Abstract
Genome-wide association studies have proved very successful in identifying novel single-nucleotide polymorphisms (SNPs) associated with disease or traits, but the related, functional SNP is usually unknown. In this paper, we describe a methodology to locate and validate candidate functional SNPs using lipoprotein lipase (LPL), a gene previously associated with triglyceride levels, as an exemplar. Two thousand seven hundred and eighty-six healthy middle-aged men from the NPHSII UK prospective study (with up to six measures of plasma lipid levels) were genotyped for 20 LPL tagging (t)SNPs using Illumina Bead technology. Using model-selection procedures and haplotypes, we identified eight SNPs that consistently maximized the fit of the model to the phenotype. Fifteen SNPs in high linkage disequilibrium with these were identified, and functional assays were carried out on all 23 SNPs. Electrophoretic mobility shift assay (EMSA) was used to identify SNPs that had the potential to alter DNA–protein interactions, reducing the number to eight possible candidate SNPs. These were examined for ability to alter expression using a luciferase reporter assay, and two regulatory SNPs, showing genotype differences, rs327 and rs3289, were identified. Finally, multiplexed-competitor-EMSA (MC-EMSA) and supershift EMSA identified FOXA2 to rs327T, and CREB-binding protein (CBP) and CCAAT displacement protein (CDP) to rs3289C as the factors responsible for transcription binding. We have identified two novel candidate functional SNPs in LPL and presented a procedure aimed to efficiently detect SNPs potentially causal to genetic association. We believe that this methodology could be successfully applied to future re-sequencing data.
INTRODUCTION
With the increasing use of genome-wide association studies (GWAS), the number of single nucleotide polymorphisms (SNPs) associated with disease or intermediate phenotypes is rapidly mounting (1). Despite this increase in signals of association, the discovery of the functional changes responsible for those signals is still a laborious and slow task. The discrepancy between the two is predominantly attributable to two factors: linkage disequilibrium (LD) between SNPs and the location of many of the SNPs in non-coding regions. LD, the violation of statistical independence between genetic loci, causes all SNPs in LD with the functional locus to carry some, or all, of the association with the trait of interest even if they have no relevant function. The second problem is that although we can, with some accuracy, predict the effect that a SNP will have on an amino acid change and thus protein function, it is becoming clear that the majority of genetic markers associated with traits or disease are present in non-coding regions and are therefore likely to affect gene regulation or pre-mRNA splicing or mRNA stability, rather than protein function (2). To achieve the goal of using genetics in disease prevention and treatment, it is important to identify these functional genetic variants, which will help disentangle the complexity evident in common diseases and allow the development of new targeted medication.
Coronary heart disease (CHD) is a complex disease that develops as a result of both environmental and genetic contributing factors. Elevated triglyceride (TG) levels in blood has been shown to be an epidemiological risk factors for CHD, where it is suggested that it increases the development of atherosclerosis, by promoting the production of pro-atherogenic small dense LDL particles (3). The risk associated with a 1 mmol difference in TG levels is 1.32 (1.26–1.39) in men and 1.72 (1.5–2.07) in women (4). Recently, several GWAS reported associations of TG with a number of loci (5–9), including the lipoprotein lipase gene (LPL), a ‘candidate’ gene that has previously demonstrated a strong and consistent association with TG levels and with CHD risk in prospective and case–control studies (10). LPL is primarily expressed in adipose tissues and muscle, where, attached to the capillary endothelium, it hydrolyses TG-rich lipoprotein particles, although expression also occurs in the heart, liver, lungs, nervous system, macrophages and pancreatic islet cells (11). LPL functions as a homodimer and catalyses the rate-limiting step in TG hydrolysis and also acts as a ligand/bridging factor for the receptor-mediated uptake of lipoproteins (12). Severe mutations that result in LPL deficiency lead to familial hyperchylomicronaemia (13), whereas common non-synonymous variants rs1801177 (D9N) and rs328 (S447X) have been consistently associated with higher and lower risk of CHD, respectively (10). Recent GWAS on lipoprotein phenotypes identified several LPL variants associated with effects on TG levels: rs10096633 (5), rs12678919 (6), rs17482753 (7), rs17410914 (8), rs6993414 (9) and rs328 (14). However, LD between neighbouring SNPs, insufficient coverage of the locus and the use of univariate statistical methods hinder the identification of the true functional change.
Here, using LPL and its association with TG as an example, we present statistical and laboratory procedures to identify candidate functional SNPs from dense tagging SNPs (tSNPs). We give examples of statistical tests available for the analysis of association between SNPs and continuous variables. Popular model selection criteria are calculated for the data and their results are compared, while haplotypes are used both as mediators of association and as an additional method of model selection. Having determined the SNPs with the highest likelihood of marking a functional variant, all SNPs in strong LD with these were examined for functional effects. Electrophoretic mobility shift assays (EMSAs) were carried out on these variant sequences, and those SNPs that showed differences in EMSA were further examined in reporter gene expression studies. For the SNPs that also demonstrated expression differences, the identification of potential DNA-binding factors mediating this effect was carried out using multiplexed-competitor EMSAs (MC-EMSAs) and supershift EMSAs.
RESULTS
Table 1 shows the basic characteristics of the genotyped subjects as well as a summary of all six TG measures available. From the 22 SNPs successfully genotyped in LPL, four SNPs were out of Hardy–Weinberg equilibrium (HWE). For two SNPs, rs1800590 and LPL-95, this was strongly significant (P < 0.0001 for both) and they were subsequently dropped from the analysis, whereas rs3779788 and rs7016529 were retained, being only moderately out of HWE (P = 0.0187 and 0.0018, respectively). Of the remaining 20 SNPs, 14 were significantly associated with levels of TG. Estimates of effect size both as beta coefficients and R2, together with levels of TG per genotype can be seen in Table 2, and the results of alternative strategies for statistical analysis are shown in the Supplementary Material. To resolve problems due to rare alleles, a permutational test of 10 000 repeats was also run with the results shown in Supplementary Material, Table S2. Adjustment for age and practice centre did not materially change the statistical significance of the results (Supplementary Material, Table S2).
Table 1.
Characteristics of the NPHSII men with complete genotype data. All six measures of TG are shown for baseline and five annual visits
| Variable | Observations | Mean | SD |
|---|---|---|---|
| Age (years) | 2385 | 55.90 | 3.37 |
| BMI (kg/m2) | 2382 | 26.48 | 3.47 |
| Total cholesterol (mmol/l) | 2370 | 5.74 | 1.00 |
| LDL-cholesterol (mmol/l) | 2068 | 3.08 | 1.00 |
| HDL-cholesterol (mmol/l) | 2054 | 1.72 | 0.59 |
| Baseline TGs (mmol/l) | 2372 | 2.07 | 1.25 |
| TGs phase 1 (mmol/l) | 2204 | 1.98 | 1.14 |
| TGs phase 2 (mmol/l) | 2097 | 1.97 | 1.17 |
| TGs phase 3 (mmol/l) | 2023 | 1.97 | 1.19 |
| TGs phase 4 (mmol/l) | 1805 | 2.12 | 1.21 |
| TGs phase 5 (mmol/l) | 1689 | 2.15 | 1.22 |
| TGs mean (mmol/l) | 2383 | 2.05 | 1.04 |
| Log TGs mean (log(mmol/l)) | 2383 | 0.61 | 0.45 |
Table 2.
