Abstract
Image enhancement is of great importance in medical imaging where image resolution remains a crucial point in many image analysis algorithms. In this paper, we investigate brain hallucination (Rousseau, 2008), or generating a high-resolution brain image from an input low-resolution image, with the help of another high-resolution brain image. We propose an approach for image super-resolution by using anatomical intermodality priors from a reference image. Contrary to interpolation techniques, in order to be able to recover fine details in images, the reconstruction process is based on a physical model of image acquisition. Another contribution to this inverse problem is a new regularization approach that uses an example-based framework integrating non-local similarity constraints to handle in a better way repetitive structures and texture. The effectiveness of our approach is demonstrated by experiments on realistic Brainweb Magnetic Resonance images and on clinical images from ADNI, generating automatically high-quality brain images from low-resolution input.
Keywords: Inverse problem, Super-resolution, Non-local approach
1. Introduction
Image processing algorithm performances are often limited by the image resolution. Resolution is for instance a key point in brain segmentation in Magnetic Resonance (MR) imaging for which partial volume effect is a limiting factor for fine image analysis. Then, it clearly appears that improving image resolution is still one of the main challenges in medical image processing. In medical imaging, a so-called low-resolution (LR) 3D image is usually a stack of 2D thick slices. As a result, 3D data are generally not isotropic. Fig. 1 shows for instance a high-resolution (HR) T1-weighted image and a LR T2-weighted image of the same patient (in this case, these images come from the Brainweb database Cocosco et al., 1997). In many cases, it may be necessary to put HR and LR images into a same coordinate system. In medical imaging, this is usually done by applying interpolation techniques (Lehmann et al., 1999). Image interpolation is a very common image processing technique in medical imaging pipelines and it may have a strong impact on other processing steps such as segmentation or registration. Interpolation methods can be divided into two groups: scene-based and object-based methods (Grevera and Udupa, 1998). Scene-based approaches use only image intensities to determine the interpolated intensity (for instance: nearest-neighbor, linear interpolation, spline-based interpolation). Such scene-based methods produce perceptually unsatisfactory results with blurred edges and textures. Many edge-preserving interpolation techniques have been reported to handle this problem. However, these techniques rely on accurate edge information that is not obtainable from coarse data. In order to guide the interpolation process, object-based methods make use of additional information extracted from images. As an example, non-rigid registration techniques have been used to drive interpolation methods between adjacent slices (Frakes et al., 2008; Penney et al., 2004). However, scene-based and object-based techniques do not take advantage of a model of the imaging process.
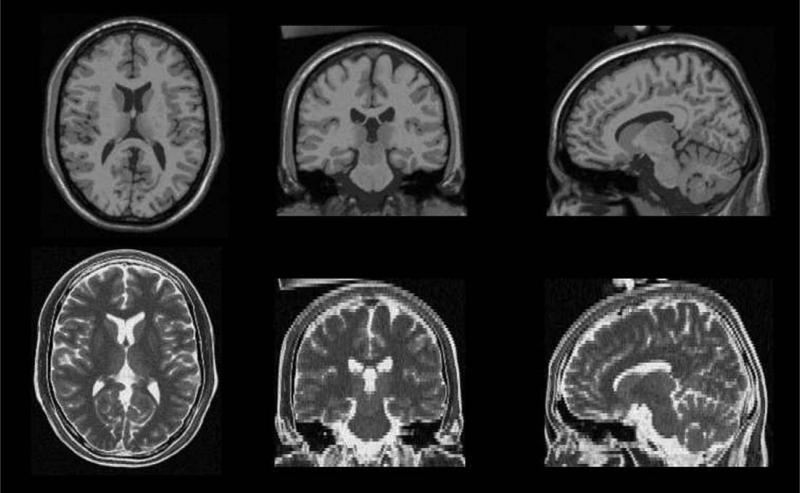
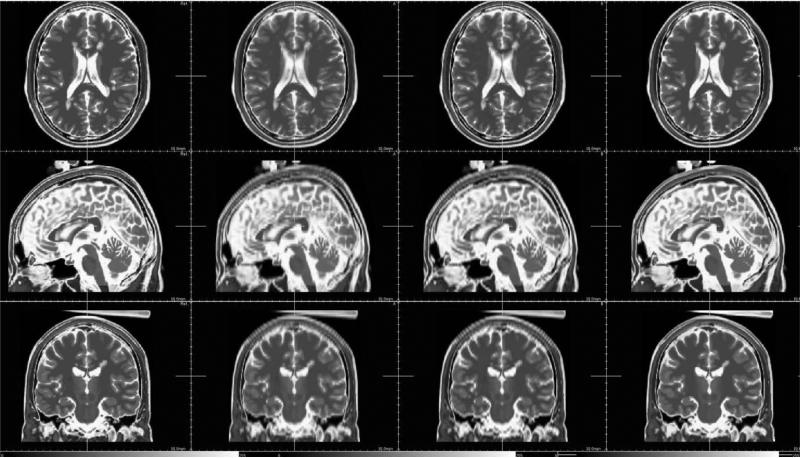
Fig. 1.
Example of MR Brainweb data (Cocosco et al., 1997). First row: high resolution T1-weighted image (1 × 1 mm2 in-plane resolution, 1 mm slice thickness). Second row: low-resolution T2-weighted image (1 × 1 mm2 in-plane resolution, 3 mm slice thickness). The purpose of the proposed approach is to reconstruct a high-resolution T2-weighted image using information from the T1-weighted image.
Super-resolution (SR) is another model-based technique which relies on modeling the imaging processes and using regularization methods describing a priori constraints. The principle of this approach is usually combining LR images to produce an image that has a higher spatial resolution than the original images (Bose et al., 2004). In medical imaging, several SR methods have been proposed to combine LR images to reconstruct one HR image (Peeters et al., 2004; Carmi et al., 2006; Rousseau et al., 2006) (usually by modifying the acquisition protocol). SR is a large research field encompassing many applications. The work we present in this paper is related to the single-frame SR framework (van Ouwerkerk, 2006), meaning that only one LR image is used to generate an HR image. More specifically, we focus on studies involving MR imaging for which an anatomical HR image and several other LR images are acquired to keep acquisition time at an acceptable level for the patient (see Fig. 1). This is the case for routinely performed clinical MR acquisitions such as follow-up of neurodegenerative diseases, brain tumor evolution or diffusion tensor imaging. Typically, one isotropic HR T1-weighted image and several anisotropic LR images (such as T2-weighted, FLAIR or proton density images) are acquired. One can note that this issue of relative difference in image resolution may also occur in other clinical settings. For instance, patient follow-up may require an analysis of scans taken at different magnetic field strength, potentially at different clinical sites.
In such context, we propose a new approach for image SR by using information from an HR MR image to drive the image reconstruction of the LR MR image. The general framework developed in this paper can be applied to other domains where image resolution is an important issue, such as remote sensing. This paper is built on previously published work (Rousseau, 2008). Many experiments have been added in order to provide further insight to the proposed approach. In Section 2, we present the image SR problem using a model-based framework and some recently proposed example-based approaches. Section 3 details our non-local approach for image SR using an HR reference MR image. In Section 4, results obtained on realistic brainweb images are presented and discussed.
