubling time of the lineage. Third, a cell must be able to halt progression through the cell cycle if any problems arise in the processes of DNA synthesis or mitosis. For example, if DNA is damaged by radiation or chemical mutagens in G1, a cell will delay entry into S phase until the damage is repaired. If completion of DNA synthesis is delayed for any reason, a cell will not enter mitosis prematurely. If replicated chromosomes have trouble aligning on the mitotic spindle, cohesins will not be cleaved prematurely. These ‘checkpoint’ mechanisms are essential in maintaining the integrity of the genome from one generation to the next.
An overriding property of this control system is robust operation in the face of molecular noise. Cell-cycle events are controlled by a network of genes, mRNAs and proteins that react with one another within the small confines of a single cell. A yeast cell, with a volume measured in femtoliters (10−15 l) contains, typically, one copy of the gene for each element of the network, a handful of mRNA molecules transcribed from that gene, and a few hundreds to thousands of protein molecules carrying out the gene's function. With such small numbers of molecules, considerable fluctuations in molecule numbers are unavoidable. The control machinery must carry out its three crucial functions reliably and robustly in the face of this noise.
Although cell-cycle progression is robust in respect to its fundamental properties, it is sloppy in many other respects. Yeast cells vary considerably in age and size at division; apparently it does not matter so much how long the cycle takes or how big the cell is when it finally divides, as long as its DNA is accurately replicated and precisely partitioned during mitosis. Variability in age and size at division is due partly to intrinsic noise in the molecular regulatory system and partly to extrinsic noise such as variability in the cell's local environment and inequities in the cell-division process.
Experiments can delineate precisely how variable are measurable properties of the cell cycle, but they cannot fully explain the molecular basis of robust-yet-variable progression through the cell cycle. To that end, one must build a biophysical model that accurately accounts for both intrinsic and extrinsic sources of noise. Of the two, intrinsic noise is more difficult to model adequately. The problem is that the molecular regulatory network is complicated and its components are present in widely different abundances. Of the many ways to describe molecular fluctuations, it is not entirely clear which theoretical approach to choose. Sveiczer et al (2001) added extrinsic noise by introducing sloppiness in the division process in a deterministic cell-cycle model. Steuer (2004) added Gaussian white noise to a deterministic model in order to explore the role of intrinsic noise in the cell cycle. Zhang et al (2006), Braunewell and Bornholdt (2007), Okabe and Sasai (2007) and Ge et al (2008)have developed stochastic models of the yeast cell cycle based on a deterministic Boolean model proposed by Li et al (2004). These models explored the robustness of the cell-cycle control network under the influence of stochastic fluctuations, but they were not compared quantitatively to experimentally observed statistics of the yeast cell cycle.
A major obstacle to building a quantitatively accurate stochastic model of the cell cycle is choosing rate laws for the reactions of the regulatory network. Cell-cycle regulation relies on positive feedback loops that create bistable switches and negative feedback loops that flip the switches on and off (Tyson and Novak, 2008). To generate these dynamic control properties, the reaction network must exhibit a certain amount of ‘nonlinearity’ in the reaction kinetics. The source of this nonlinearity is unclear. Many modelers, Novak and Tyson chief among them, have eschewed mass-action kinetics and used highly nonlinear, phenomenological rate laws like Hill kinetics or zero-order ultrasensitivity (Goldbeter and Koshland, 1981). These phenomenological rate laws violate the assumptions underlying Gillespie's stochastic simulation algorithm (SSA), which is widely used to predict the effects of molecular noise when molecule numbers are low (Gillespie, 1976). (Gillespie's algorithm assumes that reaction rates are described by mass-action kinetics.)
In this situation, we have two options: (1) to use SSA with propensities computed from phenomenological rate laws instead of mass-action kinetics, to gain some impression of the effects of molecular noise on the control system, even though this approach may not produce quantitatively accurate results or (2) to replace the phenomenological rate laws by more complex sets of pseudo-elementary (mass-action) reactions that can be reliably simulated by SSA. Mura and Csikasz-Nagy (2008) have chosen the first option, applying SSA to a comprehensive deterministic model of the budding yeast cell cycle proposed by Chen et al (2004). Sabouri-Ghomi et al (2008) and later Kar et al (2009) have taken the second approach, exploring the roles of noise in a ‘toy’ model of the budding yeast cell cycle (Tyson and Novak, 2001), which can be reformulated in pseudo-elementary reactions. In particular, Kar et al (2009) studied the effects of intrinsic and extrinsic noise on the statistics of size and cycle time distributions in yeast cell populations. They concluded that intrinsic noise makes a considerably greater contribution to cell-cycle variability than extrinsic noise, and that mRNA abundances are the major source of intrinsic noise. To keep variability within observed bounds, Kar et al had to assume that mRNA molecules are more abundant and less stable than implied by recent high-throughput studies of budding yeast mRNAs (Arava et al, 2003; http://web.wi.mit.edu/young/expression/halflife2.html).
We set out to extend these results by developing a more comprehensive model of the cell-cycle control network, based only on mass-action kinetics. We did not follow the approach of Sabouri-Ghomi et al (2008) and Kar et al (2009), who ‘unpacked’ the Michaelis–Menten kinetics in earlier models. Instead, we have followed the lead of Qu et al (2003), who proposed that multisite phosphorylation of target proteins by cyclin-dependent kinase (CDK) proteins is the likely source of nonlinear kinetic effects in cell-cycle control mechanisms. This idea has been clearly explicated by Kapuy et al (2009), who noted that multisite phosphorylation sequences may be modeled by mass-action rate laws that are suitable for either deterministic simulation (by stiff integrators) or stochastic simulation (by SSA). In this paper, we implement a generic model of cell-cycle controls (Tyson and Novak, 2008), using multisite phosphorylation sequences wherever appropriate. Using realistic estimates of mRNA and protein abundances, we carry out exact stochastic simulations of noise in various phases of the cell cycle and compare our results to recent experimental measurements (Di Talia et al, 2007) of variability in progression through G1 phase in budding yeast cells.
The budding yeast cell cycle
In order to place our results in context, we briefly summarize some specific details of the physiology and molecular biology of the budding yeast cell cycle (for more details see Pringle and Hartwell, 1981; Nasmyth, 1996; Lew et al, 1997; Mendenhall and Hodge, 1998). Saccharomyces cerevisiae has an unusual style of growth and division. Mother cells produce buds that balloon out from their sides. As the bud grows, the mother cell replicates its chromosomes. Mitosis occurs in the neck between mother and bud. At anaphase, one set of sister chromatids goes to the mother cell, and the other set goes to the bud. The cell divides at the neck to produce a large mother cell and a small daughter cell. Soon after birth, the mother cell repeats the process. The daughter cell, on the other hand, has a long G1 period before producing her first bud and entering S phase. Years ago, Hartwell et al (1974) identified this characteristic commitment step in the budding yeast cell cycle (bud initiation, onset of DNA synthesis and spindle pole body duplication) and called it ‘START.’
In budding yeast, the central regulator of the cell cycle is a cyclin-dependent protein kinase (Cdc28) encoded by the CDC28 gene. The activity of Cdc28 depends on the availability of a regulatory partner, a cyclin molecule of type Cln1–3 or Clb1–6. When associated with cyclin, Cdc28 phosphorylates different target proteins and thereby triggers crucial events of the cell cycle. Right after birth, in early G1 phase, only Cln3 is available to partner with Cdc28. When enough of this particular dimer is formed (Polymenis and Schmidt, 1997), it activates two transcription factors, SBF, a heterodimer of Swi4 and Swi6 (Tyers et al, 1993), and MBF, a heterodimer of Mbp1 and Swi6 (Wijnen et al, 2002). These transcription factors drive production of Cln1,2 and Clb5,6 proteins (Dirick and Nasmyth, 1991; Koch et al, 1993). In early G1 phase, SBF is not active because it is sequestered by Whi5 (de Bruin et al, 2004; Costanzo et al, 2004). As Cln3–Cdc28 complex accumulates beyond a threshold level, it phosphorylates Whi5 multiple times (there are 12 consensus CDK phosphorylation sites in Whi5 and 10 are phosphorylated in vivo; Wagner et al, 2009). Phosphorylated Whi5 translocates from nucleus to cytoplasm, where it is no longer available to bind to and inhibit SBF (de Bruin et al, 2004; Costanzo et al, 2004). How MBF activity is regulated is still poorly understood; it may be through phosphorylation of Swi6 by Cln3-dependent kinase (Wijnen et al, 2002). The four ‘start cyclins’ (Cln1,2 and Clb5,6) transcribed by SBF and MBF initiate the processes of budding, spindle pole body duplication and DNA replication.
