Abstract
Penicillin-binding proteins (PBPs) are the molecular target for the widely used β-lactam class of antibiotics, but how these compounds act at the molecular level is not fully understood. We have determined crystal structures of E. coli PBP5 as covalent complexes with imipenem, cloxacillin and cefoxitin. These antibiotics exhibit very different second order rates of acylation for the enzyme. In all three structures, there is excellent electron density for the central portion of the β-lactam, but weak or absent density for the R1 or R2 side chains. Areas of contact between the antibiotics and PBP 5 do not correlate with the rates of acylation. The same is true for conformational changes because although shift of a loop leading to an electrostatic interaction between Arg248 and the β-lactam carboxylate, which occurs completely with cefoxitin, partially with imipenem and is absent with cloxacillin, is consistent with the different rates of acylation, mutagenesis of Arg248 only decreased cefoxitin acylation two fold. Together, these data suggest that structures of post-covalent complexes of PBP 5 are unlikely to be useful vehicles for design of new covalent inhibitors of PBPs. Finally, superimposition of the imipenem-acylated complex with PBP5 in complex with a boronic acid peptidemimetic shows that the position corresponding to the hydrolytic water molecule is occluded by the ring nitrogen of the β-lactam. Since the ring nitrogen occupies a similar position in all three complexes, this supports the hypothesis that deacylation is blocked by the continued presence of the leaving group after opening of the β-lactam ring.
Penicillin-binding proteins (PBPs) are so named because they are the targets for the well-known and widely used class of β-lactam antibiotics. Many PBPs are transpeptidases (TPases) that catalyze the formation of cross-links between peptides on adjacent strands of glycan during the final stages of peptidoglycan synthesis in bacteria, thereby conferring strength to the cell wall against osmotic pressure. Other PBPs are carboxypeptidases (CPases), which remove the terminal d-Ala of the pentapeptidyl substrate, or endopeptidases, which cleave existing peptide cross-links. All are characterized by a set of three conserved motifs in the active site: SxxK, SxN, and KTG, where x is any residue (1). β-lactams inhibit PBPs by reacting with a serine nucleophile in the active site of the enzyme, forming a long-lived acyl-enzyme covalent complex. Whilst the active site remains so occupied, the enzyme is unavailable to react with its peptide substrate, and because cell wall autolytic activity continues, the result is a ruptured bacterial membrane and cell death (for reviews of PBPs see (2–4)).
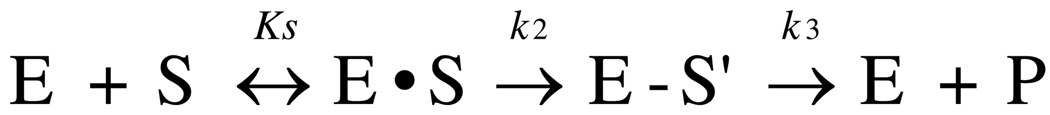
Since the discovery of β-lactam antibiotics, there has been a major effort to understand their mechanism of action. This is all the more important given the widespread development of resistance that has diminished their value as treatments for bacterial infections. Tipper and Strominger first proposed that penicillin is a structural analog of the d-Ala-d-Ala terminus of the acetylmuramyl-pentapeptide substrate (5) and this was later supported by the demonstration that penicillin and the depsipeptide substrate, diacetyl-l-lysyl-d-Ala-d-Lac, both react with the same active site serine in two d-Ala CPases (6). Accordingly, the kinetics of the reactions of PBPs with β-lactams and their peptide substrates can be described in the same way, comprising three steps: Michaelis-Menten binding (k−1/k+1 i.e. KS), acylation (k2) and deacylation (k3) (Scheme 1) (7). Acylation occurs by nucleophilic attack of a serine residue onto the carbonyl carbon of the β-lactam ring or of the penultimate d-Ala of the pentapeptide substrate to form a covalent acyl-enzyme complex. The second order rate constant of acylation, i.e. k2/KS, is widely used as a measure of the potency of an antibiotic (8–11). Deacylation occurs when an incoming water molecule (CPase) or an amino group of a neighboring peptide (TPase) attacks the acylenzyme complex, thus breaking the acyl-enzyme bond and allowing release of the product. The kinetics of deacylation (described by k3) differ between peptide and β-lactam: whereas deacylation is relatively fast for the PBP-peptidyl complex, irrespective of whether for a TPase (aminolysis) or a CPase (hydrolysis), it is slow for a PBP-β-lactam complex. Hence, to understand how β-lactams inhibit PBPs, it is necessary to determine the exact molecular mechanism that underlies the lowered rate of deacylation when the enzyme is covalently bound by a β-lactam versus a similarly bound peptide substrate.
Scheme 1.
where E•S is the non-covalent Michaelis-Menten complex, E-S’ is the covalent acyl-enzyme complex, and P is the released hydrolyzed β-lactam.
A further goal is to understand how β-lactams differ in their relative potencies toward a particular PBP. β-lactams, which vary in their R1 and R2 substituents, can exhibit significantly different rates of acylation against a specific PBP (10, 12). Whilst it might be assumed that, like non-covalent inhibitors, β-lactams with the highest affinity and therefore with the highest complementarity with the active site of a PBP would be the most potent inhibitors, PBPs do not bind β-lactams more tightly than peptide substrates (13), making it unlikely that the non-covalent affinity is the primary parameter that dictates the efficacy of enzyme inactivation. In support of this, there is a lack of correlation between the non-covalent interaction energy (a measure of complementarity) of a β-lactam complex of E. coli PBP 5 and the k2/KS value of that β-lactam against the same PBP (12). A related issue is whether induced fit contributes to the inhibition of PBPs by β-lactams because the highest extent of interaction might occur in the acylation transition state. In order to address these questions, it is necessary to determine to what degree β-lactams contact their PBP targets after acylation and to map any conformational changes that occur during this reaction.
Here, we report the crystal structures of PBP 5 from E. coli as acyl-enzyme complexes with three β-lactams, imipenem, cloxacillin and cefoxitin. In these post-covalent states, the extent of enzyme-antibiotic interaction does not correlate with the second order rate of acylation, suggesting that potency of a particular β-lactam antibiotic is only partly determined by the target enzyme. The observed conformational changes are different for each antibiotic and, at first sight, do correlate with the rates of acylation, but this is not supported by mutagenesis studies. Finally, superimpositions with the previously determined structure of PBP 5 in complex with a peptidyl boronic acid (14) supports the idea that the ring amine of the β-lactam is responsible for blocking deacylation of a β-lactam-PBP complex.
Materials and Methods
Crystal Soaking
Soluble wild-type PBP 5 protein was purified and crystallized as described previously (15). These crystals belong to the C2 space group and have cell dimensions a=109.4 Å, b=50.3 Å, c=84.5 Å and b=120.9° (15). Crystal-soaking experiments were performed with four β-lactam antibiotics: cefoxitin, penicillin G, and moxalactam (all obtained from Sigma-Aldrich Co.) and imipenem (Merck & Co., Inc). Prior to collecting diffraction data, each crystal was cryoprotected in a solution comprising stabilizing solution (8% PEG 400, 50 mM Tris-HCl, pH 8.0) and glycerol. The concentration of glycerol was gradually increased over a period of several hours to a final concentration of 25 %, after which the β-lactam was added to the solution. The concentrations of compounds and the lengths of soaking times were varied to maximize incorporation of ligand with minimal detriment to the crystal. The condition of the crystals was assessed visually and, in instances where the crystals cracked or dissolved during the soak, this “threshold” was used as an upper limit for subsequent soaking experiments. The degree of incorporation was assessed by examination of the electron density maps after collection of trial data sets. Typically, concentrations of 10–20 mM and soak times of 2–4 hours maximized the binding of the antibiotic. All attempts to produce a structure containing moxalactam (whether by crystal soaking or co-crystallization) were unsuccessful. Docking of the compound into the active site of PBP 5 suggests there is sufficient room to accommodate this antibiotic and the solvent channels in the crystal lattice appear large enough for the compound to access the active site (not shown), so the reason for the failure of moxalactam to react is unknown.
Data collection and structure determination
Data for the complexes of PBP 5 with cefoxitin and cloxacillin were collected on an R-AXIS IV++ image plate detector mounted on an RU-H3R rotating anode X-ray generator fitted with Osmic Confocal Optics (Rigaku MSC). The crystal-to-plate distance was 150 mm, and 0.5° oscillations were collected with an exposure time of 5 min. per frame. 210° and 250° of data were collected for cefoxitin- and cloxacillin-soaked crystals, respectively. These data were processed with d*Trek (16). Data for the imipenem complex were collected at the SER-CAT ID22 beam line of the Advanced Photon Source (APS) on a Mar 300 CCD detector at a wavelength of 1.00 Å. The crystal-to-plate distance was 300 mm, and 180 frames per wavelength were collected in 0.5° oscillations with a 3 sec exposure time per frame. These data were processed using HKL2000 (17).
Structure Determination and Refinement
All in cases, the starting structure for refinement was the structure of wild-type PBP 5 (15) stripped of all water molecules and a single molecule of β-mercaptoethanol. After an initial round of refinement using CNS (18), an electron density difference map (|Fobs|-|Fcalc|) was calculated and displayed using the graphics program O (19). The coordinates for each antibiotic were built in the Monomer Library Sketcher module of REFMAC5 (20) and manually positioned into the positive density in the active site of the enzyme using O. Thereafter, each model was refined using REFMAC5 alternating with rounds of manual revision. Water molecules with appropriate hydrogen bond distances were included in later rounds of refinement. Multiple conformations for residues were modeled as necessary, particularly for the complex with imipenem, which was solved at higher resolution. The stereochemistries of the final models were evaluated using PROCHECK (21).
