Abstract
Although autism spectrum disorders (ASDs) have a substantial genetic basis, most of the known genetic risk has been traced to rare variants, principally copy number variants (CNVs). To identify common risk variation, the Autism Genome Project (AGP) Consortium genotyped 1558 rigorously defined ASD families for 1 million single-nucleotide polymorphisms (SNPs) and analyzed these SNP genotypes for association with ASD. In one of four primary association analyses, the association signal for marker rs4141463, located within MACROD2, crossed the genome-wide association significance threshold of P < 5 × 10−8. When a smaller replication sample was analyzed, the risk allele at rs4141463 was again over-transmitted; yet, consistent with the winner's curse, its effect size in the replication sample was much smaller; and, for the combined samples, the association signal barely fell below the P < 5 × 10−8 threshold. Exploratory analyses of phenotypic subtypes yielded no significant associations after correction for multiple testing. They did, however, yield strong signals within several genes, KIAA0564, PLD5, POU6F2, ST8SIA2 and TAF1C.
INTRODUCTION
A portion of the genetic roots of autism trace to rare de novo and inherited copy number variants (CNVs), many of which hit genes that encode proteins affecting neuronal development, especially formation of synapses (1). These findings make sense in the context of autism, a neurodevelopmental disorder arising in childhood that is characterized by impairments in social communication and a pattern of repetitive behavior and restricted interests (2,3).
Autism, the prototypical autism spectrum disorder (ASD), is diagnosed in ∼15–20 per 10 000 people (4). The broader ASD category affects at least 60 in 10 000 children (5), but may be as high as 100 in 10 000 (6). Consistent with substantial heritability of ASD, risk to siblings of a proband with autism is 5–10%, substantially higher than population prevalence (7). A spectrum of severity is plausible due to the distribution of milder phenotypes in relatives of probands (8,9).
As yet, however, only rare de novo and inherited variants are soundly established genetic risk factors for ASD, and thus far these only account for a small proportion of the total genetic risk. Autism is a possible manifestation of single-gene disorders, such as those due to mutations in FMR1, TSC1, TSC2, MECP2 and PTEN. Some chromosomal rearrangements appear causal, with the most common being maternal duplication of 15q11–q13. Mutations of high penetrance for ASD have been identified in synaptic genes, including NLGN3, NLGN4X and SHANK3 (10,11). Rare deletion CNVs of SHANK3 and the surrounding 22q13.33 region have also been found in individuals with an ASD. In this regard, genome-wide microarray studies have implicated a substantial number of other individually rare submicroscopic CNV loci, including hemizygous deletions and duplications of 16p11.2, NRXN1 and PTCHD1 (12–18).
These microscopic and submicroscopic CNVs are presumed to have a major and sometimes causal impact on risk for ASD. In contrast, common variants rarely have such an impact on risk for any disorder, especially one like ASD that is known to diminish reproductive success. Nonetheless, even if a common variant has only a small impact on individual risk, its population attributable risk could be substantial because it is carried by many individuals. To date, studies identifying common variants affecting ASD risk have met with limited success. In addition to candidate-gene association studies, in which some genes garner supporting evidence (19), genome-wide association (GWA) studies have highlighted two ASD risk loci: 5p14.1, between the neuronal cadherin loci CDH9 and CDH10 (20), and 5p15.2, between the semaphorin (SEMA5A) and bitter taste receptor (TAS2R1) genes (21).
To search for additional common variation contributing to ASD susceptibility, the AGP conducted high-resolution genotyping to examine >1500 families. Our principal GWA analysis uses an additive model and our principal partitions of the data split along two axes: all ancestry versus European; and inclusive spectrum versus strict diagnostic groups. In exploratory analyses, we used additional phenotypic dimensions of ASD to localize susceptibility loci.
RESULTS
ASD families and genotyping
The AGP Consortium, which represents more than 50 centers in North America and Europe, collected data from 1558 ASD families (4712 subjects) for this study (Supplementary Material, Table S1). Both Autism Diagnostic Interview-Revised, ADI-R (2), and Autism Diagnostic Observation Schedule, ADOS (3), were used for research diagnostic classification. Nested research classification of subjects into ‘strict’ or ‘spectrum’ (i.e. encompasses strict) was developed based on ADI-R and ADOS classification. Subjects with known karyotypic abnormalities, fragile X mutations or other genetic disorders were excluded. Genotyping was performed by using the Illumina Human 1M-single Infinium BeadChip array. A total of 1369 ASD families comprising 1385 ASD probands (Table 1) passed quality control (QC) filters (Supplementary Material, Table S1). Counting up to third-degree relatives in the 1369 families, 43.6% had two or more ASD children (multiplex), 42.4% had one affected child (simplex) and 14% were unknown (extended family not evaluated); note, however, that we typically genotyped only one proband per family, as well as parents, even if the family were multiplex. Proband distribution by gender was 84% male and 16% female; 58.6% attained a strict research diagnosis of autism; and, based on genetic analysis, 88% of subjects were of European ancestry (Supplementary Material, Fig. S1).
Table 1.
Number of families (number of probands) used for analysis
| Group | AGP Discovery | AGRE | Combined |
|---|---|---|---|
| Primary analysis | |||
| Spc|All | 1369 (1385) | 595 (1086) | 1887 (2394) |
| Str|All | 809 (812) | 431 (687) | 1181 (1440) |
| Spc|Eur | 1217 (1230) | 440 (783) | 1603 (1959) |
| Str|Eur | 718 (720) | 311 (485) | 984 (1160) |
| Exploratory analyses | |||
| Spc|Verbal | 897 (909) | 476 (702) | 1314 (1552) |
| Spc|Non-Verbal | 453 (454) | 295 (375) | 731 (812) |
| IQ > 80 | 561 (564) | —a | — |
| IQ < 70 | 279 (281) | — | — |
Spc, spectrum; Str, strict; All, all ancestries; Eur, European ancestry.
aWe could not derive an assessment of IQ for the AGRE data that would be comparable to that from the AGP.
Genome-wide SNP association: primary analyses
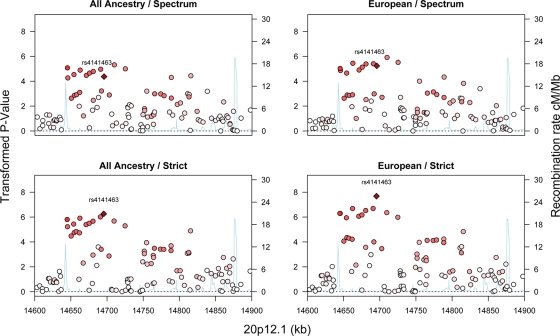
A priori we planned and conducted four nonindependent GWA analyses corresponding to data partitions along axes of diagnosis and ancestry: spectrum versus strict and European versus all ancestries (Table 2, Supplementary Material, Table S1–3, Fig. S2). Q–Q plots (Supplementary Material, Fig. S3) show that the distributions of observed test statistics are only modestly different from their expected distributions under the null hypothesis of no association. Largest associations arise in a 300 kb intronic region of MACROD2 for the most homogeneous samples, strict diagnosis and European ancestry (Fig. 1 and Table 2). The most noteworthy association occurs at rs4141463, P = 2.1 × 10−8, which falls below a commonly used GWA significance threshold of 5 × 10−8.
