Abstract
Human pluripotent stem cells (hPSCs) hold significant promise for use in regenerative medicine, or as a model to understand human embryo development. However, the basic mechanisms required for proliferation and self-renewal of hPSCs have not been fully uncovered. Proliferation in all eukaryotes is dependent upon highly regulated expression of the histone H3 variant Centromere protein A (CENP-A). In the current study, we demonstrate that hPSCs have a unique messenger ribonucleic acid (mRNA) reserve of CENP-A not found in somatic fibroblasts. Using short hairpin RNA technology to reduce but not ablate CENP-A, we show that CENP-A-depleted hPSCs are still capable of maintaining a functional centromeric mark, whereas fibroblasts are not. However, upon induction of differentiation or DNA damage, hPSCs with depleted CENP-A arrest in G2/M and undergo apoptosis. Analysis of CENP-A dynamics following DNA damage in hPSCs reveals that 60 min after irradiation, CENP-A is found in multiple small nuclear foci that are mutually exclusive to γH2AX as well as CENP-C. Furthermore, following irradiation, hPSCs with depleted CENP-A mount a normal apoptotic response at 6 h; however at 24 h, apoptosis is significantly increased in CENP-A-depleted hPSCs relative to control. Taken together, our results indicate that hPSCs exhibit a unique mechanism for maintaining genomic integrity by possessing the flexibility to reduce the amount of CENP-A required to maintain a functional centromere under self-renewing conditions, and maintaining a reserve of CENP-A mRNA to rebuild the centromere following differentiation or DNA damage.
INTRODUCTION
Maintaining the genomic integrity of human pre-implantation embryos and human pluripotent stem cells (hPSCs) is of considerable importance to human reproduction and regenerative medicine. Aneuploidy in human pre-implantation embryos is a common occurrence with an estimated 30% of all human embryos failing to progress at implantation due to chromosome structural defects (1,2). Furthermore, hPSCs derived from the inner cell mass of human blastocysts, which are called human embryonic stem cells (hESCs), acquire aneuploidies after culture in suboptimal conditions, continuous culture and adaptation or passaging as single cells without Rock inhibitor or neurotrophins (3–8). However, not all aneuploidies are tolerated in hPSCs as derivation of hESC lines from embryos diagnosed as being monosomic after pre-implantation genetic screening are selected against during the derivation procedure (9).
Chromosome segregation defects occur due to abnormal centromeric chromatin, abnormal construction of the kinetochore or defects in the activity of the spindle assembly checkpoint during mitosis. Centromeric chromatin in metazoans is uniquely identified by the incorporation of the histone H3 variant centromere protein A (CENP-A) in alpha satellite DNA repeats which act to promote centromere identity and assembly of the kinetochore protein complex required for cell division (10–16). Centromeric chromatin is surrounded by peri-centromeric heterochromatin which is hypothesized to act as a barrier to prevent CENP-A spreading (reviewed in 17–20). Conversely, when the number of CENP-A containing nucleosomes is reduced, heterochromatin spreading is hypothesized to cause dissolution of the centromeric mark (21). A complete loss of CENP-A in mitotic cells including pluripotent cells of the pre-implantation mouse embryo is lethal (11). In the complete absence of CENP-A or when CENP-A has been depleted to below the threshold required to create an epigenetic centromeric mark, cells exhibit a transient mitotic delay followed by apoptosis (22–25). In contrast, overexpression of CENP-A causes random incorporation of CENP-A into chromatin, and the creation of functional ectopic kinetochores (10). Overexpression of CENP-A has been described in human cancer cells, where it is hypothesized to cause aneuploidy (26). Taken together, the levels of CENP-A in a given cell are highly regulated to ensure appropriate functional activity and mitotic fidelity.
Microarray analysis comparing human oocytes, hESCs and somatic cells have revealed that CENP-A messenger ribonucleic acid (mRNA) is highly expressed in both oocytes and hESCs (27–31). High expression levels of CENP-A in oocytes most likely act as a maternal reserve to support the small number of mitotic pluripotent cell divisions (cleavages) that occur prior to embryonic genome activation (11). However, the purpose of the relatively high CENP-A mRNA levels in hESCs is unclear. Furthermore, it is also not known whether high CENP-A mRNA levels are found in hPSCs generated by induced reprogramming, which creates human-induced pluripotent stem (hIPS) cells, or whether hPSCs have an increase in CENP-A protein load that correlates with the increased levels of CENP-A mRNA.