These start cyclins also inactivate Sic1 and Cdh1 (Schneider et al, 1996; Zachariae et al, 1998), which are inhibitors of Clb1-6 in G1 phase. Sic1 is a stoichiometric inhibitor of Clb–Cdc28 complexes (Schwob et al, 1994). Start cyclins phosphorylate Sic1 multiple times, and once Sic1 is hyperphosphorylated, it is degraded by SCF-mediated proteolysis. Degradation of Sic1 leads to increasing levels of Clb5 and -6, which initiate DNA synthesis in budding yeast (Verma et al, 1997). Cdh1, in conjunction with the anaphase promoting complex (APC), polyubiquitinates Clb proteins, causing them to be rapidly degraded by proteasomes (Visintin et al, 1997; Schwab et al, 1997). Start cyclins phosphorylate Cdh1 multiple times (there are 11 CDK phosphorylation sites in Cdh1), preventing its binding with Clb1 and -2. Inactivation of Cdh1 allows Clb1,2 levels to rise to initiate mitosis.
After Cdh1 is depressed and Sic1 is degraded, the rising activity of Clb1,2–Cdc28 initiates mitotic events and also inactivates SBF (Amon et al, 1993; Koch et al, 1996), causing a loss of Cln1,2 from the cell. Clb1,2-kinase also inhibits MBF activity, but complete inactivation of MBF requires a corepressor, Nrm1 (de Bruin et al, 2006). As the level of start cyclins drops, Cdh1 does not reactivate, because Clb1,2-dependent kinase keeps Cdh1 phosphorylated and inactive. At the metaphase–anaphase transition, when all chromosomes are aligned on the metaphase plate, another ancillary protein of the APC, Cdc20, is activated. Among the substrates of APC-Cdc20 are Pds1 (an inhibitor of the protease that cleaves cohesin) and Clb1-6 (Lim et al, 1998; Shirayama et al, 1999; Yeong et al, 2000). Clb1,2 protein stores are only partially depleted by Cdc20-mediated proteolysis in anaphase. Their complete removal in telophase requires activation of Cdh1, which is the job of a crucial phosphatase, Cdc14 (Visintin et al, 1998). During most of the cell cycle, Cdc14 is sequestered in the nucleolus, bound to Net1 in a complex known as regulator of nucleolar silencing and telophase (RENT) (Shou et al, 1999; Visintin et al, 1999). At anaphase, Clb1,2-kinase phosphorylates Net1 multiple times (there are six CDK phosphorylation sites in Net1; Azzam et al, 2004), resulting in the release of Cdc14 from RENT. Cdc14 moves into both nucleus and cytoplasm, where it promotes re-establishment of G1 phase by two mechanisms: (1) it dephosphorylates and activates Cdh1 (Jaspersen et al, 1999) and (2) it activates Sic1 by promoting its synthesis and inhibiting its degradation (Visintin et al, 1998). Active Cdh1 finishes the destruction of Clb1 and -2, and Sic1 inhibits any remaining Clb-dependent kinase activity. As a result, cell division is triggered, and the mother and daughter cells are returned to G1 phase. The only cyclin that remains in early G1 is Cln3.
The model
Di Talia et al (2007) made careful observations of single budding yeast cells to measure the variability of specific cell-cycle events and to investigate the mechanisms controlling the START transition. (They defined START as the activation of SBF in late G1 phase.) By mathematical modeling of the molecular events controlling the yeast cell cycle, we seek to reconcile the variability observed by Di Talia et al in cell size and age at START and at the G1-S transition with the variability expected of the macromolecular regulatory network in a yeast-sized cell, with ∼10 mRNA molecules and ∼1000 protein molecules per gene involved in the network.
The model we propose is based on a general theory of cell-cycle organization presented in Chen et al (2004) and Tyson and Novak (2008). In their view, the mitotic B-type cyclins (Clb1,2) are involved in a power struggle with G1-phase stabilizers (Cdh1 and Sic1). As described in the previous section, Cdh1 degrades Clb1,2 and Sic1 inhibits Clb1,2-kinase activity, whereas the kinases phosphorylate both Cdh1 and Sic1 and nullify their effects. The mutual antagonism between these two sets of proteins creates a bistable switch, with an ‘OFF’ state (low Clb-dependent kinase activity) corresponding to G1 phase of the cell cycle, and an ‘ON’ state (high Clb-dependent kinase activity) corresponding to S-G2-M phases of the cell cycle. The START transition is driven by upregulation of Cln1,2 and Clb5,6 production, because these cyclins, by opposing Cdh1 and Sic1, flip the switch from OFF to ON. As mitotic Clb-dependent kinases rise, they downregulate the production of Cln1,2 and Clb5,6, which have already served their purpose (this is a negative feedback loop). Exit from mitosis, on the other hand, is accomplished by a different negative feedback loop. High activity of mitotic Clb-kinase promotes the activation of Cdc20 (which degrades Clb1,2) (Zhu et al, 2000; Rudner and Murray, 2000) and Cdc14 (which activates Cdh1 and Sic1) (Azzam et al, 2004), thereby flipping the switch from ON to OFF. As mitotic cyclins are degraded, Cdc14 returns to its inactive state. In the next paragraphs we provide more details about how these feedback loops are implemented in our mathematical model.
To build a mathematical model of any regulatory network, one must introduce certain assumptions and simplifications to limit the complexity of the model while still capturing the molecular details that are relevant to the experiments under consideration. In this paper, we are interested primarily in the statistical properties of populations of budding yeast cells as they were observed to progress, one-by-one, through the cell-division cycle by Di Talia et al (2007, 2009), with special emphasis on a critical event in G1 phase when the cell-cycle inhibitor, Whi5, is phosphorylated and transported out of the nucleus. To this end, we must include the interactions among Whi5, SBF, and the start cyclins (Cln1,2 and Clb5,6) whose levels of expression are controlled by SBF. In this model (Figure 1), we have chosen to combine the start cyclins into a single variable, called ‘ClbS,’ and to ignore the role of MBF, a second transcription factor whose mode of regulation is still poorly understood. A crucial role of the start cyclins, in addition to initiating bud emergence and DNA synthesis, is to flip the bistable switch (described in the previous paragraph) from OFF to ON. To model the bistable switch, we combine the mitotic cyclins into a single variable ‘ClbM’ and the G1-stabilizers (Cdh1 and Sic1) into a single component with the properties of Cdh1.
Figure 1.
Molecular mechanism for cell-cycle control in budding yeast. A solid arrow represents a chemical reaction and a dotted arrow represents an enzymatic activity on a reaction. Dashed arrows represent multisite phosphorylation chains. A T-shaped arrow with balls on the cross bar indicates a reversible binding reaction. Parameter values for multisite phosphorylation/dephosphorylation of regulatory proteins: for Whi5, N=6 and q=2; for SBF, N=4 and q=0; for Cdh1, N=10 and q=0; for Net1, N=8 and q=5. Hi5, Hbf and Ht1 are three unregulated phosphatases, which oppose cyclin-dependent kinases on the Whi5, SBF and Net1 phosphorylation chains, respectively. The synthesis and degradation reactions of Whi5, SBF, Cdh1, Net1, Hbf, Hi5 and Ht1 are not shown here. The diagram includes synthesis and degradation of MbS, the mRNA for ClbS, which is the only regulated mRNA species in the model. The mRNAs encoding all other proteins in the model are assumed to be synthesized and degraded by simple birth-death processes.
As is commonly done in this field, we assume that Cdc28 subunits are always available to bind to any cyclin partners that accumulate in the cell. Hence, the model focuses on the synthesis and degradation of cyclins and ignores fluctuations in Cdc28 level.
Clb3 and -4 may be involved in DNA replication and help to promote the G2/M transition (Richardson et al, 1992), but cells deleted of both CLB3 and CLB4 genes are viable with no detectable phenotype (Fitch et al, 1992). As not much is known about their regulation (Mendenhall and Hodge, 1998), we do not include them in this model of cell-cycle controls.
In order for our model cells to exit mitosis and return to G1, we must propose a simplified version of the ‘mitotic exit network’ (Azzam et al, 2004; Stegmeier and Amon, 2004). To this end we focus attention on the inhibition of Cdc14 by Net1. Mitotic exit in this model (Figure 1) is driven by phosphorylation of Net1, which causes Net1 to release Cdc14, allowing free Cdc14 to flip the bistable switch from ON to OFF. The phosphorylation of Net1 during anaphase and telophase is a complex process involving several kinases (Clb2, Cdc5 and Cdc15) and phosphatases (Cdc14 and Cdc55), an E3-ubiquitin ligase (APC-Cdc20) and a protease (Esp1), and a G-protein signaling network (Tem1) (Queralt et al, 2006). We have reduced this complex machinery to a simple antagonism between ClbM (phosphorylating Net1) and a constitutively active phosphatase (Ht1) whose job is to activate Net1. This highly simplified exit module is sufficient to return mitotic cells to G1 phase, so that we can study the START transition in the next cell cycle.