As a guide of the relative degree of contact made by each antibiotic, the solvent accessible area of PBP 5 occluded by each antibiotic was calculated with the CCP4 program AREAIMOL (22) using a probe radius of 1.4 Å. This program calculates the total area difference (in Å2) between ligand-bound and unbound protein. Atoms of each antibiotic that were not visible in the electron density were excluded from these calculations.
Determination of kinetic constants
R248K and R248A mutants of PBP 5 were constructed using overlap-extension PCR (23), cloned into pT7-7K, and expressed and purified as described for wild-type PBP 5 (15). k2/KS constants were determined from first order rates of acylation of wild-type and mutant PBP 5 by [14C]penicillin G (Moravek, Brea, CA) as described previously (15). The k2/Ks values of non-radioactive cefoxitin was measured in a competition experiment with [14C]penicillin G. All experiments were done at low concentrations of [14C]penicillin G to ensure that the reaction occurred at sub-saturating conditions (i.e. below the noncovalent dissociation constant). Carboxypeptidase (CPase) activities of the PBP 5 variants were measured using the experimental substrate, diacetyl-l-Lys-d-Ala-d-Ala (Ac2-KAA), as described (24). Hydrolysis of the PBP 5-[14C]penicillin G covalent complex was measured according to Nicholas et al (15). t1/2 values were determined from semi-log plots of percentage of [14C]penicillin G-bound PBP 5 remaining versus time and were converted to k3 values by dividing the values in seconds into 0.693.
Results
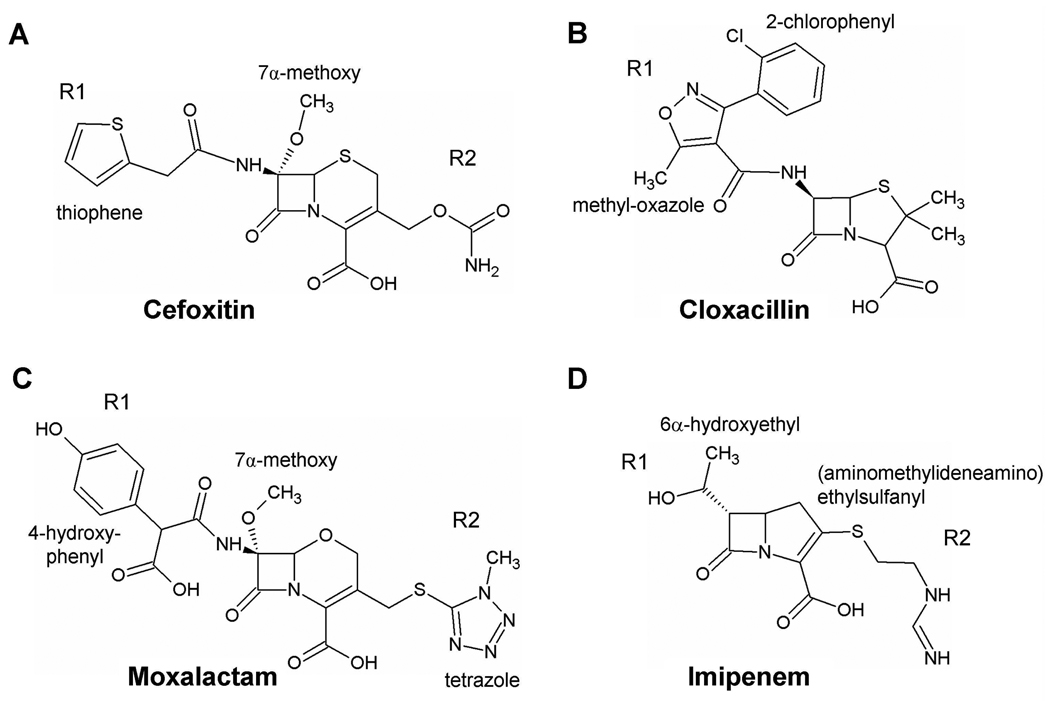
The four antibiotics chosen for this study (Fig. 1) were the same ones used to measure the non-covalent interaction energies of PBP 5 when bound with each of these antibiotics (12). Cefoxitin is a third-generation cephalosporin that contains a thiophene R1 group, a 7α methoxy group and a carbamoyloxymethyl R2 group. The latter substituent is lost during acylation, leaving an exomethylene group at C-3’ in the acyl-enzyme complex (25, 26). Moxalactam is the largest of the antibiotics used in the study and has similarities with third-generation cephalosporins. Both of its side chains are large and bulky groups: a 4-hydroxyphenyl R1 group and a tetrazolethiol R2 group (also a leaving group during acylation). Cloxacillin is a penicillin characterized by a large R1 side chain comprising methyl-oxazole and 2-chlorophenyl rings. Imipenem is a carbapenem with an extended R2 side chain [an amino(methylideneamino)ethylsulfanyl group] and a bulky 6α-hydroxyethyl group.
Fig. 1.
Molecular structures of the β-lactam antibiotics used in this study.
Description of the three structures
Imipenem
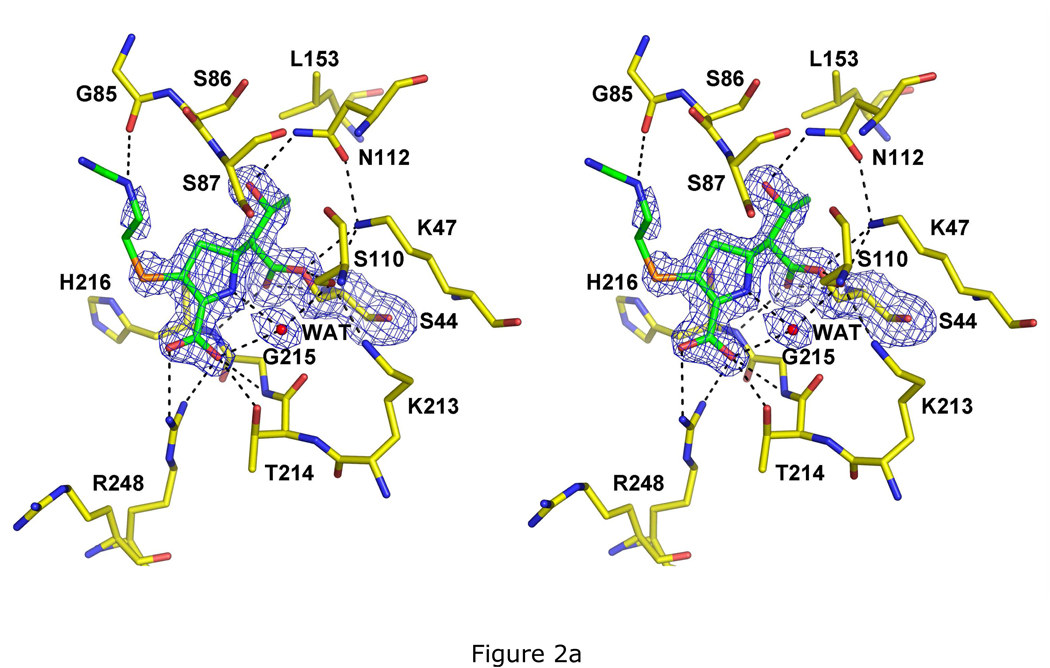
The PBP 5-imipenem structure is the first structure of a PBP in complex with this antibiotic and, at 1.5 Å, is the highest resolution of an E. coli PBP 5 structure to date. For the majority of the imipenem molecule, the electron density is well resolved (Fig. 2A). Strong density is visible between the Oγ of Ser44 and C4 of imipenem, confirming this as an acyl-enzyme complex. The carbonyl oxygen of the β-lactam, which forms an oxyanion during both the acylation and deacylation steps of the reaction, is within hydrogen-bonding distance of the amides of His216 and Ser44 (the ‘oxyanion’ hole of PBP 5) (14). A hydrogen bond between the Nδ atom of Asn112 and the 6α-hydroxyethyl group is the sole hydrogen bond involving the R1 group of imipenem. An ordered water molecule is observed in the active site, which occupies a space between the ring nitrogen and carboxylate of imipenem, the carbonyl of Thr214 and the Oγ of Ser110. This is interesting because Ser110 is believed to be involved in activating or positioning the deacylating water molecule (14) and such a water may represent the initial position of the deacylating water prior to its movement along a trajectory toward the carbonyl carbon. The R2 side chain exhibits weak density, presumably due to its disorder, and there are no contacts between imipenem and enzyme in this region, except for a possible hydrogen bond with the carbonyl of Gly85.
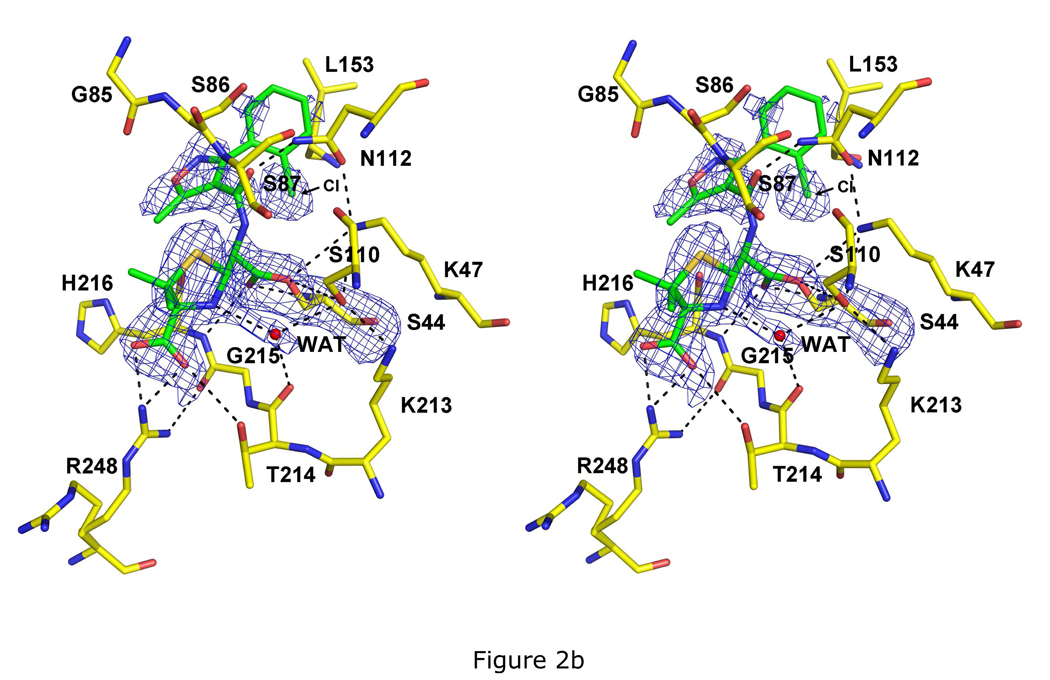
Fig. 2.