2. Image super-resolution
In this section, we present the model-based framework for image SR which leads to an ill-posed inverse problem. Then, recently proposed regularization approaches are described.
2.1. Model-based framework
Contrary to interpolation approaches, model-based approaches use a generic observation model such as:
| (1) |
where y denotes the LR image, x is the HR image, n represents observation noise, D is the sub-sampling matrix, B a blur matrix, W is the geometric transformation.
The three operators can be combined into a single matrix H: H = DBW. The matrix H thus incorporates motion compensation, degradation effects, and sub-sampling for the LR image y. In this paper, we assume that the degradation operator H and the noise characteristics are known and we focus only on the reconstruction step.
Based on this observation model, the SR image can be estimated by minimizing a least-square cost function such as:
| (2) |
For such an inverse problem, some form of regularization plays a crucial role and must be included in the cost function to stabilize the problem or constrain the space of solutions (Idier, 2008). Thus, the HR image is computed by considering the following equation:
| (3) |
is a data fidelity term related to the physical model that penalizes inconsistency between the estimated HR image x and the observed LR image y. This term depends on the observation model used. In our case, the data fidelity term is simply defined as following:
| (4) |
is a regularization term. A common approach for regularization is to take explicitly into account the image geometry and to introduce a global weight λ that balances the contribution of prior smoothness terms and a fidelity term.
2.2. Image regularization
Examples of popular pixel-based regularizers are:
Tikhonov regularization: where cp are positive coefficients, Ω is the domain of integration (all the voxels) and x(p) is pth-order derivative of x.
Variational approach: where q is different from 2 (q = 2 is the Tikhonov regularization).
Markov random field a priori image model: where ϕ can be a L2L1 function for instance.
However, the use of such a regularization term assumes a specific image model: for instance, variational approaches can be based on the assumption that images are made of smooth regions separated by sharp edges. Such prior regularization approach introduces then strong constraints into the reconstruction process.
Explicit models for the many regularities and geometries seen in local patterns are needed to develop better image reconstruction algorithms. In contrast to the pixel-based regularization approach discussed above, example-based methods consists of modeling non-local pairwise interactions from training data or a library of image patches. The principle of example-based SR methods is to add a similarity constraint between the voxels of HR image and the nearest examples present in the learning database (Baker and Kanade, 2002; Datsenko and Elad, 2007). Such an approach suggests that the reconstructed image should locally look like examples existing in the learning database. The regularization term can then be defined in a general way as follows:
| (5) |
where f(x(v)) is an operator on the HR image x at the voxel v, Ω(v) is a neighborhood of v; is the learning database, is a element of related to v and wv,k is a local weight. It is important to note that in example-based methods the regularization term depends on the learning database . For instance (see Baker and Kanade, 2002), f(x(v)) can stand for the gradient value of the SR image x at the voxel v and would be the gradient value of the best matching pixel with respect to v in the learning database.
Although the possibility to introduce new image priors makes the example-based approach very attractive, the key point (which can be a major drawback depending on the application) is the need of a relevant learning database.
In this work, we proposed to take advantage of the existence of an HR reference image in the MR imaging context and to investigate the use of a patch-based approach without any learning database by assuming that there exist related patterns in the LR anisotropic image and an HR isotropic image of the same patient. From the point of view of example-based techniques, the HR iso-tropic image can be seen as a specific learning image database since information present in the HR image is used to constrain the ill-posed reconstruction problem.
3. Proposed approach
3.1. Overview
The main idea of this work is to reconstruct an HR image using one LR image and intermodality priors from another HR image (see Fig. 2). In this context, we propose to use a non-local patch-based approach to define the regularization term in order to take into account complex spatial interactions within images. Moreover, in contrast to example-based approaches for image modeling, the proposed method is unsupervised and thus uses no image patch learning database and no computationally intensive training algorithms.
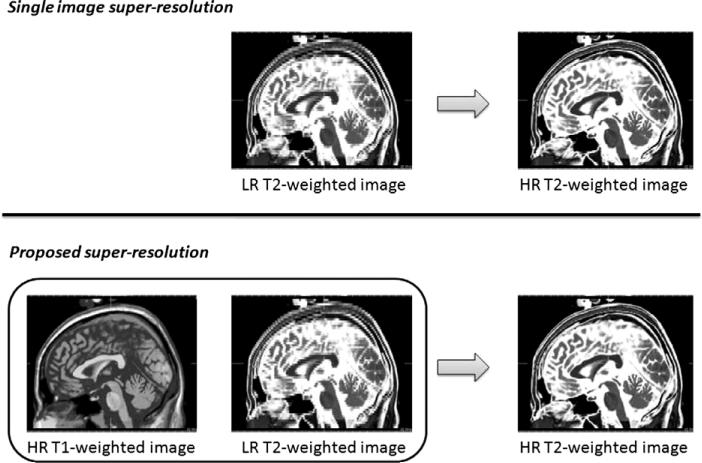
Fig. 2.
Comparison of single image SR framework and the proposed SR approach. Single image SR techniques aim at estimating an HR image using only one LR image as input. In the proposed strategy, the SR estimation is done using jointly the LR image and a reference HR image.
The three key points of the proposed approach are:
a non-local regularization functional which can handle repetitive patterns in the image domain,
the assumption that an HR reference image of a patient may contain relevant examples which should be used to reconstruct an HR image x from a LR image y of the same patient,
no use of external learning database.
The principle of non-local techniques is briefly described in Section 3.2. Section 3.3 presents how the patterns of the reference image are used to drive the reconstruction process of x. An overview of the algorithm is provided in Section 3.4.
3.2. A non-local approach
Recently, Buades et al. (2005) have proposed a very efficient denoising algorithm relying on a non-local framework. Since then, this non-local strategy has been studied and applied in several image processing applications such as non-local regularization functionals in the context of inverse problems (Rousseau, 2008; Mignotte, 2008; Kinderman et al., 2005; Gilboa and Osher, 2007; Peyré et al., 2008).
Let us consider a weighted graph w that links together voxels v, k over the image domain with a weight w(v, k). This weighted graph w is a representation of non-local interactions between image elements. In Buades et al. (2005), the graph w is used for denoising purpose using a non-local neighborhood averaging strategy (called non-local means (NLM)):
| (6) |
where dNLM(x(v)) is a denoised version of x(v). The computation of the weighted graph is described in Appendix A.
Buades et al. have shown that, for 2D natural images, the NLM filter outperforms state-of-the-art denoising methods such as the Rudin–Osher–Fatemi Total Variation minimization scheme or the Perona–Malik Anisotropic diffusion. The NLM method tries to take advantage of the high degree of redundancy of any natural image and appears to be an unsupervised example-based denoising method.
