Abstract
Transcription, mRNA decay, translation and protein degradation are essential processes during eukaryotic gene expression, but their relative global contributions to steady-state protein concentrations in multi-cellular eukaryotes are largely unknown. Using measurements of absolute protein and mRNA abundances in cellular lysate from the human Daoy medulloblastoma cell line, we quantitatively evaluate the impact of mRNA concentration and sequence features implicated in translation and protein degradation on protein expression. Sequence features related to translation and protein degradation have an impact similar to that of mRNA abundance, and their combined contribution explains two-thirds of protein abundance variation. mRNA sequence lengths, amino-acid properties, upstream open reading frames and secondary structures in the 5′ untranslated region (UTR) were the strongest individual correlates of protein concentrations. In a combined model, characteristics of the coding region and the 3′UTR explained a larger proportion of protein abundance variation than characteristics of the 5′UTR. The absolute protein and mRNA concentration measurements for >1000 human genes described here represent one of the largest datasets currently available, and reveal both general trends and specific examples of post-transcriptional regulation.
Keywords: gene expression regulation, protein degradation, protein stability, translation
Introduction
Proteins and their absolute concentrations determine the physiological state of a cell. Transcription regulation, albeit extremely important, is insufficient by itself to completely describe protein abundance (MacKay et al, 2004). Each gene also has many features and regulatory elements that modulate translation and protein degradation, and their actions impact the steady-state protein abundance, on top of transcription and mRNA decay (Hieronymus and Silver, 2004; Mata et al, 2005). Such extensive post-transcriptional regulation leads to generally low correlation between mRNA and protein concentrations. For many prokaryotic and eukaryotic organisms, only 50% or less of variation in protein abundance is explained by variations in mRNA concentrations (de Sousa Abreu et al, 2009).
Eukaryotic translation influences protein abundance in multiple ways through initiation, elongation and termination, each requiring a number of specialized factors. Translation initiation mostly occurs in a cap-dependent manner, but exceptions exist, for example Internal Ribosome Entry Sites (Filbin and Kieft, 2009). The translation start codon is normally placed within a highly conserved short sequence, also known as Kozak sequence (Kozak, 1987). Translation initiation can be influenced by several features and specific sequences in the 5′ untranslated region (UTR), for example secondary structures, sub-optimal initiation sites or upstream Open Reading Frames (uORFs). Secondary structures and uORFs can slow down the passage of the ribosome and subsequently reduce translation of the main ORF (Calvo et al, 2009).
In addition to initiation, the efficiency of translation elongation influences steady-state protein abundance. It is thought that frequent codons have more tRNAs available than infrequent codons; as a result, codon usage and tRNA adaptation may impact elongation rates and have been used as proxies of translation efficiency, in particular in bacteria and single-cellular eukaryotes (Ermolaeva, 2001). However, recent work in bacteria suggests that codon usage may have a different function than assumed so far (Kudla et al, 2009; Welch et al, 2009), a topic of active debate (Tuller et al, 2010; Waldman et al, 2010). In eukaryotes, mRNA processing and modification, such as poly-adenylation, influences mRNA stability and translation: the length of the poly(A) tail generally correlates with translation efficiency (Preiss and Hentze, 1998). Finally, the action of cis-regulators influence translation: RNA-binding proteins and miRNAs recognize specific binding motifs and modulate the interaction of members of the translation machinery with the mRNA (Abaza and Gebauer, 2008). The human genome encodes, for example, ∼600 RNA-binding proteins (Wilson et al, 2009) many of which may have regulatory functions.
Likewise, protein degradation impacts protein abundance. During ubiquitin-proteasome-mediated proteolysis, target proteins are initially ubiquitinated and then degraded by the proteasome. Regulation takes place during poly-ubiquitinylation; the most important event is dictated by E3 ubiquitin ligases that specifically recognize degradation or destruction signals (degrons) on target proteins and promotes the attachment of a poly-ubiquitin chain (Ang and Wade Harper, 2005; Ravid and Hochstrasser, 2008). The protein's sequence can contain several degradation signals. For example, N-degrons relating the identity of the N-terminal residue to the protein half-life (N-end rule; Bachmair et al, 1986), or PEST sequences, named after richness in proline, glutamic acid, serine and threonine (Rogers et al, 2008). PEST sequences lead to rapid protein turnover by directing a protein to the ubiquitin-proteasome pathway (Rechsteiner and Rogers, 1996; Spencer et al, 2004). Intrinsically, unstructured protein regions, that is regions that do not assume a particular three-dimensional structure, can also destabilize a protein (Dyson and Wright, 2005; Gsponer et al, 2008; Tompa et al, 2008).
Given such plethora of regulatory mechanisms that modify cellular protein abundance, defining the relative contributions of each feature is still a challenging task (de Sousa Abreu et al, 2009), and has so far been possible only for bacteria and yeast (Nie et al, 2006; Brockmann et al, 2007; Tuller et al, 2007; Wu et al, 2008). Here, we present the first comprehensive measurement of the influence of measures of translation and protein degradation on protein abundance variation in a human cellular system. We experimentally measure absolute protein and matching mRNA concentrations for >1000 genes in the Daoy medulloblastoma cell line, using shotgun proteomics and microarrays, respectively (Figure 1). These data comprise one of the largest such sets available today for human (de Sousa Abreu et al, 2009). We analyze ∼200 sequence features including length, nucleotide composition and structure of the coding sequence and UTRs, composition of the translation initiation site, presence of uORFs, putative target sites of miRNAs, codon usage, amino-acid composition and protein degradation signals. We identify sequence characteristics, which have dominant functions in the regulation of translation and protein degradation. Our combined model including mRNA and sequence features can explain 67% of the variation of protein abundance in this system—and thus has the highest predictive power for human protein abundance achieved so far.
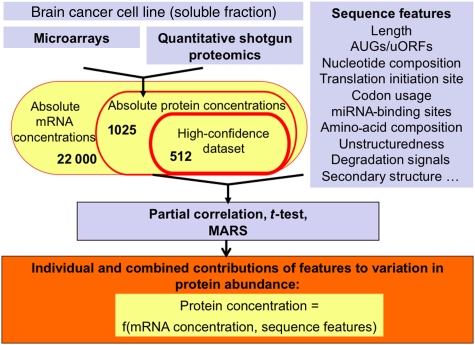
Figure 1.