CENP-A localization in hPSCs is known to be unique relative to somatic cells. For example, in hPSCs, CENP-A centromeric foci occupy a central position in the nucleus (32–36). However, upon differentiation and accumulation of heterochromatin, CENP-A is redistributed to the heterochromatic nuclear periphery and perinucleolar regions (33). The relationship between CENP-A levels and accumulation of heterochromatin upon hPSC differentiation is unknown (35,37). Furthermore, it is also unknown whether dynamic relocalization of CENP-A only occurs during hPSC differentiation, or whether it can be induced under alternative circumstances such as DNA damage where CENP-A has been shown to accumulate at foci of DNA damage in somatic cells (38,39). Given these intriguing unanswered questions, in the current study we sought to evaluate the unique biology of CENP-A in hPSC self-renewal, differentiation and DNA damage.
RESULTS
A high transcriptional load of CENP-A is a general property of hPSCs
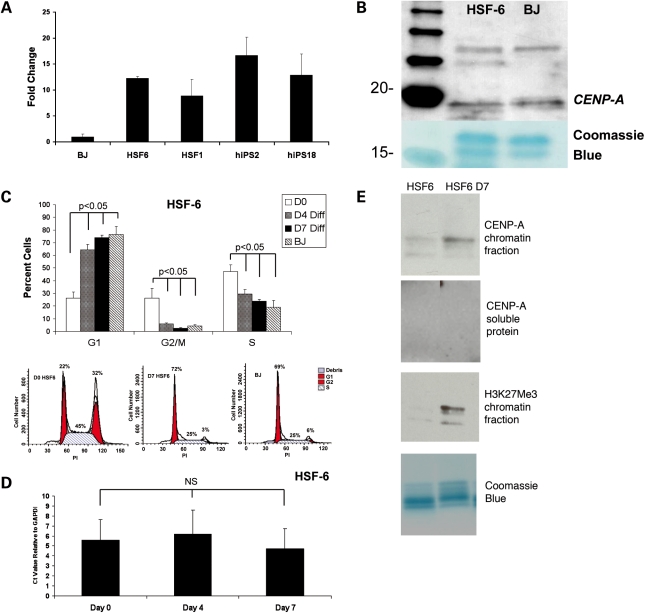
In order to determine whether elevated levels of CENP-A mRNA are a property of hPSCs derived from the inner cell mass of blastocysts (hESCs), relative to hPSCs derived by induced reprogramming from skin fibroblasts (hIPS cells), we performed real-time PCR for CENP-A mRNA in triplicate for two hESC lines (HSF-6 and HSF-1) and two hIPS cell lines (hIPS2 and hIPS18) (40), and compared this with a primary skin fibroblast cell line called BJ (Fig. 1A). We found that hPSCs derived from the inner cell mass of blastocysts or by induced reprogramming have on average 10–15-fold higher levels of CENP-A mRNA relative to somatic BJ cells. Therefore, elevated levels of CENP-A mRNA are a general property of pluripotent stem cells regardless of origin, and are not a historical remnant of ESCs derived from the inner cell mass. Next, we used western blot of purified chromatin to evaluate whether elevated levels of CENP-A mRNA correlate with elevated levels of CENP-A protein in undifferentiated hPSCs (shown is HSF-6) (Fig. 1B). Our results showed that the expression of CENP-A protein is equivalent between BJ cells and undifferentiated hPSCs.
Figure 1.
hPSCs have a reserve of CENP-A mRNA but not protein when compared with fibroblasts. (A) Quantitative real-time PCR for CENP-A normalized to GAPDH (n = 3 replicates). (B) Western blot of purified chromatin. (C) Cell cycle analysis in undifferentiated and differentiated HSF-6 and BJ cells (n = 3 replicates for each). A representative flow plot of each is also shown. (D) Quantitative real-time PCR for CENP-A mRNA comparing undifferentiated and differentiated cells (n = 3 replicates). (E) Chromatin and soluble fraction of undifferentiated and 7 days differentiated HSF-6 hPSCs (Diff, differentiation; D0, undifferentiated; D4, day 4 of differentiation; D7, day 7 of differentiation).