Means and SDs of genotype distribution and TG for the 20 genotyped LPL SNPs. Beta coefficients and R2of the additive model for log mean of TG together with the observed counts, means, SDs and P-values for the 20 successfully genotyped LPL SNPs for each SNP of LPL. SNPs ordered by chromosome position
| SNP rs number | Beta coefficienta | R2 | Common/rare allele | Common homozygote |
Heterozygote |
Rare homozygote |
P-value | HW equilibrium | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | Mean TG levels | SD | n | Mean TG levels | SD | n | Mean TG levels | SD | ||||||
| rs17410577 | 0.041* | 0.0009 | G/C | 1547 | 1.994 | 0.945 | 728 | 2.145 | 1.173 | 105 | 2.150 | 1.316 | 0.0103 | 0.0999 |
| rs1534649 | −0.015 | 0.0028 | G/T | 737 | 2.062 | 0.994 | 1145 | 2.058 | 1.085 | 496 | 1.999 | 0.998 | 0.2322 | 0.1691 |
| rs3779788 | −0.073*** | 0.0006 | C/T | 1693 | 2.103 | 1.093 | 648 | 1.910 | 0.886 | 40 | 1.907 | 0.837 | 0.0001 | 0.0187* |
| rs7016529 | 0.103* | 0.0065 | T/C | 2310 | 2.04 | 1.038 | 70 | 2.302 | 1.088 | 3 | 1.893 | 0.491 | 0.0395 | 0.0018** |
| rs1121923 | 0.016 | 0.0018 | G/A | 2247 | 2.049 | 1.051 | 133 | 2.017 | 0.827 | 3 | 1.976 | 0.053 | 0.6665 | 0.4800 |
| rs248 | −0.049* | 0.0001 | G/A | 2035 | 2.068 | 1.066 | 337 | 1.921 | 0.852 | 11 | 2.013 | 1.015 | 0.0469 | 0.4612 |
| rs249 | −0.025 | 0.0017 | A/G | 2020 | 2.059 | 1.046 | 352 | 1.979 | 1.006 | 11 | 2.109 | 0.734 | 0.3020 | 0.2910 |
| rs253 | −0.032* | 0.0004 | C/T | 725 | 2.108 | 1.048 | 1164 | 2.050 | 1.078 | 489 | 1.949 | 0.925 | 0.0129 | 0.5677 |
| rs264 | −0.082**** | 0.0026 | G/A | 1707 | 2.112 | 1.095 | 630 | 1.884 | 0.860 | 45 | 1.902 | 0.923 | <0.0001 | 0.1683 |
| rs268 | 0.127** | 0.0084 | T/C | 2284 | 2.034 | 1.031 | 97 | 2.379 | 1.177 | 2 | 1.336 | 0.777 | 0.0045 | 0.3575 |
| rs270 | 0.022 | 0.0034 | G/T | 1711 | 2.017 | 0.965 | 598 | 2.146 | 1.242 | 67 | 1.964 | 0.867 | 0.2090 | 0.0973 |
| rs283 | 0.030 | 0.0007 | C/T | 1584 | 2.014 | 1.005 | 701 | 2.113 | 1.083 | 98 | 2.117 | 1.243 | 0.0648 | 0.0703 |
| rs301 | −0.079**** | 0.0014 | T/C | 1367 | 2.142 | 1.124 | 878 | 1.931 | 0.902 | 133 | 1.825 | 0.828 | <0.0001 | 0.6622 |
| rs316 | −0.058** | 0.0112 | C/A | 1852 | 2.076 | 1.068 | 495 | 1.965 | 0.935 | 36 | 1.669 | 0.757 | 0.0036 | 0.6508 |
| rs328 | −0.081*** | 0.0036 | G/C | 1889 | 2.092 | 1.081 | 467 | 1.870 | 0.830 | 26 | 1.940 | 1.046 | 0.0001 | 0.7598 |
| rs10099160 | 0.003 | 0.0063 | A/C | 1406 | 2.048 | 1.021 | 839 | 2.030 | 1.064 | 134 | 2.144 | 1.086 | 0.8651 | 0.5542 |
| rs3289 | 0.113** | 0.0000 | A/G | 2255 | 2.035 | 1.032 | 123 | 2.221 | 1.106 | 4 | 3.568 | 1.636 | 0.0037 | 0.0915 |
| rs13702 | −0.063**** | 0.0035 | T/C | 1212 | 2.144 | 1.143 | 984 | 1.954 | 0.908 | 184 | 1.894 | 0.891 | <0.0001 | 0.4602 |
| rs4921684 | −0.043* | 0.0079 | C/T | 1730 | 2.072 | 1.068 | 598 | 2.001 | 0.966 | 53 | 1.756 | 0.825 | 0.0190 | 0.8673 |
| rs2197089 | 0.037** | 0.0023 | A/G | 695 | 1.962 | 0.941 | 1175 | 2.060 | 1.057 | 509 | 2.134 | 1.120 | 0.0048 | 0.7497 |
aAsterisks denote level of significance: *0.05, **0.01, ***0.001, ****<0.0001.
Model selection methods
The SNPs considered were not completely independent of each other, with LD ranging from an r2 value of 3.53 × 10−7 to 0.74 (Supplementary Material, Fig. S1), and so the respective P-values for their association with TG would also be non-independent. A model using all of the SNPs as explanatory variables will account for the between-SNP associations, but would also lead to overfitting. To find the smallest set of SNPs accounting for the association of the LPL with TG, we used a number of criteria of fit, with both stepwise and best-subset methods. Table 3 shows all the criteria used and the SNPs in the best model in each case. As expected, from all the possible models, the Bayesian information criterion (BIC) chose the simplest model with only rs301, whereas the Akaike information criterion, Mallows Cp and a Cross-validation scheme all selected a model containing five SNPs: rs17410577, rs264, rs268, rs301 and rs3289, all significant in the univariate analysis. Other criteria used were the residual mean square, which left out of the best model SNPs rs17410577 and rs264, and the adjusted R2 measure, which was the most permissive, allowing, in addition to the previous five SNPs, rs10099160 which is non-significant in the univariate analysis. A rank of P-values is also shown in Table 3, with only the top-ranking SNP (rs301) found in all selected models, while the second-ranking SNP is in four and the third-ranking SNP in none of the six selected best models. Stepwise regression using AIC stopped at the same five-SNP model as the best-subset method, whereas when a P-value of 0.1 was used as a cut-off for removal from the model, the stepwise regression chose an alternative five-SNP model with rs17410577, rs268, rs301, rs10099160 and rs3289.
Table 3.
Comparison of different model selection approaches. All the methods and criteria used to select SNPs most likely to be tightly associated with a functional variant
| SNP rs number | Mean of logarithm of TG for additive modela | Rank of P-value | Adj. R2 | Mallows Cp | RMS | AIC | BIC | Cross-validation | Stepwise regression using AIC | Stepwise regression using P = 0.1 |
|---|---|---|---|---|---|---|---|---|---|---|
| rs17410577 | 0.0103* | 9 | × | × | × | × | 0.130399 | 0.015 | ||
| rs1534649 | 0.2322 | 19 | ||||||||
| rs3779788 | 0.0001*** | 4 | ||||||||
| rs7016529 | 0.0395* | 13 | ||||||||
| rs1121923 | 0.6665 | 20 | ||||||||
| rs248 | 0.0469* | 15 | ||||||||
| rs249 | 0.3020 | 17 | ||||||||
| rs253 | 0.0129* | 12 | ||||||||
| rs264 | <0.0001**** | 2 | × | × | × | × | 0.069453 | |||
| rs268 | 0.0045** | 6 | × | × | × | × | × | 0.005754 | 0.005 | |
| rs270 | 0.2090 | 16 | ||||||||
| rs283 | 0.0648 | 14 | ||||||||
| rs301 | <0.0001**** | 1 | × | × | × | × | × | × | 0.004087 | <0.001 |
| rs316 | 0.0036** | 8 | ||||||||
| rs328 | 0.0001*** | 5 | ||||||||
| rs10099160 | 0.8651 | 18 | × | 0.060 | ||||||
| rs3289 | 0.0037** | 7 | × | × | × | × | × | 0.016188 | 0.020 | |
| rs13702 | <0.0001**** | 3 | ||||||||
| rs4921684 | 0.0190* | 11 | ||||||||
| rs2197089 | 0.0048** | 10 |
For additive model and stepwise regression with a P-value cut-off point, P-values are shown. For the stepwise AIC the AIC is given. For the best-subset models, cross (×) denotes inclusion in the best model.
aAsterisks denote level of significance: *0.05, **0.01, ***0.001, ****<0.0001.