Flowchart of methods. We measured absolute mRNA and protein concentrations in cellular lysate from the Daoy medulloblastoma cell line. We integrated transcript level information with data on sequence characteristics to explain variation in protein abundance. All sequence characteristics analyzed are listed in the Supplementary information. MARS, Multivariate Adaptive Regression Splines.
Results and discussion
A large-scale dataset on absolute mRNA and protein concentrations
We measured absolute mRNA and matching protein concentrations for >1000 genes, describing the average concentration of each mRNA or protein across a population of Daoy medulloblastoma cells. The data are presumed to reflect the population's steady state, as cells were harvested during logarithmic growth (at 80–90% confluency) and were neither under nutrient deprivation or other stressors. Concentration measurements are estimated to be accurate to within two- to three-fold on average (Vogel and Marcotte, 2008) (Supplementary Figure S5), with both mRNA and protein concentrations spanning four orders of magnitude (Supplementary Figures S2 and S8). We extracted a high-confidence dataset of 512 genes, which we examine more closely; however, all general trends hold true for the entire dataset (Supplementary information).
Steady-state protein concentrations are the combined result of cellular processes that impact mRNA (transcription or RNA decay) and protein (translation and degradation) expression. First, we evaluated individual correlations of sequence features with protein abundance, accounting for what can be explained by variation in mRNA abundance already. Second, we combined information on mRNA abundance and sequence features to derive a model predicting steady-state protein abundance variation.
We observe a significant positive correlation between mRNA and protein concentrations (Figure 2; Spearman's rank correlation Rs=0.46 (P-value<2E−16), Pearson's correlation of log-transformed abundances R2=0.29, R=0.54, P-value<2E−16)—larger than many previous measurements in mammalian systems (de Sousa Abreu et al, 2009). We also estimated the correlation coefficient corrected for errors in the underlying protein and mRNA measurements, using Rcorr=RPR/sqrt(rPP × rRR) (Spearman, 1904, 1910), in which RPR is the Pearson's correlation coefficient between logged protein and mRNA abundances, and rPP and rRR are the reliabilities of the protein and mRNA measurements, respectively. Measurement reliabilities can be estimated from the Pearson's correlation coefficient between technical replicates in test–retest experiments, as is shown in Supplementary Figures S1B and S3. Thus, Rcorr=0.54/sqrt(0.92 × 0.97)=0.57 and Rcorr2=0.32. This estimate implies that the correlation coefficient between perfectly measured protein and mRNA concentrations is very similar to the observed one, and measurement reliabilities are of minor influence.
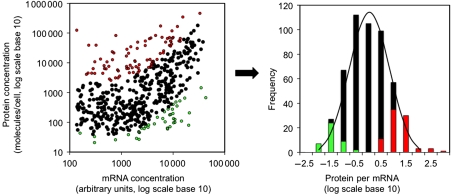
Figure 2.
Human protein and mRNA concentrations. Left: protein and mRNA concentrations correlate significantly at a log–log scale (N=512, R2=0.29, Rs=0.46 with P-value<2.2e−16). Right: genes with extremely high (red) or low (green) protein-per-mRNA ratios are likely regulated at the level of translation or protein stability. Source data is available for this figure at www.nature.com/msb.
The relationship between protein and mRNA concentration is non-linear, but can be approximated by a piece-wise linear function (Supplementary Figure S12). The protein-per-mRNA ratio is approximately log-normally distributed (Figure 2). Log-normal distributions are, in general, the result of multiplicative independent random variables, in the same way as normal distributions are the result of additive independent random variables. In many biological and physical processes, independent effects act in a multiplicative manner and produce log-normal distributions (Koch, 1966). We identified two populations of extreme protein-per-mRNA ratios—the genes in these populations are likely subject to stronger translation or protein degradation regulation (Figure 2). A gene with a large protein-per-mRNA ratio may be very efficiently translated and/or may encode a very stable protein; a gene with few protein molecules per mRNA may be subject to the opposite regulation.
Deviations from the correlation between protein and mRNA concentration result from regulation at the level of translation and protein degradation, and the relationship between protein and mRNA in our data (Figure 2) implies that >70% of the variation in protein abundance can be attributed to some combination of these processes and biological and measurement noise. To evaluate the contribution of translation and protein degradation to gene expression, we examined sequence features related to translation and protein degradation in their ability to explain these remaining 70% of variation in protein expression.
We conducted three types of tests: (a) we examine partial Spearman's rank correlation of numerical features (e.g. length, uORFs) with protein concentration, accounting for variation in mRNA concentrations; (b) for numerical and categorical features (e.g. function), we compare two extreme populations with Welch's t-test and (c) using a Multivariate Adaptive Regression Splines (MARS) model, we analyze the combined contributions of mRNA expression and sequence features to protein abundance variation (Figure 1). To account for the non-linearity of many relationships (e.g. Figure 2), we use non-parametric approaches throughout the analysis (Supplementary Sections S2 and S4).
Individual correlations
We tested ∼200 sequence features for their impact on protein abundance, that is the remaining variation in steady-state protein concentrations after accounting for the variation that can be explained by mRNA expression levels (Figure 1). We correlated each individual feature with protein concentrations, accounting for variation of mRNA expression (partial correlation). In this manner, we explicitly focused on translation and protein degradation. For example, mRNA concentrations alone, representing combined effects of transcription and mRNA stability, correlate weakly with coding sequence length (Rs=−0.22, P-value=9E−7). In comparison, when examining sequence length and protein abundance through partial correlation and factoring out the effects of variation in mRNA concentration, the negative correlation strengthens considerably (RS=−0.53, P-value=5E−46; Table I). Translation and/or protein degradation are strongly inversely correlated with protein sequence length, and more so than transcription and mRNA decay. As partial rank correlation is sensitive to measurement noise, we consider this part of the analysis exploratory rather than confirmatory.