Given the unique cell cycle properties of undifferentiated hPSCs in which a significant fraction of cells occupy S phase and G2/M, we next tested whether CENP-A mRNA levels change upon differentiation for 7 days in the presence of retinoic acid where the cell cycle normalizes to that of a somatic cell. Our results show that by day 7 of differentiation, the proportion of cells occupying G1 is significantly increased, and the proportion of cells in G2/M and S is significantly reduced to become equivalent to cycling BJ fibroblast cells (Fig. 1C, Supplementary Material, Fig. S1). Despite this change in cell cycle dynamics, the levels of CENP-A mRNA remain equivalent to the levels of undifferentiated hPSCs (Fig. 1D). In order to determine whether CENP-A protein levels change with differentiation, we evaluated CENP-A in the chromatin and soluble fractions of hPSCs (shown is HSF-6), and compared this to cells differentiated for 7 days. Our data show that CENP-A protein is exclusive to the chromatin fraction of hPSCs, and that the levels of CENP-A in chromatin increase with hPSC differentiation. Furthermore, we found that increased levels of CENP-A protein correlate with an increase in the amount of histone H3 lysine 27 (H3K27me3), which is known to occur as cells transition from the self-renewing pluripotent state to differentiation (Fig. 1E).
Significant depletion of CENP-A in pluripotent cells under self-renewing conditions is not lethal
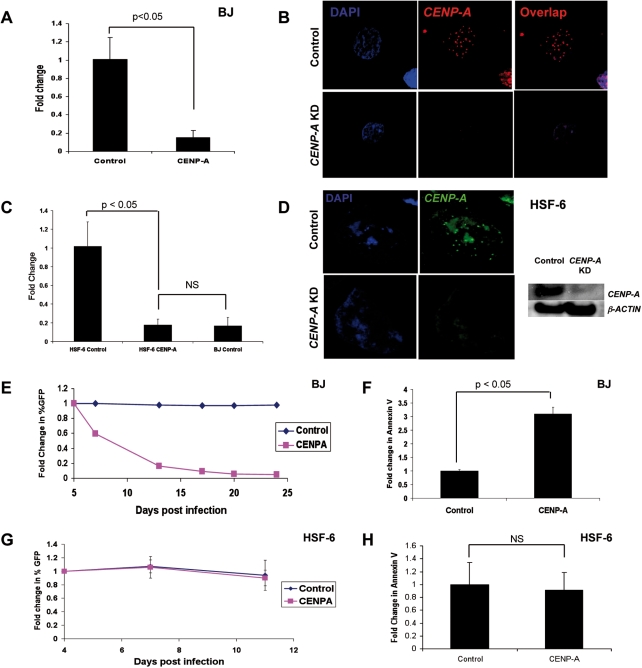
Given the high mRNA reserve of CENP-A in undifferentiated hPSCs relative to fibroblasts, we were next interested in determining whether modulating the levels of CENP-A in hPSCs had an effect on undifferentiated hPSC self-renewal. In this experiment, our goal was to reduce but not ablate the levels of CENP-A in undifferentiated hPSCs. To achieve this, we used short hairpin RNA (shRNA) technology and assayed pluripotent stem cells in parallel with somatic BJ fibroblasts. We initially tested three independent shRNAs in fibroblasts and found one which consistently resulted in >90% knockdown (data not shown). This shRNA was used for the remainder of the study. Transduction of control or CENP-A shRNA lentiviral vectors into BJ fibroblasts resulted in 90% knockdown of CENP-A mRNA levels relative to control following 5 days selection in hygromycin (Fig. 2A). Using immunofluorescence with an antibody against CENP-A, we determined that this level of knockdown significantly reduced the CENP-A centromeric foci in knockdown cells relative to cells transduced with the control vector (Fig. 2B) suggesting that the centromeric mark is compromised in the knockdown cells. Transduction of HSF-6 hPSCs with control or CENP-A shRNA lentivirus resulted in an 80% reduction in CENP-A mRNA levels compared with control cells after 5 days of selection (Fig. 2C). This knockdown reduced the CENP-A mRNA load to an equivalent level found in fibroblasts under unmanipulated conditions. However, using immunofluorescence and western blot comparing control and CENP-A shRNA knockdown cultures CENP-A shRNAs also caused a significant reduction in CENP-A protein, with loss of CENP-A foci by immunofluorescence and a significant reduction in protein levels by western blot (Fig. 2D).