Haplotype approaches
For the 20 SNPs, 242 haplotypes were identified (220 possible) of which 21 were present at a frequency higher than 1% capturing 87.1% of the population, and with just 4 above 5% accounting for only 52.9% of haplotypic variability. To capture 90% of all the haplotypes, 26 were needed with a minimum frequency of 0.52%, whereas for 99% coverage, 101 haplotypes were required with a minimum relative frequency of 0.021% (Supplementary Material, Table S3). Using the frequency cut-off of 1%, three haplotypes had TG levels significantly lower than those of the most common haplotype in the population, with a single haplotype being associated with higher TG levels, when additivity of haplotypes was assumed (Supplementary Material, Table S4). Aligning the five haplotypes SNP by SNP as shown in Supplementary Material, Table S5, we can see that eight SNPs were common in all haplotypes making them unlikely to be responsible for the lowering of TG observed. It is expected that functional variants, or SNPs in LD with them, will be the same in the three lowering haplotypes and differ between them and the most common and raising haplotypes. Three SNPs—rs301, rs13702 and rs2197089—followed this pattern, with the LD r2 ranging from 0.23 to 0.33 between them. For the TG-raising haplotype, only SNP rs17410577 was consistent with the phenotypic change (Supplementary Material, Table S5).
We repeated the entire haplotype procedure including only the five SNPs (rs17410577, rs264, rs268, rs301 and rs3289) chosen by the model selection step described earlier. Twenty haplotypes were found, 7 with a frequency of more than 1% accounting for 98.2% of the population (Supplementary Material, Table S6). Three haplotypes were found to have statistically different TG levels from the most common haplotype, 2 of them associated with lower and 1 with higher TG levels (Supplementary Material, Table S7). Again, aligning the haplotypes revealed that rs301 was consistent with the pattern of the lowering haplotypes, whereas, in contrast to what was seen earlier, the rs268 SNP was now characterizing the haplotype of higher TG levels (Supplementary Material, Table S8).
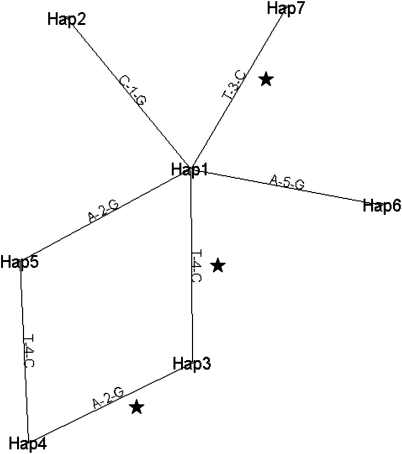
Using the 21 common haplotypes obtained for all 20 LPL SNPs, we constructed a haplotypic tree showing their sequence relatedness (Supplementary Material, Fig. S2). Using TreeScan, all the branches were tested for association with TG levels. After 10 000 simulations using the analysis of variance tests, the only evidence for association with the phenotype was the change between Haplotype (Hap)10 (10th most common haplotype) and the node between Hap11 and Hap9, with a permutational P-value (after monotonicity is enforced) of 0.053. This branch of the tree was associated with the change of the SNP rs301 (Supplementary Material, Fig. S2). We repeated the tree inference and analysis using only the five SNPs selected earlier, and the results are presented in Figure 1. The tree has a very pronounced loop between Hap1, Hap3, Hap4 and Hap5. For the benefit of analysis, we assumed that common haplotypes were giving rise to haplotypes of smaller frequency. Thus, the two possible alternatives of the tree is either a cut of the loop between Hap4 and Hap5 or between Hap5 and Hap1. Testing both alternatives, we found that there were three branches associated with TG levels. The strongest association (P < 0.0001) was between Hap1 and Hap3 because of a change in the rs301 SNP. The branches between Hap3 and Hap4 (P = 0.0010), and between Hap1 and Hap7 (P = 0.0310) due to SNPs rs264 and rs268, respectively, were also significantly associated with the phenotype.
Figure 1.
Haplotypic tree for five selected SNPs. Haplotypes are ordered by frequency with Hap1 the most common. Numbers on branches stand for the specific SNP change between the haplotypes with SNP 1 rs17410577, SNP 2 rs264, SNP 3 rs268, SNP 4 rs301 and SNP 5 rs3289. When each change in the tree was tested for association with TG, the changes between Hap1 and Hap7, Hap1 and Hap3, and Hap3 and Hap4 were found to be statistically significant.
Association of minimal SNP set with other lipid traits
To explore the effect of the selected set of LPL SNPs on other CHD traits, the association of both the SNPs and their haplotypes was examined. As shown in Supplementary Material, Table S9, in addition to TG, LPL was also associated with changes in HDL-cholesterol (HDL-C) levels, though with more modest effects. Apolipoprotein (apo) AI and apo B also showed signals of association with LPL with three and one SNPs, respectively, although, owing to the number of tests performed and the P-values obtained, these associations do not permit safe conclusions to be drawn for these associations. Using the haplotypes of the five SNPs of the reduced model to test for association with other lipid markers, we found that haplotype 4 was associated with an increase in both ApoAI and HDL-C levels when compared with the most common haplotype (Supplementary Material, Table S10).
Selection of putative SNPs that marks functional variants
From the statistical tests of best model selection, five SNPs were deemed as the most likely candidates: rs17410577, rs264, rs268, rs301 and rs3289. These five SNPs as well as rs10099160 (selected from the R2 criterion), rs13702 and rs2197089 (suggested from the haplotypic association) were examined for functionality. SNPs in high LD with these eight SNPs (r2 > 0.8) were identified from the Genome Variation Server database (http://gvs-p.gs.washington.edu/GVS/). A total of 23 SNPs were selected from the original eight, and the LD between the SNPs is shown in Supplementary Material, Figure S3.
Functional assessment of LPL SNPs
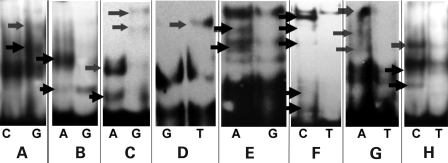
To investigate the effect of the putative functional LPL SNPs on DNA-binding proteins, EMSAs were performed using probes of ∼30 bp sequences that encompassed the common or rare variant of each of the 23 SNPs. Nuclear extracts from Huh7 (human liver cell line) and human smooth muscle cells were used in the assays to include a wide range of possible DNA-binding proteins that may be involved in LPL regulation. Initial EMSA analysis revealed potential binding differences between eight wild-type and variant alleles (rs17410577, rs268, rs327, rs326, rs331, rs3289, rs3208305 and rs2197089; Fig. 2) using the Huh7 cell line, and all these were followed-up for further analysis. Using nuclear extract from human smooth muscle cells did not provide any further allele-specific bands to those found using the Huh7 cell line (data not shown). The probes that did not bind any proteins by EMSAs, or those that the common and rare variants were bound by the same intensity, and therefore likely to bind outside the SNP location, were rejected from further analysis. The relative location of SNPs examined and those that showed allele-specific protein binding are shown in Supplementary Material, Figure S4.
Figure 2.
Representative EMSA binding using probes surrounding SNPs associated with TG levels. Red arrows indicate the presence of an allele-specific DNA–protein complex: (A) rs17410577; (B) rs268; (C) rs326; (D) rs327; (E) rs331; (F) rs3289; (G) rs3208305; (H) rs2197089.