In Rousseau (2008) and Mignotte (2008), a non-local regularization functional for inverse problems has been proposed relying on a NLM denoised version of the reconstructed image:
| (7) |
Such a regularization functional has the advantage over popular variational regularization functionals to handle in a better way repetitive structures and texture without incorporating strong priors on the reconstructed image.
3.3. A non-local regularization functional using intermodality priors
We propose to introduce into the regularization term a similarity constraint between local patterns of the reconstructed HR image x and the HR reference image .
Let the weighted graph estimated on the HR reference image and wx the weighted graph estimated on the HR reconstructed image x.
A naive approach to use intermodality priors for SR would be to directly compute the weighted graph and to estimate the SR image using the regularization functional defined in Eq. (7). In this case, the denoised version of the reconstructed HR image x would be computed using the weighted graph in Eq. (6):
| (8) |
This simple strategy relies on the assumption that local patterns in the HR reference image could be directly used as examples to regularize the reconstructed HR image x.2 However, there are some cases where this assumption may not hold. In the context of MR imaging, images and y are not acquired with the same MR sequence. Multimodal MR data do not reveal the same tissue specificity; for instance, Multiple Sclerosis (MS) lesions are clearly visible in T2-weighted images but not in T1-weighted images (see Fig. 3). In this case, the HR T1-weighted image may not be a relevant candidate to guide the reconstruction process. In order to handle the case of possible outliers, we propose to modify the regularization term by locally analyzing the correlation between the two graphs wx and .
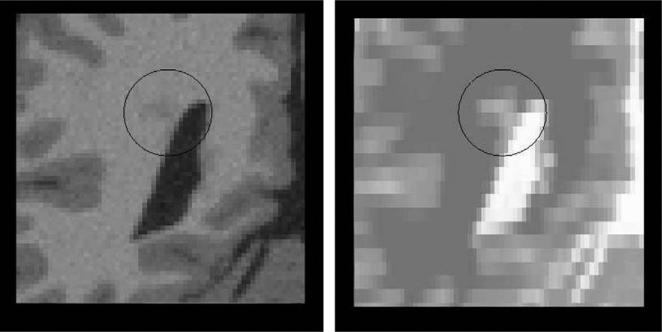
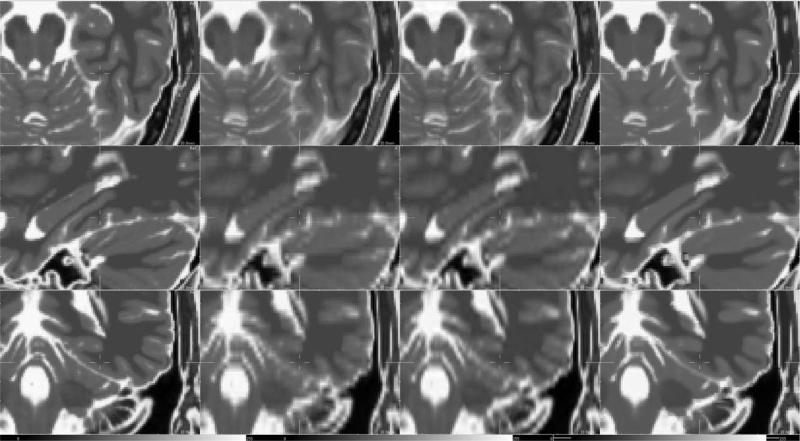
Fig. 3.
Illustration of possible outliers. The MS lesion (in the circle) is less visible in the HR T1-weighted image than in the LR T2-weighted image. In this case, the assumption that local patterns in the HR image could be used as examples to regularize the reconstructed HR image x does not hold.
For each voxel v, we define a scalar α as the correlation between the two weighted graphs wx and . To handle outliers (such as MS lesions), we propose an adaptive scheme by modifying the way to compute the denoised version of the HR reconstructed image x:
| (9) |
is now a weighted average of two denoised versions of x(v). The first term is the denoised version of x(v) computed by using the HR reference image and the second term is a denoised version of x(v) obtained with the current estimate of the reconstructed HR image x. If the two weighted graphs are correlated, the HR reference image is likely to be a relevant candidate to guide the reconstruction process and thus, α is close to 1. If the weighted graphs are uncorrelated, the presence of outliers is detected and α is close to 0. Defining in such way allows us to choose the best examples to regularize the reconstructed HR image x.
We define then a regularization functional taking advantage of intermodality priors as follows:
| (10) |
where is a denoised version of x(v) using the HR reference image (see Eq. (9)).
3.4. Overview of the algorithm
Require: LR image y, starting estimate of HR image x, HR reference image
Compute the weighted graph
repeat
Compute the weighted graph wx
Update α
Estimate
Update x by minimizing
until convergence
4. Results
To explore the ability to reconstruct high-resolution images of realistic typical anatomical brain structures, we applied the algorithm on MRI images of Brainweb (Cocosco et al., 1997) and real images (ADNI data).
4.1. Brainweb images
Brainweb is a simulated brain database which is often used as a gold standard for the analysis of in vivo MR data. Using the HR image provided by Brainweb, we have generated LR images with an in-plane resolution of 1 mm × 1 mm and a slice thickness of 3 mm, using the observation model described by Eq. (1). As in the Brainweb simulator, the point spread function is modeled with a boxcar function. Intensities of HR Brainweb images have been normalized between 0 and 255.
The database contains simulated brain MRI data based on two anatomical models: normal and multiple sclerosis (MS lesions have been extracted from real MRI data). We also have investigated the influence of Gaussian noise (in this case, both LR images and HR reference images are corrupted with Gaussian noise). The proposed methodology has been implemented on four cases:
normal anatomical model without noise,
multiple sclerosis anatomical model without noise,
normal anatomical model with Gaussian noise (σ = 10),
multiple sclerosis anatomical model with Gaussian noise (σ = 10),
To investigate the influence of the parameters, we use the PSNR in decibels (dB) to quantify the quality of reconstructed images:
| (11) |
where MSE is the mean square error and d is the dynamic range of the image x(d = max(x) – min(x) = 255).
In all experiments, a gradient descent method is used to optimize the cost function. The parameter λ is set to 0.01 (this value has been estimated in order to maximize the PSNR of the recovered image on Brainweb database).
By comparing the ground truth image with the reconstructed HR brain images, the Brainweb dataset allows us to evaluate the influence of each parameter of the proposed approach.