Table 1. Individual correlations.
| Partial correlation: Spearman's Rs to protein abundance (given mRNA abundance variation) | Rs to mRNA abundance | |
|---|---|---|
| Variation in human steady-state protein abundance is dominated by measures of sequence length, protein and mRNA decay, amino-acid composition and translation initiation (upstream Open Reading Frames). The table lists significant sequence features, as well as some other sequence features of interest and experimental data for comparison. Spearman's rank correlation with protein abundance is calculated as the partial correlation between protein concentration and the feature of interest, accounting for variation in mRNA concentration. P-values are the result of testing of the hypothesis of zero partial correlation with protein expression (given mRNA abundance variation). Spearman's rank correlation with mRNA abundance is calculated as direct correlation between mRNA concentration and the feature of interest. Rs—Spearman's rank correlation. Significance: *P-value⩽0.01, **P-value⩽0.001, ***P-value⩽0.0001. Stability of the secondary structures in the 5′UTR is defined as the difference between the lowest free energy calculated for a segment of the real RNA sequence and the average of the lowest free energies of a large number of randomized segments with the same base composition and the same size divided by the s.d. of the free energies from the random sample (Supplementary Table S2). | ||
| Sequence lengths | ||
| Coding sequence length | −0.53*** | −0.22*** |
| 5′UTR length | −0.10 | −0.10 |
| 3′UTR length | −0.19*** | −0.26*** |
| Nucleotide frequencies and properties | ||
| Local secondary structures at the 3′ end of the 5′UTR (window size 40 or 60, significance score) | 0.20*** | 0.06 |
| AUG frequency in 5′UTR | −0.21*** | −0.17*** |
| uORFs frequency in 5′UTR | −0.18*** | −0.14* |
| Amino-acid frequencies and properties | ||
| Serine | −0.24*** | −0.30*** |
| Glutamine | −0.18*** | −0.14* |
| Leucine | −0.18*** | −0.13* |
| Glycine | 0.17*** | 0.05 |
| Polar amino acids | −0.18*** | −0.04 |
| PEST region frequency | −0.37*** | −0.18*** |
| Intrinsic protein unstructuredness | −0.18* | −0.12 |
| Experimental data | ||
| Polysomes (rank ordered) (Mazan-Mamczarz et al, 2005) | 0.19* | 0.17* |
| Phosphorylation (Bodenmiller et al, 2008) | 0.06 | 0.18*** |
| Protein stability index (PSI) (Yen et al, 2008) | 0.09 | 0.15 |
| mRNA decay rate (Yang et al, 2003) | −0.37*** | −0.32*** |
| Other features (not significant) | ||
| miRNAs per 3′UTR (TargetScan90) | −0.08 | −0.03 |
| Polyadenylation sites/3′UTR | 0.02 | −0.11 |
| Codon bias index | 0.08 | 0.12* |
| G+C (total, in coding strand) | 0.04 | 0.11* |
| G+C (third codon position) | 0.06 | 0.15** |
Table I lists a subset of sequence features with the strongest correlation with protein abundance (each with P-value ⩽0.0001), as well as additional tested features of biological interest. Complete results are in Supplementary Section 3 (Supplementary information). Measures of mRNA sequence length are the strongest correlates: expression is significantly lower among proteins with long coding and 3′UTR sequences than among proteins with short sequences (Table I). Correct protein folding is a major determinant of expression levels (Drummond et al, 2005), and the ability to fold fast and correctly may decrease with sequence length, rendering the inverse correlation plausible. In addition, ribosome density decreases in long yeast sequences (Ingolia et al, 2009), resulting in lower translation rates. Short mRNAs tend to be more stable than long mRNAs (Feng and Niu, 2007), are more efficiently translated (Lackner et al, 2007) and tend to have higher transcript levels (Coghlan and Wolfe, 2000). Other evidence suggests a decrease of translation initiation in long sequences (Arava et al, 2003, 2005; Lackner and Bahler, 2008). Indeed, the length of the 5′UTR and its folding energy are strongly correlated (Supplementary information). In comparison, short 3′UTRs have on average fewer binding sites for potential repressor molecules, such as miRNAs and RNA-binding proteins, than long 3′UTRs, and thus a short sequence may be advantageous for high protein abundance (Sandberg et al, 2008; Mayr and Bartel, 2009; Santhanam et al, 2009).
A second set of significant correlations arises from frequencies and properties of amino acids (Table I). These compositional biases can have several origins: (i) amino acids have different costs associated with their use in the cell, that is essential amino acids may be depleted in highly expressed proteins; (ii) the amino-acid sequence may influence a protein's folding and stability; (iii) some amino acids are post-translationally modified and thus not detectable by mass spectrometry and (iv) some peptide sequences are more easily ionizable and hence observable by electrospray mass spectrometry than others, and the differential ionization impacts the observed protein concentration.
Mechanism (iv) is accounted for by the quantitative proteomics method (Lu et al, 2007) (Supplementary Section 1.2; Supplementary Figures S6 and S7). We could not find biases regarding essential amino acids (mechanism i, not shown). The other reasons can be evaluated by concurrent correlations of other features. For example, protein phosphorylation does not correlate with expression (mechanism iii), whereas intrinsic protein unstructuredness, that is the fraction of intrinsically unstructured protein regions, and the presence of PEST motifs correlate strongly and negatively with protein abundance (Table I). These findings suggest a dominant influence of protein degradation (mechanism ii) on biases in the amino-acid composition of proteins with different expression levels. This interpretation is supported by a weak positive correlation with experimental protein degradation data (Yen et al, 2008).
A third set of features relates to translation initiation (Table I). The more structured the 5′UTR of an mRNA is, the more difficult it is for scanning ribosomes to reach the translation start site, suppressing translation. Indeed, we observe a significant positive correlation between local mRNA stability of the 5′UTR (measured by a negative score) and protein abundance (Rs=0.20, P-value<2.5E−5). Recent work in Escherichia coli and yeast confirms the function of secondary structures in protein expression regulation, in particular in the 5′ end of the mRNA (Ringner and Krogh, 2005; Kudla et al, 2009; Gu et al, 2010; Tuller et al, 2010). In addition, we observe an enrichment in upstream start codons (AUG) and uORFs in the 5′UTRs of genes with low protein abundance, suggesting ribosome stalling at these secondary initiation sites and lowered translation of the main open reading frame (Rs=−0.21, P-value=2E−6 and Rs=−0.18, P-value=6E−5, respectively). Concordantly, the translation initiation site is marked by a position-specific nucleotide composition (Kozak, 1987): we find weak differences between the two extreme populations at the nucleotide positions −5 and +4 (Supplementary Figure S9), suggesting a sub-optimal translation initiation for genes with few protein molecules observed per mRNA. Secondary structures of the 3′UTR do not have significant individual effects on protein abundance in our dataset, although they have recently been shown to positively influence translation as measured by association with the translation initiation factor eIF4E (Santhanam et al, 2009).