Figure 1 provides additional details about the production of ClbS at START in our simplified model. In G1 phase, Whi5 binds to and inhibits the transcription factor SBF that controls expression of CLBS. In a newborn cell, Cln3 is the only cyclin available to pair with Cdc28 and phosphorylate Whi5. Cells need a threshold amount Cln3-kinase activity to counteract the effect of a phosphatase (Hi5) that keeps Whi5 in its active (unphosphorylated) form. Newly divided mother cells have enough Cln3 to phosphorylate Whi5 and to start a new cell cycle promptly. (Later we will describe a genetic mechanism that biases mother cells to contain more Cln3 molecules than daughter cells.) Newborn daughter cells are too small to execute START (Whi5 phosphorylation and SBF activation). As the cell grows, enough Cln3 accumulates to prime the phosphorylation of Whi5. Multiply phosphorylated Whi5 moves from the nucleus to the cytoplasm, releasing some SBF to upregulate the synthesis of ClbS. Then ClbS-kinase promotes further phosphorylation of Whi5 and additional release of SBF. This auto-enhancement by ClbS of its own production was recently shown by Skotheim et al (2008) to have a significant function in the START transition.
The cell-division cycle in budding yeast is coupled to cell growth at the START transition (Hartwell and Unger, 1977; Johnston et al, 1977; Lord and Wheals, 1980), primarily through the action of Cln3-kinase (Miller and Cross, 2001; Cross et al, 2002). In order for the level of Cln3 protein to be indicative of cell growth, it must be produced at an accelerated rate relative to other regulatory proteins (in particular, the phosphatase Hi5 in Figure 1). In our model equations (Table I), the synthesis rate of Cln3 protein increases quadratically with cell volume (V2), whereas the synthesis rates of other proteins, for example, ClbS, ClbM, and Cdc14, are proportional to V. This assumption is supported by the fact that mRNAs (like CLN3) with low intrinsic initiation rates experience a proportionally greater enhancement of translation with increasing availability of ribosomal precursors (Lodish, 1974). Furthermore, the translation of CLN3 mRNA molecules is affected by a specific 5′ leader sequence that is sensitive to cell growth rate (Polymenis and Schmidt, 1997). Newcomb et al (2002) showed that the transcription factor AZF is a glucose-dependent positive regulator of CLN3; in the presence of glucose, CLN3 mRNA level rises higher than other mRNAs (e.g. CLN2 mRNA). The V2 factor is meant to reflect these transcriptional/translational controls over Cln3 production rate.
Table 1. Dynamical equations of the model.

|
The bistable switching behavior that underlies progression through the cell cycle in our model relies on the mutual antagonism between ClbM and Cdh1 and on a requirement that the phosphorylation of Cdh1 depends ultrasensitively on the ratio of ClbM kinase activity to Cdc14 phosphatase activity. In earlier models of the yeast cell cycle (Chen et al, 2000, 2004), we relied heavily on zero-order ultrasensitivity (Goldbeter and Koshland, 1981), which depends on saturation of enzyme by substrate in Michaelis–Menten kinetics. In that case, the signaling enzyme (say, the kinase) will be all tied up in the enzyme–substrate complex and will be unavailable to phosphorylate other target proteins. An alternative mechanism to achieve ultrasensitivity is multisite phosphorylation (Gunawardena, 2005; Salazar and Hofer, 2007; Kapuy et al, 2009), which works well in the opposite limit, where enzyme binds only transiently to substrate. The enzyme quickly carries out phosphorylation (or dephosphorylation) and dissociates from the substrate. Multisite phosphorylation can be classified as either processive (each enzyme–substrate encounter results in multiple phosphorylations of the substrate) or distributive (each enzyme–substrate encounter results in a single phosphorylation event). Multiple phosphorylations can happen in a specific order (‘ordered’ phosphorylation) or in a random order (‘disordered’ phosphorylation).
Kapuy et al (2009) have shown that an ordered, distributive, multisite phosphorylation mechanism can produce very robust, ultrasensitive, signal-response curves and bistable switching behavior, and so we adopt this mechanism in our model. Furthermore, we assume a ‘threshold rule’ on the activity of multiphosphorylated proteins, that is, the first q phosphorylation states (0, 1, … q) are identically active (or inactive) and the higher phosphorylation states (q+1, q+2, … N) are identically inactive (or active). Our assigned values of N and q for each multiphosphorylated protein are given in the legend to Figure 1. For example, for Whi5 (N=6, q=2) we assume that the unphosphorylated form (Whi5) and the first two phosphorylated forms (Whi5P1 and Whi5P2) are ‘active,’ that is, they bind to the unphosphorylated form of SBF to form the complexes Cmp, CmpP1 and CmpP2 (the phosphorylation state of Cmp refers to its Whi5 component) and thereby inhibit CLBS transcription. Higher phosphorylation states of the complex do not exist, because, we assume, Whi5P3,…Whi5P6 do not bind to SBF. For SBF (N=4, q=0), which is phosphorylated by ClbM, we assume only the free, unphosphorylated form is an active transcription factor, whereas SBFP1, …, SBFP4 are all inactive forms, as well as SBF molecules in the complexes Cmp, CmpP1 and CmpP2.
The dynamic properties of the model are governed by 60 differential equations (Table I) involving 71 kinetic parameters (Table II). Verbal descriptions of the variables and parameters are provided in Supplementary Tables S1 and S2. The kinetic-parameter values have been estimated from experimental measurements as best we are able. The degradation rate constants for all proteins are calculated from protein half-life measurements done by Belle et al (2006). We set the synthesis rate constant for each protein so that the average number of protein molecules calculated for an asynchronous population of cells closely matches the experimentally measured values in Ghaemmaghami et al (2003). For phosphorylation and dephosphorylation reactions, we do not have experimentally measured rate constants so we assigned what we consider to be realistic values for those parameters, so that the model compares well with the observations of Di Talia et al (2007). We assume that cell size (V) grows exponentially, as indicated in Di Talia et al (2007), with specific growth rate (α=0.007 min−1) corresponding to a mass-doubling time (MDT=ln(2)/α) of about 100 min.
Table 2. Parameter values.
| Parameter | min−1 | Parameter | min−1 | Parameter | min−1 |
|---|---|---|---|---|---|
| α | 0.007 | k s,mbS | 11.5 | k d,c | 0.2 |
| k d,g | 12.0 | k ′ d,h1 | 1.65 | k d,r | 0.075 |
| γbM | 0.03 | γbS | 0.1 | γn3 | 0.14 |
| γbf | 0.01 | γhbf | 0.01 | γc14 | 0.02 |
| γhi5 | 0.01 | γt1 | 0.015 | γi5 | 0.01 |
| γh1 | 0.01 | γmn3 | 1.0 | γht1 | 0.01 |
| γmbS | 0.25 | γmc14 | 0.7 | γmbM | 0.7 |
| γmi5 | 0.7 | γmhi5 | 0.7 | γmhbf | 0.7 |
| γmh1 | 0.7 | γmht1 | 0.7 | γmt1 | 0.7 |
| Parameter | fL molec−1 min−1 | Parameter | fL molec−1 min−1 | Parameter | fL−1 min−1 |
| k d,bMi | 1.7 × 10−5 | k d,bMa | 0.025 | k s,bM | 0.27 |
| k a,c | 0.0417 | k a,r | 0.042 | k s,bS | 0.3 |
| k a,g | 0.25 | k p,h1 | 0.033 | k s,i5 | 0.715 |
| k ′ p,h1 | 0.18 | k p,bf | 0.0625 | k s,h1 | 0.275 |
| k p,i5 | 0.1835 | k ′ p,i5 | 0.0475 | k s,t1 | 0.725 |
| k p,cm | 0.1835 | k ′ p,cm | 0.0475 | k s,c14 | 0.38 |
| k p,t1 | 0.042 | k p,nt | 0.042 | k s,hi5 | 0.1275 |
| k d,h1 | 0.134 | k d,bf | 4.6 × 10−3 | k s,hbf | 0.1275 |
| k ′ d,bf | 0.0165 | k d,i5 | 9.6 × 10−3 | k s,ht1 | 0.102 |
| k ′ d,i5 | 0.033 | k d,cm | 9.6 × 10−3 | ||
| k ′ d,cm | 0.033 | k d,t1 | 3.3 × 10−3 | ||
| k d,nt | 3.3 × 10−3 | ||||
| Parameter | molec min−1 | Parameter | molec min−1 | Parameter | Dimensionless |
| k s,mn3 | 7.5 | k s,mbM | 5.25 | P | 1.0 |
| k s,mc14 | 5.25 | k s,mhbf | 7.0 | f | 0.4 |
| k s,mi5 | 5.25 | k s,mhi5 | 7.0 | ||
| k s,mt1 | 5.25 | k s,mh1 | 5.25 | ||
| k s,mht1 | 7.0 | ||||
| Parameter | molec fL−1 min−1 | Parameter | fL−2 min−1 | ||
| k s,bf | 1.53 | k s,n3 | 0.0024 |
We model mRNA turnover by a simple birth–death process, although in reality mRNA production and destruction likely involve complex processes of gestation and senescence (Pedraza and Paulsson, 2008). As a single mRNA molecule may be translated into 10–100 protein molecules, fluctuations at the mRNA level are amplified at the level of expressed proteins. For a simple birth–death process of mRNA turnover, the half-life of mRNA must be very short (∼1 min) in order to keep protein-level fluctuations within reasonable bounds (Swain et al, 2002; Pedraza and Paulsson, 2008; Kar et al, 2009). As we show in Supplementary Appendix 1, the longer mRNA half-lives reported in the website (http://web.wi.mit.edu/young/expression/halflife2.html) can be accommodated by a more complex (and more realistic) model of mRNA gestation and senescence. To do so would roughly double the number of variables and reactions in our model, whose only roles would be to track fluctuations in mRNA species that are currently unobservable. Hence, we have chosen to model mRNA turnover by a simple birth–death process with a short ‘effective’ half-life for each mRNA species, in order to simulate correctly the expected fluctuations at the protein level.