4.2. Influence of the smoothing parameter β
b
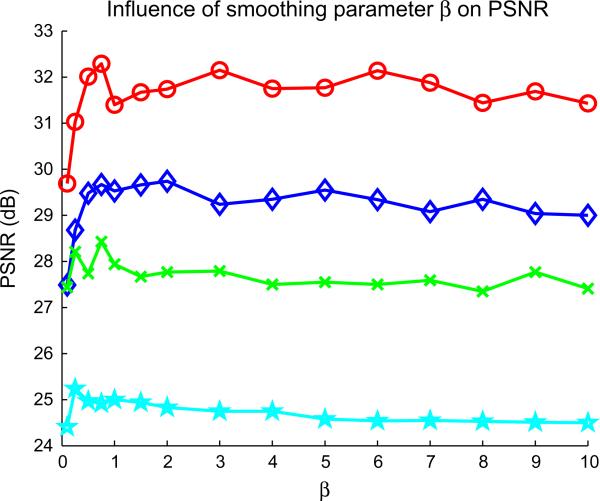
Fig. 4 shows the influence of the smoothing parameter β for the computation of non-local weights wNLM. For denoising purpose, Buades et al. (2005) show that this parameter should be set to 1. Using the Brainweb image ground truth for normal and pathological cases, we investigate different values of β. Based on this experiment, it appears that a small value of β (such as 0.1 or 0.25) may significantly decrease the quality of the reconstructed image. However, for higher values of β, the quality of the reconstructed image does not seem to be strongly affected which leads to the assumption that the proposed non-local approach is not very sensitive to this setting.
Fig. 4.
Influence of the smoothing parameter β on the PSNR computed with the reconstructed image compare the Brainweb ground truth. ○, Non-pathological noise free Brainweb image; ◇, multiple sclerosis noise free Brainweb image; ×, non-pathological noisy Brainweb image; ☆, multiple sclerosis noisy Brainweb image.
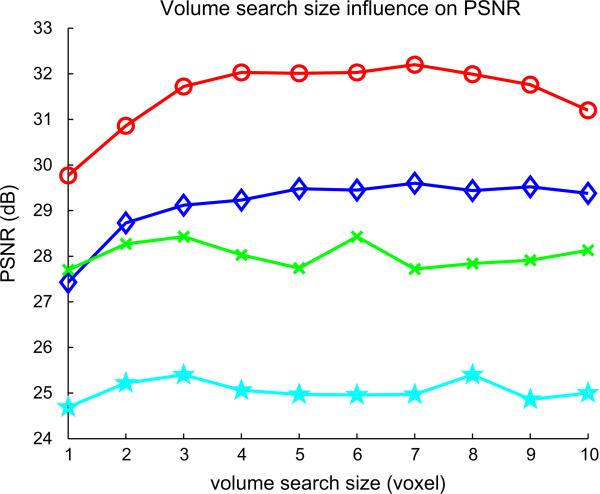
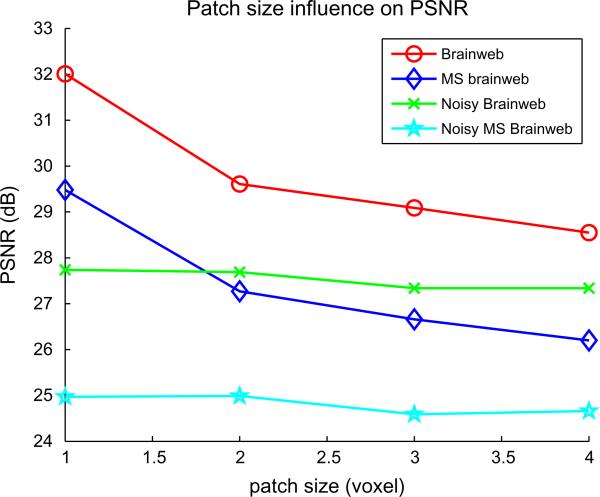
4.3. Influence of the size of the search volume and the neighborhood
Figs. 5 and 6 show the influence of the size of the search volume and the local neighborhood. Increasing the number of voxels in the search volume does not seem to affect the PSNR when the half-size of the search volume is greater than 3. Practically, using a small value for search volume size prevents useless computations. Concerning the patch size, it clearly appears that a patch of 3 × 3 × 3 is of optimal size with respect to the PSNR and the computation time.
Fig. 5.
Influence of the size of the search volume on the PSNR computed with the reconstructed image compare the Brainweb ground truth. ○, Non-pathological noise free Brainweb image; ◇, multiple sclerosis noise free Brainweb image; ×, non-pathological noisy Brainweb image; ☆, multiple sclerosis noisy Brainweb image.
Fig. 6.
Influence of the size of the neighborhood on the PSNR computed with the reconstructed image compare the Brainweb ground truth. ○, Non-pathological noise free Brainweb image; ◇, multiple sclerosis noise free Brainweb image, × : non-pathological noisy Brainweb image, ☆: multiple sclerosis noisy Brainweb image.
4.4. Influence of the image correlation
Fig. 7 shows the correlation map computed between the HR T1-weighted image and the HR T2-weighted image for the multiple sclerosis Brainweb case. This figure shows that for regions containing outliers (i.e. MS lesions), α is low. For these regions, the final image is only reconstructed using information of LR T2-weighted image. Moreover, we also report results obtained with fixed values of α in Table 1 (α = 0 or 1). When α = 0 (which corresponds to the case of SR single-frame with non-local regularization), the quality of the reconstructed image is better than using interpolation methods. However, better results are obtained using an HR reference image during the reconstruction process (adaptive α using graph correlation or α = 1).
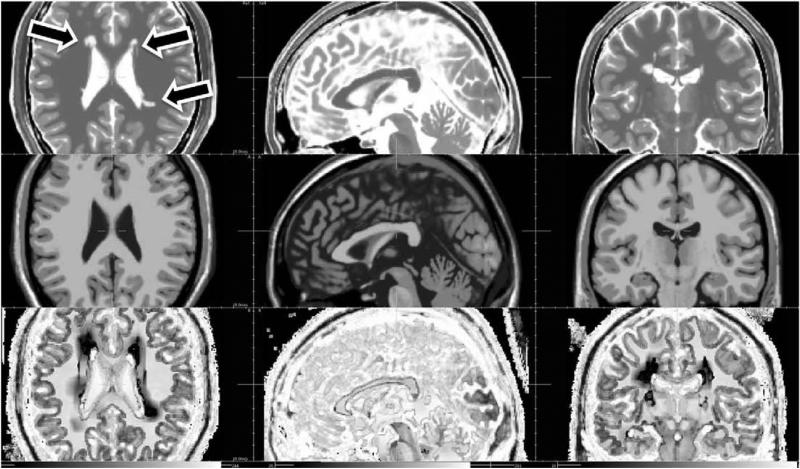
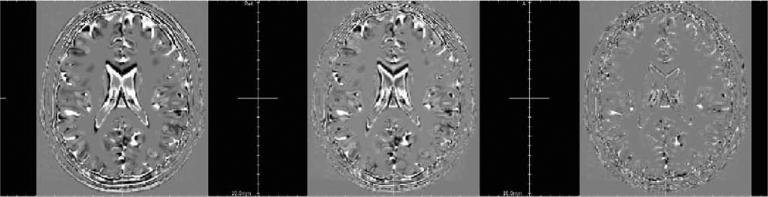
Fig. 7.
Estimated correlation map for the multiple sclerosis Brainweb image. First row: HR T2-weighted image. Second row: HR T1-weighted image. Third row: correlation map (values of the parameter a. A linear intensity scale between 0 and 1 is used to display α.) Three arrows show MS lesion locations (axial view).
Table 1.