Figure 2.
hPSCs with depleted CENP-A undergo normal self-renewal. (A) Quantitative PCR for CENP-A mRNA in BJ cells (n = 5 replicates). (B) Immunofluorescence of BJ cells for CENP-A knockdown and control conditions. (C) Quantitative PCR for CENP-A in control and knockdown hPSCs (HSF-6) compared with BJ (n = 6 replicates). (D) Immunofluorescence and western blot comparing control and knockdown hPSCs (HSF-6). (E) Competition assay in BJ cells knockdown compared with control transduced. A representative graph from n = 3 replicates is shown. (F) Annexin V staining of BJ cells (n = 3 replicates). (G) Competition assay in CENP-A knockdown cells compared with control hPSCs (HSF-6). A representative graph from n = 3 replicates is shown. (H) Annexin V staining of hPSCs (HSF-6) (n = 6 replicates).
Next, we evaluated the functional consequence of CENP-A knockdown in BJ cells and hPSCs (Fig. 2E–H, Supplementary Material, Fig. S2). Knockdown of CENP-A in BJ cells caused rapid out-competition of the GFP positive knockdown cells by wild-type cells, with a 40% reduction in the percentage of GFP positive CENP-A knockdown cells within 2 days of the assay, and a 90% loss in GFP positive CENP-A knockdown cells after 15 days (Fig. 2E). Control GFP positive cells and wild-type cells maintained a constant ratio over this period demonstrating that the vector was not silenced. Phenotypically, knockdown of CENP-A in BJ fibroblasts caused a significant increase in apoptosis relative to cells transduced with the control vector (Fig. 2F) and this was morphologically associated with a significant reduction in cell number after 5 days of selection when starting with equivalent seeding densities (Supplementary Material, Fig. S2b). In contrast, knockdown of CENP-A in undifferentiated hPSCs resulted in no change in morphology by phase contrast (Supplementary Material, Fig. S2a), or proliferation rate relative to cells transduced with the control vector as measured using the competition assay (Fig. 2G). Furthermore, knockdown of CENP-A in undifferentiated hPSCs did not cause a significant change in apoptosis relative to control (Fig. 2H, Supplementary Material, Fig. S3a). Analysis of basic self-renewal markers including stage-specific embryonic antigen 4 (SSEA4) by flow cytometry, and OCT4, NANOG and SOX2 mRNA by real-time PCR revealed no difference in levels when compared to cells transduction with the control vector (Supplementary Material, Figs S2c–d and S3b). Taken together, these results suggest two alternative hypotheses; (i) knocking down CENP-A in undifferentiated hPSCs has no effect on the integrity of the centromere resulting in normal mitosis, or (ii) hPSCs with significantly reduced levels of CENP-A are proliferating, but accumulating considerable aneuploidies due to relaxed cell cycle checkpoints.
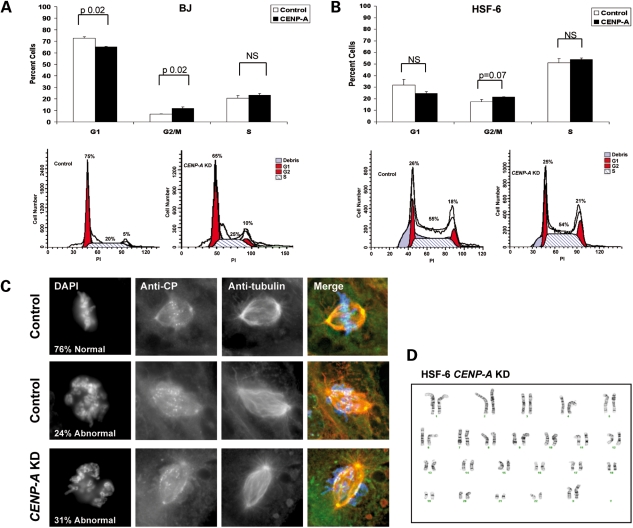
To address this, we evaluated the cell cycle of BJ fibroblasts and undifferentiated hPSCs transduced with control and CENP-A shRNA vectors. Our results show that knockdown of CENP-A in BJ cells caused a delay in G2/M and a loss of cells from G1 as expected for cells arresting in mitosis due to segregation defects (Fig. 3A). However, knockdown of CENP-A in hPSCs resulted in a trend towards a G2/M delay, but no statistically significant effect on cell cycle phasing (Fig. 3B). To determine whether knockdown of CENP-A in hPSCs caused chromosomal alignment defects, we evaluated 50–55 mitotic cells from control and CENP-A knockdown cultures 5 days after transduction and selection (Fig. 