Table 2.
Results from primary analyses of AGP Discovery, AGRE, and SAGE data sets
| Features |
SNPs associated in discovery seta |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| rs6731562, 2q31.1 | rs10258862, 7p14.1 | rs6557675, 8p21.3 | rs4078417, 14q22.1 | rs7142002, 14q32.31 | rs17284809, 16p13.11 | rs205409, 16p11.2 | rs4150167b, 16q24.1 | rs4141463c, 20p12.1 | ||
| Gene | HAT1 | POU6F2 | – | – | PPP2R5C | MYH11 | GSG1L | TAF1C | MACROD2 | |
| Minor allele frequency | 0.34 (G) | 0.33 (G) | 0.31 (A) | 0.34 (C) | 0.06 (G) | 0.04 (A) | 0.43 (G) | 0.02 (A) | 0.43 (A) | |
| Group | Str|Eur | Spc|All | Str|All | Spc|Eur | Spc|Eur | Spc|All | Spc|Eur | Spc|Eur | Str|Eur | |
| AGP discovery | ORd | 1.64 | 1.41 | 0.61 | 1.43 | 0.54 | 0.52 | 0.69 | 0.38 | 0.56 |
| 95% CI | 1.35–1.99 | 1.23–1.61 | 0.51–0.72 | 1.25–1.64 | 0.41–0.70 | 0.39–0.69 | 0.60–0.79 | 0.24–0.58 | 0.47–0.67 | |
| P | 4.7 × 10−6 | 3.7 × 10−6 | 2.2 × 10−7 | 4.8 × 10−6 | 1.9 × 10−6 | 1.7 × 10−6 | 1.1 × 10−6 | 1.0 × 10−6 | 2.1 × 10−8 | |
| AGRE | OR | 1.08 | 0.85 | 1.00 | 1.13 | 0.73 | 0.88 | 1.21 | 0.71 | 0.84 |
| 95% CI | 0.86–1.37 | 0.73–0.99 | 0.83–1.22 | 0.95–1.34 | 0.52–1.02 | 0.41–1.88 | 1.03–1.43 | 0.44–1.12 | 0.67–1.04 | |
| P | 5.2 × 10−1 | 4.6 × 10−2 | 1.0 × 10−0 | 1.8 × 10−1 | 4.1 × 10−2 | 7.1 × 10−1 | 2.9 × 10−2 | 1.0 × 10−0 | 1.3 × 10−1 | |
| AGP + AGRE | OR | 1.38 | 1.13 | 0.77 | 1.29 | 0.62 | 0.56 | 0.86 | 0.48 | 0.65 |
| 95% CI | 1.19–1.61 | 1.03–1.25 | 0.68–0.88 | 1.16–1.44 | 0.50–0.76 | 0.43–0.72 | 0.77–0.95 | 0.35–0.66 | 0.57–0.75 | |
| P | 8.5 × 10−5 | 2.3 × 10−2 | 2.3 × 10−4 | 2.7 × 10−5 | 2.1 × 10−6 | 4.2 × 10−6 | 9.3 × 10−3 | 7.0 × 10−7 | 4.7 × 10−8 | |
| AGP + SAGE | OR | 1.32 | 1.24 | 0.76 | 1.24 | 0.59 | 0.59 | 0.80 | 0.45 | 0.69 |
| 95% CI | 1.15–1.52 | 1.12–1.38 | 0.66–0.87 | 1.11–1.38 | 0.47–0.75 | 0.46–0.76 | 0.73–0.89 | 0.31–0.66 | 0.60–0.79 | |
| P | 8.8 × 10−5 | 3.8 × 10−5 | 8.5 × 10−5 | 7.5 × 10−5 | 7.5 × 10−6 | 1.4 × 10−5 | 2.6 × 10−5 | 1.7 × 10−5 | 8.1 × 10−8 | |
| AGP + AGRE + SAGE | OR | 1.25 | 1.09 | 0.84 | 1.19 | 0.64 | 0.63 | 0.91 | 0.54 | 0.73 |
| 95% CI | 1.11–1.41 | 1.00–1.18 | 0.76–0.93 | 1.10–1.30 | 0.53–0.78 | 0.50–0.79 | 0.84–0.99 | 0.40–0.73 | 0.66–0.82 | |
| P | 2.0 × 10−4 | 4.6 × 10−2 | 1.0 × 10−3 | 5.6 × 10−5 | 2.9 × 10−6 | 5.7 × 10−5 | 2.8 × 10−2 | 2.1 × 10−5 | 3.7 × 10−8 | |
Spc, spectrum; Str, strict; All, all ancestries; Eur, European ancestry.
aReporting restricted to SNPs with statistics meeting or falling below the threshold of P < 5 × 10−6. See Supplementary Material for description of all SNPs with statistics meeting or falling below the threshold of P ≤ 5 × 10−4.
bBold emphasis denotes SNPs discussed.
cTop association signal reported only; additional signal at reporting threshold for markers in strong LD include rs6079536, rs6079537, rs6079540, rs6079544, rs6074787, rs10446030, rs6079553, rs4814324, rs6074798, rs980319 (Supplementary Material, Table S2).
dOdds ratio based on the minor allele.
Figure 1.
Association results, presented as the −log(base 10) of the P-values, for an intronic region of MACROD2 (20p12.1). The panels show the combinations of two diagnostic levels, strict versus spectrum and any versus European ancestry of the subjects. Recombination rates were calculated using Release 22 of the HapMap CEU Panel.
We sought support for the results obtained from the primary analyses by two approaches. First, we analyze independent ASD families from the Autism Genetics Resource Exchange (AGRE) database, combining AGP trios with AGRE simplex/multiplex families to perform a ‘mega-analysis’ in 2179 families for the four primary analyses. This ‘mega-analysis’, performed on all markers, did not add much additional support (Table 2), in terms of more significant association signals identified at the discovery phase, and no new loci emerged for their association with ASD (see Supplementary Material). For example, at rs4141463 (in MACROD2), the estimated odds ratio changed from 0.56 to 0.65 for the strict diagnosis (Table 2), European ancestry, resulting in a P-value of 4.7 × 10−8. An observation merits consideration for this and related results. Although the common allele is over-transmitted in both the original and AGRE data sets, the differential transmission as measured by the odds ratio is notably smaller for the latter, a result that is consistent with the winner's curse (22). Thus, if rs4141463 truly confers risk, the estimate from the AGRE data is a more realistic estimate of risk.