Performances of image reconstruction methods (PSNR in dB) using noise free Brainweb images. The proposed method is compared with several interpolation methods. The influence of the correlation map (value of the parameter α) on the reconstruction results is also reported.
| Method | Non-pathological image | Multiple sclerosis image |
|---|---|---|
| Nearest-neighbor interpolation | 23.74 | 20.89 |
| Trilinear interpolation | 24.65 | 21.61 |
| Third-order B-spline interpolation | 24.76 | 21.69 |
| Fifth-order B-spline interpolation | 24.73 | 21.67 |
| Proposed method | 32.20 | 29.48 |
| Proposed method with α = 0 | 28.30 | 26.05 |
| Proposed method with α = 1 | 32.09 | 29.16 |
4.5. Quantitative comparison with interpolation methods on Brainweb images
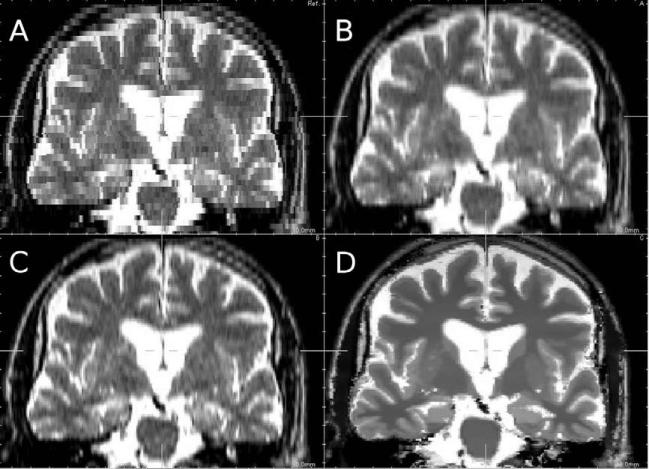
We have reported PSNR results obtained with the different methods in Table 1. Results obtained with the proposed approach compare favorably with B-spline interpolation. Figs. 8 and 9 show the results for non-pathological and pathological MR Brainweb images for axial, coronal and sagittal views. Fig. 10 is a zoom in on reconstruction results and allows a better visual comparison between interpolation method (trilinear and cubic B-spline) and the proposed SR method. It can be seen that details have been recovered and contrast between structures has been improved. Typical interpolation artefacts (see the skull for instance) are less visible on images reconstructed using the proposed method.
Fig. 8.
Reconstruction results (non-pathological case). From left to right: ground truth, trilinear interpolation, third-order B-spline interpolation, the proposed approach.
Fig. 9.
Reconstruction results in presence of MS lesions. From left to right: ground truth, trilinear interpolation, third-order B-spline interpolation, the proposed approach.
Fig. 10.
Zoom on reconstruction results. From left to right: ground truth, trilinear interpolation, third-order B-spline interpolation, the proposed approach.
Fig. 11 shows the difference between the T2-weighted ground truth image and the reconstructed images (non-pathological case). It highlights the fact that the proposed framework provides a better reconstructed image, specifically at boundaries of brain structures. As pointed out in Section 3.3, the presence of brain lesions might introduce bias in reconstructed images using intermodality priors. Fig. 12 shows results in MS lesion areas. Again, it appears that the proposed method using intermodality priors compares favorably with interpolation methods. More specifically, no spurious artefacts have been detected in reconstructed images. These results tend to show that our reconstruction method is robust to outliers such as MS lesions.
Fig. 11.
Difference between ground truth image and reconstruction results (non-pathological case, axial view). From left to right: trilinear interpolation, third-order B-spline interpolation, the proposed approach. The same linear intensity scale is used for the display of image differences.
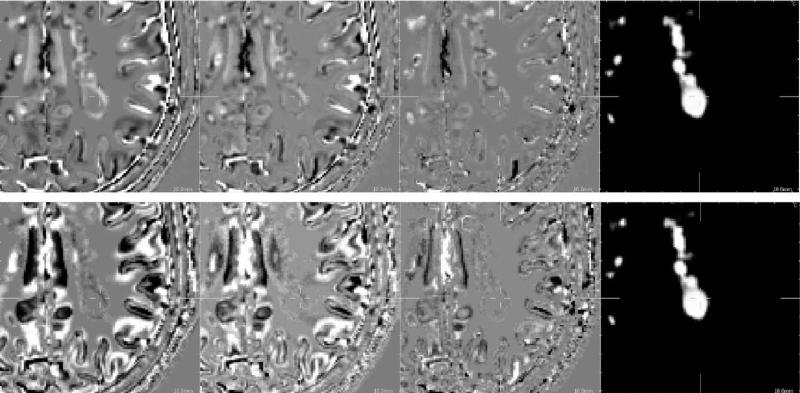
Fig. 12.
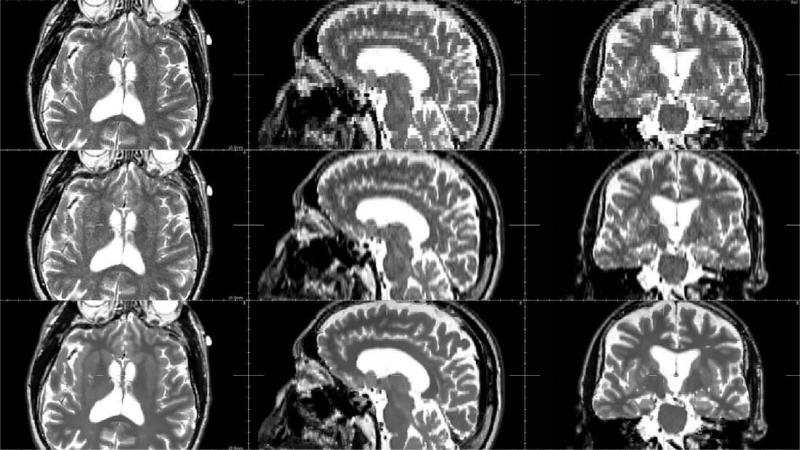
Difference between ground truth image and reconstruction results (pathological case). First row: reconstruction of a T2-weighted image. Second row: reconstruction of a T1-weighted image. From left to right: trilinear interpolation, third-order B-spline interpolation, the proposed approach, MS lesion mask. The same linear intensity scale is used for the display of image differences.
In MS, lesions are usually less visible in T1-weighted images than in T2-weighted images. To verify that our algorithm does not add any reconstruction artefacts, an HR T1-weighted image has been reconstructed using an HR T2-weighted image as reference image in a pathological case. Fig. 12 shows details of reconstructed images in a MS lesion area. It seems that the proposed method does not add “hallucinated” (or false) lesion. This particular point is discussed in Section 5.