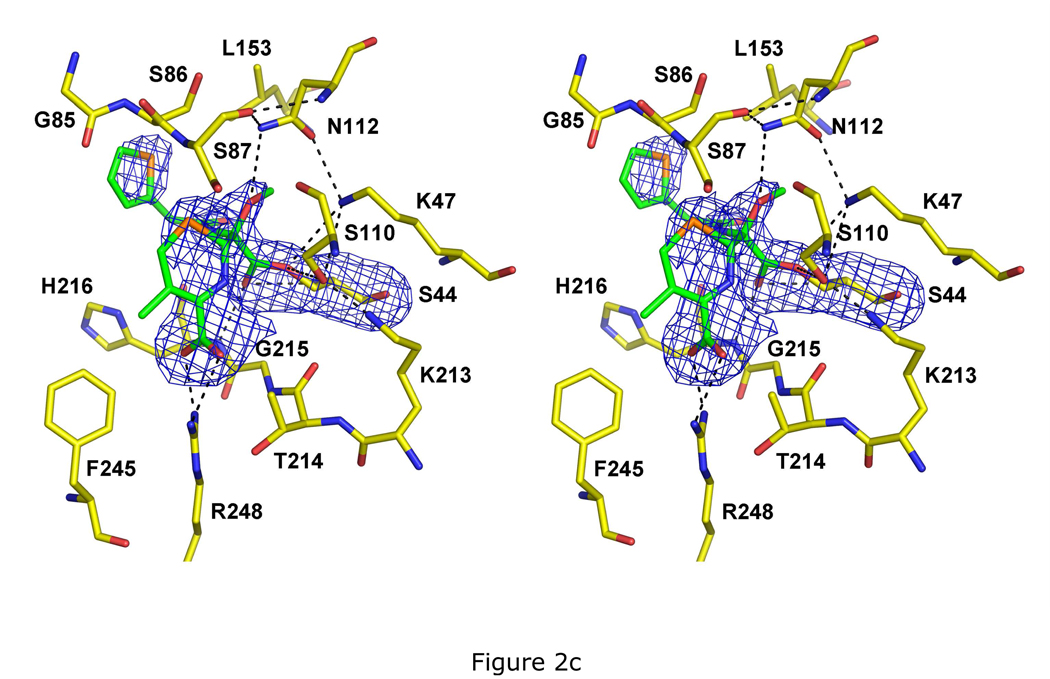
Crystal structures of PBP 5 as acyl-enzyme complexes with three β-lactam antibiotics. All three structures are shown as stereoviews of the active site region where the covalently bound β-lactam is displayed in green bonds and the amino acids are colored with yellow bonds. The density shown is difference |Fo|-|Fc| electron density calculated after a refinement in which the atoms corresponding to the β-lactam and to Ser44 were excluded. Potential hydrogen bonds are shown as dashed lines. The three structures are acyl-enzyme complexes of: A, imipenem at 1.5 Å resolution and contoured at 2.8 sigma; B, cloxacillin at 1.9 Å resolution and 2.5 sigma; and C, cefoxitin at 2.1 Å resolution and 2.5 sigma.
Interestingly, Arg248 exhibits two conformations in the electron density: one that contacts the carboxylate and the other that occupies the same position as observed in the wild-type structure of PBP 5. Compared to the structure of wild-type PBP 5, the guanidinium group of Arg248 has moved over 6 Å to contact the carboxylate. This interaction was unexpected because we anticipated previously that Arg198 in PBP 5 would contact the carboxylate of β-lactams (15). A similar arrangement is seen in the complex with cloxacillin, whereas in the cefoxitin-bound structure only the conformer that contacts the carboxylate is observed (see below).
Cloxacillin
The structure of PBP 5 in complex with cloxacillin was determined at 1.9 Å resolution (Table 1). Most of the density corresponding to the thiazolidine ring and carboxylate of cloxacillin is clear except for one of the two methyl groups at position 2 (which may be due to multiple conformations of the thiazolidine caused by ring puckering) (Fig. 2B). Again, the side chain of Arg248 is present in two conformations, one the same as in the native enzyme and the other in contact with the cloxacillin carboxylate. Most of the R1 side chain exhibits weak density, especially the 2-chlorophenyl ring. The latter was positioned (tentatively) using the few peaks of positive density present, including a stronger peak that might correspond to the chloro group. In this position, the ring lies within a pocket formed by Leu153, Ser86 and Asn112. The weak density is probably caused by rotation about the carbon-carbon bond connecting to the oxazole ring. Such a rotation would explain the relatively stronger density observed for the oxazole because, as this ring rotates, it would remain in essentially the same position, whereas there would be multiple positions for the 2-chlorophenyl ring. Consistent with this idea, there are some peaks of positive electron density near Asp154 that might be the 2-chlorophenyl ring occupying an alternative conformation. This latter position is similar to the one observed in the structure of the Class C AmpC β-lactamase in complex with cloxacillin (27) (see supplemental Fig. 1 for superimposition). Since the 2-chlorophenyl ring cannot be modeled without ambiguity in the crystal structure, its atoms have been modeled with zero occupancy.
Table 1.
Data collection and refinement statistics for the three PBP 5 structures
| Antibiotic | Cloxacillin | Cefoxitin | Imipenem |
|---|---|---|---|
| Molarity of soak (mM) | 10 | 10 | 10 |
| Time of soak (mins) | 60 | 60 | 90 |
| Wavelength (Å) | 1.5418 | 1.5418 | 1.0000 |
| Resolution range (Å) | 54.0–1.9 (1.97–1.90) |
54.0–2.1 (2.18–2.10) |
33.0–1.50 (1.55–1.50) |
| Rmerge (%) | 7.3 (33.6) | 9.5 (37.1) | 5.5 (48.8) |
| Redundancy | 4.9 (3.6) | 4.3 (4.1) | 6.8 (3.9) |
| Completeness (%) | 97.5 (78.5) | 100.0 (100.0) | 96.6 (76.3) |
| <I>/<σI> | 7.0 (2.0) | 5.2 (1.9) | 40.1 (1.8) |
| Number of glycerol molecules | 1 | 4 | 1 |
| Number of water molecules | 309 | 118 | 369 |
| Crystallographic R factor (%) | 21.6 | 22.8 | 18.9 |
| R work (%) | 21.4 | 22.5 | 18.8 |
| R free (%) | 25.7 | 28.1 | 21.2 |
| RMS deviations from ideal stereochemistry:- | |||
| bond lengths (Å) | 0.011 | 0.012 | 0.009 |
| bond angles (°) | 1.37 | 1.49 | 1.23 |
| Mean B factors (Å2): | |||
| all atoms | 25.7 | 28.6 | 22.9 |
| main chain | 24.6 | 27.8 | 20.7 |
| side chain | 25.8 | 28.7 | 22.5 |
| Waters | 30.7 | 29.8 | 32.7 |
| Antibiotic | 41.5 | 43.1 | 28.7 |
| Ramachandran plot: | |||
| residues in most favored region (%) | 92.2 | 91.1 | 92.8 |
| residues in additionally allowed regions (%) | 7.5 | 8.2 | 6.8 |
| residues in generously allowed regions (%) | 0.3 | 0.3 | 0.3 |
| residues in disallowed regions (%) | 0.0 | 0.3 | 0.0 |
| RMSD of Cα atom with native structure* (Å) | 0.56 | 0.47 | 0.61 |
| PDB code | 3MZD | 3MZE | 3MZF |
where Rmerge = ∑| Ii-Im | / ∑ Ii where Ii is the intensity of the measured reflection and Im is the mean intensity of all symmetry-related reflections. Figures within brackets are for the outer resolution shell.
calculated using all common main chain atoms between the two structures.
Most of the potential hydrogen bonds between cloxacillin and PBP 5 are similar to those in the imipenem-bound structure. The same water molecule between the ring nitrogen, Thr214 and Ser110 is also observed (although it is relatively farther away from the ring carboxylate) and the side chain of Thr214 has also rotated to contact the carboxylate. In addition, a hydrogen bond between the Nδ2 atom of Asn112 and the carbonyl of the amido group of cloxacillin is equivalent to that between the same atom and the hydroxyl of the 6α–hydroxyethyl in imipenem.
Cefoxitin
The structure of PBP 5 in complex with cefoxitin was solved at 2.1Å resolution (Table 1). The density is clear and unambiguous for the covalent linkage, the 7α-methoxy group, and most of the dihydrothiazine ring except for the C3 carbon and its substituent methylene group (Fig. 2C). A similar lack of density at C3 was also observed in the crystal structure of PBP 5 in complex with a designed cephalosporin (28) and, as for cloxacillin, it may be due to ring puckering. The dihydrothiazine ring is packed on one side by the hydrophobic portion of the side chain Arg198 and to a lesser extent by the thiophene ring on the other side. Phe245 also contributes to the hydrophobicity of the binding pocket and has undergone a conformational change to move closer to the active site (see below). In a similar fashion to the cloxacillin-bound structure, the R1 side chain of cefoxitin shows weak density and makes only minimal contact with the enzyme.