We also combined the results from the family-based analysis with allele frequencies from control data from the Study on Addiction: Genetics and Environment (SAGE), also genotyped with the Illumina Human 1M-single Infinium BeadChip (23). Combining the AGP family-based transmission data with control data also yielded no new loci (see Supplementary Material). The peak association for MACROD2 remained at rs4141463 (Table 2 and Supplementary Material), but the P-value for association increased to 8.1 × 10−8 (strict diagnosis, European ancestry). For the loci identified by primary analyses of AGP data, the AGP, AGRE and control data taken together (Table 2) had little effect on the significance level for rs4141463 (P = 3.7 × 10−8 for strict diagnosis, European ancestry). In fact, the combined AGP, AGRE and control analyses showed similar results to those from the combined AGP and AGRE analysis (see Table 2), with the exception of rs4150167; the P-value for this SNP rises to 2.1 × 10−5. Looking over the entire genome, analysis of the combined data did not reveal compelling new loci (see Supplementary Material).
Genome-wide SNP association: exploratory analyses
To examine whether greater phenotypic homogeneity within ASD could help identify common risk variants, we performed a number of exploratory analyses examining specific sub-groups of the ASD sample. In this study, we report in detail two categorical variables: verbal status and IQ; see Methods for description of exploratory categories. We also evaluated parental origin effects through parental transmission. Sample sizes are given in Table 1; results are given in Supplementary Material, Table S4. None of our exploratory analyses detected association below the threshold of 5 × 10−8 in the AGP discovery sample alone. We do observe signals that are close to the threshold (P < 1 × 10−7, chosen strictly for heuristic purposes) in the discovery sample in PLD5, POU6F2 and an intergenic region on 8p21.3. Moreover, in a combined analysis of the AGP and AGRE data, we observe three associations that cross the P-value threshold: for verbal individuals, for SNPs rs3784730 (in ST8SIA2) and rs2196826 (in PLD5); and, for maternal parent of origin, rs9532931 (in a gene for an uncharacterized predicted protein KIAA0564). Importantly, these findings would not be significant after correction for multiple testing of diagnostic groups and sub-phenotypes. A summary of all association signals at P < 1 × 10−6 in the exploratory analyses are detailed in Supplementary Material, Tables S3 and S4); see Supplementary Material for results for SNPs with association (P ≤ 5 × 10−4).
Level of function, as measured by IQ, has been assumed to be a major source of etiological heterogeneity for autism. When we explored the impact of IQ on GWA results by splitting the sample by probands with IQ >80 and those with IQ <70, no P-value exceeded the threshold for GWA significance and none met criterion P < 1 × 10−6.
Genome-wide SNP association: candidate loci
We compared our data with replicated candidate-gene studies, which were derived from (19), as well as the recent GWA reports that implicated intergenic intervals at the 5p14.1 CDH9–CDH10 and 5p15.2 SEMA5A–TAS2R1 loci, respectively (20,21,24) (Supplementary Material, Tables S5 and S6). Because the estimated effect sizes for these studies typically fall in the range of 1.1–1.3, our power to replicate these findings was low (Supplementary Material, Fig. S4) and some of the prior candidate-gene studies made use of markers not well tagged by SNPs in our study. We found some support for several prior candidate loci, including CNTNAP2, RELN and SLC25A12 (P < 10−4), but our analysis did not garner additional evidence for either of the top findings from the prior GWA studies.
DISCUSSION
After testing ∼1 million SNPs for association with ASD, we identified in one of our set of four primary analyses one SNP, rs4141463, in MACROD2 crossing a preset threshold of P < 5 × 10−8. Three other SNPs crossed this threshold in the context of exploratory analyses, making their interpretation more difficult due to multiple testing. All of these results spring from a relatively small sample size for GWA studies (n ≤ 1369 families), limiting both our power to detect association and the certainty of the associations detected. Unbiased estimates of odds ratios detected by GWA studies are typically in the range of 1.1–1.3; to have good power to detect such effect sizes requires many thousands of samples, which is beyond the reach of the autism genetics community at the moment. This issue could at least partially explain why most genomic regions with prior evidence of SNP associations for ASD risk garner little support from our data (Supplementary Material, Table S6). Moreover, the winner's curse and shrinkage to the mean (22,25,26) could explain the smaller odds ratios that we estimated from the replication data (Table 2).
Keeping these caveats in mind, several results from our study are potentially relevant to autism risk. The function of MACROD2 (previously c20orf133) is largely unknown. The protein contains a MACRO domain which is a high-affinity ADP-ribose-binding domain that is important in multiple biological processes. Recent genome-wide studies have highlighted copy number variation at MACROD2 in an individual with schizophrenia (27), brain infarct (28) and brain volume in multiple sclerosis (29). Also ∼500 kb from the association signal observed for ASD is FLRT3, which is embedded in MACROD2. FLRT3 is a cell adhesion molecule with functions in neuronal development.
It is interesting to consider that, while rs4141463 falls in a MACROD2 intron, the precise location could be irrelevant to its possible functional impact on ASD risk. Recent evidence (30), yet to be corroborated, suggests that this SNP or one of many correlated SNPs in this region (Fig. 1) acts to regulate expression of PLD2. The observation becomes more noteworthy in light of the fact that our exploratory analyses also identify PLD5 as another locus possibly associated with autism. PLD proteins could play an important role in risk for autism. The protein derived from PLD2 has been shown to regulate axonal outgrowth (31) and metabotropic glutamate receptor signaling (32).
A second association signal of interest from the primary analyses (Table 2 and Supplementary Material, Table S2) involves a missense variation in the TAF1C gene (rs4150167; G523R; P = 1.0 × 10−6). TAF1C (TATA box-binding protein-associated factor 1C) is involved in the initiation of transcription by RNA polymerase I. This process requires the formation of a complex composed of the TATA-binding protein (TBP) and three TBP-associated factors (TAFs) specific for RNA polymerase I. TAF1C and its complex are displaced by PTEN (33). Mutations in PTEN have been highlighted in a number of cases of autism and related disorders (34–39). A caveat about the data for this SNP is worth noting: visual inspection revealed typical genotype clusters, yet the relatively common allele (≈0.98) is over-transmitted, a pattern consistent with poor genotyping quality.