We examined a number of other features of which many had a surprisingly small contribution (Supplementary information). For example, the presence of putative miRNA-binding sites only shows very weak negative impact on protein abundance in the dataset, confirming recent evidence that miRNAs fine-tune expression regulation, rather than affect gross changes in protein concentrations (Baek et al, 2008; Selbach et al, 2008). Further, codon bias index does not correlate significantly with human protein abundance, in agreement with recent observations in E. coli (Kudla et al, 2009) (Table I). It is, however, a selected feature in the combined model (see below). The codon bias index is weakly positively correlated with mRNA concentration (Rs=0.12, P-value<0.01). Measures of GC content display positive correlations with mRNA, but not protein abundance. As high GC content positively correlates with translation initiation (Santhanam et al, 2009), mRNAs may be stabilized and translated more efficiently, but the overall protein production per mRNA may not be affected. Correspondingly, we find codon usage biases in some amino acids, which could be explained by differences in the GC content, but not by the number of available tRNA genes (Supplementary Figures S10 and S11). Thus, weak codon usage preferences in our dataset may be a by-product of the observed biases in nucleotide composition and mRNA secondary structure.
Finally, we observe a significant enrichment in glycolytic enzymes (P-value<0.05) among genes with high numbers of proteins produced per mRNA, for example MDH1, PKM2, DLD, PGK1, TPI1, LDHB, LDHA, TXN, ETFA, MDH2 and PDHB (Supplementary Table S3). The essential functions of these enzymes concur with their high expression levels. Some translation initiation factors (EIF3C, D, F, M and EIF4B) have extremely low protein-per-mRNA ratios, although this bias is weak (P-value<0.05).
Explaining two-thirds of the variation in protein abundance
In addition to examination of individual correlations, we assessed the combined contributions of mRNA expression and sequence features to protein steady-state abundance. Our model uses MARS, approximating non-linear relationships with continuous piece-wise linear functions. The MARS model analysis differs from the individual partial correlations described above in that features are selected successively based on their additional contribution to explaining variation in protein concentration. Using the full MARS model, we are able to account for two-thirds (67%; Figure 3A; Supplementary Figure S13) of the variation in protein abundance across the proteins using 25 sequence features (Supplementary Table S4). In a pruned model, the top 11 features combined with mRNA expression explain 57%. These results apply specifically to our dataset; when generalizing the model, we can explain ∼30–60% of protein abundance variation (Supplementary Section 4.4). Compared with mRNA data or sequence length alone (Figure 2; Supplementary Figure S15), we can thus more than double the amount of variance explained in protein abundance by using additional sequence information.
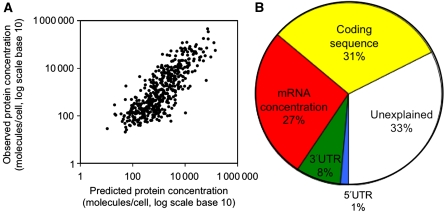
Figure 3.
Combined contributions. (A) Predicted protein abundance using the entire, combined MARS model, R2=0.67 (log scale, P-value<0.001). (B) Contributions of different feature groups to explanation of protein abundance variation. Yellow, green, blue: length, composition, structure and other characteristics of the coding sequence, 5′UTR and 3′UTR, respectively. Details are provided in Supplementary Section S4. See Supplementary Figure S14 for a different feature grouping. Source data is available for this figure at www.nature.com/msb.
Although the order and relative contributions of the individual features may vary from dataset to dataset, we attempted to extract general trends on the types of features that explain variation in protein abundance (Figure 3B). When grouping features of similar types, we observe that characteristics of the coding sequence are the largest contributors, explaining 30% of protein abundance in addition to what can be accounted for by mRNA concentration, that is transcription and mRNA decay. These features include length, nucleotide and amino-acid composition, as well as other characteristics. Codon bias again has only a minor function (2%). Characteristics of the 3′UTR and of the 5′UTR, that is lengths, nucleotide composition and secondary structures, describe another ∼9% of the variation, leaving 33% expression variation unexplained (Figure 3B). The unexplained fraction may be accounted for by mechanisms not considered in this analysis (e.g. regulation by RNA-binding proteins or gene-specific structural motifs), as well as expression and measurement noise. Measurement noise arises, for example, from batch and sampling effects both in the RNA and protein analyses. Overall, these results suggest that the contributions of translation and protein degradation regulation to protein abundance are comparable with those of transcription and mRNA decay.
Summary and conclusions
We present a comprehensive characterization of determinants of human protein abundance, based on large-scale measurement of absolute protein and mRNA concentrations in a medulloblastoma cell line. We show that the contribution of translation and protein degradation is at least as important as the contribution of mRNA transcription and stability to the abundance variation of the final protein products—a finding that may be surprising given that it is commonly assumed that the first step in expression, that is transcription, is the major target of regulation. Protein and matching mRNA concentrations correlate significantly, with variation in mRNA expression explaining ∼25–30% of the variation in protein abundance. Another 30–40% of the variation can be accounted for by characteristics of the sequences, which we identified in a comparative assessment of global correlates. Among these characteristics, sequence length, amino-acid frequencies and also nucleotide frequencies are of strong influence.