Results and discussion
Deterministic calculations
Before carrying out stochastic simulations, we first characterize the deterministic properties of the reaction network. In Figure 2A, we show a one-parameter bifurcation diagram that plots steady-state and oscillatory ClbM-dependent kinase activity as a function of cell volume. In this calculation, we treat V as a parameter by removing the dynamical equation for V(t). For small V, the only stable state of the control system is a steady state of low ClbM-kinase activity, representing the G1 phase of the cell cycle. If V were to increase slowly, the cell would remain in G1 until V exceeds 32.3 fL (SN2), where the stable G1 steady state is destroyed by coalescing with an unstable saddle point. Beyond this size, ClbM level increases abruptly as Cdh1 is inactivated. However, the system cannot attain a stable G2/M state of steady high ClbM level, because this steady state is unstable. The stable, large amplitude limit cycle oscillations, for V>32.3 fL, are created by the negative feedback loop: ‘ClbM inhibits Net1 inhibits Cdc14 activates Cdh1 destroys ClbM.’ SN2 is a special type of bifurcation where the saddle and the node coalesce and give birth to a large amplitude, infinite period, stable limit cycle (Figure 2B). As we have explained elsewhere (Tyson et al, 2002), a bifurcation diagram with these features is ideal for governing cell cycles with the three fundamental properties mentioned in the introduction.
Figure 2.
One-parameter bifurcation diagrams. (A) Number of ClbM molecules per cell as a function of (fixed) cell volume. Black line: stable steady state; red line: unstable steady state; blue lines: maximum and minimum levels during a stable limit cycle oscillation. The steady-state curve shows a region of multiple steady states between two saddle-node (SN) bifurcations at V=23.8 and 32.3 fL, and a Hopf bifurcation (HB) at V=23.9 fL. (B) Period of limit cycle oscillation (dashed line) is overlaid on top of bifurcation diagram.
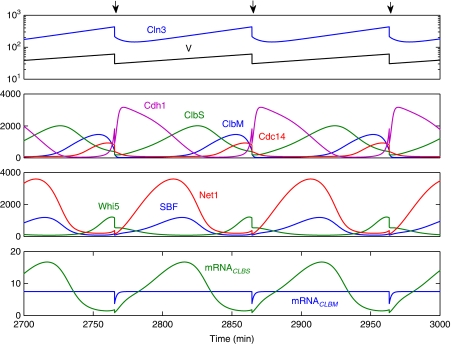
In Figure 3, we show a deterministic simulation of the cell-cycle model for a lineage of cells that grow exponentially and divide evenly when ClbM concentration drops below 12.5 nM (∼300 molecules per cell) as the cell exits mitosis. At cell division, we partition all molecular species (proteins and mRNAs) equally between the two daughter cells. In this simulation, the various protein species oscillate exactly as expected from our earlier description of the control system. In Figure 3, the bottom panel shows the time courses of CLBS and CLBM mRNAs. The CLBS transcript oscillates because CLBS is transcriptionally regulated, whereas every other mRNA is maintained at a constant number of molecules per cell. (At cell division, there is a brief, two-fold drop in the numbers of mRNA molecules per cell, but these numbers quickly return to their steady-state values.)
Figure 3.
Deterministically simulated time courses of some key regulatory species in the cell-cycle control system. Top panel: blue curve is the number of Cln3 molecules per cell and black curve is cell volume in fL. Arrows indicate times of cell division. Other panels plot numbers of molecules per cell for the indicated proteins.
Stochastic simulations
We use Gillespie's SSA to simulate the stochastic firing of the 176 elementary reactions comprising our model. In these stochastic simulations, we use a pseudo-steady state approximation for activation of the CLBS gene,
under the assumption that the activation and inactivation of the CLBS gene responds rapidly to changes in the concentration of free SBF. (We are assuming that SBF binds to and dissociates from the CLBS gene very rapidly. Were this not the case, then transcriptional noise would likely be too large for reliable operation of the cell-cycle control system.)
In glucose medium, the ratio of average volumes at birth of daughter and mother cells was observed by Di Talia et al (2007) to be 40:60. In these simulations, we take into account the asymmetry of budding yeast division, by assigning a fraction f of the dividing cell's volume to the daughter cell and 1−f to the mother cell, where f=0.4. At division, we partition mRNA and protein molecules to mother and daughter cells according to their volume, except for Cln3. Laabs et al (2003) have shown that the CLN3 gene is downregulated in daughter cells relative to mother cells, resulting in a daughter-specific delay of START. Recently, Di Talia et al (2009) have found that, in a population of newborn daughter cells, the CLN3 gene is ∼3 times less expressed compared with a population of newly divided mother cells. In our simulations, we have modeled this observation by partitioning Cln3 molecules asymmetrically: we give 25% of Cln3 to the daughter cell and 75% to the mother cell at division.
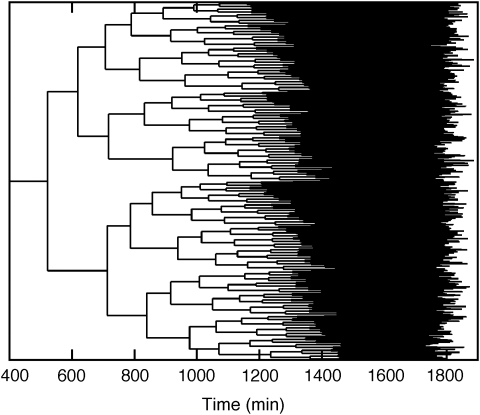
In the stochastic simulation procedure, we generate entire trees of budding yeast cells that derive from a single progenitor (see Figure 4). We trigger cell division (as in deterministic simulations) when ClbM concentration drops below 12.5 nM both for daughter and mother cells. In Figure 5A, we present numbers of various proteins per cell from birth to division. In Figure 5B, we superimpose ‘cell-cycle trajectories’ of several daughter and mother cells on the bifurcation diagram (Figure 2).
Figure 4.
Cell-cycle pedigree generated from a representative stochastic simulation.
Figure 5.
Stochastic simulation of the budding yeast cell cycle. (A) Time courses of some key regulatory proteins. (B) Cell-cycle trajectories (blue line) superimposed on the bifurcation diagram (Figure 2). Arrows show the direction of progress through the cell cycle and the abrupt jump from right to left indicates a cell-division event. A cell-cycle trajectory is the locus of [ClbM](t) (the blue line in panel A) versus V(t), plotted parametrically in time from cell birth to cell division. A newborn cell starts at the left end of a jump, and follows the blue trajectory until it reaches cell division, at the right end of the next jump. At each division, the newborn daughter cell receives 40% of the size of the dividing mother cell.
Next, we simulate experiments of Di Talia et al (2007) that address the question of size control over G1 events. Under the assumption of exponential growth, size at birth (Vbirth) can be related to size at budding (Vbud) by
where α is the specific growth rate of the culture and TG1 is the duration of G1 phase. (Bud emergence and initiation of DNA synthesis occur simultaneously in wild-type budding yeast cells.) If START is controlled by cell size, then the duration of G1 phase will be perfectly correlated with volume at birth, and the slope of αTG1 versus ln(Vbirth) will be −1. On the other hand, if START is controlled by a timer rather than a sizer, then the slope will be 0. Di Talia et al (2007) measured birth sizes and G1 durations of single cells by time lapse fluorescence microscopy and found that, for daughter cells, the scatter plot of αTG1 versus ln(Vbirth) is best fit by two straight lines with a break point at intermediate birth size. Smaller cells are associated with the larger slope (−0.7), indicating a strong size control over small cells. The smaller slope (−0.3) associated with larger cells indicates that timer control dominates the sizer mechanism when cells are large at birth. A similar analysis of mother cells shows no evidence of size control in the experimental data.
Our simulations of these experiments are shown in Figure 6. Following the lead of Di Talia et al, we average αTG1 over all cells in small intervals of Vbirth (bins) to get the red data points in Figure 6. We suppose that budding and S phase begin when ClbS level increases above a threshold (37.5 nM, which is ∼50% of the maximum ClbS level). For daughter cells (Figure 6B), the binned data are well fit by a two-slope (bilinear) model, with slopes −0.59 (for small cells) and −0.25 (for large cells), similar to the experimental measurements. On the other hand, as observed, mother cells (Figure 6D) are controlled by a timer (slope=−0.08) because at birth they are already larger than the critical size required for START. The smallest quartile of mother cells show evidence of weak size control (slope=−0.36).
Figure 6.