From the exploratory analyses (Supplementary Material, Table S3), we identify a number of loci as having noteworthy association. One of the most appealing genes for risk for autism is ST8SIA2, coding for a protein expressed very highly throughout the mammalian brain (expression level or density >90 for 14 out of 17 brain regions assessed in the Allen Brain Atlas, http://www.brain-map.org/). Mice without polysialyltransferases ST8SiaII and ST8SiaIV, which modify neural cell adhesion molecule (NCAM1), show malformations of major brain axon tracts (40). Loss of either ST8 protein alone results in milder phenotypes. Inactivation of ST8SiaII in mice alters axonal targeting, involving hippocampal infrapyramidal mossy fibers, and the mice show increased exploration and diminished fear (40,41), behaviors of potential relevance to autism. Learning and memory, mediated through morphological synaptic plasticity, are also critically dependent on NCAM polysialylation status but in a complex way (42). Further studies are needed to determine the relevance of these neurodevelopmental results to the genetics of autism and identify the genetic variation affecting expression or function of ST8SIA2. With regard to genetic variation, in addition to the results found in our study, variation in ST8SIA2 has been associated with risk for schizophrenia in Asian populations (43,44).
While we and others (20,21) find limited evidence that common alleles affect risk for autism, the number of families studied is still relatively small. Our findings appear to rule out a common allele increasing relative risk by 2-fold or more. Much larger samples will be required to detect subtle effects on relative risk (e.g. 1.2), which is more typical of risk loci for common diseases. With such low relative risk, replication of true positive findings is further complicated by chance findings, as well as differences in ascertainment. These challenges are not unique to common variants. The same challenges are faced when searching for rare sequence mutations and CNVs affecting risk for ASD. Moreover, our ultimate goal is to integrate results across the range of rare to common variation, thereby describing the genetic architecture of autism. This will require larger cohorts comprised of individuals exhibiting the relatively stringent ASD phenotype of this study, as well as an unselected group more representative of the general ASD population, both being examined at the highest resolution for CNVs, rare sequence variation and common alleles. The heterogeneity of ASD will continue to complicate ameliorative opportunities; however, the identification of risk variants could reveal target gene pathways amenable for therapeutic intervention.
MATERIALS AND METHODS
Sample collection and ascertainment
Diagnostic classes
For these analyses, we primarily grouped families into two nested diagnostic classes (strict and spectrum ASD) based on proband diagnostic measures (45). To qualify for the strict class, affected individuals met the criteria for autism on both primary diagnostic instruments, the ADI-R (2) and the ADOS (3). In addition to individuals meeting criteria for autism, a spectrum class included all individuals who were classified as ASD on both the ADI-R and ADOS or who were not evaluated on one of the instruments but were diagnosed with autism on the other instrument.
AGRE cohort
One additional family-based autism cohort was evaluated in this study. The AGRE sample consists of families in which a proband and often one or more siblings are diagnosed with ASD (46). A total of 595 families (1086 probands) that were shown to be independent of the AGP sample were identified for replication.
SAGE control cohort
A control group, namely subjects from the Study on Addiction: Genetics and Environment (SAGE), was chosen because it was genotyped with Illumina Human 1M-single Infinium BeadChip (23). This cohort consisted of 1965 control subjects (from the larger SAGE case–control study). The consented sample included 31% males and 69% females, with mean age of 39.2 (SD 9.1), and 73% subjects self-identified as European-American (Caucasian), 26% as African-American and 1% as other (http://zork.wustl.edu/gei/study_description.htm). Both raw intensities and genotypes were available through NHGRI–dbGaP (http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000092.v1.p1). The SAGE control subjects have had exposure to alcohol (and possibly to other drugs), but did not meet the criteria for any illicit drug dependence.
Genotyping
Samples were genotyped using the Illumina Human 1M-single Infinium BeadChip. We performed stringent, uniform QC procedures on the resulting data. The Illumina Human 1M-single Infinium BeadChip contains a total of 1 072 820 markers (50-mer probes) for SNP and CNV analyses. Samples were processed using the manufacturer's recommended protocol with no modifications for Infinium II arrays, and BeadChips were scanned on the Illumina BeadArray Reader using default settings. Analysis and intra-chip normalization were performed using Illumina's BeadStudio software v.3.3.7, with a GenCall cutoff of 0.1. Built-in controls, both sample independent (including staining controls, extension controls, target removal controls and hybridization controls) and sample dependent (including stringency controls, nonspecific binding controls and nonpolymorphic controls), were inspected to assess the quality of the experiment. For genotype calling, we followed the manufacturer's protocols and used technical controls. Trios consisting of an affected offspring and both parents were genotyped, and in total genotyping was completed for 4683 individuals from 1558 families. For the control sample, 1880 individuals were genotyped on the Illumina Human 1M-single Infinium BeadChip, as described elsewhere (23).
The AGRE sample was genotyped on the Illumina HapMap550 array (20), which yields genotypes for roughly 550 000 SNPs of the 1 million contained on the 1M chip. To make these data comparable with the 1M platform, we inferred the missing SNP genotypes by using three sources of information: haplotypes called from trios genotyped on the 1M, haplotypes called from the HapMap550 genotypes and a small set of 105 samples that were genotyped on both platforms and yielded high-quality genotypes. This smaller set of overlapping samples was used to evaluate the accuracy of inferred genotypes. We used Beagle 3.0.1 to call haplotypes and infer missing genotypes (47). Because Beagle 3.0.1 allows only families with a single offspring, we created trios from multiplex families; inferred missing genotypes; then, after putting family data back together, resolved inconsistencies when possible and ‘zeroed’ inconsistencies otherwise. Using the 105 samples genotyped on both platforms, we assessed imputation accuracy; imputed genotypes for an SNP were retained only when none of the called genotypes were discrepant with the 1M genotypes. Following this QC process, each sample from the AGRE data set contained genotypes from the HapMap550 and 248 642 additional imputed genotypes for subsequent analysis.
Association analysis
Genetic QC for association analysis
As a first QC step prior to GWA analysis, probands from 80 families were removed because they either carried chromosomal abnormalities, exhibited chromosomal cell line artifacts or, based on literature reports, had highly penetrant ASD CNVs. In broad outline, subsequent QC for association analyses was performed at family and individual levels, followed by QC for individual SNPs.
We first assessed gender miscalls based on X chromosome genotypes and allele calls for Y, adjusting gender when appropriate (e.g. miscoding) and dropping samples (e.g. Klinefelter syndrome) or genotypes (e.g. loss of X in cell line) from the X chromosome. We searched the database for duplicate samples using a subset of 5254 SNPs that were independent and had a >99.9% completion rate for genotypes at this QC stage. Duplicates from four families were removed. Data were subsequently checked for Mendelian errors, and 19 families with large numbers of errors were removed from the analysis. In all other cases of Mendelian inheritance errors, the SNPs were set to missing in the family exhibiting the error. The fraction of complete genotypes per individual was required to be ≥95% over the autosomes; 27 samples fell below this criterion. Following this QC step, 4304 genotyped individuals were retained for 1445 pedigrees.