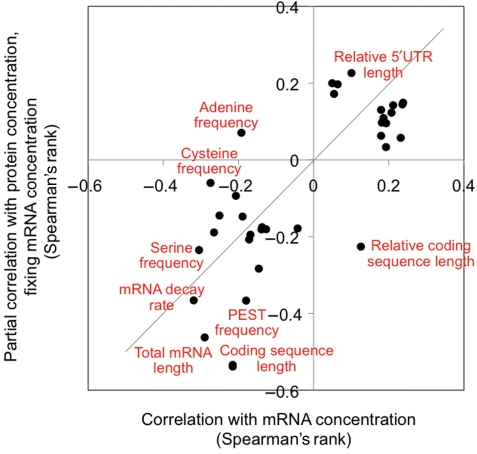
Most of the sequence features in our analysis correlate both with protein and mRNA abundance (Figure 4), underlined by the fact that sequence features alone can explain >50% of protein abundance variation. Some features, for example amino-acid frequencies and secondary structures in the 5′UTR, appear specific to expression regulation at the level of translation and protein degradation, as they correlate more strongly with protein abundance than with mRNA abundance. This means that during evolution, the overall trends in expression regulation are concordant between human protein expression, that is translation and protein degradation, and mRNA expression, that is transcription and mRNA decay, similar to what has been observed for yeast (Garcia-Martinez et al, 2007; Lackner et al, 2007). The bulk of protein expression regulation in our dataset is explained by features of the coding or protein sequence, and not by features of the UTRs. Fine regulation of gene expression may occur through transcription regulation, but also through the action and interactions of dynamic post-transcriptional regulators such as miRNAs and RNA-binding proteins.
Figure 4.
Concordance of mRNA and protein expression regulation. The figure shows the correlation coefficients for features listed in Table I. All correlations are listed in the Supplementary information; x axis: Spearman's rank correlation between the respective feature, and the mRNA concentration, which is the combined outcome of transcription and mRNA decay; y axis: partial Spearman's rank correlation between the respective feature and the protein concentration, fixing variation in mRNA concentration, which describes the combined outcome of translation and protein degradation.
The correlations between mRNA and protein concentrations are typically low (although significant) and variable across organisms, with most R2 values between 0.30 and 0.50 (reviewed in de Sousa Abreu et al, 2009). Several large-scale analyses in the bacterium Desulfovibrio vulgaris (Nie et al, 2006) and baker's yeast Saccharomyces cerevisae (Brockmann et al, 2007; Wu et al, 2008) have attempted to quantify the impact of post-transcriptional regulation on protein expression levels, but the set of features as well as direct measurements of protein concentrations were often limited. Nie et al (2006) investigated only sequence features related to translation initiation (e.g. Shine–Dalgarno sequences), elongation (e.g. codon usage) and termination (e.g. stop codon identity). Similarly, Brockmann et al (2007) analyzed only factors contributing to translational activity, for example ribosome occupancy and density, and the codon adaptation index. Wu et al (2008) included properties related to translation and also an estimate of protein half-life. These studies identified features that explained ∼15–33% of the variation in protein concentrations, in addition to the contribution of mRNA concentrations.
Our study provides one of the first large-scale measurements of absolute mRNA and protein concentrations in a human cell line and assesses the relative importance of ∼200 features describing protein translation, post-translational modification and protein degradation. In comparison with previous studies, which primarily used linear regression, we use non-parametric methods throughout our work. Although the exact extent of individual feature contributions differs across systems, the strong function of amino-acid composition and protein degradation on expression level regulation has also been observed in bacteria and yeast, respectively (Nie et al, 2006; Wu et al, 2008). Thus, it may be a universal characteristic. Amino acid and nucleotide composition (e.g. codon usage) in the coding region relate to elongation, which has been identified as an important contributor of protein translation efficiency in bacteria (Nie et al, 2006) and yeast (Brockmann et al, 2007; Wu et al, 2008). We detect some compositional biases in human sequences, but codon usage only has minor impact.
Regulation of translation initiation has a large function, for example through the flanking 5′ and 3′UTRs, and we find some contribution of the nucleotide composition and the resulting secondary structures of UTRs to protein abundance variation. Concurrently, in yeast, 1.2% of the variation in protein concentration can be explained by the minimum free energy of the 5′UTR-predicted structure, possibly by influencing ribosome scanning (Wu et al, 2008). In human, secondary structures just after the stop codon correlate with expression levels of the translation initiation factor eIF4E (Santhanam et al, 2009). The strong impact of uORFs on protein abundance (Table I) has, to our knowledge, not been observed before. The inverse relationship between the lengths (coding sequence, UTRs) and expression levels or translation activity has been eluded to directly or indirectly in recent work (de Sousa Abreu et al, 2009; Santhanam et al, 2009).
About 33% of the variation in protein abundance cannot be explained by our model (Figure 3B). This unexplained variation may be accounted for by measurement reliability (as addressed above and in Supplementary Figures S1B and S3) and measurement accuracy. With respect to the latter, our control experiments indicate that measurements of protein concentrations are ∼84% accurate (Supplementary Figure S5), suggesting that we cannot explain at least 16% of the expression variation unless our methods improve significantly. In other words, we achieve already ∼80% of our maximal predictability (67% out of 84%).
The remaining, unexplained protein abundance variation may also be explained by gene expression noise (Raser and O’Shea, 2005) and by sequence features that are not included in this study. For example, genomic rearrangements such as chromosomal duplication should mainly not only affect transcription regulation, but may also impact the relationship between protein and mRNA production. Furthermore, the human cells were not synchronized and expression values of cell cycle genes represent their average across both the population and the cell cycle—and this averaging may account for some lack in correlation between mRNA and protein concentrations.
Our study has some restrictions, which will be addressable in future. We analyzed a dataset of limited size and focused on soluble proteins in the cell lysate. Although our combined model uses cross-validation to show the generality of the findings, it remains to be shown how trends hold true for membrane proteins or for other cellular systems. Indeed, analyses similar to ours may be used to describe cellular systems and to quantify molecular ‘expression states’ through a comparison of the contributions of mRNA concentration and sequence features to protein abundance. Further, our results may prove useful for parameter optimization during heterologous protein expression optimization—a field of ongoing scientific debate and investigation (Welch et al, 2009).