Compared to the structures in complex with imipenem and cloxacillin, the side chain of Arg248 fully occupies the position that contacts the carboxylate of the antibiotic and both nitrogens of the guanidinium group are within hydrogen bonding distance of the carboxylate, thus forming a bidentate interaction. Interestingly, the side chain of Thr214 does not appear to have rotated to contact the carboxylate. It is possible that the stronger interaction between the carboxylate group and Arg248 in this structure negates the necessity for the interaction with the threonine hydroxyl. Another difference is the lack of an ordered water molecule near the ring nitrogen (although there is a hint of such a water in the |Fo|-|Fc| difference electron density map and its relative weakness may simply be due to the lower resolution of this structure). These differences aside, the hydrogen bonding is very similar to the imipenem- and cloxacillin-bound structures, including a hydrogen bond (albeit relatively longer) between Asn112 and the 7α-methoxy of cefoxitin (equivalent to the 6α-hydroxyethyl in imipenem).
Superimposition of the three structures
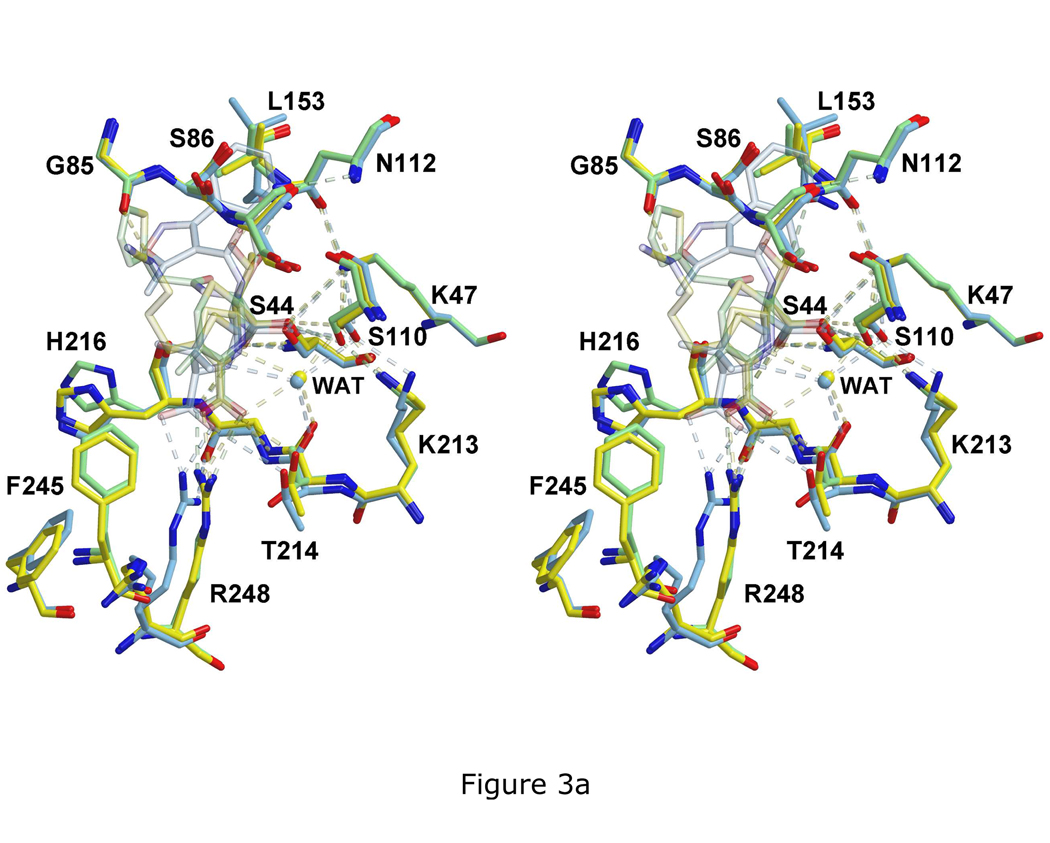
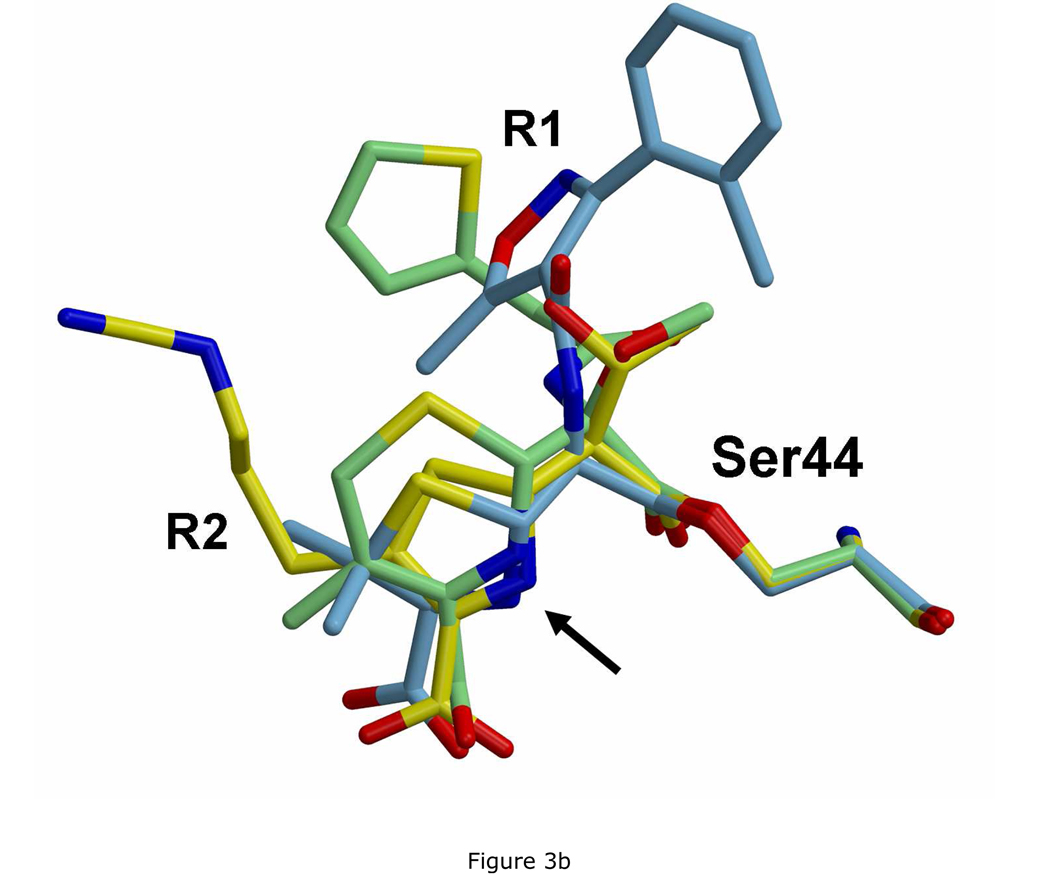
The overall conservation of the hydrogen-bonding network with the actives sites of the three acyl-enzyme complexes is most evident when the three structures are superimposed (Fig. 3a). The intra-enzyme hydrogen bonds between Ser110 and Lys213, Lys47 and Ser44, and Lys47 and Asn112 are essentially the same as those observed in the structure of wild-type PBP 5 (15). The superimposition also shows the gradual divergence in the positions of the antibiotics in the three structures when moving away from Ser44 (Fig. 3a and 3b). There appears to be a large amount of space available in the active site of PBP 5 to accommodate the R1 group, which might explain its weak density and variable positions in the cloxacillin- and cefoxitin-bound structures and makes the failure to obtain a structure in complex with moxalactam somewhat surprising.
Fig. 3.
Overlay of the three structures showing the divergence in the positions of R1 or R2 groups. A. In this stereoview of the superimposed active sites, the imipenem complex is colored yellow, the cloxacillin complex is blue and the cefoxitin complex is green. For clarity, atoms of the β-lactams have been rendered with a degree of transparency. Note the conservation of the hydrogen bonding network, shown as dashed lines. B. The same overlay, but showing only the three antibiotics. The arrow marks the position of the ring nitrogen.
Enzyme-antibiotic complementarity
One goal of this study was to determine whether the degree of complementarity between antibiotic and enzyme correlates with the second order rates of acylation rates of the same antibiotics against PBP 5. Excluding atoms that were not visible in the electron density (i.e. were modeled with zero occupancy), the area occluded by antibiotic in each of the three complexes was calculated and compared with the second order rate constant of acylation and the non-covalent interaction energy for the same antibiotics, as determined by Beadle et al. (12) (Table 2). Even though cloxacillin occludes the greatest surface area of PBP 5 (258 Å2), it exhibits the lowest rate of acylation with PBP 5. Overall, there is no correlation with acylation rate and this agrees with Beadle et al., who found a similar lack of correlation between acylation rate and the non-covalent interaction energy (12).
Table 2.