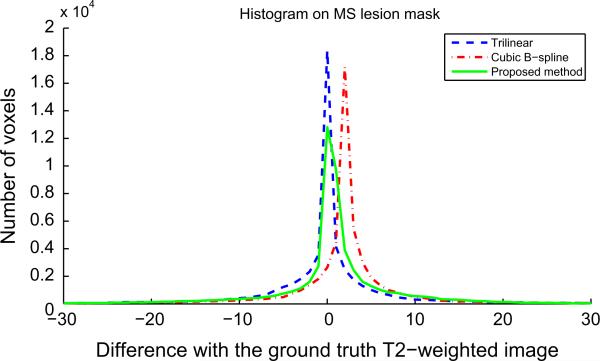
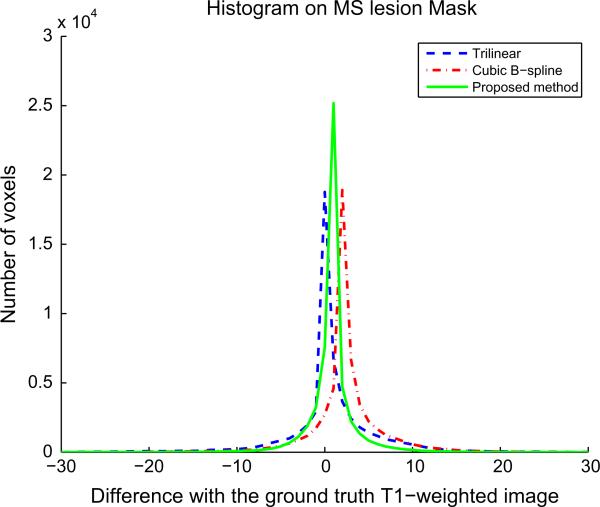
In addition to PSNR values and visual assessment, an error analysis has been done in MS lesion areas. Figs. 13 and 14 show the histograms of error reconstruction for a reconstructed T2-weighted image (using an HR T1-weighted image as reference) and a reconstructed T1-weighted image (using an HR T2-weighted image as reference). In both cases, the standard deviation of the error reconstruction distribution is smaller for our method than for interpolation method. This tends to confirm that the proposed method allows a better reconstruction (specifically at boundaries) and that it is robust to outliers such as MS lesions.
Fig. 13.
Histogram of error reconstruction (difference between ground truth T2-weighted image and reconstruction results) in MS lesion mask (pathological case). Mean and standard deviation of these distributions are: trilinear (0.32; 97.62), cubic B-spline (2.10; 71.17), proposed method (0.74; 67.72).
Fig. 14.
Histogram of error reconstruction (difference between ground truth T2-weighted image and reconstruction results) in MS lesion mask (pathological case). Mean and standard deviation of these distributions are: trilinear (0.64; 27.68), cubic B-spline (2.34; 18.10), proposed method (0.83; 7.71).
4.6. Results on ADNI images
Data used in the preparation of this article were obtained from the Alzheimer's Disease Neuroimaging Initiative (ADNI) database (http://www.loni.ucla.edu/ADNI). The ADNI was launched in 2003 by the National Institute on Aging (NIA), the National Institute of Biomedical Imaging and Bioengineering (NIBIB), the Food and Drug Administration (FDA), private pharmaceutical companies and non-profit organizations, as a $60 million, 5-year public–private partnership. The primary goal of ADNI has been to test whether serial magnetic resonance imaging (MRI), positron emission tomography (PET), other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of mild cognitive impairment (MCI) and early Alzheimer's Disease (AD). Determination of sensitive and specific markers of very early AD progression is intended to aid researchers and clinicians to develop new treatments and monitor their effectiveness, as well as lessen the time and cost of clinical trials. The Principle Investigator of this initiative is Michael W. Weiner, M.D., VA Medical Center and University of California, San Francisco. ADNI is the result of efforts of many co-investigators from a broad range of academic institutions and private corporations, and subjects have been recruited from over 50 sites across the US and Canada. The initial goal of ADNI was to recruit 800 adults, ages 55–90, to participate in the research – approximately 200 cognitively normal older individuals to be followed for 3 years, 400 people with MCI to be followed for 3 years, and 200 people with early AD to be followed for 2 years. For up-to-date information see http://www.adni-info.org.
The image size and resolution are, respectively, 160 × 192 × 192 voxels and 1.2 mm × 1.250 mm × 1.250 mm for the T1-axial T2-weighted image. The weighted image, and 228 × 256 × 55 voxels and 0.938 mm × 0.938 mm × 3.000 mm for the LR T1-weighted image is first registered to the LR T2-weighted image using a rigid transform and then the proposed SR technique is applied. Figs. 15 and 16 show typical results obtained on the ADNI data using a T1-weighted image as the reference image to drive the reconstruction process of an axial T2-weighted image (the resolution of the reconstructed T2-weighted image is then: 0.938 mm × 0.938 mm × 0.938 mm). We can observe on these figures that for the reconstructed image using the proposed approach, the contrast of brain structures is higher than using interpolation techniques. Typical interpolation artefacts (like staircase artefact) are less visible on the reconstructed image. Moreover, it can be noticed that the non-local regularization framework allows to jointly denoise and reconstruct the T2-weighted image.
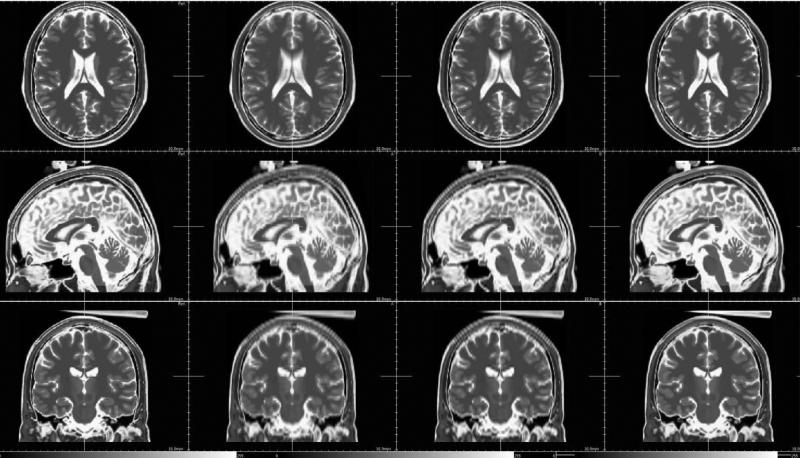
Fig. 15.
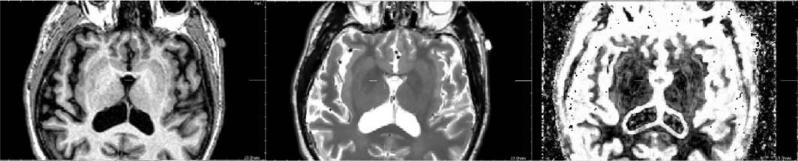
Reconstruction results on ADNI data (the LR T2-weighted image is firstly registered on the HR T1-weighted image). First row: nearest-neighbor interpolation. Second row: third-order B-spline interpolation. Third row: the proposed approach.
Fig. 16.
Zoom on reconstruction results on ADNI data (coronal view). (A) Nearest-neighbor interpolation, (B) trilinear interpolation, (C) third-order B-spline interpolation, (D) proposed approach.