We then removed monomorphic SNPs or those with a genotyping completion rate <95%. After this step, 991 221 SNPs were retained. We note that using a genotyping completion rate of 95% or more allows some SNPs of poor genotyping quality to enter the analysis; the alternative is to use a more stringent completion rate, such as 99%, which has the advantage of removing low-quality SNPs at the cost of removing some high-quality SNPs. Recognizing the tradeoff, we chose to use the less stringent criterion for association analysis, and follow up SNPs with small P-values, by manual inspection of genotype clusters. However, for the figures in the manuscript, we use the more stringent criterion for genotyping completion rate, which more accurately reflects the final results after manual inspection of genotype clusters.
Ancestry was then determined for the proband by using 5239 widely spaced, independent SNPs that had a genotype completion rate of ≥99.9%. The software used was SpectralGEM (48), which estimated five significant dimensions of ancestry (Supplementary Material, Fig. S1). Subsequent clustering on the dimensions of ancestry resulted in six clusters: three clusters of European ancestry, with n = 824, 353 and 87; and three clusters reflecting other major ancestral groups, with n = 68, 54 and 35 (e.g. African and Asian); see also Supplementary Material, Figure S1. The major European cluster (n = 824) was used to determine minor allele frequencies (MAFs), to evaluate Hardy–Weinberg equilibrium (HWE) and Wright's Fst among the genotyping sites. (Note, however, that all three European clusters were used for the ultimate association analyses of this ancestry.) Specific SNPs were eliminated on the basis of the genotypes in this homogeneous European cluster for the following reasons: 5499 were monomorphic; 2102 for completion rate <95%; 132 894 for MAF <0.01; 7734 for HWE P < 0.005; and 89 for Fst > 0.02. Following this QC step, there were 842 348 SNPs available for association analysis. In this final SNP-based edit, six more individuals had a genotype completion rate of <95%. Merging diagnostic information with genotype information to determine informative families yielded 1369 families with complete genotype data for parents and offspring; with at least one genotyped offspring carrying an ASD diagnosis (16 families had more than one genotyped, affected child).
Genetic association analyses
Family-based analyses were first performed using FBAT, which allows for rapid calculation of statistics under additive, dominant and recessive genetic models. We do not present, here, the results for the dominant or recessive models because they did not contribute meaningfully to the results. However, to implement more flexible analyses, such as parent-of-origin analyses, we also used an in-house program written for family-based association (49) that implements methods described by Cordell et al. (50). Comparisons of the in-house program to FBAT results, when appropriate, yielded excellent agreement (unpublished data). A priori, we planned four principal analyses, all under the additive model: spectrum versus strict diagnosis by all ancestries versus European ancestry. These four analyses covered the extremes of phenotype and ancestry. Numerous exploratory analyses were also performed under an additive genetic model: parent of origin analysis considering paternal- and maternal-specific transmissions for both strict and spectrum diagnostic classes; and for the largest, spectrum sets, we stratified by proband's verbal/non-verbal status (51).
To determine whether the level of cognitive function, as measured by IQ, was an important covariate for heterogeneity, we split probands according to IQ into four groups: (i) those with IQ > 80; (ii) those with 80 ≥ IQ ≥ 70; (iii) those with 70 > IQ > 25; and those with IQ ≤ 25. For GWA analyses, we used only Groups (i) and (iii), which had the largest sample size. IQ was measured in various ways by the different recruitment centers, but for our purposes we used verbal, non-verbal (performance) and full-scale IQ assessments. If an individual's score was >80 for any of these three measures, the proband was classified into the above 80 group; otherwise, providing IQ was evaluated on at least two measures and none were ≥70, the proband was classified into one of the below 70 groups. Sample sizes for principal analyses are given in Table 1.
To enhance power for GWA tests, two additional data sets were combined with the AGP data. We analyzed the AGRE data using family-based analyses and the AGRE and AGP data combined using mega-analyses. All primary analyses were performed with both data sets. We limited exploratory analyses to these nine: broad diagnostic group; verbal and nonverbal status by the diagnostic groups and by ancestry. For the primary analyses, we also analyzed two other sets of combined samples: AGP trios together with AGRE families; AGP trios together with unrelated SAGE controls; and all three data sets. The method to analyze control and family-based data (49) builds on two related ideas: matched case–control analysis using conditional logistic regression (e.g. 52) and the natural connection between family-based analysis and conditional logistic regression of alleles found in probands (the transmitted alleles) and matched pseudo-controls (formed from transmitted and un-transmitted alleles) (49). Unrelated SAGE controls were matched by genetic ancestry to probands and combined with the ‘pseudo-controls’ produced by the family-based analysis: first, by spectral analysis, we estimated the genetic ancestry of probands and unrelated controls (48); then, using the optimal matching algorithm, we formed genetically homogeneous strata (52), each consisting of a single proband and one or more unrelated controls. In those strata where a single control was matched with more than one proband, the control was paired with the best match in the stratum and the remaining probands each form their own stratum. Finally, within each stratum, we contrasted the genotype of the proband with the genotypes of the matched controls and pseudo-controls via conditional logistic regression.