Lack of correlation between enzyme-β-lactam complementarity and second order rate of acylation
| Antibiotic | Total area difference (Å2) |
Non- covalent interaction energy (kcal/mol)* |
k2/KS (M−1/sec−1)* |
|---|---|---|---|
| Imipenem | 166 | 2.6 ± 0.4 | 790 ± 170 |
| cloxacillin | 258 | 2.7 ± 0.2 | 104 ± 30 |
| cefoxitin | 193 | 2.0 ± 0.4 | 2750 ± 590 |
data from (12)
Conformational changes
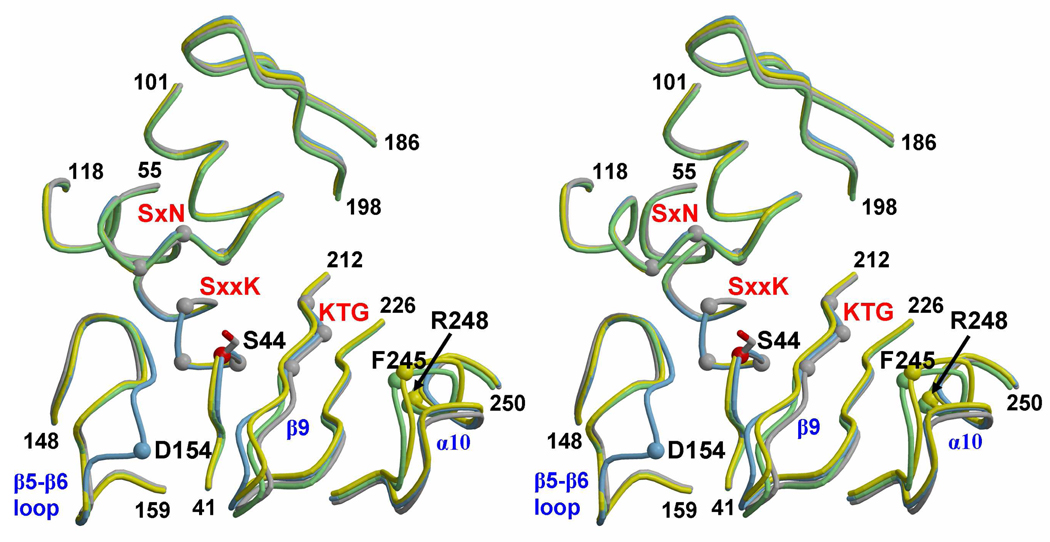
The absence of a relationship between contact surface area and acylation rate led us to examine whether conformational changes that occur in PBP 5 in response to acylation by a specific β-lactam might correlate with the observed rates of acylation. When comparing the three antibiotic-bound and wild-type structures of PBP 5, two of the three conserved motifs of the active site (SxxK and SxN) overlap almost exactly, whereas there are minor differences in the KTG motif. The 74–90 region, which is disordered in the structure of a deacylation-defective mutant of PBP 5 but ordered in wild type PBP 5 (15, 29), is unchanged in conformation. The structural differences occur in three regions of PBP 5 near the active site (β9, β5-β6 loop and α10) and are different for each antibiotic (Fig. 4).
Fig. 4.
Mapping of conformational changes that occur in the three acyl-enzyme complexes of PBP 5. This stereoview shows the superimposed backbones of the following structures: wild-type, grey; imipenem, yellow, cloxacillin, blue; and cefoxitin, green. The three conserved motifs that are the hallmark of PBPs are labeled in red and the Cα atoms of these residues are displayed in grey CPK format. The Cα atoms of Arg248 and Phe245 in the imipenem complex are shown in green and for Asp154 in the cloxacillin complex in blue. Specific elements of secondary structure are labeled blue. Residue numbers at the beginning and end of each segment displayed are marked. (For description of the secondary structure see (29)).
β9 (which contains the KTG motif) and the β9-β10 loop are both shifted toward the active site (to various degrees) in all three structures. The largest shift is observed in the cloxacillin- and imipenem-bound structures and may be due to the contact between Thr214 and ring carboxylate, which is not observed in the cefoxitin-bound structure. This shift is often observed in acyl-enzyme complexes of PBPs because this β strand (usually called β3 in other PBPs) provides a main-chain amide that helps form the oxyanion hole and a threonine or serine that contacts the β-lactam carboxylate, e.g. (30–33).
One structural difference that occurs only in the cloxacillin-bound structure is the shift toward the active site of residues 152–155 in the β5-β6 loop. At first sight, this shift is surprising because, even though it has brought Asp154 closer to the active site, there is no direct contact of this residue with cloxacillin. However, the loop may have moved in response to one of the apparent alternative conformations of the 2-chlorophenyl ring (described above), as suggested by some positive difference density near Asp154. Interestingly, a similar shift of these residues occurred in the crystal structure of PBP 5 bound to a boronic acid compound (14), but in two equally occupied conformations. Small peaks of positive electron density indicate that the wild-type like conformation is present in the cloxacillin-bound structure, but at very low occupancy.
The most pronounced structural difference between wild-type and acylated structures is observed in the cefoxitin-bound enzyme in which a shift of residues 242–250, comprising the N-terminal region of α10, brings both Phe245 and Arg248 closer to the active site. Arg248 contacts the carboxylate (as mentioned above) whilst Phe245 lies close to the methylene group of the dihydrothiazine ring. This same loop exhibits apparent flexibility in the imipenem-bound structure, with two apparent routes for residues 242–249. (Additional peaks in the electron density suggest a third position, but not one that can be modeled). By contrast, this loop does not shift in the cloxacillin-bound structure. The complete shift of this loop, together with the specific nature of the contact made by Arg248, may explain why cefoxitin acylates PBP 5 at a higher rate than either cloxacillin or imipenem. In fact, whether by coincidence or not, the degree of differences with wild-type PBP 5 of this region does correlate with k2/KS, as discussed below.
Mutations at position 248 lower CPase activity
To determine whether the different positions of Arg248 in the acylated complexes of PBP 5 correlate with the kinetics of acylation, we determined k2/KS for both penicillin G and cefoxitin of wild-type PBP 5 and of two mutants: Arg248Lys and Arg248Ala (Table 3). For penicillin G and cefoxitin, the rates of acylation were lowered for both mutants, but by only 2- to 4-fold. Acylation constants for cefoxitin, though, were not as affected as those for penicillin, which suggests that the movement of the N-terminal end of α10 does not promote acylation of PBP 5 by an induced-fit mechanism. The mutations did not affect deacylation of the penicillinoyl complex either, with only a slight lowering of k3 for R248K, compared to wild type, and no difference in that for R248A. The carboxypeptidase activity of both mutants, however, was severely reduced: the kcat/Km was lowered by 20-fold for R248K and 40-fold for R248A. This suggests an important role for Arg248 in CPase activity and one probable function of this residue is positioning of the carboxylate of the terminal d-Ala during acylation, as suggested by the crystal structure of PBP 6 in complex with a synthetic muramyl-pentapeptide substrate, in which the equivalent to Arg248 (Arg244) contacts the carboxylate of the d-Ala via a bridging water molecule (34).
Table 3.
Kinetic constants of Arg248 mutants of PBP 5. WT=wild-type. The number of measurements are given in parentheses.
|
kcat/Km (M−1 s−1) |
k2/KS (M−1s−1) |
t1/2 (min) |
k3 (s−1 × 10−5) |
||
|---|---|---|---|---|---|
| Substrate | Ac2-KAA | Penicillin G | Cefoxitin | Penicillin G | Penicillin G |
| WT | 47.0 ± 1.9 (n=4) |
450 ± 70 (n=6) |
3410 ± 20 (n=3) |
14.9 ± 1.6 (n=2) |
78.3 ± 8.5 |
| R248K | 2.3 ± 0.9 (n=4) |
134 ± 20 (n=7) |
1760 ± 80 (n=3) |
25.9 ± 1.7 (n=3) |
44.8 ± 2.8 |
| R248A | 1.2 ± 0.5 (n=3) |
125 ± 14 (n=5) |
1550 ± 150 (n=3) |
14.3 ± 0.4 (n=3) |
80.8 ± 2.1 |
Superimposition with PBP 5 bound with a boronic acid inhibitor
Previously, we determined the structure of PBP 5 in complex with a tripeptide boronic acid inhibitor, which is a mechanistic probe of deacylation (14). Of the two hydroxyls bonded to the boron, one (O2) mimics the oxyanion of the tetrahedral intermediate for deacylation and lies within the oxyanion hole comprising the amides of Ser44 and His216, and the other (O3) mimics the incoming water that hydrolyzes the ester linkage (the hydrolytic or deacylating water).
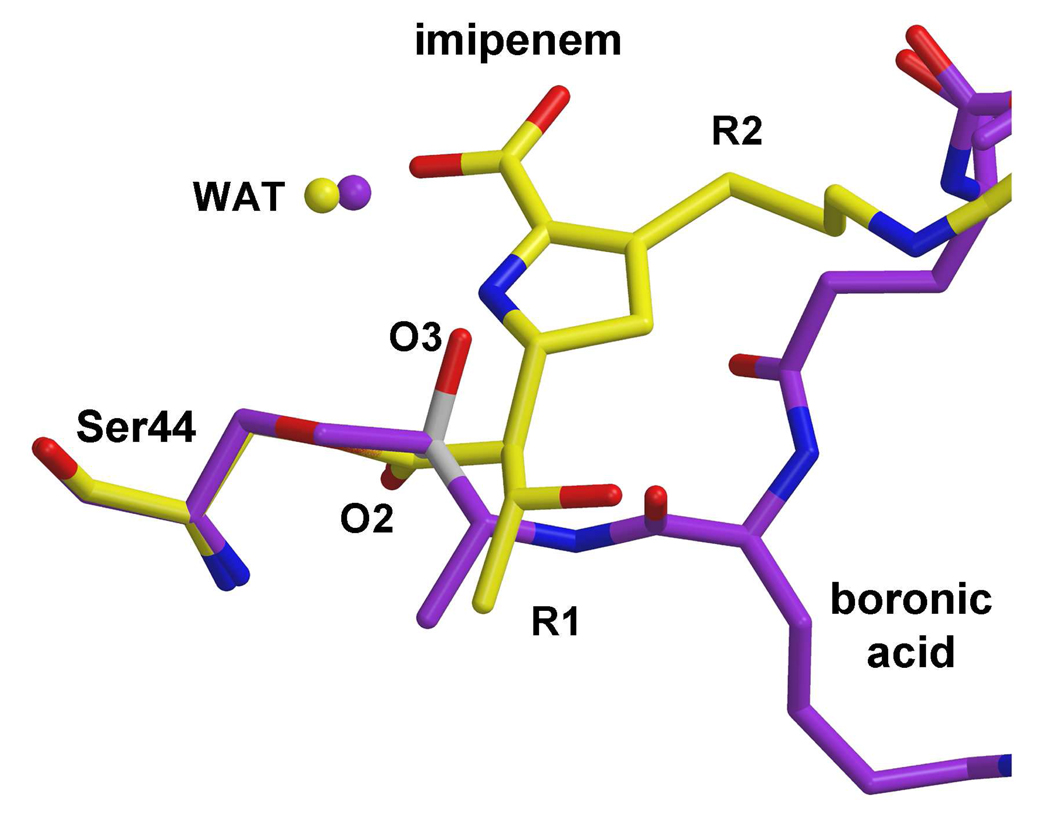
The structure of the boronic acid complex was compared to the imipenem-bound structure of PBP 5 in order to examine why the rate of deacylation is lowered for a β-lactam-bound complex (the imipenem-bound structure was chosen because it is the highest resolution of the three structures). The superimposition shows that, beyond the vicinity of Ser44, there is little overlap between the boronic acid and imipenem (Fig. 5), although the R2 group extends generally in the direction of the l-lysyl side chain of the inhibitor, and were an R1 group present, it would overlap approximately with the main chain of the peptidyl region of the inhibitor. Critically, the O3 oxygen atom lies very close to the ring nitrogen of imipenem (1.4 Å) in support of a mechanism where deacylation is inhibited because the ring nitrogen of the β-lactam blocks access of the water molecule to the ester linkage. The close overlap of the ring nitrogen in all three antibiotics (marked by arrow in Fig. 3b) suggests the water molecule would also be blocked by the other two β-lactams.