Materials and methods
The medulloblastoma Daoy cell line was obtained from American Type Culture Collection. Cells were cultured, harvested and prepared for protein and mRNA analysis as described before (Ramakrishnan et al, 2009). Briefly, total mRNA was extracted using the Trizol method, and analyzed on NimbleGen Homo sapiens 4-Plex (HG18 60mer expr 4plex) arrays using the Agilent Microarray Scanner G2565AA. Soluble proteins were extracted from lysed cells, enzymatically digested into tryptic peptides and analyzed 10 times through LC–MS/MS on a Thermo Electron LTQ-Orbitrap Classic (Ramakrishnan et al, 2009). We re-analyzed the respective raw datasets published before (Ramakrishnan et al, 2009) to obtain estimates of absolute mRNA and protein concentrations.
Estimates of absolute mRNA concentrations
Gene expression values were generated using NimbleScan expression Robust Multi-array Analysis (Irizarry et al, 2003). Quantile normalization and background subtraction were performed across replicate arrays. Report files were further analyzed in the R Project for Statistical Computing using the Bioconductor packages including limma and arrayQuality. We assessed array quality by evaluating diagnostic plots and hierarchical clustering plots. One microarray was eliminated from the study because of the lack of quality. To achieve the same empirical distribution on the single-channel microarray intensities of the two biological replicates, we performed a second quantile normalization. Finally, we averaged gene expression values to obtain the final values. Supplementary Figure S1A addresses the accuracy of the concentration estimates. The data is deposited under the NCBI accession number GSE20492.
Estimates of absolute protein concentrations
Each of the LC–MS/MS runs was analyzed independently with Bioworks (Thermo Fisher Scientific), searching a database of the respective amino-acid sequences (ENSEMBL H. sapiens v. 47.36). The database of protein sequences was made non-redundant with respect to alternative splice variants: we used only the longest sequence to represent each protein. The search results were combined for analysis by PeptideProphet (Keller et al, 2002) and ProteinProphet (Nesvizhskii et al, 2003), and post-processed in the APEX pipeline (Lu et al, 2007; Vogel and Marcotte, 2008) to estimate absolute protein abundance based on weighted spectral counts. We accepted proteins as confidently identified if their ProteinProphet probability was above a cutoff corresponding to <5% global FDR.
We estimate absolute protein concentrations for 1025 proteins, scaling to units of molecules/cell by assuming an average of 8000 molecules/protein. This assumption is based on the findings in yeast and E. coli of ∼4000 and ∼500 molecules/cell, respectively (Lu et al, 2007). All results presented here are valid independent of the precise number of molecules per cell. Control experiments to assess measurement accuracy were performed on protein mixes of known concentrations (Supplementary Figure S5; Supplementary Table S1).
Raw and post-processed data files are provided at http://marcottelab.org/MSdata/, dataset 05.
Data integration
Using reference sequence (Refseq, ENSEMBL) identifiers, we integrated absolute levels of steady-state mRNA and protein concentrations obtained by microarrays and MS-based shotgun proteomics. We found a match for 1025 data points; their UTRs and coding sequences (FASTA format) were obtained from NCBI36 (Ensembl v.44.36f). We filtered the data to construct a high-confidence dataset of 512 genes with information on mRNA and protein concentration. These filters removed genes with <7 arbitrary units (mRNA concentration based on a frequency distribution plot, see Supplementary Figure S2); genes with transmembrane helices (as the sample was cytosolic); genes with ambiguous identifiers and/or gene predictions and genes lacking sequence information (3′ or 5′UTR).
Sequence features
We selected a set of ∼200 sequence features that are likely to have dominant functions in translation and protein degradation regulation: composition of the translation initiation site (Kozak sequence); length and composition of the UTRs and coding sequence; presence and arrangement of uORFs; over-represented motifs that might function as regulatory sequences (PEST); putative-binding sites for miRNAs and secondary structure of the UTRs (Supplementary information; Supplementary Table S2). To account for high cross-correlation among the sequence features, we combined any set of the features with Spearman's correlation coefficients between features ∣Rs∣⩾0.90 to a single feature. For comparison, we also included in the analysis complementary experimental data for mRNA and protein degradation and ribosome attachment from references (Yang et al, 2003; Mazan-Mamczarz et al, 2005; Yen et al, 2008). Details of the source and use of each feature are described in the Supplement information.
Comparisons
We calculated Spearman's rank correlation coefficients between mRNA expression levels and each individual feature. We also calculated partial correlation coefficients (Spearman's rank) of each feature with protein concentrations fixing for the variation introduced by mRNA concentrations, in this way, focusing on effects specifically relevant to translation and protein degradation regulatory processes.
In addition, we compared features of sets of genes with extremely low or high protein-per-mRNA ratios (highlighted in green and red, respectively, in Figure 2), using Welch's t-tests. This adaptation of Student's t-test is intended for use with samples, which may have unequal variances. Genes of extreme protein-per-mRNA ratios were selected according to the following method: assuming a non-linear relationship between mRNA and protein levels, the protein level was modeled as a smooth function of mRNA protein level, that is protein=f(mRNA), where f is a smooth function. We used robust local polynomial regression fit in estimating function f(x) (Cleveland et al, 1992). The outliers or extreme values were then selected as the points in the sample that outer-mostly deviated from the fitted curve f(x), which is measured as the length of the standardized residual. As a result, genes were selected with extremely small or large numbers of protein molecules per mRNA (as highlighted in Figure 2).
In the Supplement information, we comment on the use of partial correlation tests and of protein-per-mRNA ratios to analyze types of data similar to ours (Supplementary Section S2).
Multiple regression (MARS)
To describe the non-linear relationship between protein abundance and biological sequence features and mRNA abundance and the combined contributions of all features, we used MARS. The MARS models were fitted in R (Team, 2004) using functions contained in the ‘earth’ library (Milborrow, 2009). In this model, non-linear responses between protein abundance and biological factors (variables, features) are described by a series of linear segments of differing slope, each of which is fitted using a basis function (Friedman and Roosen, 1995; Hastie et al, 2001). The MARS model uses generalized cross-validation to choose a best set of variables and their functional forms and is particularly useful for the automatic selection of a best set of variables (features) out of all variables in our data. We discuss details of the MARS models, its generality and performance/advantages compared with alternative models (linear regression, principal component analysis) in Supplementary Section S4; Supplementary Figure S16; Supplementary Tables S5, S6, S7 and S8.