SUPPLEMENTARY MATERIAL
FUNDING
This research was primarily supported by Autism Speaks (USA), the Health Research Board (HRB, Ireland), The Medical Research Council (MRC; UK); Genome Canada/Ontario Genomics Institute and the Hilibrand Foundation (USA). Additional support for individual groups was provided by the US National Institutes of Health [HD055751, HD055782, HD055784, HD35465, MH52708, MH55284, MH057881, MH061009, MH06359, MH066673, MH077930, MH080647, MH081754, MH66766, NS026630, NS042165, NS049261]; the Canadian Institutes for Health Research (CIHR), Assistance Publique - Hôpitaux de Paris (France), Autistica, Canada Foundation for Innovation/Ontario Innovation Trust, Deutsche Forschungsgemeinschaft (grant: Po 255/17-4) (Germany), EC Sixth FP AUTISM MOLGEN, Fundação Calouste Gulbenkian (Portugal), Fondation de France, Fondation FondaMental (France), Fondation Orange (France), Fondation pour la Recherche Médicale (France), Fundação para a Ciência e Tecnologia (Portugal), GlaxoSmithKline-CIHR Pathfinder Chair (Canada), the Hospital for Sick Children Foundation and University of Toronto (Canada), INSERM (France), Institut Pasteur (France), the Italian Ministry of Health [convention 181 of 19.10.2001], the John P. Hussman Foundation (USA), McLaughlin Centre (Canada), Netherlands Organization for Scientific Research [Rubicon 825.06.031], Ontario Ministry of Research and Innovation (Canada), Royal Netherlands Academy of Arts and Sciences [TMF/DA/5801], the Seaver Foundation (USA), the Swedish Science Council, The Centre for Applied Genomics (Canada), the Utah Autism Foundation (USA) and the Wellcome Trust core award [075491/Z/04 UK]. We wish to acknowledge SAGE as part of this study. Funding support for the Study of Addiction: Genetics and Environment (SAGE) was provided through the NIH Genes, Environment and Health Initiative [GEI] (U01 HG004422). SAGE is one of the genome-wide association studies funded as part of the Gene Environment Association Studies (GENEVA) under GEI. Assistance with phenotype harmonization and genotype cleaning, as well as with general study coordination, was provided by the GENEVA Coordinating Center (U01 HG004446). Assistance with data cleaning was provided by the National Center for Biotechnology Information. Support for collection of datasets and samples was provided by the Collaborative Study on the Genetics of Alcoholism (COGA; U10 AA008401), the Collaborative Genetic Study of Nicotine Dependence (COGEND; P01 CA089392), and the Family Study of Cocaine Dependence (FSCD; R01 DA013423). Funding support for genotyping, which was performed at the Johns Hopkins University Center for Inherited Disease Research, was provided by the NIH GEI (U01 HG004438), the National Institute on Alcohol Abuse and Alcoholism, the National Institute on Drug Abuse, and the NIH contract ‘High throughput genotyping for studying the genetic contributions to human disease’ (HHSN268200782096C). Funding to pay the Open Access Charge was provided by Autism Speaks.
Supplementary Material
ACKNOWLEDGEMENT
The authors gratefully acknowledge the families participating in the study.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Cook E.H., Jr, Scherer S.W. Copy-number variations associated with neuropsychiatric conditions. Nature. 2008;455:919–923. doi: 10.1038/nature07458. [DOI] [PubMed] [Google Scholar]
- 2.Lord C., Rutter M., Couteur A. Autism Diagnostic Interview-Revised: a revised version of a diagnostic interview for caregivers of individuals with possible pervasive developmental disorders. J. Autism Dev. Disord. 1994;24:659–685. doi: 10.1007/BF02172145. doi:10.1007/BF02172145. [DOI] [PubMed] [Google Scholar]
- 3.Lord C., Risi S., Lambrecht L., Cook E.H., Jr, Leventhal B.L., DiLavore P.C., Pickles A., Rutter M. The autism diagnostic observation schedule-generic: a standard measure of social and communication deficits associated with the spectrum of autism. J. Autism Dev. Disord. 2000;30:205–223. doi:10.1023/A:1005592401947. [PubMed] [Google Scholar]
- 4.Fombonne E. Epidemiology of pervasive developmental disorders. Pediatr. Res. 2009;65:591–598. doi: 10.1203/PDR.0b013e31819e7203. doi:10.1203/PDR.0b013e31819e7203. [DOI] [PubMed] [Google Scholar]
- 5.Fernell E., Gillberg C. Autism spectrum disorder diagnoses in Stockholm preschoolers. Res. Dev. Disabil. 2010;31:680–685. doi: 10.1016/j.ridd.2010.01.007. [DOI] [PubMed] [Google Scholar]
- 6.Baron-Cohen S., Scott F.J., Allison C., Williams J., Bolton P., Matthews F.E., Brayne C. Prevalence of autism-spectrum conditions: UK school-based population study. Br. J. Psychiatry. 2009;194:500–509. doi: 10.1192/bjp.bp.108.059345. doi:10.1192/bjp.bp.108.059345. [DOI] [PubMed] [Google Scholar]
- 7.Bailey A., Le Couteur A., Gottesman I., Bolton P., Simonoff E., Yuzda E., Rutter M. Autism as a strongly genetic disorder: evidence from a British twin study. Psychol. Med. 1995;25:63–77. doi: 10.1017/s0033291700028099. doi:10.1017/S0033291700028099. [DOI] [PubMed] [Google Scholar]
- 8.Hurley R.S., Losh M., Parlier M., Reznick J.S., Piven J. The broad autism phenotype questionnaire. J. Autism Dev. Disord. 2007;37:1679–1690. doi: 10.1007/s10803-006-0299-3. doi:10.1007/s10803-006-0299-3. [DOI] [PubMed] [Google Scholar]
- 9.Constantino J.N., Todd R.D. Intergenerational transmission of subthreshold autistic traits in the general population. Biol. Psychiatry. 2005;57:655–660. doi: 10.1016/j.biopsych.2004.12.014. doi:10.1016/j.biopsych.2004.12.014. [DOI] [PubMed] [Google Scholar]