Correlation of αTG1 with ln(Vbirth) for (A, B) daughter cells and (C, D) mother cells. Vbirth is normalized by average volume of daughter cells at birth. (A, C) Black dots are raw data from simulation. Red circles represent data binned in 2 fL intervals. (B, D) Binned data (filled black circles) are fitted well by two straight lines (red).
Experiments of Di Talia et al (2007) suggest that Whi5 phosphorylation and export from the nucleus is the key event that is correlated to cell size. After Whi5 is translocated to the cytoplasm, SBF activity drives CLBS expression, and downstream events follow in due course. Inspired by their experiment, we calculate the time interval, denoted T1, between cell birth and SBF activation (when SBF level increases above 15 nM). The correlation of αT1 to ln(Vbirth) is plotted for daughter cells in Figure 7A. As before, we bin the data and fit with two straight lines (Figure 7B). The correlation of αT1 with ln(Vbirth) behaves identically with that of αTG1 (Figure 8). The offset between these two lines, αT2=α(TG1−T1), is roughly constant (T2≈15 min), independent of size at birth. These properties of the model agree nicely with the experiments of Di Talia et al (2007). Analogous simulations for mother cells match the experimental results as well (Supplementary Figure S1). Also, in the model as in experiments, there is no correlation between T2 and size at birth (not shown).
Figure 7.
Correlation of αT1 with ln(Vbirth) for daughter cells, as in Figure 6. (A) Raw data. (B) Binned data (2fL intervals).
Figure 8.
Simulated values of αTG1 (blue circles) and αT1 (green circles) for daughter cells are binned and plotted against ln(Vbirth). Solid lines represent best bilinear fits.
In Table III, we show that the statistical properties of our simulated cell populations are in excellent agreement with measured means and s.d. of budding yeast cell-cycle properties. (Relevant histograms are provided in Supplementary Figures S2 and S3.) In particular, the time period (T1) from cell division to SBF activation is the noisiest part of the daughter cell cycle (CV=74%), because during this time the number of molecules of all molecular species are small and highly fluctuating. In our simulated mother cells, T1 is very short (∼6 min) and highly variable (CV=115%), which is consistent with the observation of Di Talia et al (2007) that the T1 duration of mother cells is too short to measure experimentally. Because of the strong size control operating in daughter cells, the coefficient of variation in volume at division is less than the coefficient of variation in cycle time. But for mother cells, which have only weak size control at best, the CVs are almost identical.
Table 3. Mean and coefficient of variation for cell cycle properties, calculated from the model; experimental values (in parentheses) are from Di Talia et al (2007) .
| Quantity | Daughter cells |
Mother cells |
||
|---|---|---|---|---|
| Mean (min) | CV | Mean (min) | CV | |
| CV=coefficient of variation=mean/(s.d.). Age@division=cell cycle time=TG1SG2M. TG1=T1+T2. | ||||
| Age @ div | 114.25 (112) | 0.2242 (0.22) | 89.35 (87) | 0.1990 (0.14) |
| T G1 | 40.66 (37) | 0.4834 (0.50) | 20.73 (16) | 0.4815 (0.50) |
| T SG2M | 73.59 (76) | 0.2231 (0.20) | 68.62 (72) | 0.1958 (0.17) |
| T 1 | 23.64 (20) | 0.7404 | 6.34 (∼2) | 1.15 |
| T 2 | 17.02 (17) | 0.5131 | 14.38 (14) | 0.3967 |
| Mean (fL) | Mean (fL) | |||
| V birth | 27.52 (28) | 0.2253 (0.20) | 41.27 (40) | 0.2253 (0.18) |
| V division | 60.80 | 0.1919 | 76.64 | 0.1930 |
The average numbers of transcripts and proteins per gene per cell, computed from asynchronous populations of mother and daughter cells are recorded in Table IV. Protein abundances are in rough agreement with measured quantities (Ghaemmaghami et al, 2003), except for Cdc14. (We do not understand how the reported abundance of Cdc14 can be five-fold larger than that of Net1. The abundance of Cdc14 may have been overestimated.) In addition, we plot histograms of some representative mRNAs and proteins (Figure 9) for an asynchronous population of mother cells. Recently, Zenklusen et al (2008) have used fluorescent in situ hybridization in budding yeast cells to measure abundances of selected mRNAs for both regulated and constitutively expressed genes. Although they did not measure any of the mRNAs we are interested in, the shapes of their histograms are similar to the histograms we have calculated.
Table 4. Average number of transcripts and protein molecules in an asynchronous population of cells.
| Cell | Gene |
||||||
|---|---|---|---|---|---|---|---|
| CLN3 | CLBS | CLBM | CDC14 | CDH1 | NET1 | WHI5 | |
| NM, not measured. | |||||||
| aExperimental values from Ghaemmaghami et al (2003). CLBS=CLN2+CLB5, CLBM=CLB1+CLB2. | |||||||
| Transcript | |||||||
| Daughter | 7 | 8 | 7 | 7 | 7 | 7 | 7 |
| Mother | 7 | 10 | 7 | 7 | 7 | 7 | 7 |
| Protein | |||||||
| Daughter | 207 | 913 | 364 | 225 | 1515 | 1809 | 433 |
| Mother | 397 | 1409 | 564 | 341 | 1684 | 1791 | 307 |
| Protein | |||||||
| Experimenta | NM | 1800 | 600 | 8550 | NM | 1590 | 1440 |
Figure 9.
Histograms of representative mRNAs (A–C) and proteins (D–I) in mother cells.
In Figure 10, we plot the effects of ploidy on the CVs of various cell-cycle properties. Our simulations are in good quantitative accord with the measurements in Supplementary Table S8 of Di Talia et al (2007), except for the CV of age at division for mother cells. In general, in our simulations, CV decreases with increasing ploidy by the expected inverse-square-root dependence. In Supplementary Table S3, we record how the means of these properties depend on ploidy.
Figure 10.
Effect of ploidy on noise. Plot of coefficient of variation (CV) for various cell-cycle properties as a function of ploidy. Blue squares: simulated data; red circles: experimental data, from Supplementary Table S8 of Di Talia et al (2007). Black lines represent expected dependence of CV on  ploidy. Left panel: daughter cells; right panel: mother cells, except for last row.
ploidy. Left panel: daughter cells; right panel: mother cells, except for last row.
In Table V, we compute the contributions of size-dependent and size-independent noise to the variability of TG1. These quantities are calculated exactly as described in Di Talia et al (2007). Our simulated values for daughter cells agree quite well with the measured values in Table I of Di Talia et al (2007). For mother cells, the agreement is less good: our model seems to attribute more mother-cell variability to size-dependent noise than is warranted by experiments. This discrepancy is related to the fact that, in our simulation (Figure 6D), small mother cells show some size control of TG1, which is not observed in Figure 2D of Di Talia et al (2007).
Table 5. Contributions of size-dependent and size-independent noise to the coefficient of variation of G1 duration.
| Ploidy | Daughter cell |
Mother cell |
||||
|---|---|---|---|---|---|---|
| Noise in TG1 | Size-dependent noise | Size-independent noise | Noise in TG1 | Size-dependent noise | Size-independent noise | |
| aExperimental values of CV given inside the parentheses are from Table I of Di Talia et al (2007). | ||||||
| Haploid | 0.4834 (0.55)a | 0.3184 (0.31) | 0.3684 (0.45) | 0.4815 (0.50) | 0.3130 (0.20) | 0.3681 (0.46) |
| Diploid | 0.38 (0.42) | 0.2682 (0.28) | 0.2681 (0.31) | 0.3711 (0.39) | 0.2496 (0.13) | 0.2551 (0.37) |
| Tetraploid | 0.3157 (0.24) | 0.2535 (0.15) | 0.1857 (0.19) | 0.2928 (0.26) | 0.2333 (0.09) | 0.1759 (0.24) |
In Figure 11, we show how the CV of protein abundance varies with mean protein abundance. This graph has been simulated by a method similar to the ‘gating’ used by Newman et al (2006) to reduce the effects of external noise. For an asynchronous population of simulated cells, we selected a small sample of cells from a narrow window around the mean cell size of the population. From this sample, we calculated the mean and CV of the number of molecules of every protein in our model, doing so for all three ploidy levels (haploid, diploid, tetraploid). The graph shows the expected linear dependence (slope=−1) of log(CV2) on log(mean). For low-abundance proteins, our simulations are in good quantitative agreement with the measurements in Figure 2G of Newman et al (2006). At high abundance, Newman's measurements level off at ∼10%, due presumably to sources of extrinsic noise that are not included in our model. In Supplementary Figure S4, we plot CV versus mean abundance for specific proteins in our model. We observe the expected dependence of CV on mean−1/2, except for ClbS and ClbM, the two proteins in our model whose total abundances oscillate dramatically during the cell cycle.
Figure 11.