Fig. 5.
Superimposition of the imipenem- and peptidyl boronyl-complexes of PBP 5. The imipenem molecule is colored yellow and the boronic acid is purple. The conserved water molecules are shown as spheres of the same respective colors.
Discussion
In this paper, we present the crystal structures of the acylated complexes of E. coli PBP 5 with cefoxitin, cloxacillin and imipenem. The goal of this work was to see whether the structures explain why these antibiotics exhibit such dramatically different rates of acylation against PBP 5 and also to examine how deacylation is blocked in complexes with β-lactams but not peptides. The weak or absent electron density corresponding to the R1 and R2 side chains for each of the antibiotics suggests that these substituents do not participate in interactions with PBP 5, at least in the acylated complex, and the area of contact of the antibiotics with PBP 5 does not correlate with their rates of acylation of the enzyme. We also investigated whether a conformational rearrangement of a loop resulting in an electrostatic interaction of Arg248 and the β-lactam carboxylate of the antibiotics might explain the specificity of these antibiotics for inhibition of the enzyme, but mutagenesis of Arg248 had only minor effects on β-lactam binding. In contrast, the Arg248 mutants had a major effect on carboxypeptidase activity, suggesting that this residue plays an important role in enzymatic activity with peptide substrates. Finally, these structures show that the amino group generated upon acylation of the β-lactam is likely to block the deacylating water molecule, providing more support for the hypothesis that β-lactam antibiotics block hydrolysis of the acylated complex due to the continued presence of the leaving group. The implications of these structures on both the acylation and deacylation steps of the reaction are discussed below.
Lack of electron density for β-lactam side chains
One of the surprises of the structures of PBP 5 in complex with the three β-lactams is the lack of electron density around the R1 and R2 groups. In each structure, the strongest density is focused around the central core of the antibiotic, with weaker or absent density for the R1/R2 groups. A similar distribution of electron density was also observed in crystal structures of PBP 5 in complex with peptidoglycan-mimetic β-lactams (28). This contrasts with acyl-enzyme complexes of other PBPs where the electron density typically covers the entire antibiotic molecule (PBP 2a (31), PBP 2X (35), PBP 5fm (36), PBP 4 (37), PBP4 from H. influenzae (38) and R61 (39)). The paucity of electron density for the side chains is not merely an unusual property of the chosen antibiotics because all three show good density when bound in crystal structures of β-lactamases (i.e. cloxacillin in AmpC (class C β-lactamase) (27); cefoxitin in the CTX-M extended-spectrum β-lactamase (40) and BS3 (class A β-lactamase) (41); and imipenem in AmpC (42) and TEM-1 (class A β-lactamase) (43)).
The reason for the lack of density for the R1/R2 groups is not obvious. One possibility is that the soluble construct used to crystallize PBP 5 differs structurally in the presumed binding sites for the β-lactam side chains compared to the protein in vivo, perhaps due to the loss of association with the cell membrane or a protein-binding partner. This has been proposed as one possible reason why PBPs in vitro catalyze cross-linking reactions at rates much slower than would be required for bacterial growth (4, 44). A second possibility is that the active site of PBP 5 is distorted due to crystal contacts. However, since the acyl-enzyme complexes were generated by soaking of PBP 5 crystals, it appears that the active site within the crystal is competent for reactivity with β-lactams. In regard to both of these possibilities, it is pertinent to note that, compared to other PBPs, PBP 5 has a wider active site cavity, as measured by Asn112-Gly115 and Ser44-Gly115 distances (E. coli PBP 5 numbering)(28), and this relative openness may reduce binding interactions with the β-lactam side chains. A third explanation for the lack of electron density for R1/R2 side chains is that such contacts occur only in the pre-covalent state and/or in the acylation transition state, and not in the post-covalent state that is observed in the crystal structures (discussed below).
What determines the acylation rate of β-lactams?
Our observation that the interaction surfaces between PBP 5 and β-lactam antibiotic in the three complexes do not correlate with the second order rates of acylation (Table 2) is similar to that of Beadle et al (12) who, by measuring the non-covalent interaction energies of the same antibiotics when bound covalently to PBP 5, also observed a lack of correlation between complementarity and acylation. This led us to examine whether specific structural changes that occur during acylation by β-lactams might instead be responsible. Central amongst these is the shift of the N-terminal end of α10, which brings Phe245 and Arg248 toward the active site. This displacement appears to correlate with k2/KS because it is largest in the cefoxitin-bound structure, with a partial shift in the imipenem complex and is absent for the cloxacillin complex. The driving force for this shift appears to be the strong electrostatic linkage between Arg248 and the β-lactam carboxylate. Where this interaction is weaker, as in the imipenem-bound complex, or absent, as in the cloxacillin-bound complex, Thr214 rotates to contact the carboxylate. Interesting, a similarly large shift of Phe245 and Arg248 was observed in another acyl-enzyme complex of PBP 5 with a designed cephalosporin, but not with a similar penicillin (28). Again, this shift appeared as if it might correlate with the kinetics because the cephalosporin inhibited PBP 5 by a two-fold higher rate than penicillin G (44). Mutation of Arg248 to Lys or Ala, however, lowered the k2/KS for penicillin and cefoxitin by 4- and 2-fold, respectively. The relatively minor effects of the mutations suggest that the conformational shift observed is not particularly critical for acylation by β-lactams, and it may instead be a consequence of the carboxylate occupying slightly different positions in the three structures.
So what factors influence the different rates of acylation of β-lactams toward PBP 5? Answering such a question is obviously important for the design of new antimicrobials directed against PBPs. The significantly different rates of acylation observed between the three antibiotics suggests that the R1 or R2 groups play a major role in acylation, but, given the lack of enzyme-antibiotic contacts in PBP 5 beyond the vicinity of the β-lactam ring, their contribution to rate enhancement is unclear (A possible exception here is cefoxitin because loss of the leaving group at C-3’ and concurrent changes in the thiazine geometry may both impact acylation rate). One immediate explanation is that the structures observed are of the post-covalent states; thus, any contacts present during the Michaelis-Menten interaction that contribute to the rate of acylation are not observed. Arguing against this idea, however, is the relatively small magnitude of the conformational differences between bound and unbound structures of PBP 5 (see Fig. 4). Moreover, with the exception of the 152–156 loop that moves in response to the binding of cloxacillin, the observed shifts bring the enzyme into closer contact with groups that are common to β-lactams, i.e. the acyl-enzyme linkage and the carboxylate, and not the R1 or R2 groups, which suggests that any induced fit mechanism is the same for the three antibiotics. The number and strength of the molecular interactions present is another potential factor, but again these are very similar in the three complexes (Fig. 3a).
Taken together, it seems that the central β-lactam ring is primarily responsible for reactivity with PBP 5, in keeping with the idea that β-lactams acylate PBPs due to their resemblance to the tetrahedral intermediates or transition states of the reaction with peptide (5, 45). The R1/R2 side chains, however, must exert some influence and, although they make only minimal binding contributions in the acylated complexes of PBP 5, their size and shape may limit the number of bound conformations available to the antibiotic during acylation and this may explain the very different rates. Irrespective of the exact mechanism, it seems that post-covalent complexes of PBPs with β-lactams do not provide relevant information regarding the relative potencies of antibiotics, and thus may not be optimal vehicles for the design of new β-lactam inhibitors of PBPs (12).
Implications for the mechanism of deacylation
Previously, by determining the structure of PBP 5 as a tetrahedral complex of a tripeptide boronic acid inhibitor, we established the position of the hydrolytic water in the active site of PBP 5 (14). In that structure, the hydroxyl in the boronic acid compound that mimics the hydrolytic water molecule (O3 in Fig. 5) was hydrogen bonded to a water molecule, which in turn was hydrogen bonded to the carbonyl of Thr214 and the side chain hydroxyl of Ser110 (14). Based on this, we proposed a mechanism of hydrolysis comprising a polarizing network linking the hydrolytic water molecule to Lys213 via the bridging water molecule and Ser110. A recent study has argued, however, that Lys213 does not act in deacylation, but rather serves as an electrostatic anchor for the carboxylate of the d-Ala leaving group and the β-lactam carboxylate, and Lys47 was proposed instead to activate the hydrolytic water molecule (46). On two counts, such a scenario is unlikely. Firstly, in the boronic acid-PBP 5 complex, Lys47 does not appear well placed to orient a water molecule occupying the same position as O3 of the boronic acid. Secondly, the idea of Lys213 acting as electrostatic anchor for the β-lactam carboxylate is not supported by our structures because, in all three cases, this lysine is too far away and, notably, in all crystal structures of PBP 5 solved to date, irrespective of whether these are of the native enzyme or of acyl-enzyme complexes, this residue is always hydrogen bonded to Ser110, which is consistent with the idea of a polarization network involving these residues. Moreover, in structures of other PBPs and of Class A β-lactamases, the equivalent lysine also does not contact the carboxylate (e.g. (36, 37, 47)). Hence, the structures of PBP 5 in complex with the β-lactams provide no compelling reason to alter our existing hypotheses regarding the catalytic mechanism of deacylation in this enzyme.