Supplementary Material
Supplementary text, Supplementary figures S1–16, Supplementary tables S1–8
Acknowledgments
We thank Jörg Gsponer for help with Disopred predictions, Barron Blackman for help with PEST predictions and Marcelo Bento Soares for ALU predictions. We thank Joshua Plotkin for helpful discussions. This work was supported by the Children's Cancer Research Institute (to RSA and LOP), the NIH (GM076536, GM67779, GM088624 to EMM), NIH/NINDS (NS60658 to DK) and NSF (0640923 to EMM); the Welch (F-1515, to EMM), Tengg (to LOP) and Packard (to EMM) Foundations and the International Human Frontier Science Program (to CV). This research was supported (in part) by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Footnotes
The authors declare that they have no conflict of interest.
References
- Abaza I, Gebauer F (2008) Trading translation with RNA-binding proteins. RNA 14: 404–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang XL, Wade Harper J (2005) SCF-mediated protein degradation and cell cycle control. Oncogene 24: 2860–2870 [DOI] [PubMed] [Google Scholar]
- Arava Y, Boas FE, Brown PO, Herschlag D (2005) Dissecting eukaryotic translation and its control by ribosome density mapping. Nucleic Acids Res 33: 2421–2432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arava Y, Wang Y, Storey JD, Liu CL, Brown PO, Herschlag D (2003) Genome-wide analysis of mRNA translation profiles in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 100: 3889–3894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmair A, Finley D, Varshavsky A (1986) In vivo half-life of a protein is a function of its amino-terminal residue. Science 234: 179–186 [DOI] [PubMed] [Google Scholar]
- Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP (2008) The impact of microRNAs on protein output. Nature 455: 64–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodenmiller B, Campbell D, Gerrits B, Lam H, Jovanovic M, Picotti P, Schlapbach R, Aebersold R (2008) PhosphoPep—a database of protein phosphorylation sites in model organisms. Nat Biotechnol 26: 1339–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockmann R, Beyer A, Heinisch JJ, Wilhelm T (2007) Posttranscriptional expression regulation: what determines translation rates? PLoS Comput Biol 3: e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo SE, Pagliarini DJ, Mootha VK (2009) Upstream open reading frames cause widespread reduction of protein expression and are polymorphic among humans. Proc Natl Acad Sci USA 106: 7507–7512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland WS, Grosse E, Shyu WM (1992) Local regression models. In Statistical Models in S. New York: Wadsworth & Brooks/Cole, Chapman & Hall [Google Scholar]
- Coghlan A, Wolfe KH (2000) Relationship of codon bias to mRNA concentration and protein length in Saccharomyces cerevisiae. Yeast 16: 1131–1145 [DOI] [PubMed] [Google Scholar]
- de Sousa Abreu R, Penalva LO, Marcotte E, Vogel C (2009) Global signatures of protein and mRNA expression levels. Mol Biosyst 5: 1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond DA, Bloom JD, Adami C, Wilke CO, Arnold FH (2005) Why highly expressed proteins evolve slowly. Proc Natl Acad Sci USA 102: 14338–14343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson HJ, Wright PE (2005) Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol 6: 197–208 [DOI] [PubMed] [Google Scholar]
- Ermolaeva MD (2001) Synonymous codon usage in bacteria. Curr Issues Mol Biol 3: 91–97 [PubMed] [Google Scholar]
- Feng L, Niu DK (2007) Relationship between mRNA stability and length: an old question with a new twist. Biochem Genet 45: 131–137 [DOI] [PubMed] [Google Scholar]
- Filbin ME, Kieft JS (2009) Toward a structural understanding of IRES RNA function. Curr Opin Struct Biol 19: 267–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman JH, Roosen CB (1995) An introduction to multivariate adaptive regression splines. Stat Methods Med Res 4: 197–217 [DOI] [PubMed] [Google Scholar]
- Garcia-Martinez J, Gonzalez-Candelas F, Perez-Ortin JE (2007) Common gene expression strategies revealed by genome-wide analysis in yeast. Genome Biol 8: R222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gsponer J, Futschik ME, Teichmann SA, Babu MM (2008) Tight regulation of unstructured proteins: from transcript synthesis to protein degradation. Science 322: 1365–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Zhou T, Wilke C (2010) A universal trend of reduced mRNA stability near the translation-initiation site in prokaryotes and eukaryotes. PLoS Comput Biol 6: e1000664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastie T, Tibshirani RJ, Friedman JH (2001) The Elements of Statistical Learning: Data Mining, Inference and Prediction. New York: Springer Verlag [Google Scholar]
- Hieronymus H, Silver PA (2004) A systems view of mRNP biology. Genes Dev 18: 2845–2860 [DOI] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS (2009) Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324: 218–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP (2003) Summaries of affymetrix GeneChip probe level data. Nucleic Acids Res 31: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller A, Nesvizhskii AI, Kolker E, Aebersold R (2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem 74: 5383–5392 [DOI] [PubMed] [Google Scholar]
- Koch AL (1966) The logarithm in biology. 1. Mechanisms generating the log-normal distribution exactly. J Theor Biol 12: 276–290 [DOI] [PubMed] [Google Scholar]
- Kozak M (1987) An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res 15: 8125–8148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudla G, Murray AW, Tollervey D, Plotkin JB (2009) Coding-sequence determinants of gene expression in Escherichia coli. Science 324: 255–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackner DH, Bahler J (2008) Translational control of gene expression from transcripts to transcriptomes. Int Rev Cell Mol Biol 271: 199–251 [DOI] [PubMed] [Google Scholar]
- Lackner DH, Beilharz TH, Marguerat S, Mata J, Watt S, Schubert F, Preiss T, Bahler J (2007) A network of multiple regulatory layers shapes gene expression in fission yeast. Mol Cell 26: 145–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu P, Vogel C, Wang R, Yao X, Marcotte EM (2007) Absolute protein expression profiling estimates the relative contributions of transcriptional and translational regulation. Nat Biotechnol 25: 117–124 [DOI] [PubMed] [Google Scholar]