3C). To detect the centromere in these studies, we used CREST antisera commonly used to identify active centromeres. Unlike staining with CENP-A, CREST foci could be identified in the CENP-A knockdown cells (Fig. 3C), confirming that reducing the levels of CENP-A by 80% is not sufficient to induce complete loss of the centromere under self-renewing conditions. Analysis of the metaphase plate showed 38/50 or 76% of cells transduced with the control vector exhibited normal chromosomal alignment in metaphase (Fig. 3C). However, under control conditions, we also identified 12 metaphases (24%) with morphologically abnormal alignment. Similarly, in the CENP-A knockdown cells, 69% of metaphases were scored as normal, whereas 17/55 (31%) of metaphases exhibited abnormal alignment (Fig. 3C). In order to determine whether this small increase in chromosome misalignment translates to an increase in aneuploidy, we performed karyotype analysis of the CENP-A knockdown cells 10 days after transduction (Fig. 3D). Our results showed that knockdown of CENP-A under self-renewing conditions does not result in aneuploidy within the first passage.
Figure 3.
Significantly reducing CENP-A levels in hPSCs does not cause segregation defects. (A) Cell cycle profile of CENP-A knockdown BJ cells relative to control after 5 days of selection with hygromycin (n = 3 replicates). A representative flow plot of each is also shown. (B) Cell cycle profiles of control and CENP-A knockdown hPSCs (HSF-6) (n = 3 replicates). A representative flow plot of each is shown. (C) Immunofluorescence (IF) of control and CENP-A knockdown hPSCs with anti-tubulin, anti-CREST and DAPI. (D) Karyotype of CENP-A knockdown HSF-6 hPSCs 10 days after transduction with lentivirus.
Taken together, our results indicate that the CENP-A mRNA reserve found in undifferentiated PSCs is responsible for sustaining the CENP-A protein load, as loss of the mRNA reserve results in a significant reduction in CENP-A protein. However, unlike fibroblasts, depleting CENP-A protein levels has no impact on proliferation and self-renewal. Therefore, our data point to the idea that under self-renewing conditions, the amount of CENP-A protein necessary to generate a functional epigenetic centromeric mark in hPSCs is considerably less than the amount of CENP-A protein required to create a functional centromere in fibroblasts.
CENP-A depleted pluripotent stem cells cannot support the transition from pluripotency to lineage commitment
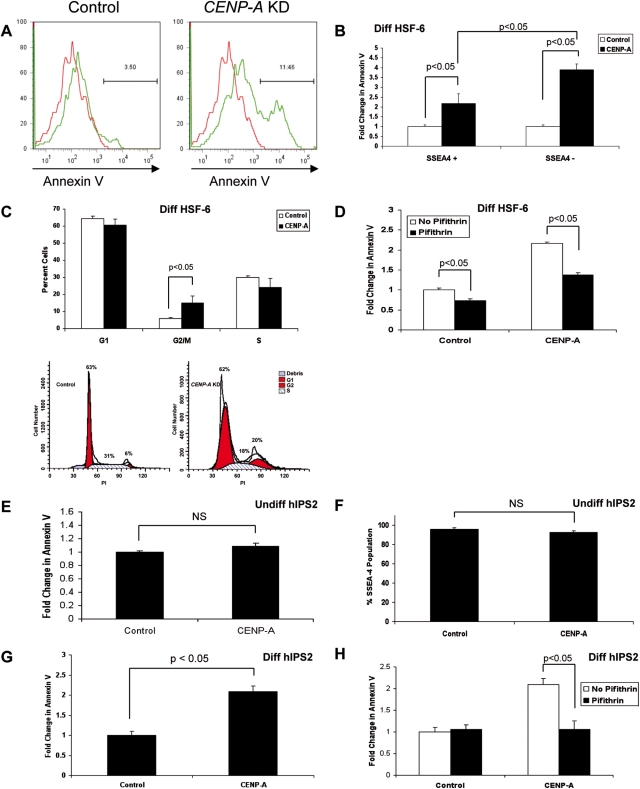
Next, we investigated whether reducing CENP-A has an effect on centromeric maintenance when hPSCs are induced to differentiate and accumulate increased amounts of H3K27me3 (Fig. 1E). To achieve this, we transduced hPSCs with either CENP-A shRNA lentivirus or control lentivirus under self-renewing conditions, allowed the cells to recover for 72 h in selection media and then induced differentiation in the presence of retinoic acid for 4 days. Our results show that differentiation of cells with reduced levels of CENP-A caused in a significant increase in the proportion of cells undergoing apoptosis relative to control (Fig. 4A). This was observed in HSF-6 (Fig. 4A) as well as HSF-1 hPSC lines (Supplementary Material, Fig. S3c). Furthermore, apoptosis on day 4 of differentiation was accompanied by accumulation of cells in the G2/M phase of the cell cycle (Fig. 4C), indicating that the apoptosis is due to cells with segregation defects. In order to determine whether apoptosis could be attributed to all differentiated cells or whether there was a quantitative difference between cells with more stem-like characteristics (retaining SSEA4 positive surface staining), we analyzed Annexin V staining in the SSEA4 positive and SSEA4 negative fractions at day 4 of differentiation (Fig. 4B). Our results showed that apoptosis is enriched in the SSEA4 negative (differentiated) fraction of CENP-A knockdown cells, confirming that apoptosis was more tightly coupled with the process of differentiation in cells with reduced levels of CENP-A.