Variation of protein noise with mean abundance. From an asynchronous population of mother and daughter cells, we picked a sample of cells from a narrow range of volumes around the mean cell size (mean±1 fL). This procedure mimics the ‘gating’ method that Newman et al (2006) used to reduce extrinsic noise. We calculated the mean and coefficient of variation of every protein in this sample of cells. For proteins with multiple phospho-forms (SBF, Whi5, Cdh1 and Net1) and/or bound in complexes (Cdc14, Net1, SBF and Whi5), we computed the mean and CV for the total population of each protein. We include data for haploid, diploid and tetraploid cells in the same plot.
Finally, we have investigated the role of asymmetric CLN3 expression in mother and daughter cells just after cell division. As explained in Di Talia et al (2009), two transcription factors, Ace2 and Ash1, accumulate specifically in daughter cells where they repress the expression of CLN3 relative to mother cells. Consequently, in early G1, daughter cells contain significantly less Cln3 protein than mother cells, beyond the difference expected on the basis of their smaller cell size. Di Talia et al studied double-mutant cells, denoted ACE2* ASH1*, for which Ace2 and Ash1 accumulate symmetrically in mother and daughter cells. To simulate their mutant cells, we repeated our simulations with Cln3 equally distributed between mother and daughter cells, instead of the 75:25 split we have used so far. In Figure 12, we plot αT1 as a function of log(Vbirth) for both wild-type and ACE2* ASH1* cells, in the same format used by Di Talia et al in their Figures 2A and G. The agreement between model and experiments is excellent.
Figure 12.
Correlation of αT1 with log(Vbirth) for mother (red) and daughter (blue) cells. Left panels: at division, Cln3 is distributed 25:75 to the daughter and mother cells, which is representative of the situation in wild-type (WT) cells. Right panels: at division, Cln3 is distributed 50:50 to daughter and mother cells, which is representative of the situation in ACE2* ASH1* mutant cells (Di Talia et al, 2009). In the lower panels, we replot the data by randomly picking 50 data points (both for daughter and mother cells) from the respective upper panel plots, so that the correlation can be compared more easily with the experimental plots (Figure 2A and G) in Di Talia et al (2009). In our simulations, the average T1 delay between daughter and mother cells of the same birth size is 11.5 min in WT cells and 1.14 min in mutant cells. In the experiments of Di Talia et al, these delays are 8 and 1.3 min, respectively.
Conclusion
We have presented a new model of the regulation of CDK activity in budding yeast cells, based on multisite phosphorylation of CDK target proteins. Our model displays the three fundamental properties of cell-cycle regulation: alternating phases of DNA synthesis and mitosis, balanced growth and division, and functional checkpoints (in this case, a critical-size checkpoint in G1 phase). These desirable features of the model depend in subtle ways on the fact that most CDK targets have multiple CDK-phosphorylation sites. Multisite phosphorylation appears to be crucial to the dynamics of cell-cycle regulation. Deterministic analysis of the model (bifurcation theory) shows how these properties arise from the regulatory network, and stochastic simulations of the model show that these essential features are robust to molecular fluctuations of the magnitude expected in yeast cells.
The major sources of noise in protein regulatory networks of a yeast cell are related to gene transcription and the small number of unique mRNA transcripts. As each mRNA molecule may instruct the synthesis of dozens of protein molecules, the coefficient of variation (CVP) of molecular fluctuations at the protein level may be dominated by fluctuations at the mRNA level, as expressed in the formula, derived by Swain et al (2002) and Pedraza and Paulsson (2008):
 |
where NM, NP denote the number of mRNA and protein molecules, respectively, and ρ=τM/τP is the ratio of half-lives of mRNA and protein molecules. For a yeast cell, typical values of NM and NP are 8 and 800, respectively (Ghaemmaghami et al, 2003; Zenklusen et al, 2008). If ρ=1, then CVP≈25%. Such large fluctuations in protein levels are inconsistent with the observed variability of size and age at division in yeast cells, as shown in the simplified cell-cycle model of Kar et al (2009) and as we have confirmed with our more realistic model.
Based on Equation (3), there seem to be only two ways out of this impasse. Either NM for cell-cycle genes is larger than expected on the basis of the measurements of Zenklusen et al (2008), or τM is smaller than expected from the measurements reported in http://web.wi.mit.edu/young/expression/halflife2.html. To get reasonable agreement between their model and experimental observations, Kar et al (2009) used a combination of larger NM and smaller τM values than warranted by the literature.
However, there is a third possibility. As stressed by Pedraza and Paulsson (2008), Equation (3) is based on a simple, one-stage, birth–death model of mRNA turnover. If mRNA production and destruction are more complex, with several ‘gestational’ intermediates and ‘senescent’ forms, then CVP can be much smaller than predicted by Equation (3). In Supplementary Appendix 1, we show that a model of transcription–translation coupling with 10 stages each of mRNA gestation and senescence gives reasonable fluctuations at the protein level (CVP≈5%), with an mRNA half-life of 10 min. By comparison, a one-stage model with τM=10 min gives CVP≈25%, as expected from Equation (3). A one-stage model with τM=1 min gives similar fluctuations (CVP≈5%) to a 10-stage model with τM=10 min. In Table SA1.1 of Supplementary Appendix 1, we report trial results of our cell-cycle model with five stages of gestation and senescence for each type of mRNA and with a 10-min half-life for each type, except CLN2 and CLN3 mRNAs (half-life=5 min). The means and CVs of cell-cycle properties of mother and daughter cells are comparable to experimental observations and to the results of a one-stage model of mRNA turnover with half-life=1 min.
From these considerations, we conclude that a yeast cell, because it is so small and has so few mRNA transcripts per protein-coding gene, must do a certain amount of ‘time averaging’ over the processes of gene expression and mRNA processing in order to keep protein-level fluctuations at reasonable levels. In our stochastic simulations, we have chosen to implement this time averaging by two assumptions: (1) transcription factor binding to regulated genes (CLBS in our example) is rapidly reversible, so that mRNA production is not too bursty in time and (2) the ‘effective’ half-life of mRNA molecules is short, so that large fluctuations in mRNA numbers do not persist for lengthy intervals of time. The short half-life that we use (∼1 min) is a consequence of our decision to model mRNA turnover by a simple birth–death process. If we include intermediate stages of mRNA gestation and senescence, then mRNA half-lives can be considerably longer, but the model must keep track of many mRNA forms that are currently unobservable. In any case, it appears that a full understanding of the mechanisms by which yeast cells limit the magnitude of protein fluctuations must await further theoretical developments and experimental measurements.
The alternation of S and M phases in the eukaryotic cell cycle is accomplished by switching the CDK control system back and forth between alternative stable states (Nasmyth, 1996; Tyson and Novak, 2008): a G1 state, with low CDK activity, and an S/G2/M state, with high CDK activity. Proper functioning of this switch requires a degree of nonlinearity in the kinetics that is provided by distributive multisite phosphorylation (Kapuy et al, 2009). This type of mechanism requires that CDK binds only transiently to its target proteins, quickly phosphorylating a site, dropping off and searching for another substrate to attack. Such a strategy is ideal for a kinase such as CDK, which has many substrates to phosphorylate and which cannot afford to spend too much time tied up in complexes with any one of them. It is also suitable for creating the kind of kinetic nonlinearities required for the reaction network to exhibit bistability and hysteresis. From a modeling point of view, multisite phosphorylation is convenient because the reaction mechanism can be described by mass-action rate laws, which are easy to simulate either deterministically or stochastically.
We have tested the adequacy of the model by carrying out extensive stochastic simulations of recent experiments by Di Talia et al (2007) on size control of the START transition in single cells of budding yeast, Saccharomyces cerevisiae. They were able to identify two subphases (T1 and T2) of the unbudded phase (G1 phase) of the yeast cell cycle. The statistics of T1 and T2 are quite different, and our model is in excellent quantitative agreement with the experimental distributions.
Although the model has all the basic features of cell-cycle control outlined by Tyson and Novak (2008), it is still incomplete as a realistic model of the budding yeast cell cycle. It lacks several regulatory proteins (MBF, Clb5, Sic1, Cdc20 and Cdc5) that have important functions in the budding yeast cell cycle. The model will be extended to these components in a future publication.
Materials and methods
The wiring diagram in Figure 1 was translated into a set of ordinary differential equations (Table I), representing the dynamical evolution of the proteins and mRNAs in the model. Parameter values used for our simulations are given in Table II.
Deterministic simulations were carried out in MATLAB using the ode15s solver. For the purpose of bifurcation analysis, we removed the dynamical equation for V(t) from the set of differential equations and treated V as a fixed parameter. Bifurcation diagrams were drawn using XPP-Aut, available from http://www.math.pitt.edu/~bard/xpp/xpp.html.
In deterministic simulations, we triggered cell division whenever ClbM concentration level drops below 12.5 nM as the cell exits mitosis. At cell division, all molecular species (proteins and mRNAs) were partitioned equally between the two newborn cells.