- MacKay VL, Li X, Flory MR, Turcott E, Law GL, Serikawa KA, Xu XL, Lee H, Goodlett DR, Aebersold R, Zhao LP, Morris DR (2004) Gene expression analyzed by high-resolution state array analysis and quantitative proteomics: response of yeast to mating pheromone. Mol Cell Proteomics 3: 478–489 [DOI] [PubMed] [Google Scholar]
- Mata J, Marguerat S, Bahler J (2005) Post-transcriptional control of gene expression: a genome-wide perspective. Trends Biochem Sci 30: 506–514 [DOI] [PubMed] [Google Scholar]
- Mayr C, Bartel DP (2009) Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell 138: 673–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazan-Mamczarz K, Kawai T, Martindale JL, Gorospe M (2005) En masse analysis of nascent translation using microarrays. Biotechniques 39: 61–62, 64, 66–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milborrow S (2009) earth: Multivariate Adaptive Regression Spline Models. R Software Package (http://cran.r-project.org/web/packages/earth/index.html)
- Nesvizhskii AI, Keller A, Kolker E, Aebersold R (2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem 75: 4646–4658 [DOI] [PubMed] [Google Scholar]
- Nie L, Wu G, Zhang W (2006) Correlation of mRNA expression and protein abundance affected by multiple sequence features related to translational efficiency in Desulfovibrio vulgaris: a quantitative analysis. Genetics 174: 2229–2243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preiss T, Hentze MW (1998) Dual function of the messenger RNA cap structure in poly(A)-tail-promoted translation in yeast. Nature 392: 516–520 [DOI] [PubMed] [Google Scholar]
- Ramakrishnan SR, Vogel C, Prince JT, Li Z, Penalva LO, Myers M, Marcotte EM, Miranker DP, Wang R (2009) Integrating shotgun proteomics and mRNA expression data to improve protein identification. Bioinformatics 25: 1397–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raser JM, O’Shea EK (2005) Noise in gene expression: origins, consequences, and control. Science 309: 2010–2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravid T, Hochstrasser M (2008) Diversity of degradation signals in the ubiquitin-proteasome system. Nat Rev Mol Cell Biol 9: 679–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rechsteiner M, Rogers SW (1996) PEST sequences and regulation by proteolysis. Trends Biochem Sci 21: 267–271 [PubMed] [Google Scholar]
- Ringner M, Krogh M (2005) Folding free energies of 5′-UTRs impact post-transcriptional regulation on a genomic scale in yeast. PLoS Comput Biol 1: e72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers JT, Bush AI, Cho HH, Smith DH, Thomson AM, Friedlich AL, Lahiri DK, Leedman PJ, Huang X, Cahill CM (2008) Iron and the translation of the amyloid precursor protein (APP) and ferritin mRNAs: riboregulation against neural oxidative damage in Alzheimer's disease. Biochem Soc Trans 36: 1282–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB (2008) Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science 320: 1643–1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santhanam AN, Bindewald E, Rajasekhar VK, Larsson O, Sonenberg N, Colburn NH, Shapiro BA (2009) Role of 3′UTRs in the translation of mRNAs regulated by oncogenic eIF4E—a computational inference. PLoS One 4: e4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N (2008) Widespread changes in protein synthesis induced by microRNAs. Nature 455: 58–63 [DOI] [PubMed] [Google Scholar]
- Spearman C (1904) The proof and measurement of association between two things. Am J Psychol 15: 72–101 [PubMed] [Google Scholar]
- Spearman C (1910) Correlation calculated from faulty data. Br J Psychol 3: 271–295 [Google Scholar]
- Spencer ML, Theodosiou M, Noonan DJ (2004) NPDC-1, a novel regulator of neuronal proliferation, is degraded by the ubiquitin/proteasome system through a PEST degradation motif. J Biol Chem 279: 37069–37078 [DOI] [PubMed] [Google Scholar]
- Team RDC (2004) R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing ISBN 3-900051- 07-0: http://www.R-project.org [Google Scholar]
- Tompa P, Prilusky J, Silman I, Sussman JL (2008) Structural disorder serves as a weak signal for intracellular protein degradation. Proteins 71: 903–909 [DOI] [PubMed] [Google Scholar]
- Tuller T, Kupiec M, Ruppin E (2007) Determinants of protein abundance and translation efficiency in S. cerevisiae. PLoS Comput Biol 3: e248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuller T, Waldman YY, Kupiec M, Ruppin E (2010) Translation efficiency is determined by both codon bias and folding energy. Proc Natl Acad Sci USA 107: 3645–3650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel C, Marcotte EM (2008) Calculating absolute and relative protein abundance from mass spectrometry-based protein expression data. Nat Protoc 3: 1444–1451 [DOI] [PubMed] [Google Scholar]
- Waldman YY, Tuller T, Shlomi T, Sharan R, Ruppin E (2010) Translation efficiency in humans: tissue specificity, global optimization and differences between developmental stages. Nucleic Acids Res 38: 2964–2974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch M, Govindarajan S, Ness JE, Villalobos A, Gurney A, Minshull J, Gustafsson C (2009) Design parameters to control synthetic gene expression in Escherichia coli. PLoS ONE 4: e7002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson D, Pethica R, Zhou Y, Talbot C, Vogel C, Madera M, Chothia C, Gough J (2009) SUPERFAMILY—sophisticated comparative genomics, data mining, visualization and phylogeny. Nucleic Acids Res 37: D380–D386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Nie L, Zhang W (2008) Integrative analyses of posttranscriptional regulation in the yeast Saccharomyces cerevisiae using transcriptomic and proteomic data. Curr Microbiol 57: 18–22 [DOI] [PubMed] [Google Scholar]
- Yang E, van Nimwegen E, Zavolan M, Rajewsky N, Schroeder M, Magnasco M, Darnell JE Jr (2003) Decay rates of human mRNAs: correlation with functional characteristics and sequence attributes. Genome Res 13: 1863–1872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen HC, Xu Q, Chou DM, Zhao Z, Elledge SJ (2008) Global protein stability profiling in mammalian cells. Science 322: 918–923 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary text, Supplementary figures S1–16, Supplementary tables S1–8