- 10.Jamain S., Quach H., Betancur C., Rastam M., Colineaux C., Gillberg I.C., Soderstrom H., Giros B., Leboyer M., Gillberg C., et al. Mutations of the X-linked genes encoding neuroligins NLGN3 and NLGN4 are associated with autism. Nat. Genet. 2003;34:27–29. doi: 10.1038/ng1136. doi:10.1038/ng1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Durand C.M., Betancur C., Boeckers T.M., Bockmann J., Chaste P., Fauchereau F., Nygren G., Rastam M., Gillberg I.C., Anckarsäter H., et al. Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nat. Genet. 2006;39:25–27. doi: 10.1038/ng1933. doi:10.1038/ng1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sebat J., Lakshmi B., Malhotra D., Troge J., Lese-Martin C., Walsh T., Yamrom B., Yoon S., Krasnitz A., Kendall J., et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. doi:10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marshall C.R., Noor A., Vincent J.B., Lionel A.C., Feuk L., Skaug J., Shago M., Moessner R., Pinto D., Ren Y., et al. Structural variation of chromosomes in autism spectrum disorder. Am. J. Hum. Genet. 2008;82:477–488. doi: 10.1016/j.ajhg.2007.12.009. doi:10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weiss L.A., Shen Y., Korn J.M., Arking D.E., Miller D.T., Fossdal R., Saemundsen E., Stefansson H., Ferreira M.A., Green T., et al. Association between microdeletion and microduplication at 16p11.2 and autism. N. Engl. J. Med. 2008;358:667–675. doi: 10.1056/NEJMoa075974. doi:10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- 15.Kumar R.A., KaraMohamed S., Sudi J., Conrad D.F., Brune C., Badner J.A., Gilliam T.C., Nowak N.J., Cook E.H., Jr, Dobyns W.B., Christian S.L. Recurrent 16p11.2 microdeletions in autism. Hum. Mol. Genet. 2008;17:628–638. doi: 10.1093/hmg/ddm376. doi:10.1093/hmg/ddm376. [DOI] [PubMed] [Google Scholar]
- 16.Fernandez B.A., Roberts W., Chung B., Weksberg R., Meyn S., Szatmari P., Joseph-George A.M., Mackay S., Whitten K., Noble B., et al. Phenotypic spectrum associated with de novo and inherited deletions and duplications at 16p11.2 in individuals ascertained for diagnosis of autism spectrum disorder. J. Med. Genet. 2010;47:195–203. doi: 10.1136/jmg.2009.069369. doi:10.1136/jmg.2009.069369. [DOI] [PubMed] [Google Scholar]
- 17.Glessner J.T., Wang K., Cai G., Korvatska O., Kim C.E., Wood S., Zhang H., Estes A., Brune C.W., Bradfield J.P., et al. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature. 2009;459:569–573. doi: 10.1038/nature07953. doi:10.1038/nature07953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pinto D., Pagnamenta A.T., Klei L., Anney R., Merico D., Regan R., Conroy J., Magalhaes T., Correia C., Abrahams B.S., et al. Functional impact of global rare copy number variation in autism. Nature. 2010;466:368–372. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abrahams B.S., Geschwind D.H. Advances in autism genetics: on the threshold of a new neurobiology. Nat. Rev. Genet. 2008;9:341–355. doi: 10.1038/nrg2346. doi:10.1038/nrg2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang K., Zhang H., Ma D., Bucan M., Glessner J.T., Abrahams B.S., Salyakina D., Imielinski M., Bradfield J.P., Sleiman P.M., et al. Common genetic variants on 5p14.1 associate with autism spectrum disorders. Nature. 2009;459:528–533. doi: 10.1038/nature07999. doi:10.1038/nature07999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weiss L.A., Arking D.E., Daly M.J., Chakravarti A. Gene Discovery Project of Johns Hopkins & the Autism Consortium. A genome-wide linkage and association scan reveals novel loci for autism. Nature. 2009;461:802–808. doi: 10.1038/nature08490. doi:10.1038/nature08490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Göring H.H., Terwilliger J.D., Blangero J. Large upward bias in estimation of locus-specific effects from genomewide scans. Am. J. Hum. Genet. 2001;69:1357–1369. doi: 10.1086/324471. doi:10.1086/324471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bierut L.J., Agrawal A., Bucholz K.K., Doheny K.F., Laurie C., Pugh E., Fisher S., Fox L., Howells W., Bertelsen S., et al. A genome-wide association study of alcohol dependence. Proc. Natl Acad. Sci. USA. 2010;107:5082–5087. doi: 10.1073/pnas.0911109107. doi:10.1073/pnas.0911109107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma D., Salyakina D., Jaworski J.M., Konidari I., Whitehead P.L., Andersen A.N., Hoffman J.D., Slifer S.H., Hedges D.J., Cukier H.N., et al. A genome-wide association study of autism reveals a common novel risk locus at 5p14.1. Ann. Hum. Genet. 2009;73:263–273. doi: 10.1111/j.1469-1809.2009.00523.x. doi:10.1111/j.1469-1809.2009.00523.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhong H., Prentice R.L. Correcting ‘winner's curse’ in odds ratios from genomewide association findings for major complex human diseases. Genet. Epidemiol. 2010;34:78–91. doi: 10.1002/gepi.20437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun L., Bull S.B. Reduction of selection bias in genomewide studies by resampling. Genet. Epidemiol. 2005;28:352–367. doi: 10.1002/gepi.20068. doi:10.1002/gepi.20068. [DOI] [PubMed] [Google Scholar]
- 27.Xu B., Woodroffe A., Rodriguez-Murillo L., Roos J.L., van Rensburg E.J., Abecasis G.R., Gogos J.A., Karayiorgou M. Elucidating the genetic architecture of familial schizophrenia using rare copy number variant and linkage scans. Proc. Natl Acad. Sci. USA. 2009;106:16746–16751. doi: 10.1073/pnas.0908584106. doi:10.1073/pnas.0908584106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Debette S., Bis J.C., Fornage M., Schmidt H., Ikram M.A., Sigurdsson S., Heiss G., Struchalin M., Smith A.V., van der Lugt A., et al. Genome-wide association studies of MRI-defined brain infarcts: meta-analysis from the CHARGE Consortium. Stroke. 2010;41:210–217. doi: 10.1161/STROKEAHA.109.569194. doi:10.1161/STROKEAHA.109.569194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baranzini S.E., Wang J., Gibson R.A., Galwey N., Naegelin Y., Barkhof F., Radue E.W., Lindberg R.L., Uitdehaag B.M., Johnson M.R., et al. Genome-wide association analysis of susceptibility and clinical phenotype in multiple sclerosis. Hum. Mol. Genet. 2009;18:767–778. doi: 10.1093/hmg/ddn388. doi:10.1093/hmg/ddn388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Duan S., Huang R.S., Zhang W., Bleibel W.K., Roe C.A., Clark T.A., Chen T.X., Schweitzer A.C., Blume J.E., Cox N.J., Dolan M.E. Genetic architecture of transcript-level variation in humans. Am. J. Hum. Genet. 2008;82:1101–1113. doi: 10.1016/j.ajhg.2008.03.006. doi:10.1016/j.ajhg.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kanaho Y., Funakoshi Y., Hasegawa H. Phospholipase D signalling and its involvement in neurite outgrowth. Biochim. Biophys. Acta. 2009;1791:898–904. doi: 10.1016/j.bbalip.2009.03.010. [DOI] [PubMed] [Google Scholar]