Luciferase reporter assays
To determine the possible transcriptional effect that the transcription factor (TF)-binding sites identified by EMSAs may be having on LPL expression, a luciferase reporter system was created, whereby the LPL promoter (from −724 to +39) was inserted upstream of luciferase, and 100 bp sequences encompassing each SNP allele were individually placed downstream of the SV40 late poly(A) signal for the luc+ reporter, and therefore able to act as an enhancer for gene expression (Supplementary Material, Fig. S5). No SNPs located in the LPL promoter itself were identified by statistical analysis, and so inserting SNP sequences directly upstream of luciferase was not appropriate in this model.
Transfection of LPL constructs was carried out in Huh7 and smooth muscle cells, using a Renilla-containing vector to control for transfection efficiency. Figure 3A and B shows expression levels in Huh7 and smooth muscle cells, respectively, relative to expression of the pGL3 basic vector containing the LPL promoter only. In Huh7 cells, expression from rs327 G construct was 1.7-fold higher than that from the rs327 T construct (P < 0.001). Expression from the rs3289 C construct was 1.6-fold higher than that from the rs3289 T construct (P < 0.01). There were no other significant differences in reporter expression between alleles. The differences in expression were not observed when transfected into human smooth muscle cells, indicating that the enhancer/silencer elements are likely to be tissue-specific, as is the case with the majority of distal regulatory elements.
Figure 3.
Results of luciferase reporter assay showing relative expression of LPL-luciferase-enhancer constructs relative to the LPL-luciferase construct (promoter only). (A) Transfection of a Huh7 cell line. Significant differences in expression are seen in rs327, where expression from the G allele is 1.7 times that of the T allele. Similarly, in rs3289, expression from the C allele is 1.6 times that shown of the T allele. (B) Transfection of a smooth muscle vascular cell line. There were no significant differences in expression between alleles for this cell line.
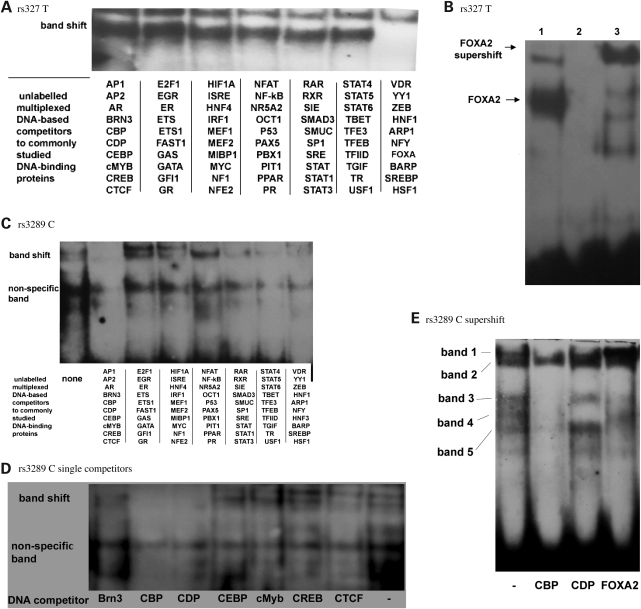
MC-EMSA and supershift assay
To identify the TFs involved in binding to the LPL variants, which also conferred differential expression in luciferase reporter assays, competition assays were carried out using MC-EMSAs (15). In this procedure, excess unlabelled competitor dsDNA consensus sequences for well-characterized TFs were multiplexed and included in the EMSA-binding reaction. Elimination of the bandshift for a particular set of TFs is caused by the sequestration of the causal TF by the unlabelled competitor probe. When used on the sequence containing the rs327 T allele, one set of MC sequences eliminated the bandshift (Fig. 4A). When individual competitors from this set were examined in a further EMSA (data not shown), this indicated that the additional band produced by rs327 T compared with G was due to a FOXA family TF. A supershift assay confirmed this to be FOXA2 (Fig. 4B). Similarly, MC-EMSAs suggested that the additional bands produced by rs3289 T were from a heterodimer of TFs, CREB-binding protein (CBP) and CCAAT displacement protein (CDP) (Fig. 4C–E), although without a clear supershift this could not be conclusively proved.
Figure 4.
(A) MC-EMSA using cocktails of unlabelled DNA competitors to 70 well-characterized DNA-binding proteins, using the rs327 T allele probe and Huh7 nuclear extract. The final column of multiplexed competitors prevents binding of rs327 T DNA to the protein. (B) Supershift EMSA on rs327 T probe. The single competitors from the final column in 7(a) were run individually in a further EMSA, showing FOXA competitor resulted in competition (data not shown). To confirm FOXA binding, a supershift assay was carried out using an antibody to FOXA2. Lane 1 represents the shift produced by the FOXA2–DNA interaction, lane 2 includes unlabelled consensus DNA for FOXA competition, lane 3 demonstrates a supershift with the addition of a FOXA2 monoclonal antibody, confirming binding of FOXA2. (C) MC-EMSA using cocktails of unlabelled DNA competitors to 70 well-characterized DNA-binding proteins, using the rs3289 C allele probe and Huh7 nuclear extract. The first column of multiplexed competitors prevents binding of rs3289 C DNA to the protein. (D) The single competitors from the first column in 7(c) were run individually in a further EMSA, showing both CBP and CDP competitors resulted in competition. (E) Supershift against rs3289 C allele probe using antibodies for CBP, CDP and FOXA2 (as a negative control). Addition of antibody to CBP results in the elimination of bands 1, 3 and 4. Addition of antibody to CDP results in the elimination of band 4. A supershift is not observed, possibly due to hindrance from heterodimeric binding.
DISCUSSION
Using 20 dense tSNPs for LDL, we identified a smaller subset of SNPs likely to be associated with DNA changes affecting TG levels. The selection was based on best-subset and stepwise regression techniques using a number of criteria of fit together with an analysis of haplotypes both before and after model selection. These SNPs, in addition to SNPs in strong LD with them, were investigated further in functional studies. We performed an EMSA for each SNP and those that showed differential binding were subsequently cloned into a reporter construct to study their effect on expression. We identified two SNPs that showed novel functionality, through a mechanism affecting gene expression. Finally, we identified the binding TF using MC-EMSA and supershift EMSA methodologies.
In terms of the statistical analyses, in the supplementary section we have presented a detailed walkthrough of a number of alternative approaches for the analysis of genotypic data in association studies. We showed that a power transformation is better in achieving a distribution close to the normal than the logarithmic transformation commonly used. Nevertheless, we chose to present the latter, because effect size measures on a logarithmic scale have an intuitive meaning of multiplication, which the more complex power transformation is unable to convey. We also presented both a longitudinal model through all multiple measurements of TG as well as a much simpler model where the mean TG levels over 6 years was used as the response variable. Again, a trade-off between simplicity and accuracy was evident. The longitudinal model is better suited for the available data but it lacks the user-friendliness of the simple model. In this case, the choice between them does not change our conclusions; thus simplicity was favoured. Similarly, the more elaborate genotypic model, making no assumption for the dominance between the two alleles, is usually a better approximation of the truth, since only rarely will the heterozygous trait mean level be situated exactly between the two homozygotes, as is the assumption of the additive model, though the genotypic model will have a degree of freedom more than the corresponding additive model, lowering the power of the test. We tested the dominance deviation for all the SNPs and we found that only one of the SNPs showed significant deviation from additivity, thus making the use of the assumption-free model unnecessary, although, as a general rule, we suggest that both models are considered initially.
Model selection
It is evident that model selection is an important tool in distinguishing overlapping genetic effects when dense SNPs are considered. With most of the criteria used, we ended up with the same or similar subset of SNPs. SNPs rs3289, identified as functional, and rs301, in strong LD with the functional SNP rs327, were in all selected models, except BIC which selected only rs301. In contrast, we were unable to find any functional change associated with SNP rs268, which was also considered a very good candidate and had similar statistical evidence with the other two. It is difficult to say which criterion of fit is the most appropriate for all cases but we can see that, at least in our case, BIC was the most conservative. Cross-validation, Mallows Cp and AIC are identical but AIC is the easiest of the three to calculate and has been implemented in a wide range of statistical packages. However, the commonly used method of considering the importance of SNPs based on their P-values was not supported from our results, with the first and seventh ranked SNPs found to be associated with changes of LPL transcription. We are currently working on a simulation study to assess the efficiency of each selection criterion in different scenarios of LD and the number of functional SNPs.