To understand how the HR T1-weighted is used to drive the reconstruction process, the correlation map is displayed in Fig. 17. It appears that the parameter α has a high value near to the edges which means that the HR reference image is mainly used for brain region with high contrast (interface between white matter and grey matter for instance). Conversely, uniform and/or textured regions such as caudate nucleus or putamen are only reconstructed using information existing in the LR T2-weighted image.
Fig. 17.
Estimated correlation map for ADNI images. First row: HR T1-weighted image. Second row: reconstructed T2-weighted image. Third row: correlation map (values of the parameter α. A linear intensity scale between 0 and 1 is used to display α).
Looking at Fig. 15, one can argue that the proposed reconstructed framework leads to an over smooth image which may not be realistic. As Fig. 18 shows the reconstructed T2-weighted image has indeed a very similar aspect to the HR T1-weighted image denoised using the Non-Local Mean algorithm (Buades et al., 2005). Therefore, this property of the proposed algorithm appears to be beneficial since the proposed processing leads to a denoised HR T2-weighted MR image.
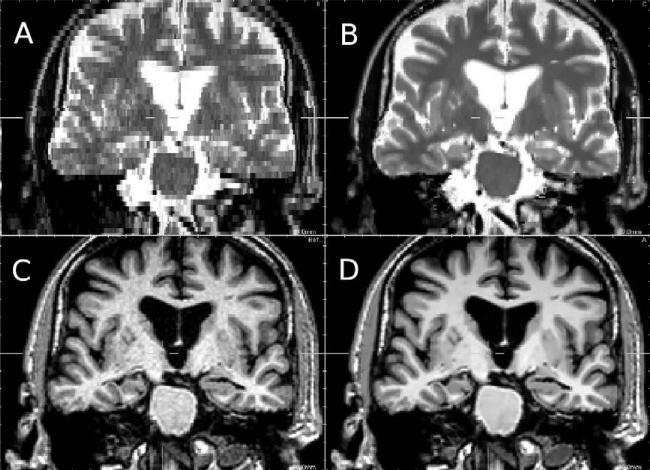
Fig. 18.
Zoom on reconstruction results on ADNI data (coronal view). (A) Nearest-neighbor interpolation, (B) proposed approach, (C) T1-weighted image used as reference image, (D) denoised T1-weighted image.
5. Discussion
The first contribution of this work concerns a simple idea for image SR which is the use of an HR reference image to improve the resolution of the LR image. As previously stated, such approach is directly related to the medical context we are interested in. However, we believe that such new image SR approach may have a substantial impact in the image processing research field. Moreover, possible further work can focus on the unification of this framework with other previously proposed approaches such as object-based interpolation and registration-based interpolation.
The second contribution is about the modeling of the SR problem introducing a non-local regularization term. We have shown that example-based approaches such as non-local means can be embedded into the reconstruction process to enhance the performance of super-resolution techniques. If an HR brain image of the patient is available, LR images of the same patient can be enhanced by exploiting non-local pairwise interactions. We believe that this work is a new original way to tackle the problem of image enhancement in MR imaging by exploiting the multimodality aspect of MR data. The key point of this work is the use of a non-local approach to define a new regularization term in the reconstruction process. The proposed methodology is related to example-based SR methods such as the one developed by Baker and Kanade (2000) but in our case the learning database is reduced to one reference image. Moreover, in this work we exploit non-local interactions between voxels by defining non-local weights w (also called non-local graphs) which take into account non-local interactions in the reconstructed image and the reference image.
Experimental results show that the developed algorithm compares favorably with interpolation approaches. The two key points of the proposed approach, with respect to interpolation methods, are the use of an observation model (as in SR approaches) and the use of a reference HR image which drives the reconstruction process. Experiments on Brainweb images show that even with the presence of lesions visible only in the LR T2-weighted image, the reconstruction method we propose is able to recover such “outliers” without introducing artefacts which may come from the HR T1-weighted image (where lesions are not clearly visible). In this particular case, we would have thought that the lesions would disappear. Although our experiments tend to show that our approach is robust to such outliers, this is a crucial point that needs to be further investigated (small lesions, tumors, etc.). In general, this point raises the following question: what is the influence of an interpolation or image SR algorithm on image analysis (segmentation, detection, etc.)?
This points out that such image enhancement technique needs to be validated using physical phantoms. Moreover, an evaluation on large database is required to prove that SR based methods can have a significant impact on medical image processing pipelines (specifically for registration, segmentation or change detection). Such validation can be application dependent and thus requires a specific evaluation framework. As it has been shown in Section 4, in addition to PSNR and visual assessment, there is a clear need to develop standard criteria to measure image quality reconstruction of SR technique. This is a key point for image enhancement technique particularly in medical imaging where image artefacts have to be avoided.
On one hand, this work shows that SR based techniques provides higher MR image quality than standard interpolation algorithms. On the other hand, interpolation techniques do not require any rigid registration step and are less time consuming (SR based methods may require several hours per reconstruction). Based on our experiments (not shown in the paper), registration does not introduce artefacts in the reconstructed images using the proposed approach. If images are not correctly registered, the correlation coefficient α tends to zero which means that the reconstruction process is only driven by the T2-weighted image.
High-resolution imaging is a key point for MR brain image analysis in order to study anatomical details. Since few brain image analysis algorithms take into account an observation model such as in Eq. (1), the proposed reconstruction approach may have a substantial impact on segmentation or registration. Therefore, such model-based HR image reconstruction algorithm may represent an important step towards multimodal brain analysis at fine scale.
While in this work we are only focused on brain MR image reconstruction, the proposed approach relying intermodality priors might have potential applications to other medical image modalities. Future work would involve studying a similar method for multimodal images (computerized tomography, diffusion MRI, ultrasound, etc.). Depending on the intended medical application, such approach could allow to save time of data acquisition by enabling image quality improvement as a post-acquisition step. The corner stone would concern then the criterion for linking the different modalities (How to calculate the local correlation map? What type of information can be used between the imaging modalities?.)
Acknowledgments
The research leading to these results has received funding from the European Research Council under the European Community's Seventh Framework Programme (FP7/2007-2013 Grant Agreement No. 207667).
The author would like to thank Sylvain Faisan and Vincent Noblet from LSIIT (CNRS/University of Strasbourg) for fruitful discussions.
Data collection and sharing for this project was funded by the Alzheimer's Disease Neuroimaging Initiative (ADNI; Principal Investigator: Michael Weiner; NIH Grant U01 AG024904). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering (NIBIB), and through generous contributions from the following: Pfizer Inc., Wyeth Research, Bristol-Myers Squibb, Eli Lilly and Company, GlaxoSmithK-line, Merck & Co. Inc., AstraZeneca AB, Novartis Pharmaceuticals Corporation, Alzheimer's Association, Eisai Global Clinical Development, Elan Corporation plc, Forest Laboratories, and the Institute for the Study of Aging, with participation from the US Food and Drug Administration. Industry partnerships are coordinated through the Foundation for the National Institutes of Health. The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer's Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory of Neuro Imaging at the University of California, Los Angeles.