How β-lactams work
The molecular basis for the antibiotic action of β-lactams is the prevention of deacylation in an acyl-enzyme complex with a PBP, but how that is achieved mechanistically remains unclear. This issue, however, has been examined extensively for β-lactamases, which are evolutionarily and mechanistically related to PBPs, and a number of theories have been posited. One is that deacylation can be blocked by impeding the formation of the tetrahedral intermediate (or its transition state). For instance, in AmpC (a class C β-lactamase), it has been suggested that interactions with the R1 group of ceftazidime cause the dihydrothiazine ring to occupy a position that sterically hinders formation of the tetrahedral intermediate/transition state (48). The 7α-methoxy group of cefoxitin may also cause a similar steric hindrance in CTX-M, an extended spectrum β-lactamase (49). Neither mechanism is very likely in PBP 5, however, because very few interactions involving R1 groups are observed in the crystal structures and because the 7α-methoxy in cefoxitin and 7α-hydroxyethyl of imipenem lie on the opposite side of acyl linkage from the two hydroxyls of the boronic acid, which mimics the tetrahedral intermediate. A third mechanism for hindering the tetrahedral intermediate, the displacement of the carbonyl from the oxyanion hole, which was observed in imipenem complexes of TEM-1 (Class A β-lactamase) and AmpC (Class C) (42, 43) is also unlikely in PBP 5 because, in the complex with imipenem, the carbonyl is positioned normally within the oxyanion hole formed by the amide nitrogens of Ser44 and His216.
A second mechanism for blocking deacylation is occlusion of the deacylating water by the pendant leaving group of β-lactams (39, 50, 51) and this appears to be the case for PBP 5. If the water molecule observed in the crystal structures of the complexes with imipenem and cloxacillin (see Fig. 2), as well as in the boronyl complex (see Fig. 6), represents the initial position of the deacylating water, then its movement along a trajectory toward its final position, as represented by O3 in the boronyl complex, would be impeded by the ring nitrogen. If there is any role for the β-lactam carboxylate, it might be to restrain the water to its initial position, but it does not appear well placed to block the trajectory of the water, as has been suggested for the inhibition of AmpC by loracarbef (27). Importantly, in all three structures, the position of the ring nitrogen remains fairly constant, even though the positions of the R groups vary considerably (Fig. 3B).
This conclusion contrasts with a similar analysis of PBP6 that was reported recently. When the structure of PBP6 acylated by ampicillin was superimposed with the structure of PBP 5 in complex with the boronic acid inhibitor, it was observed that the bridging carbon of the β-lactam was the closest to the deacylating water and not the nitrogen atom (Chen, 2009). Although PBP 5 and PBP6 share very high sequence identity (61.4 %), such an analysis is likely to be more accurate when structures from the same PBP, i.e. PBP 5, are superimposed. Irrespective of whether occlusion of the hydrolytic water is mediated by the ring nitrogen or the bridging carbon of the β-lactam, it is obvious that blockage of the hydrolytic water cannot occur for a peptide substrate because the terminal d-Ala of the pentapeptide substrate has left during acylation (Chen, 2009). For a β-lactam, though, the “leaving” group does not leave because the β-lactam is cyclic and remains tethered. The ring structure of the β-lactam is, therefore, critical to its success as an inhibitor (50, 52). Examination of high-molecular weight PBPs in complex with beta-lactams suggests that the same ring nitrogen (or bridging carbon) would be blocked by the terminal amino group of an incoming acceptor peptide. If so, the same mechanism would also apply to PBPs that are transpeptidases, the clinical targets for β-lactams, thus explaining mechanistically the core antibiotic activity of β-lactams.
Supplementary Material
Acknowledgements
Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. W-31-109-ENG-38. Data were collected at Southeast Regional Collaborative Access Team (SER-CAT) 22-ID beamline at the Advanced Photon Source, Argonne National Laboratory. Supporting institutions may be found at www.ser-cat.org/members.html. The X-ray crystallography facility used for this work is supported by the Medical University of South Carolina's Research Resource Facilities program.
Abbreviations used
- CPase
carboxypeptidase
- PBP
penicillin-binding protein
- RMSD
root mean square deviation
- TPase
transpeptidase
Footnotes
This work was supported by the National Institutes of Health grants GM66861 to CD, AI36901 to RAN and AI17986 to RFP.
SUPPORTING INFORMATION AVAILABLE
A superimposition of the structures of E. coli PBP 5 and AmpC β-lactamase, showing cloxacillin bound in each active site. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Ghuysen JM. Serine beta-lactamases and penicillin-binding proteins. Annu. Rev. Microbiol. 1991;45:37–67. doi: 10.1146/annurev.mi.45.100191.000345. [DOI] [PubMed] [Google Scholar]
- 2.Waxman DJ, Strominger JL. Penicillin-binding proteins and the mechanism of action of beta-lactam antibiotics. Annu. Rev. Biochem. 1983;52:825–869. doi: 10.1146/annurev.bi.52.070183.004141. [DOI] [PubMed] [Google Scholar]
- 3.Macheboeuf P, Contreras-Martel C, Job V, Dideberg O, Dessen A. Penicillin binding proteins: key players in bacterial cell cycle and drug resistance processes. FEMS Microbiol. Rev. 2006;30:673–691. doi: 10.1111/j.1574-6976.2006.00024.x. [DOI] [PubMed] [Google Scholar]
- 4.Pratt RF. Substrate specificity of bacterial DD-peptidases (penicillin-binding proteins) Cell Mol Life Sci. 2008;65:2138–2155. doi: 10.1007/s00018-008-7591-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tipper DJ, Strominger JL. Mechanism of action of penicillins: a proposal based on their structural similarity to acyl-D-alanyl-D-alanine. Proc. Natl. Acad. Sci. U S A. 1965;54:1133–1141. doi: 10.1073/pnas.54.4.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yocum RR, Waxman DJ, Rasmussen JR, Strominger JL. Mechanism of penicillin action: penicillin and substrate bind covalently to the same active site serine in two bacterial D-alanine carboxypeptidases. Proc. Natl. Acad. Sci. U S A. 1979;76:2730–2734. doi: 10.1073/pnas.76.6.2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frere JM, Ghuysen JM, Iwatsubo M. Kinetics of interaction between the exocellular dd-carboxypeptidase- transpeptidase from Streptomyces R61 and beta-lactam antibiotics. A choice of models. Eur. J. Biochem. 1975;57:343–351. doi: 10.1111/j.1432-1033.1975.tb02307.x. [DOI] [PubMed] [Google Scholar]
- 8.Frere JM, Nguyen-Disteche M, Coyette J, Joris B. Mode of action: interaction with the penicillin-binding proteins. In: Page MI, editor. The chemistry of β-lactams. Glasgow: Chapman & Hall; 1992. pp. 148–196. [Google Scholar]
- 9.Chambers HF, Sachdeva MJ, Hackbarth CJ. Kinetics of penicillin binding to penicillin-binding proteins of Staphylococcus aureus. Biochem. J. 1994;301:139–144. doi: 10.1042/bj3010139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Graves-Woodward K, Pratt RF. Reaction of soluble penicillin-binding protein 2a of methicillin-resistant Staphylococcus aureus with beta-lactams and acyclic substrates: kinetics in homogeneous solution. Biochem. J. 1998;332:755–761. doi: 10.1042/bj3320755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu WP, Sun Y, Bauer MD, Paule S, Koenigs PM, Kraft WG. Penicillin-binding protein 2a from methicillin-resistant Staphylococcus aureus: kinetic characterization of its interactions with beta-lactams using electrospray mass spectrometry. Biochemistry. 1999;38:6537–6546. doi: 10.1021/bi990025e. [DOI] [PubMed] [Google Scholar]
- 12.Beadle BM, Nicholas RA, Shoichet BK. Interaction energies between beta-lactam antibiotics and E. coli penicillin-binding protein 5 by reversible thermal denaturation. Protein Sci. 2001;10:1254–1259. doi: 10.1110/ps.52001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ghuysen JM, Frere JM, Leyh-Bouille M, Coyette J, Dusart J, Nguyen-Disteche M. Use of model enzymes in the determination of the mode of action of penicillins and delta 3-cephalosporins. Ann. Rev. Biochem. 1979;48:73–101. doi: 10.1146/annurev.bi.48.070179.000445. [DOI] [PubMed] [Google Scholar]
- 14.Nicola G, Peddi S, Stefanova ME, Nicholas RA, Gutheil WG, Davies C. Crystal structure of Escherichia coli penicillin-binding protein 5 bound to a tripeptide boronic acid inhibitor: a role for Ser-110 in deacylation. Biochemistry. 2005;44:8207–8217. doi: 10.1021/bi0473004. [DOI] [PubMed] [Google Scholar]
- 15.Nicholas RA, Krings S, Tomberg J, Nicola G, Davies C. Crystal structure of wild-type penicillin-binding protein 5 from E. coli: Implications for deacylation of the acyl-enzyme complex. J. Biol. Chem. 2003;278:52826–52833. doi: 10.1074/jbc.M310177200. [DOI] [PubMed] [Google Scholar]