Model selection also benefited the analysis of haplotypes. Excluding SNPs not selected as contributing to the fit of the model decreased the noise associated with haplotypic analysis. In this case, the analysis of haplotypes was used as a means of identifying potentially interesting SNPs when the pattern of common lineage was accounted for rather than a test of association for a single extended haplotype. The use of haplotypic trees gave another level of information for the haplotypes, both in terms of the relationship between them, sometimes in evolutionary terms, as well as on the effect of SNPs in a context-dependent manner, as seen in the TreeScan method. We also explored the idea of using the inferred haplotypes and their respective trees as tools for finding SNPs associated with functional changes. Although we were able to identify rs301 in both cases, this method severely penalizes rare SNPs, and more work is required before we can claim that it is a successful method for further refinement of association signals. If rare functional SNPs in and around LPL are contributing to a significant extent to the association of the gene with TG levels, then rare haplotypes sharing the same lineage might prove informative, as outlined recently in a modelling paper (16). In this study, we considered HapMap CEU SNPs with frequencies higher than 4%, thus limiting our ability to identify truly rare polymorphisms, which would require re-sequencing of many subjects for a comprehensive analysis.
Functional assays of identified SNPs
Once the SNPs in strong LD with the tSNPs associated with TG levels were identified, a simple procedure to examine functionality of a large subset of SNPs was required. As the SNPs were present almost exclusively in non-coding regions, there was a high likelihood that a functional SNP would be effecting gene regulation. As a regulatory SNP would necessarily alter DNA-binding properties, EMSA analysis was the most appropriate and simplest tool to measure this possibility. Chromatin immunoprecipitation would be another potential tool, with the advantage of being an in vivo approach, but this procedure requires prior knowledge of the DNA-binding proteins involved. Once EMSAs had identified allele-specific differences in affinity for DNA-binding proteins, expression differences were then examined. Luciferase reporter assays were carried out, using the LPL promoter driving luciferase, with the potential regulatory alleles acting as an enhancer element.
The strongest statistical association with TG levels was with the tSNP rs301. One SNP in strong LD with this SNP, rs327, also affected expression levels in an enhancer-based luciferase construct driven by the LPL promoter. In this scenario, the T allele resulted in decreased expression and appears to act as a distal transcriptional silencer. An alternative explanation is that there is a transcription silencer nearby, but not including rs327, and the G allele is acting as a classical enhancer. Using MC-EMSAs, followed by supershift assay demonstrated strong binding of FOXA2 to the T allele, but no binding to the G allele. FOXA2 (previously known as hepatic nuclear factor-3β) is a member of the forkhead class of DNA-binding proteins, which acts as a transcriptional activator of liver-specific genes, and can also interact with chromatin. The LPL promoter has been well-characterized, and a study by Enerbäck et al. (17) has previously identified two elements in the promoter with DNA-binding properties similar to those of the forkhead family of TFs. These elements were able to confer differentiation-linked expression in vitro in a system that mimicked LPL expression during adipocyte differentiation. A similar system may be working at this intronic enhancer, where the T allele, but not the G allele, of rs327 may affect the differentiation-linked LPL expression. Another explanation could lie with the involvement of FOXA2 in chromatin remodelling. The FOXA proteins are able to open highly compacted chromatin in vitro through a process that does not involve the SWI/SNF chromatin remodelling complex and likely occurs through the ability of the FOXA proteins to bind the core histones H3 and H4 (18). Binding of the T allele to FOXA2 may facilitate an open chromatin structure surrounding intron 8/9, facilitating further access of DNA-binding proteins that may act as a complex transcriptional silencer. Indeed, analysis of chromatin signatures indicates the presence of a regulatory region surrounding rs327. A study into genome-wide histone methylation profiles identified an H3K4me1 (histone H3 monomethyl K4) profile surrounding rs327 (Supplementary Material, Fig. S6A and B) (19). This signature is often found downstream of transcription start sites and is associated with enhancer/silencer elements. Another genome-wide study examined nuclease-accessible sites in CD34+/− cells, which, in strong concordance with the H3K4me1 profile, demonstrates such a site in this region (Supplementary Material, Fig. S6A and B) (20). Together, these data add weight to the likelihood of this being an important regulatory region and the possibility that rs327 is an important SNP in LPL regulation.
The ‘HindIII’ polymorphism (rs320), also in strong LD with rs301, has been associated with TG levels in a number of studies (10). A study by Chen et al. (21) examined the functionality of this SNP, and EMSA analysis demonstrated the binding of TATA-binding protein to both alleles, with a marginally higher affinity to the common allele. This was complemented by a reporter assay, whereby the entire intron 8 sequence with either rs320 T or G was cloned downstream of the LPL promoter (1537 bp), driving luciferase expression. The authors found higher luciferase expression with the T allele construct. We, however, found no allelic difference in DNA binding to rs320 and did not pursue this SNP further. The model used for the reporter assay by Chen et al. (21) was for a promoter, rather than an enhancer. As the SNP is located >22 kb downstream of the transcription start site, any functional effect is likely to be through long-range interactions and acting as an enhancer/silencer element.
Another SNP which showed both differential expression and TF-binding in an enhancer construct was rs3289. Although rare (minor allele frequency < 0.05), this variant provides an additional, independent functional SNP that may regulate LPL levels. EMSA-based analysis indicated binding of both CBP and CDP to the T allele but not the C allele. CDP acts as a transcriptional repressor by binding to unique nucleotide sequence elements and is also involved in the regulation of gene transcription through nuclear matrix attachment (22). CBP is a transcription co-activator, having acetyltransferase activity with its binding partners. A study by Li et al. (23) demonstrated that CDP interacts with CBP, and the transcriptional repression is regulated by acetylation of specific lysine residues near the homeodomain of CDP (23). This mechanism may explain the reduced transcriptional activity of the construct containing the rs3289 T allele.
A recent study using the Cardiometabolic Illumina chip (24), encompassing 50 000 SNPs focusing on candidate genes for cardiovascular disease (25), included both rs327 and rs3289. There are 74 SNPs on the CVD chip in and around the LPL locus with 31 of them significant at the 10−5 cut-off point. Of the 20 SNPs we analysed, 17 were also included in the HumanCVD BeadChip (rs248, rs283 and rs4921648 only in NPHSII data) with 7 significant in both studies (rs253, rs264, rs301, rs328, rs3289, rs13702 and rs2197089). After variable selection, only rs3289 was common in the best model in both studies, consistent with our conclusion that this is a functional SNP. In contrast, rs327, although present in the second study, was dropped from the best model and two SNPs in high LD with it were included (rs331: r2 = 0.949;rs3916027: r2 = 0.899). We are unsure if the exclusion of rs327 was due to chance and the inherent uncertainty of best model selection, or it signifies that an as yet undiscovered SNP is responsible for the improvement of rs331 and rs3916027 fit beyond that of rs327. Interestingly, the well-characterized stop codon rs328 SNP, although significant in the univariate analysis of both the present study (P = 0.0001) and the HumanCVD study (P = 8.4 × 10−10), was not selected in the best model for either of them. In our data, rs328 is no longer informative when the rs301 SNP is included in the best model, with rs301 in high LD with our functional candidate SNP rs327 (r2 = 0.94). This raises the possibility that rs328, at least in Caucasians, might not itself be a functional locus, but in LD with one or more causal variants. This is further supported by the work of Deo et al. (26), which used admixture mapping in an African American sample from the Jackson Heart Study. They concluded that, in European-derived populations, rs328 is not itself the major causal variant but a marker for it, because of its effect size showing a statistically significant dependence on local ancestral background. Our candidate functional SNP, rs327, although above the statistical significance cut-off level of 0.0006 (P = 7.8 × 10−4), had a consistent effect in all subpopulations tested, more so than the rs10096633 SNP suggested by the authors as a possible functional variant. Unfortunately, the majority of studies, although identifying more than one statistical significant signal in the LPL gene (5–9,27), as we do, do not perform model selection or conditional P-value analysis in the locus in order to find which SNP is more closely associated with the functional variant. In the case of Tang et al. (28), conditional analysis of rs326 and rs13702 using rs328 as a covariate suggested that their effect is beyond what is explained by the stop codon rs328, with rs326 in high LD with our functional candidate rs327 SNP. Imputation of the SNPs previously identified as important or likely functional (rs10096633 (5), rs12678919 (6), rs17482753 (7), rs320 (10), rs328 (14)), and subsequent conditional analysis with the SNPs we identified (Supplementary Material, Table S11) revealed that rs327 can, to some degree, account for the association observed with the previously reported SNPs, suggesting that although these are significant in a univariate analysis, and are unlikely to be functional.