Appendix A. A Computation of weighted graphs wx and
wx and are computed in the same way. For the sake of clarity, we describe only the computation of wx.
The weight wx(v, k) between two voxels v and k is computed as follows (Coupé et al., 2008):
| (A.1) |
where P(x(k)) is a 3D patch of x centered in voxel k and P(x(v)) is the 3D patch of x centered in voxel v; Zv is a constant of normalization; Ni is the number of voxels of a 3D patch. The distance between the 3D patches is the sum over voxels of patches of intensity differences using the L2 norm. The specific case concerning the central point of the patch is computed as in Kinderman et al. (2005). With the assumption of Gaussian noise in images, β is set to 1 (see Buades et al., 2005 for theoretical justifications) and the standard deviation of noise is estimated via pseudo-residuals εv as defined in Gasser et al. (1986). For each voxel v of x, let us define:
| (A.2) |
where Ω(v) is the 6-neighborhood at voxel v. The standard deviation of noise is computed as the least-square estimator:
| (A.3) |
where n is the number of voxels of x.
Moreover, as suggested by Mahmoudi and Sapiro in Mahmoudi and Sapiro (2005), voxel preselection can avoid useless computation and also improve the result of denoising. The preselection of relevant voxels is based on the mean and variance of patches (Coupé et al., 2008). wx(v, k) is set to 0 if one of these conditions is not fulfilled:
| (A.4) |
| (A.5) |
with μ = 0.95 and σ2 = 0.5.
Footnotes
Data used in the preparation of this article were obtained from the Alzheimer's Disease Neuroimaging Initiative (ADNI) database (http://www.loni.ucla.edu/ADNI). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. ADNI investigators include (complete listing available at http://www.loni.ucla.edu/ADNI/Collaboration/ADNI_Manuscript_Citations.pdf).
It is important to note that in example-based SR methods, the elements of the learning database used for reconstruction are image intensities. In the proposed approach, the elements of the learning database are weighted graphs, which describe non-local interactions between image voxels.
References
- Baker S, Kanade T. Hallucinating faces. Fourth International Conference on Automatic Face and Gesture Recognition. 2000 [Google Scholar]
- Baker S, Kanade T. Limits on super-resolution and how to break them. IEEE Transactions on Pattern Analysis and Machine Intelligence. 2002;24(9):1167–1183. [Google Scholar]
- Bose NK, Chan RH, Ng MK. Special Issue: High Resolution Image Reconstruction. International Journal of Imaging Systems and Technology. 2004;14(2–3) [Google Scholar]
- Buades A, Coll B, Morel JM. A review of image denoising algorithms, with a new one. Multiscale Modeling & Simulation. 2005;4(2):490–530. [Google Scholar]
- Carmi E, Liub S, Alona N, Fiata A, Fiat D. Resolution enhancement in MRI. Magnetic Resonance Imaging. 2006;24(2) doi: 10.1016/j.mri.2005.09.011. [DOI] [PubMed] [Google Scholar]
- Cocosco CA, Kollokian V, Kwan RK-S, Evans AC. BrainWeb: online interface to a 3D MRI simulated brain database. Proceedings of Third International Conference on Functional Mapping of the Human Brain. 1997;5(4) [Google Scholar]
- Coupé P, Yger P, Prima S, Hellier P, Kervrann C, Barillot C. An optimized blockwise nonlocal means denoising filter for 3-D magnetic resonance images. IEEE Transactions on Medical Imaging. 2008;27(4):425–441. doi: 10.1109/TMI.2007.906087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko D, Elad M. Example-based single document image super-resolution: a global MAP approach with outlier rejection. Multidimensional Systems and Signal Processing. 2007;18:103–121. [Google Scholar]
- Frakes DH, Dasi LP, Pekkan K, Kitajima HD, Sundareswaran K, Yoganathan AP, Smith MJT. A new method for registration-based medical image interpolation. IEEE Transactions on Medical Imaging. 2008;27(3):370–377. doi: 10.1109/TMI.2007.907324. [DOI] [PubMed] [Google Scholar]
- Gasser T, Sroka L, Steinmetz C. Residual variance and residual pattern in nonlinear regression. Biometrika. 1986;73(3):625–633. [Google Scholar]
- Gilboa G, Osher S. UCLA CAM Report, 07-23. 2007. Nonlocal operators with applications to image processing. [Google Scholar]
- Grevera GJ, Udupa JK. An objective comparison of 3-D image interpolation methods. IEEE Transactions on Medical Imaging. 1998;17:642–652. doi: 10.1109/42.730408. [DOI] [PubMed] [Google Scholar]
- Idier J. Bayesian Approach to Inverse Problems. ISTE Ltd. and John Wiley & Sons Inc.; 2008. [Google Scholar]
- Kinderman S, Osher S, Jones PW. Deblurring and denoising of images by nonlocal functionals. Multiscale Modeling and Simulations. 2005;4(4):1091–1115. [Google Scholar]
- Lehmann T, Gonner C, Spitzer K. Survey: interpolation methods in medical image processing. IEEE Transactions on Medical Imaging. 1999;18(11):1049–1075. doi: 10.1109/42.816070. [DOI] [PubMed] [Google Scholar]
- Mahmoudi M, Sapiro G. Fast image and video denoising via nonlocal means of similar neighborhoods. IEEE Signal Processing Letters. 2005;12(12):839–842. [Google Scholar]
- Mignotte M. A non-local regularization strategy for image deconvolution. Pattern Recognition Letters. 2008;29(16):2206–2212. [Google Scholar]
- Peeters RR, Kornprobst P, Nikolova M, Sunaert S, Vieville T, Malandain G, Deriche R, Faugeras O, Ng M, Van Hecke P. The use of super-resolution techniques to reduce slice thickness in functional MRI. International Journal of Imaging Systems and Technology. 2004;14(3):131–138. [Google Scholar]
- Penney GP, Schnabel JA, Rueckert D, Viergever MA, Niessen WJ. Registration-based interpolation. IEEE Transactions on Medical Imaging. 2004;23(7):922–926. doi: 10.1109/TMI.2004.828352. [DOI] [PubMed] [Google Scholar]
- Peyré G, Bougleux S, Cohen L. European Conference on Computer Vision, Part III. LNCS 5304; 2008. Non-local regularization of inverse problems. pp. 57–68. [Google Scholar]
- Rousseau F, Glenn O, Iordanova B, Rodriguez-Carranza C, Vigneron D, Barkovich J, Studholme C. Registration-based approach for reconstruction of high-resolution in utero fetal MR brain images. Academic Radiology. 2006;13(9):1072–1081. doi: 10.1016/j.acra.2006.05.003. [DOI] [PubMed] [Google Scholar]
- Rousseau F. European Conference on Computer Vision, Part I. LNCS, 5302; 2008. Brain hallucination. [Google Scholar]
- van Ouwerkerk JD. Image super-resolution survey. Image and Vision Computing. 2006;24:1039–1052. [Google Scholar]