Stochastic simulations were performed using Gillespie's SSA (Gillespie, 1976). In stochastic simulations (as in the deterministic case), cell division occurs whenever ClbM concentration drops below 12.5 nM. To avoid multiple divisions caused by ClbM concentration fluctuating back-and-forth across 12.5 nM, we insist that ClbM concentration must increase above 20 nM before a subsequent division can be triggered by falling ClbM concentration. Following experimental evidence in Di Talia et al (2007), we partitioned 40% of total volume at division to the daughter cell and 60% to the mother cell. We partitioned all molecular species to the mother and daughter cells in 60:40 ratios, respectively, except for Cln3, which was apportioned 75% to the mother cell and 25% to the daughter cell. This rule was intended to account for the fact that Cln3 production is actively downregulated in newborn daughter cells (Laabs et al, 2003; Di Talia et al, 2009).
In stochastic simulations, we followed both mother and daughter cells to generate a complete pedigree of growing and dividing cells. For each cell, we recorded the total duration of G1 phase (from birth until ClbS rises above 37.5 nM) and the T1 duration (from birth until active SBF rises up above 15 nM).
Supplementary Material
Supplementary Figures S1–S4, Supplementary Tables S1–S3, Supplementary Appendix 1, (containing two figures and one table), Listing of source codes
Acknowledgments
This work was supported by the National Institutes of Health (1 R01 GM078989). We thank Fred Cross, Stefano Di Talia, Kathy Chen and Sandip Kar for helpful discussions and advice.
Footnotes
The authors declare that they have no conflict of interest.
References
- Amon A, Tyers M, Futcher B, Nasmyth K (1993) Mechanisms that help the yeast cell cycle clock tick: G2 cyclins transcriptionally activate G2 cyclins and repress G1 cyclins. Cell 74: 993–1007 [DOI] [PubMed] [Google Scholar]
- Arava Y, Wang Y, Storey JD, Liu CL, Brown PO, Herschlag D (2003) Genome-wide analysis of mRNA translation profile in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 100: 3889–3894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzam R, Chen SL, Shou WY, Mah AS, Alexandru G, Nasmyth K, Annan RS, Carr SA, Deshaies RJ (2004) Phosphorylation by cyclin B-CDK underlies release of mitotic exit activator Cdc14 from the nucleolus. Science 305: 516–519 [DOI] [PubMed] [Google Scholar]
- Belle A, Tanay A, Bitincka L, Shamir R, O’Shea EK (2006) Quantification of protein half-lives in the budding yeast proteome. Proc Natl Acad Sci USA 103: 13004–13009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunewell S, Bornholdt S (2007) Superstability of the yeast cell-cycle dynamics: ensuring causality in the presence of biochemical stochasticity. J Theor Biol 245: 638–643 [DOI] [PubMed] [Google Scholar]
- Chen KC, Calzone L, Csikasz-Nagy A, Cross FR, Novak B, Tyson JJ (2004) Integrative analysis of cell cycle control in budding yeast. Mol Biol Cell 15: 3841–3862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen KC, Csikasz-Nagy A, Gyorffy B, Val J, Novak B, Tyson JJ (2000) Kinetic analysis of a molecular model of the budding yeast cell cycle. Mol Biol Cell 11: 369–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M, Nishikawa JL, Tang X, Millman JS, Schub O, Breitkreuz K, Dewar D, Rupes I, Andrews B, Tyers M (2004) CDK activity antagonizes Whi5, an inhibitor of G1/S transcription in yeast. Cell 117: 899–913 [DOI] [PubMed] [Google Scholar]
- Cross FR, Archambault V, Miller M, Klovstad M (2002) Testing a mathematical model of the yeast cell cycle. Mol Biol Cell 13: 52–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruin RAM, Kalashnikova TI, Chahwan C, McDonald WH, Wohlschlegel J, Yates JI, Russell P, Wittenberg C (2006) Constraining G1-specific transcription to late G1 phase: the MPF associated corepressor Nrm1 acts via negative feedback. Mol Cell 23: 483–496 [DOI] [PubMed] [Google Scholar]
- de Bruin RAM, McDonald WH, Kalashnikova TI, Yates J III, Wittenberg C (2004) Cln3 activates G1-specific transcription via phosphorylation of the SBF bound repressor Whi5. Cell 117: 887–898 [DOI] [PubMed] [Google Scholar]
- Di Talia S, Skotheim JM, Bean JM, Siggia ED, Cross FR (2007) The effects of molecular noise and size control on variability in the budding yeast cell cycle. Nature 448: 947–951 [DOI] [PubMed] [Google Scholar]
- Di Talia S, Wang H, Skotheim JM, Rosebrock AP, Futcher B, Cross FR (2009) Daughter-specific transcription factors regulate cell size control in budding yeast. PLOS Biol 7: e1000221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirick L, Nasmyth K (1991) Positive feedback in the activation of G1 cyclins in yeast. Nature 351: 754–757 [DOI] [PubMed] [Google Scholar]
- Fitch I, Dahmann C, Surana U, Amon A, Nasmyth K, Goetsch L, Byers B, Futcher B (1992) Characterization of four B-type cyclin genes of the budding yeast S. cerevisiae. Mol Biol Cell 3: 805–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge H, Qian H, Qian M (2008) Synchronized dynamics and non-equilibrium steady states in a stochastic cell-cycle network. Math Biosci 211: 132–152 [DOI] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh W, Bower K, Howson RW, Belle A, Dephoure N, O’Shea EK, Weissman JS (2003) Global analysis of protein expression in yeast. Nature 425: 737–741 [DOI] [PubMed] [Google Scholar]
- Gillespie DT (1976) A general method for numerically simulating the stochastic time evolution of coupled chemical reactions. J Comput Phys 22: 403–434 [Google Scholar]
- Goldbeter A, Koshland DE Jr (1981) An amplified sensitivity arising from covalent modification in biological systems. Proc Natl Acad Sci USA 78: 6840–6844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardena J (2005) Multisite protein phosphorylation makes a good threshold but can be a poor switch. Proc Natl Acad Sci USA 102: 14617–14622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell LH, Culotti J, Pringle JR, Reid BJ (1974) Genetic control of the cell division cycle in yeast: a model to account for the order of cell cycle events is deduced from the phenotypes of the yeast mutants. Science 183: 46–51 [DOI] [PubMed] [Google Scholar]
- Hartwell LH, Unger MW (1977) Unequal division in Saccharomyces cerevisiae and its implications for the control of cell division. J Cell Biol 75: 422–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspersen SL, Charles JF, Morgan DO (1999) Inhibitory phosphorylation of the APC regulator Hct1 is controlled by the kinase Cdc28 and the phosphatase Cdc14. Curr Biol 11: 227–236 [DOI] [PubMed] [Google Scholar]
- Johnston GC, Pringle JR, Hartwell LH (1977) Coordination of growth with cell division in the yeast Saccharomyces cerevisiae. Exp Cell Res 105: 79–98 [DOI] [PubMed] [Google Scholar]
- Kapuy O, Barik D, Sananes MRD, Tyson JJ, Novak B (2009) Bistability by multiple phosphorylation of regulatory protein. Prog Biophys Mol Biol 100: 47–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kar S, Baumann WT, Paul MR, Tyson JJ (2009) Exploring the role of noise in the eukaryotic cell cycle. Proc Natl Acad Sci USA 106: 6471–6476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch C, Moll T, Neuberg M, Ahorn H, Nasmyth K (1993) A role for the transcription factors Mbp1 and Swi4 in progression from G1 to S phase. Science 261: 1551–1557 [DOI] [PubMed] [Google Scholar]
- Koch C, Schleiffer A, Ammerer G, Nasmyth K (1996) Switching transcription on and off during the yeast cell cycle: Cln/Cdc28 kinases activate bound transcription factor SBF (Swi4/Swi6) at Start, whereas Clb/Cdc28 kinase displace it from the promoter in G2. Genes Dev 10: 129–141 [DOI] [PubMed] [Google Scholar]
- Laabs TL, Markwardt DD, Slattery MG, Newcomb LL, Stillman DJ, Heideman W (2003) ACE2 is required for daughter cell-specific G1 delay in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 100: 10275–10280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lew DJ, Weinert T, Pringle JR (1997) Cell cycle control in Saccharomyces cerevisiae. In The Molecular and Cellular Biology of Yeast Saccharomyces, Pringle JR, Broach JR, Jones EW (eds), Vol. 3, pp 607–695. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory [Google Scholar]
- Li F, Long T, Lu Y, Ouyang Q, Tang C (2004) The yeast cell-cycle network is robustly designed. Proc Natl Acad Sci USA 101: 4781–4786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim HH, Goh P-Y, Surana U (1998) Cdc20 is essential for the cyclosome-mediated proteolysis of both Pds1 and Clb2 during M phase in budding yeast. Curr Biol 8: 231–234 [DOI] [PubMed] [Google Scholar]