Limitations
Our current use of the methodology described here, and tested on the LPL association with TG levels, has a number of limitations. TG level is a distant phenotype compared to LPL activity or mass, and other factors might also be involved in the regulation of this association; in contrast, to the measurements of the immediate protein levels, TG has the advantage of showing effects owing to both changes in expression and protein function. We cannot claim that the results presented describe a comprehensive analysis of the LPL locus. The identification of tSNPs and SNPs in LD with the ones we considered as most likely linked to a functional variantwas done predominantly through the use of HapMap. Our coverage of the area relies on the completeness of the data (29), and thus more, yet undiscovered, causal common variants might still exist in and around LPL. The 1000 Genomes Project (http://www.1000genomes.org/) and the increasing use of re-sequencing will significantly enhance our ability to test all the variations in a locus, including rare SNPs. In our case, further information was lost because of the failure in genotyping some of the tSNPs, using 20 of the original 27 considered. As with all in vitro models for in vivo processes, DNA–protein interactions in vitro may not fully represent those occurring in vivo, particularly where chromatin structure and epigenetics may play an additional and important role. Furthermore, there may be tissue-specific TFs involved in LPL expression that are not represented either in the liver or smooth muscle cells used in this study. The luciferase reporter assay is a crude model to demonstrate regulatory potential of DNA sequences, and although currently the only technique to measure allelic expression differences in vitro, suffers mostly from limitations of insert size and no consideration of chromatin structure, which may be one of the major influences on gene expression.
Conclusions
We described a process of statistical analysis and experimental follow-up aimed to identify candidate functional SNPs in and around LPL. Two novel SNPs were shown to affect regulation of LPL in vitro and these were in LD with SNPs identified as being associated with levels of TG in a sample of middle-aged healthy men. We believe that the same process can be used to easily, and cost-effectively, determine potential functional variants in a number of genes shown to be associated with intermediate phenotypes or disease. More specifically, the cost of re-sequencing long stretches of DNA is becoming more and more affordable. This will provide us with a wealth of information never seen before but will also present new challenges. Signals of association are going to be clustered in areas of LD that can extend to great distances away from the gene. Moreover, any independent signals of association within the cluster are going to be difficult to distinguish. The proposed procedures offer an efficient method in identifying the most important SNPs and scanning their most closely linked SNPs to identify candidate functional variants.
MATERIALS AND METHODS
Study design and phenotypic measures
The Northwick Park Heart Study II (NPHS-II) is a prospective study of 3012 healthy middle-aged men aged 50–64 years at recruitment, sampled from nine UK general practices between 1989 and 1994 (30). They were free from disease at the time of recruitment, and information on lifestyle habits, height, weight and blood pressure were recorded at baseline and on subsequent prospective follow-up. Venepuncture was performed in the morning, and participants had been requested to take only a light breakfast. Baseline measures of cholesterol, TG and apolipoproteins AI and B were made using standard assays (31) with up to five subsequent annually repeated measurements of total cholesterol and TG. HDL-C was measured directly in samples taken at year 4 (31). A DNA repository was established using samples from 2775 participants obtained at the time of recruitment. Full details of recruitment, measurements, follow-up and definitions of incident disease have been reported elsewhere (30).
Genotyping
A customized Illumina 768 SNP genotyping array was assembled to capture common genetic variation in more than 76 genes (32) including the gene for LPL. We selected 27 tSNPs using the Haploview implementation of Tagger on HapMap data, applying a pairwise r2 threshold of 0.8 with a minor allelic frequency threshold of 0.04. Another five (c)SNPs were previously genotyped, with three also in the new array and one in LD with them. Of the 27 SNPs included in the array, 22 were successfully genotyped, with the rest either failing the final quality control or failing genotyping altogether.
Statistical analysis
TG levels were transformed using a logarithmic transformation (see Supplementary Material for a power transformation approach). The multiple measurements of TG, in baseline and five annual visits, were analysed using the mean of all six measurements adjusting for age and practice centre in STATA (version 10, STATA Corporation; see Supplementary Material for a longitudinal mixed-effect model). In terms of dominance between the two SNP alleles, we used an additive model, where the heterozygous effect is expected to be exactly between the two homozygotes (the genotypic model is described in the Supplementary Material). The subset of SNPs providing the best fit to the data, among all the possible models, was selected using a number of criteria in R (http://www.r-project.org/, 2.9.0). Haplotypes consisting of all the LPL SNPs were inferred using Phase (33). TG levels of haplotypes with a relative frequency of more than 1% were compared with that of the most common haplotype using 300 random draws from the posterior distributions of the haplotypes as computed by Phase. Each imputed data set was then tested separately, and the results were summarized using Rubin's rule (34) in Stata with mim (35). Those showing significant differences were aligned and the SNPs identifying haplotypes with lower or higher levels of TG were found. Using TCS (36), we constructed an unrouted network of haplotypes showing the relationships between the sequences. The haplotypic tree obtained was used to search for phenotype–genotype associations with TreeScan (37). Throughout, we use a rather permissive cut-off point of 0.05. This is done to reflect our prior knowledge of the effect of LPL on TG levels and the fact that our aim was not to discover signals of association but to identify the most likely SNPs of functional variation. Imputation of additional SNPs was done using MACH 1.0.16 (http://www.sph.umich.edu/csg/abecasis/MACH/index.html) and data from the full data set of HapMap phase 1 and 2 (http://hapmap.ncbi.nlm.nih.gov/).
SNPs in high LD with those identified as potentially interesting tSNPs (r > 0.8) were obtained from the Genome Variation Server database (http://gvs-p.gs.washington.edu/GVS/) using the combined LD derived from the CELERA Collection of 653 Caucasian individuals from CEPH pedigrees, CELERA Collection of 30 unrelated Caucasian individuals and 252 genotypes from Utah residents with Northern and Western European ancestry from the CEPH collection.
Cell culture and plasmid preparation
Huh7 cells (HPACC, Porton Down, UK) were maintained in high-glucose Dulbecco's modified Eagle's medium (PAA, Yeovil, UK) supplemented with 2 mm l-glutamine (PAA) and 10% fetal bovine serum (FBS) (PAA) at 37°C, 5% CO2. Human smooth muscle cells (Lonza, Porrino, Spain) were cultured in SmGM-2 Smooth Muscle Growth Medium-2 (Clonetics), containing 5% FBS.