- 32.Dhami G.K., Ferguson S.S. Regulation of metabotropic glutamate receptor signaling, desensitization and endocytosis. Pharmacol. Ther. 2006;111:260–271. doi: 10.1016/j.pharmthera.2005.01.008. doi:10.1016/j.pharmthera.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 33.Zhang C., Comai L., Johnson D.L. PTEN represses RNA Polymerase I transcription by disrupting the SL1 complex. Mol. Cell Biol. 2005;25:6899–6911. doi: 10.1128/MCB.25.16.6899-6911.2005. doi:10.1128/MCB.25.16.6899-6911.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Butler M.G., Dasouki M.J., Zhou X.P., Talebizadeh Z., Brown M., Takahashi T.N., Miles J.H., Wang C.H., Stratton R., Pilarski R., et al. Subset of individuals with autism spectrum disorders and extreme macrocephaly associated with germline PTEN tumour suppressor gene mutations. J. Med. Genet. 2005;42:318–321. doi: 10.1136/jmg.2004.024646. doi:10.1136/jmg.2004.024646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Buxbaum J.D., Cai G., Chaste P., Nygren G., Goldsmith J., Reichert J., Anckarsäter H., Rastam M., Smith C.J., Silverman J.M., et al. Mutation screening of the PTEN gene in patients with autism spectrum disorders and macrocephaly. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2007;144B:484–491. doi: 10.1002/ajmg.b.30493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goffin A., Hoefsloot L.H., Bosgoed E., Swillen A., Fryns J.P. PTEN mutation in a family with Cowden syndrome and autism. Am. J. Med. Genet. A. 2001;105:521–524. doi: 10.1002/ajmg.1477. doi:10.1002/ajmg.1477. [DOI] [PubMed] [Google Scholar]
- 37.Herman G.E., Butter E., Enrile B., Pastore M., Prior T.W., Sommer A. Increasing knowledge of PTEN germline mutations: Two additional patients with autism and macrocephaly. Am. J. Med. Genet. A. 2007;143:589–593. doi: 10.1002/ajmg.a.31619. [DOI] [PubMed] [Google Scholar]
- 38.Orrico A., Galli L., Buoni S., Orsi A., Vonella G., Sorrentino V. Novel PTEN mutations in neurodevelopmental disorders and macrocephaly. Clin. Genet. 2009;75:195–198. doi: 10.1111/j.1399-0004.2008.01074.x. doi:10.1111/j.1399-0004.2008.01074.x. [DOI] [PubMed] [Google Scholar]
- 39.Varga E.A., Pastore M., Prior T., Herman G.E., McBride K.L. The prevalence of PTEN mutations in a clinical pediatric cohort with autism spectrum disorders, developmental delay, and macrocephaly. Genet Med. 2009;11:111–117. doi: 10.1097/GIM.0b013e31818fd762. doi:10.1097/GIM.0b013e31818fd762. [DOI] [PubMed] [Google Scholar]
- 40.Hildebrandt H., Mühlenhoff M., Oltmann-Norden I., Röckle I., Burkhardt H., Weinhold B., Gerardy-Schahn R. Imbalance of neural cell adhesion molecule and polysialyltransferase alleles causes defective brain connectivity. Brain. 2009;132:2831–2838. doi: 10.1093/brain/awp117. doi:10.1093/brain/awp117. [DOI] [PubMed] [Google Scholar]
- 41.Nacher J., Guirado R., Varea E., Alonso-Llosa G., Röckle I., Hildebrandt H. Divergent impact of the polysialyltransferases ST8SiaII and ST8SiaIV on polysialic acid expression in immature neurons and interneurons of the adult cerebral cortex. Neuroscience. 2010;167:825–837. doi: 10.1016/j.neuroscience.2010.02.067. [DOI] [PubMed] [Google Scholar]
- 42.Ter Horst J.P., Loscher J.S., Pickering M., Regan C.M., Murphy K.J. Learning-associated regulation of polysialylated neural cell adhesion molecule expression in the rat prefrontal cortex is region-, cell type- and paradigm-specific. Eur. J. Neurosci. 2008;28:419–427. doi: 10.1111/j.1460-9568.2008.06326.x. doi:10.1111/j.1460-9568.2008.06326.x. [DOI] [PubMed] [Google Scholar]
- 43.Arai M., Yamada K., Toyota T., Obata N., Haga S., Yoshida Y., Nakamura K., Minabe Y., Ujike H., Sora I., et al. Association between polymorphisms in the promoter region of the sialyltransferase 8B (SIAT8B) gene and schizophrenia. Biol. Psychiatry. 2006;59:652–659. doi: 10.1016/j.biopsych.2005.08.016. doi:10.1016/j.biopsych.2005.08.016. [DOI] [PubMed] [Google Scholar]
- 44.Tao R., Li C., Zheng Y., Qin W., Zhang J., Li X., Xu Y., Shi Y.Y., Feng G., He L. Positive association between SIAT8B and schizophrenia in the Chinese Han population. Schizophr. Res. 2007;90:108–114. doi: 10.1016/j.schres.2006.09.029. doi:10.1016/j.schres.2006.09.029. [DOI] [PubMed] [Google Scholar]
- 45.Risi S., Lord C., Gotham K., Corsello C., Chrysler C., Szatmari P., Cook E.H., Jr, Leventhal B.L., Pickles A. Combining information from multiple sources in the diagnosis of autism spectrum disorders. J. Am. Acad. Child Adolesc. Psychiatry. 2006;45:1094–1103. doi: 10.1097/01.chi.0000227880.42780.0e. doi:10.1097/01.chi.0000227880.42780.0e. [DOI] [PubMed] [Google Scholar]
- 46.Geschwind D.H., Sowinski J., Lord C., Iversen P., Shestack J., Jones P., Ducat L., Spence S.J. AGRE Steering Committee. The autism genetic resource exchange: a resource for the study of autism and related neuropsychiatric conditions. Am. J. Hum. Genet. 2001;69:463–466. doi: 10.1086/321292. doi:10.1086/321292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Browning B.L., Browning S.R. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am. J. Hum. Genet. 2009;84:210–223. doi: 10.1016/j.ajhg.2009.01.005. doi:10.1016/j.ajhg.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee A.B., Luca D., Klei L., Devlin B., Roeder K. Discovering genetic ancestry using spectral graph theory. Genet. Epidemiol. 2010;34:51–59. doi: 10.1002/gepi.20434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Crossett A., Kent B.P., Klei L., Ringquist R., Trucco M., Roeder K., Devlin B. Using ancestry matching to combine family-based and unrelated samples for genome-wide association studies. Statist. Med. 2010 doi: 10.1002/sim.4057. in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cordell H.J., Barratt B.J., Clayton D.G. Case/pseudocontrol analysis in genetic association studies: a unified framework for detection of genotype and haplotype associations, gene–gene and gene–environment interactions, and parent-of-origin effects. Genet. Epidemiol. 2004;26:167–185. doi: 10.1002/gepi.10307. doi:10.1002/gepi.10307. [DOI] [PubMed] [Google Scholar]
- 51.Liu X.Q., Paterson A.D., Szatmari P. Autism Genome Project Consortium. Genome-wide linkage analyses of quantitative and categorical autism subphenotypes. Biol. Psychiatry. 2008;64:561–570. doi: 10.1016/j.biopsych.2008.05.023. doi:10.1016/j.biopsych.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Luca D., Ringquist S., Klei L., Lee A.B., Gieger C., Wichmann H.E., Schreiber S., Krawczak M., Lu Y., Styche A., et al. On the use of general control samples for genome-wide association studies: genetic matching highlights causal variants. Am. J. Hum. Genet. 2008;82:453–463. doi: 10.1016/j.ajhg.2007.11.003. doi:10.1016/j.ajhg.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.