Figure 4.
Differentiation of CENP-A-depleted hESCs and hIPS cells causes lethal mitotic defects. (A) Annexin V staining of control and CENP-A shRNA-transduced HSF-6 cells after 4 days of differentiation with 10 µM RA (n = 3 replicates). (B) Annexin V staining in SSEA4+ versus SSEA4− populations in CENP-A shRNA-transduced HSF-6 after 4 days of differentiation (n = 3 replicates). (C) Cell cycle profiles of hPSCs (HSF-6) after 4 days differentiation (n = 3 replicates). Shown is one representative flow plot from each sample. (D) Annexin V staining of HSF-6 after 4 days of differentiation before and after treatment with Pifithrin (n = 3 replicates). (E) Annexin V staining of control and CENP-A shRNA-transduced HIPS2 cells under self-renewing conditions (n = 3 replicates). (F) SSEA4 staining of control and CENP-A knockdown hIPS2 cells under self-renewing conditions (n = 3 replicates). (G) Annexin V staining of control and CENP-A knockdown hIPS2 cells after 4 days of differentiation (n = 3 replicates). (H) Apoptosis following 4 days of differentiation in control and CENP-A knockdown hIPS2 cells before and after treatment with pifithrin (n = 3 replicates).
One of the major proteins involved in coupling cell cycle checkpoints to induction of apoptosis is p53. In order to determine whether the increased apoptosis in CENP-A knock down cells is p53 dependent, we added the p53 inhibitor pifithrin to transduced hESCs 12 h before the initiation of differentiation, and measured apoptosis at day 4 of differentiation (Fig. 4D). Our results showed that the majority of apoptosis was p53 dependent in both HSF-6 (Fig. 4D) and HSF-1 hPSCs derived from the inner cell mass of blastocysts (Supplementary Material, Fig. S3d). We also showed that pifithrin induced a small yet significant decrease in the endogenous levels of apoptosis in differentiated cells transduced with the control vector.
Given that the reduced levels of CENP-A caused a significant increase in apoptosis as hESCs transition to lineage commitment, we were next interested in evaluating whether hIPS cells exhibit the same phenotype. We first transduced hIPS2 cells with either control or CENP-A shRNA lentivirus, and evaluated apoptosis and SSEA4 expression under self-renewing conditions. Our results indicated that similar to hESCs, hIPS cells with depleted CENP-A exhibit no change in SSEA4 or apoptosis relative to control cells cultured under self-renewing conditions (Fig. 4E and F). Next, we induced differentiation, and similar to hESCs, our results showed that hIPS cells with reduced levels of CENP-A exhibit increased apoptosis relative to control (Fig. 4G), and that this apoptosis is p53 dependent (Fig. 4H). Taken together, our results indicate that even though reduced levels of CENP-A in undifferentiated hPSCs is sufficient to support a basic centromeric mark, the switch to lineage commitment increases the threshold of CENP-A required to maintain the centromeric mark and cells with depleted CENP-A undergo G2/M arrest and apoptosis.