The LPL promoter (−724 to +39 relative to the transcription start site) was amplified using standard PCR methods, and ligated into the linearized pGL3 basic vector using the Acc651 and HindIII sites. The integrity of the construct was verified by sequencing. To add the putative regulatory elements surrounding the SNPs of interest, oligonucleotides were designed with the addition of BamHI restriction sites at the 5′- and 3′-ends, and PCR amplification of common and rare homozygous individuals performed (oligos and PCR parameters are available upon request). The PCR products were digested with BamHI and ligated into the BamHI-linearized LPL–pGL3 vector, downstream of the SV40 late poly(A) signal. All sequences used in plasmid construction are available upon request.
Electrophoretic mobility shift assay
Nuclear extracts were obtained from Huh7 cells and smooth muscle cells using the NE-PER Nuclear and Cytoplasmic Extraction Reagents Kit (Pierce) as described in the manual, with the addition of Complete Protease Inhibitor (Roche) to buffers CER I and NER I. EMSA probes were designed with ∼15 bp each side of the candidate SNPs (probe sequences are available upon request). Probes were labelled using the Biotin 3′-end DNA Labelling Kit (Pierce) as described in the manual. Each binding reaction consisted of 2 µl of 10X binding buffer (100 mm Tris, 500 mm KCl; pH 7.5), 1 µg of p[dI-dC], 5 µg of nuclear extract, 200 fmol of biotin-labelled DNA, made to a total of 20 µl with H2O and incubated at 25°C for 30 min, followed by the addition of 5X loading buffer. Samples were loaded onto a 6% polyacrylamide gel and electrophoresed for 150 min at 120 V at 4°C. Transfer to positively charged nylon membrane was achieved through Southern transfer, and detection using the Chemiluminescent Nucleic Acid Detection Module (Pierce).
Luciferase reporter assay
Huh7 cells were seeded at a density of 2.5 × 104 per well in a 96-well plate format, and smooth muscle cells at a density of 1.5 × 104 per well and grown to confluence overnight in the appropriate media (described above). Cells were transfected with 200 ng of pGL3 reporter construct with 10 ng of pRLTK as a transfection control. Transfection was carried out in Opti-Mem serum-free media (Sigma) using Lipofectamine 2000 (Invitrogen) as described in the manual. Media was replaced 8 h following transfection, with serum-containing media described above, and the cells left for 2 days before harvesting. Cells were lysed using Passive Lysis Buffer (Promega), and luciferase expression was determined using the Dual-Luciferase Reporter Assay System (Promega) and measured using a Tropix TR717 Microplate Luminometer (PE Applied Biosystems). Luciferase assays were carried out in triplicate, and the mean relative expression differences between alleles were determined by t-test. Three clones for each allele were examined to ensure reproducibility.
TF identification
Identification of DNA-binding proteins was carried out using MC-EMSAs, as previously described (15). In brief, 100X unlabelled DNA competitors to 70 well-characterized DNA-binding proteins were added to the binding reaction described above, using arrays of 10 competitors per reaction, with a 30-min incubation on ice prior to the addition of labelled probe. Where a band shift was eliminated by multiplexed competition, the 10 individual competitors from the relevant array were run separately in a further EMSA. For verification of DNA-binding factor, 1 µg of monoclonal antibody (FOXA2, CDP and CBP; Abcam, UK) was used in place of the DNA competitor to create a supershift.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by the British Heart Foundation (RG05/014 and PG08/008). The NPHSII study was supported by the Medical Research Council, the US National Institutes of Health (NHLBI 33014) and Du Pont Pharma. Funding to pay the Open Access Charge was provided by British Heart Foundation.
Supplementary Material
ACKNOWLEDGEMENTS
We acknowledge the contribution of the late Professor George Miller (1939–2006) who was the PI on the NPHSII study. We also thank all the medical staff and patients who contributed to the NPHSII study and the Office for National Statistics (NHS) Central Registry for provision of mortality data.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Hindorff L.A., Junkins H.A., Mehta J.P., Manolio T.A. 2009. http://www.genome.gov/26525384 . Accessed 14 October. [DOI] [PMC free article] [PubMed]
- 2.Hardy J., Singleton A. Genomewide association studies and human disease. N. Engl. J. Med. 2009;360:1759–1768. doi: 10.1056/NEJMra0808700. doi:10.1056/NEJMra0808700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kannel W.B., Vasan R.S. Triglycerides as vascular risk factors: new epidemiologic insights. Curr. Opin. Cardiol. 2009;24:345–350. doi: 10.1097/HCO.0b013e32832c1284. doi:10.1097/HCO.0b013e32832c1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hokanson J.E., Austin M.A. Plasma triglyceride level is a risk factor for cardiovascular disease independent of high-density lipoprotein cholesterol level: a meta-analysis of population-based prospective studies. J. Cardiovasc. Risk. 1996;3:213–219. doi:10.1097/00043798-199604000-00014. [PubMed] [Google Scholar]
- 5.Aulchenko Y.S., Ripatti S., Lindqvist I., Boomsma D., Heid I.M., Pramstaller P.P., Penninx B.W., Janssens A.C., Wilson J.F., Spector T., et al. Loci influencing lipid levels and coronary heart disease risk in 16 European population cohorts. Nat. Genet. 2009;41:47–55. doi: 10.1038/ng.269. doi:10.1038/ng.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kathiresan S., Willer C.J., Peloso G.M., Demissie S., Musunuru K., Schadt E.E., Kaplan L., Bennett D., Li Y., Tanaka T., et al. Common variants at 30 loci contribute to polygenic dyslipidemia. Nat. Genet. 2009;41:56–65. doi: 10.1038/ng.291. doi:10.1038/ng.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saxena R., Voight B.F., Lyssenko V., Burtt N.P., de Bakker P.I., Chen H., Roix J.J., Kathiresan S., Hirschhorn J.N., Daly M.J., et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 8.Kooner J.S., Chambers J.C., Aguilar-Salinas C.A., Hinds D.A., Hyde C.L., Warnes G.R., Gomez Perez F.J., Frazer K.A., Elliott P., Scott J., et al. Genome-wide scan identifies variation in MLXIPL associated with plasma triglycerides. Nat. Genet. 2008;40:149–151. doi: 10.1038/ng.2007.61. doi:10.1038/ng.2007.61. [DOI] [PubMed] [Google Scholar]
- 9.Willer C.J., Sanna S., Jackson A.U., Scuteri A., Bonnycastle L.L., Clarke R., Heath S.C., Timpson N.J., Najjar S.S., Stringham H.M., et al. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat. Genet. 2008;40:161–169. doi: 10.1038/ng.76. doi:10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sagoo G.S., Tatt I., Salanti G., Butterworth A.S., Sarwar N., van Maarle M., Jukema J.W., Wiman B., Kastelein J.J.P., Bennet A.M., et al. Seven lipoprotein lipase gene polymorphisms, lipid fractions, and coronary disease: a HuGE association review and meta-analysis. Am. J. Epidemiol. 2008;168:1233–1246. doi: 10.1093/aje/kwn235. doi:10.1093/aje/kwn235. [DOI] [PubMed] [Google Scholar]
- 11.Zechner R. The tissue-specific expression of lipoprotein lipase: implications for energy and lipoprotein metabolism. Curr. Opin. Lipidol. 1997;8:77–88. doi: 10.1097/00041433-199704000-00005. doi:10.1097/00041433-199704000-00005. [DOI] [PubMed] [Google Scholar]