- Lodish HF (1974) Model for the regulation of mRNA translation applied to haemoglobin synthesis. Nature 251: 385–388 [DOI] [PubMed] [Google Scholar]
- Lord PG, Wheals AE (1980) Asymmetrical division of Saccharomyces cerevisiae. J Bacteriol 142: 808–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendenhall MD, Hodge AE (1998) Regulation of Cdc28 cyclin-dependent protein kinase activity during the cell cycle of the yeast Saccharomyces cerevisiae. Microbiol Mol Biol Rev 62: 1191–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller ME, Cross FR (2001) Mechanisms controlling subcellular localization of the G1 cyclins Cln2p and Cln3p in budding yeast. Mol Cell Biol 21: 6292–6311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan DO (2007) The Cell Cycle: Principles of Control. London, UK: New Science Press Ltd [Google Scholar]
- Mura I, Csikasz-Nagy A (2008) Stochastic Petri Net extension of a yeast cell cycle model. J Theor Biol 254: 850–860 [DOI] [PubMed] [Google Scholar]
- Murray A, Hunt T (1993) The Cell Cycle. An Introduction. New York: WH Freeman & Co [Google Scholar]
- Nasmyth K (1996) At the heart of the budding yeast cell cycle. Trends Genet 12: 405–412 [DOI] [PubMed] [Google Scholar]
- Newcomb LL, Hall DD, Heideman W (2002) AZF1 is a glucose-dependent positive regulator of CLN3 transcription in Saccharomyces cerevisiae. Mol Cell Biol 22: 1607–1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman JRS, Ghaemmaghami S, Ihmels J, Breslow DK, Noble M, DeRisi JL, Weissman JS (2006) Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature 441: 840–846 [DOI] [PubMed] [Google Scholar]
- Okabe Y, Sasai M (2007) Stable stochastic dynamics in yeast cell cycle. Biophys J 93: 3451–3459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedraza JM, Paulsson J (2008) Effects of molecular memory and bursting on fluctuations in gene expression. Science 319: 339–343 [DOI] [PubMed] [Google Scholar]
- Polymenis M, Schmidt EV (1997) Coupling cell division to cell growth by translational control of the G1 cyclin CLN3 in yeast. Genes Dev 11: 2522–2531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pringle JR, Hartwell LH (1981) The Saccharomyces cerevisiae cell cycle. In The Molecular Biology of the Yeast Saccharomyces, Strathern JN, Jones EW, Broach JR (eds), pp 97–142. Cold Spring Harbor NY: Cold Spring Harbor Laboratory [Google Scholar]
- Qu Z, Weiss JN, MacLellan WR (2003) Regulation of the mammalian cell cycle: a model of the G1-to-S transition. Am J Physiol Cell Physiol 284: C349–C364 [DOI] [PubMed] [Google Scholar]
- Queralt E, Lehane C, Novak B, Uhlmann F (2006) Downregulation of PP2ACdc55 phosphatase by separase initiates mitotic exit in budding yeast. Cell 125: 1–14 [DOI] [PubMed] [Google Scholar]
- Richardson H, Lew DJ, Henze M, Sugimoto K, Reed SI (1992) Cyclin-B homologs in Saccharomyces cerevisiae function in S phase and in G2. Genes Dev 6: 2021–2034 [DOI] [PubMed] [Google Scholar]
- Rudner A, Murray A (2000) Phosphorylation by Cdc28 activates the Cdc20-dependent activity of the anaphase promoting complex. J Cell Biol 149: 1377–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabouri-Ghomi M, Ciliberto A, Kar S, Novak B, Tyson JJ (2008) Antagonism and bistability in protein interaction networks. J Theor Biol 250: 209–218 [DOI] [PubMed] [Google Scholar]
- Salazar C, Hofer T (2007) Versatile regulation of multisite protein phosphorylation by the order of phosphate processing and protein-protein interactions. FEBS J 27: 1046–1061 [DOI] [PubMed] [Google Scholar]
- Schneider BL, Yang Q-H, Futcher AB (1996) Linkage of replication to Start by the CDK inhibitor Sic1. Science 272: 560–562 [DOI] [PubMed] [Google Scholar]
- Schwab M, Lutum AS, Seufert W (1997) Yeast Hct1 is a regulator of Clb2 cyclin proteolysis. Cell 90: 683–693 [DOI] [PubMed] [Google Scholar]
- Schwob E, Bohm T, Mendenhall MD, Nasmyth K (1994) The B-type cyclin kinase inhibitor p40sic1 controls the G1 to S transition in S. cerevisiae. Cell 79: 233–244 [DOI] [PubMed] [Google Scholar]
- Shirayama M, Toth A, Galova M, Nasmyth K (1999) APC (CDC20) promotes exit from mitosis by destroying the anaphase inhibitor Pds1 and cyclin Clb5. Nature 402: 203–207 [DOI] [PubMed] [Google Scholar]
- Shou WY, Seol JH, Shevchenko A, Baskerville C, Moazed D, Chen ZWS, Jang J, Shevchenko A, Charbonneau H, Deshaies RJ (1999) Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell 97: 233–244 [DOI] [PubMed] [Google Scholar]
- Skotheim JM, Di Talia S, Siggia ED, Cross FR (2008) Positive feedback of G1 cyclins ensures coherent cell cycle entry. Nature 454: 291–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmeier F, Amon A (2004) Closing mitosis: the function of the Cdc14 phosphatase and its regulation. Annu Rev Genet 38: 203–232 [DOI] [PubMed] [Google Scholar]
- Steuer R (2004) Effects of stochasticity in models of the cell cycle: from quantized cycle times to noise-induced oscillations. J Theor Biol 228: 293–301 [DOI] [PubMed] [Google Scholar]
- Sveiczer A, Tyson JJ, Novak B (2001) A stochastic, molecular model of the fission yeast cell cycle: role of the nucleocytoplasmic ratio in cycle time regulation. Biophys Chem 92: 1–15 [DOI] [PubMed] [Google Scholar]
- Swain PS, Elowitz MB, Siggia ED (2002) Intrinsic and extrinsic contributions to stochasticity in gene expression. Proc Natl Acad Sci USA 99: 12795–12800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyers M, Tokiwa G, Futcher B (1993) Comparison of the Saccharomyces cerevisae G1 cyclins: Cln3 may be an upstream activator of Cln1, Cln2 and other cyclins. EMBO J 12: 1955–1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyson JJ, Csikasz-Nagy A, Novak B (2002) The dynamics of cell cycle regulation. BioEssays 24: 1095–1109 [DOI] [PubMed] [Google Scholar]
- Tyson JJ, Novak B (2001) Regulation of the eukaryotic cell cycle: molecular antagonism, hysteresis and irreversible transitions. J Theor Biol 210: 249–263 [DOI] [PubMed] [Google Scholar]
- Tyson JJ, Novak B (2008) Temporal organization of the cell cycle. Curr Biol 18: R759–R768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma R, Annan RS, Huddleston MJ, Carr SA, Reynard G, Deshaies RJ (1997) Phosphorylation of Sic1p by G1 CDK required for its degradation and entry into S phase. Science 278: 455–460 [DOI] [PubMed] [Google Scholar]
- Visintin R, Craig K, Hwang ES, Prinz S, Tyers M, Amon A (1998) The phosphatase Cdc14 triggers mitotic exit by reversal of CDK-dependent phosphorylation. Mol Cell 2: 709–718 [DOI] [PubMed] [Google Scholar]
- Visintin R, Hwang ES, Amon A (1999) Cfi1 prevents premature exit from mitosis by anchoring Cdc14 phosphatase in the nucleolus. Nature 398: 818–823 [DOI] [PubMed] [Google Scholar]
- Visintin R, Prinz S, Amon A (1997) CDC20 and CDH1: a family of substrate-specific activators of APC-dependent proteolysis. Science 278: 460–463 [DOI] [PubMed] [Google Scholar]
- Wagner MV, Smolka MB, de Bruin RAM, Zhou H, Wittenberg C, Dowdy SF (2009) Whi5 regulation by site specific CDK-phosphorylation in Saccharomyces cerevisiae. PLoS ONE 4: e4300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijnen H, Landman A, Futcher B (2002) The G1 cyclin Cln3 promotes cell cycle entry via the transcription factor Swi6. Mol Cell Biol 22: 4402–4418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeong FM, Lim HH, Padmashree CG, Surana U (2000) Exit of mitosis in budding yeast: biphasic inactivation of the Cdc28-Clb2 mitotic kinase and the role of Cdc20. Mol Cell 5: 501–511 [DOI] [PubMed] [Google Scholar]
- Zachariae W, Schwab M, Nasmyth K, Seufert W (1998) Control of cyclin ubiquitination by CDK-regulated binding of Hct1 to the anaphase promoting complex. Science 282: 1721–1724 [DOI] [PubMed] [Google Scholar]
- Zenklusen D, Larson DR, Singer RH (2008) Single-RNA counting reveals alternative modes of gene expression in yeast. Nat Struct Mol Biol 15: 1263–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Qian M, Ouyang Q, Deng M, Li F, Tang C (2006) Stochastic model of yeast cell-cycle network. Physica D 219: 35–39 [Google Scholar]
- Zhu G, Spellman PT, Volpe T, Brown PO, Botstein D, Davis TN, Futcher B (2000) Two yeast forkhead genes regulate the cell cycle and pseudohyphal growth. Nature 406: 90–94 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures S1–S4, Supplementary Tables S1–S3, Supplementary Appendix 1, (containing two figures and one table), Listing of source codes