- 16.Pflugrath JW. The finer things in X-ray diffraction data collection. Acta Cryst. 1999;D55:1718–1725. doi: 10.1107/s090744499900935x. [DOI] [PubMed] [Google Scholar]
- 17.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Meths. Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 18.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Cryst. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 19.Jones TA, Zou J-Y, Cowan SW, Kjeldgaard M. Improved methods for building protein structures in electron-density maps and the location of errors in these models. Acta Cryst. 1991;A47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 20.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Cryst. 1997;D53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 21.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Cryst. 1993;26:283–291. [Google Scholar]
- 22.CCP4. The CCP4 suite: programs for protein crystallography. Acta Cryst. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 23.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 24.Stefanova ME, Davies C, Nicholas RA, Gutheil WG. pH, inhibitor, and substrate specificity studies on Escherichia coli penicillin-binding protein 5. Biochim. Biophys. Acta. 2002;1597:292–300. doi: 10.1016/s0167-4838(02)00311-4. [DOI] [PubMed] [Google Scholar]
- 25.Faraci WS, Pratt RF. Mechanism of inhibition of RTEM-2 beta-lactamase by cephamycins: relative importance of the 7 alpha-methoxy group and the 3' leaving group. Biochemistry. 1986;25:2934–2941. doi: 10.1021/bi00358a030. [DOI] [PubMed] [Google Scholar]
- 26.Faraci WS, Pratt RF. Nucleophilic re-activation of the PC1 beta-lactamase of Staphylococcus aureus and of the DD-peptidase of Streptomyces R61 after their inactivation by cephalosporins and cephamycins. Biochem. J. 1987;246:651–658. doi: 10.1042/bj2460651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patera A, Blaszczak L, Shoichet B. Crystal structures of substrate and inhibitor complexes with AmpC β-lactamase: possible implications for substrate-assisted catalysis. J. Am. Chem. Soc. 2000;122:10504–10512. [Google Scholar]
- 28.Sauvage E, Powell AJ, Heilemann J, Josephine HR, Charlier P, Davies C, Pratt RF. Crystal structures of complexes of bacterial DD-peptidases with peptidoglycan-mimetic ligands: the substrate specificity puzzle. J. Mol. Biol. 2008;381:383–393. doi: 10.1016/j.jmb.2008.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davies C, White SW, Nicholas RA. Crystal structure of a deacylation-defective mutant of penicillin-binding protein 5 at 2.3 Å resolution. J. Biol. Chem. 2001;276:616–623. doi: 10.1074/jbc.M004471200. [DOI] [PubMed] [Google Scholar]
- 30.Kelly JA, Knox JR, Zhao H, Frere JM, Ghuysen JM. Crystallographic mapping of beta-lactams bound to a d-alanyl-d-alanine peptidase target enzyme. J. Mol. Biol. 1989;209:281–295. doi: 10.1016/0022-2836(89)90277-5. [DOI] [PubMed] [Google Scholar]
- 31.Lim D, Strynadka NC. Structural basis for the beta lactam resistance of PBP2a from methicillin-resistant Staphylococcus aureus. Nat. Struct. Biol. 2002;9:870–876. doi: 10.1038/nsb858. [DOI] [PubMed] [Google Scholar]
- 32.Macheboeuf P, Di Guilmi AM, Job V, Vernet T, Dideberg O, Dessen A. Active site restructuring regulates ligand recognition in class A penicillin-binding proteins. Proc. Natl. Acad. Sci. U S A. 2005;102:577–582. doi: 10.1073/pnas.0407186102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sauvage E, Herman R, Petrella S, Duez C, Bouillenne F, Frere JM, Charlier P. Crystal structure of the Actinomadura R39 DD-peptidase reveals new domains in penicillin-binding proteins. J Biol Chem. 2005;280:31249–31256. doi: 10.1074/jbc.M503271200. [DOI] [PubMed] [Google Scholar]
- 34.Chen Y, Zhang W, Shi Q, Hesek D, Lee M, Mobashery S, Shoichet BK. Crystal structures of penicillin-binding protein 6 from Escherichia coli. J. Am. Chem. Soc. 2009;131:14345–14354. doi: 10.1021/ja903773f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamada M, Watanabe T, Baba N, Takeuchi Y, Ohsawa F, Gomi S. Crystal structures of biapenem and tebipenem complexed with penicillin-binding proteins 2X and 1A from Streptococcus pneumoniae. Antimicrob. Agents Chemother. 2008;52:2053–2060. doi: 10.1128/AAC.01456-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sauvage E, Kerff F, Fonze E, Herman R, Schoot B, Marquette JP, Taburet Y, Prevost D, Dumas J, Leonard G, Stefanic P, Coyette J, Charlier P. The 2.4 Å crystal structure of the penicillin-resistant penicillin-binding protein PBP5fm from Enterococcus faecium in complex with benzylpenicillin. Cell. Mol. Life Sci. 2002;59:1223–1232. doi: 10.1007/s00018-002-8500-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kishida H, Unzai S, Roper DI, Lloyd A, Park SY, Tame JR. Crystal structure of penicillin binding protein 4 (dacB) from Escherichia coli, both in the native form and covalently linked to various antibiotics. Biochemistry. 2006;45:783–792. doi: 10.1021/bi051533t. [DOI] [PubMed] [Google Scholar]
- 38.Kawai F, Clarke TB, Roper DI, Han GJ, Hwang KY, Unzai S, Obayashi E, Park SY, Tame JR. Crystal structures of penicillin-binding proteins 4 and 5 from Haemophilus influenzae. J. Mol. Biol. 2010;396:634–645. doi: 10.1016/j.jmb.2009.11.055. [DOI] [PubMed] [Google Scholar]
- 39.Kuzin AP, Liu H, Kelly JA, Knox JR. Binding of cephalothin and cefotaxime to d-ala-d-ala-peptidase reveals a functional basis of a natural mutation in a low-affinity penicillin-binding protein and in extended-spectrum beta-lactamases. Biochemistry. 1995;34:9532–9540. doi: 10.1021/bi00029a030. [DOI] [PubMed] [Google Scholar]
- 40.Chen Y, Delmas J, Sirot J, Shoichet B, Bonnet R. Atomic resolution structures of CTX-M beta-lactamases: extended spectrum activities from increased mobility and decreased stability. J. Mol. Biol. 2005;348:349–362. doi: 10.1016/j.jmb.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 41.Fonze E, Vanhove M, Dive G, Sauvage E, Frere JM, Charlier P. Crystal structures of the Bacillus licheniformis BS3 class A beta-lactamase and of the acyl-enzyme adduct formed with cefoxitin. Biochemistry. 2002;41:1877–1885. doi: 10.1021/bi015789k. [DOI] [PubMed] [Google Scholar]
- 42.Beadle BM, Shoichet BK. Structural basis for imipenem inhibition of class C beta-lactamases. Antimicrob Agents Chemother. 2002;46:3978–3980. doi: 10.1128/AAC.46.12.3978-3980.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maveyraud L, Mourey L, Kotra L, Pedelacq JD, Guillet V, Mobashery S, Samama JP. Structural basis for the clinical longevity of carbapenem antibiotics in the face of challenge by the common class A beta-lactamases from the antibiotic-resistant bacteria. J. Am. Chem. Soc. 1998;120:9748–9752. [Google Scholar]
- 44.Josephine HR, Charlier P, Davies C, Nicholas RA, Pratt RF. Reactivity of penicillin-binding proteins with peptidoglycan-mimetic beta-lactams: what's wrong with these enzymes? Biochemistry. 2006;45:15873–15883. doi: 10.1021/bi061804f. [DOI] [PubMed] [Google Scholar]
- 45.Lee B. Conformation of penicillin as a transition-state analog of the substrate of peptidoglycan transpeptidase. J. Mol. Biol. 1971;61:463–469. doi: 10.1016/0022-2836(71)90393-7. [DOI] [PubMed] [Google Scholar]
- 46.Zhang W, Shi Q, Meroueh SO, Vakulenko SB, Mobashery S. Catalytic mechanism of penicillin-binding protein 5 of Escherichia coli. Biochemistry. 2007;46:10113–10121. doi: 10.1021/bi700777x. [DOI] [PubMed] [Google Scholar]
- 47.Gordon E, Mouz N, Duee E, Dideberg O. The crystal structure of the penicillin-binding protein 2x from Streptococcus pneumoniae and its acyl-enzyme form: implication in drug resistance. J. Mol. Biol. 2000;299:477–485. doi: 10.1006/jmbi.2000.3740. [DOI] [PubMed] [Google Scholar]
- 48.Powers RA, Caselli E, Focia PJ, Prati F, Shoichet BK. Structures of ceftazidime and its transition-state analogue in complex with AmpC beta-lactamase: implications for resistance mutations and inhibitor design. Biochemistry. 2001;40:9207–9214. doi: 10.1021/bi0109358. [DOI] [PubMed] [Google Scholar]
- 49.Chen Y, Shoichet B, Bonnet R. Structure, function, and inhibition along the reaction coordinate of CTX-M beta-lactamases. J. Am. Chem. Soc. 2005;127:5423–5434. doi: 10.1021/ja042850a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pratt RF. Functional evolution of the serine β-lactamase active site. J. Chem. Soc., Perkin Trans. 2002;2:851–861. [Google Scholar]
- 51.Silvaggi NR, Josephine HR, Kuzin AP, Nagarajan R, Pratt RF, Kelly JA. Crystal structures of complexes between the R61 DD-peptidase and peptidoglycan-mimetic beta-lactams: a non-covalent complex with a "perfect penicillin". J. Mol. Biol. 2005;345:521–533. doi: 10.1016/j.jmb.2004.10.076. [DOI] [PubMed] [Google Scholar]
- 52.Sheehan JC. The Enchanted Ring: The Untold Story of Penicillin. Massachusetts: MIT Press, Cambridge; 1982. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.