- 12.Stein Y., Stein O. Lipoprotein lipase and atherosclerosis. Atherosclerosis. 2003;170:1–9. doi: 10.1016/s0021-9150(03)00014-5. doi:10.1016/S0021-9150(03)00014-5. [DOI] [PubMed] [Google Scholar]
- 13.Merkel M., Eckel R.H., Goldberg I.J. Lipoprotein lipase: genetics, lipid uptake, and regulation. J. Lipid. Res. 2002;43:1997–2006. doi: 10.1194/jlr.r200015-jlr200. doi:10.1194/jlr.R200015-JLR200. [DOI] [PubMed] [Google Scholar]
- 14.Kathiresan S., Melander O., Guiducci C., Surti A., Burtt N.P., Rieder M.J., Cooper G.M., Roos C., Voight B.F., Havulinna A.S., et al. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat. Genet. 2008;40:189–197. doi: 10.1038/ng.75. doi:10.1038/ng.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith A.J., Humphries S.E. Characterization of DNA-binding proteins using multiplexed competitor EMSA. J. Mol. Biol. 2009;385:714–717. doi: 10.1016/j.jmb.2008.11.035. doi:10.1016/j.jmb.2008.11.035. [DOI] [PubMed] [Google Scholar]
- 16.Dickson S.P., Wang K., Krantz I., Hakonarson H., Goldstein D.B. Rare variants create synthetic genome-wide associations. PLoS Biol. 2010;8:12. doi: 10.1371/journal.pbio.1000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Enerback S., Ohlsson B.G., Samuelsson L., Bjursell G. Characterization of the human lipoprotein-lipase (LPL) promoter - evidence of 2 cis-regulatory regions, Lp-alpha and Lp-beta, of importance for the differentiation-linked induction of the LPL gene during adipogenesis. Mol. Cell. Biol. 1992;12:4622–4633. doi: 10.1128/mcb.12.10.4622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cirillo L.A., Lin F.R., Cuesta I., Friedman D., Jarnik M., Zaret K.S. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol. Cell. 2002;9:279–289. doi: 10.1016/s1097-2765(02)00459-8. doi:10.1016/S1097-2765(02)00459-8. [DOI] [PubMed] [Google Scholar]
- 19.Bernstein B.E., Kamal M., Lindblad-Toh K., Bekiranov S., Bailey D.K., Huebert D.J., McMahon S., Karlsson E.K., Kulbokas E.J., Gingeras T.R., et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. doi:10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 20.Gargiulo G., Levy S., Bucci G., Romanenghi M., Fornasari L., Beeson K.Y., Goldberg S.M., Cesaroni M., Ballarini M., Santoro F., et al. NA-Seq: A discovery tool for the analysis of chromatin structure and dynamics during differentiation. Dev. Cell. 2009;16:466–481. doi: 10.1016/j.devcel.2009.02.002. doi:10.1016/j.devcel.2009.02.002. [DOI] [PubMed] [Google Scholar]
- 21.Chen Q., Razzaghi H., Demirci F.Y., Kamboh M.I. Functional significance of lipoprotein lipase HindIII polymorphism associated with the risk of coronary artery disease. Atherosclerosis. 2008;200:102–108. doi: 10.1016/j.atherosclerosis.2007.12.011. doi:10.1016/j.atherosclerosis.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu J.Q., Barnett A., Neufeld E.J., Dudley J.P. Homeoproteins CDP and SATB1 interact: potential for tissue-specific regulation. Mol. Cell. Biol. 1999;19:4918–4926. doi: 10.1128/mcb.19.7.4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li S.D., Aufiero B., Schiltz R.L., Walsh M.J. Regulation of the homeodomain CCAAT displacement/cut protein function by histone acetyltransferases p300/CREB-binding protein (CBP)-associated factor and CBP. Proc. Natl Acad. Sci. USA. 2000;97:7166–7171. doi: 10.1073/pnas.130028697. doi:10.1073/pnas.130028697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keating B.J., Tischfield S., Murray S.S., Bhangale T., Price T.S., Glessner J.T., Galver L., Barrett J.C., Grant S.F., Farlow D.N., et al. Concept, design and implementation of a cardiovascular gene-centric 50K SNP array for large-scale genomic association studies. PLoS ONE. 2008;3:e3583. doi: 10.1371/journal.pone.0003583. doi:10.1371/journal.pone.0003583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Talmud P.J., Drenos F., Shah S., Shah T., Palmen J., Verzilli C., Gaunt T.R., Pallas J., Lovering R., Li K., et al. Gene-centric association signals for lipids and apolipoproteins identified via the HumanCVD BeadChip. Am. J. Hum. Genet. 2009;85:628–642. doi: 10.1016/j.ajhg.2009.10.014. doi:10.1016/j.ajhg.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deo R.C., Reich D., Tandon A., Akylbekova E., Patterson N., Waliszewska A., Kathiresan S., Sarpong D., Taylor H.A., Jr, Wilson J.G. Genetic differences between the determinants of lipid profile phenotypes in African and European Americans: the Jackson Heart Study. PLoS Genet. 2009;5:e1000342. doi: 10.1371/journal.pgen.1000342. doi:10.1371/journal.pgen.1000342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lanktree M.B., Anand S.S., Yusuf S., Hegele R.A. and the SHARE Investigators. Replication of genetic associations with plasma lipoprotein traits in a multiethnic sample. J. Lipid Res. 2009;50:1487–1496. doi: 10.1194/jlr.P900008-JLR200. doi:10.1194/jlr.P900008-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tang W.M.D.P., Apostol G.M.D.M.S., Schreiner P.J.P., Jacobs D.R.J.P., Boerwinkle E.P., Fornage M.P. Associations of lipoprotein lipase gene polymorphisms with longitudinal plasma lipid trends in young adults: the Coronary Artery Risk Development in Young Adults (CARDIA) Study. Circulation: Cardiovasc. Genet. 2010;3:179–186. doi: 10.1161/CIRCGENETICS.109.913426. doi:10.1161/CIRCGENETICS.109.913426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Altshuler D., Brooks L.D., Chakravarti A., Collins F.S., Daly M.J., Donnelly P. International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cooper J.A., Miller G.J., Bauer K.A., Morrissey J.H., Meade T.W., Howarth D.J., Barzegar S., Mitchell J.P., Rosenberg R.D. Comparison of novel hemostatic factors and conventional risk factors for prediction of coronary heart disease. Circulation. 2000;102:2816–2822. doi: 10.1161/01.cir.102.23.2816. [DOI] [PubMed] [Google Scholar]
- 31.Talmud P.J., Hawe E., Miller G.J., Humphries S.E. Nonfasting apolipoprotein B and triglyceride levels as a useful predictor of coronary heart disease risk in middle-aged UK men. Arterioscler. Thromb. Vasc. Biol. 2002;22:1918–1923. doi: 10.1161/01.atv.0000035521.22199.c7. doi:10.1161/01.ATV.0000035521.22199.C7. [DOI] [PubMed] [Google Scholar]
- 32.Drenos F., Talmud P.J., Casas J.P., Smeeth L., Palmen J., Humphries S.E., Hingorani A.D. Integrated associations of genotypes with multiple blood biomarkers linked to coronary heart disease risk. Hum. Mol. Genet. 2009;18:2305–2316. doi: 10.1093/hmg/ddp159. doi:10.1093/hmg/ddp159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stephens M., Smith N.J., Donnelly P. A new statistical method for haplotype reconstruction from population data. Am. J. Hum. Genet. 2001;68:978–989. doi: 10.1086/319501. doi:10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rubin D.B. Multiple imputation for nonresponse in surveys. New York: John Wiley & Sons, Inc.; 1987. [Google Scholar]
- 35.Royston P., Carlin J.B., White I.R. Multiple imputation of missing values: new features for mim. Stata J. 2009;9:252–264. [Google Scholar]
- 36.Clement M., Posada D., Crandall K.A. TCS: a computer program to estimate gene genealogies. Mol. Ecol. 2000;9:1657–1659. doi: 10.1046/j.1365-294x.2000.01020.x. doi:10.1046/j.1365-294x.2000.01020.x. [DOI] [PubMed] [Google Scholar]
- 37.Posada D., Maxwell T.J., Templeton A.R. TreeScan: a bioinformatic application to search for genotype/phenotype associations using haplotype trees. Bioinformatics. 2005;21:2130–2132. doi: 10.1093/bioinformatics/bti293. doi:10.1093/bioinformatics/bti293. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.