CENP-A-depleted ESCs undergo significant apoptosis when challenged with DNA damage
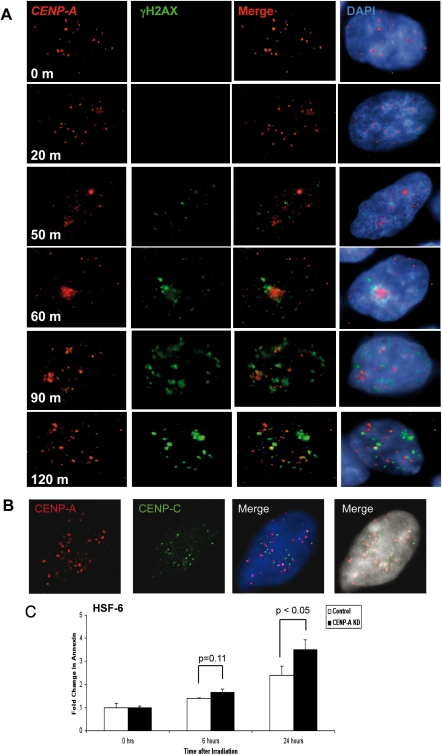
A role for CENP-A was recently identified in human somatic cells following DNA damage (38). Given that significantly reducing the levels of CENP-A in undifferentiated hPSCs still supports a functional centromere, we were next interested in evaluating whether the DNA damage response is affected in hPSCs with significantly reduced levels of CENP-A mRNA and protein. To achieve this, we first evaluated the kinetics of CENP-A localization in hPSCs (HSF-6) over the course of 2h following DNA damage with 5 Gy of irradiation (Fig. 5A). Our results show that within 50 min of DNA damage, CENP-A protein is identified as numerous small nuclear foci rather than large centromeric dots. The generation of the numerous small CENP-A foci was associated with an increase in γH2AX signal indicating initiation of DNA repair, however CENP-A and γH2AX do not co-localize. At 50–90 min, a major proportion of the hESC nuclei reorganize, resulting in clustering of the majority of CENP-A foci. Clustered and non-clustered CENP-A foci continue to exhibit minimal to no overlap with γH2AX. By 120 min, CENP-A foci had returned to the central nuclear position in most cells, however the intense staining of γH2AX indicates that repair is still ongoing. Furthermore, similar to the proceeding time points, γH2AX staining remained mostly mutually exclusive from the foci of CENP-A. Given the dynamic remodeling of CENP-A foci following irradiation, we next evaluated whether during the clustering phase 60 min after DNA damage, the CENP-A foci localized with a second constitutive centromeric protein (CENP-C) (Fig. 5B). CENP-C was chosen because centromere localization and function is mutually dependent upon both CENP-C and CENP-A (reviewed in 17,18). Our results clearly show that at 60 min following irradiation damage, CENP-C foci also rearrange and cluster in the nucleus, however CENP-A and CENP-C foci rarely co-localize.
Figure 5.
The dynamics and role of CENP-A following DNA damage. (A) IF of unmodified HSF-6 hPSCs with CENP-A and γH2AX localization after DNA damage by 5 Gy IR. (B) IF of unmodified HSF-6 hPSCs with CENP-A (red) and CENP-C (green) 60 min following 5 Gy IR. (C) Annexin V staining of control- and CENP-A shRNA-transduced HSF-6 with and without DNA damage with 5 Gy IR (n = 6 replicates).
In order to evaluate the effects of reduced levels of CENP-A mRNA and protein during DNA damage, we knocked down CENP-A in undifferentiated hPSCs (HSF-6) and after 5 days of selection we exposed the cells to 5 Gy of irradiation. Our functional read-out was to measure apoptosis at 6 and 24 h after irradiation damage. Our results showed that in cells transduced with the control vector, a 2–3-fold increase in apoptosis was identified on 24 h of irradiation damage. In contrast, hPSCs transduced with CENP-A shRNA vectors exhibited a statistically significant increase in apoptosis above the control cells at 24 h (Fig. 5C).
DISCUSSION
In the current study, we show that the mRNA levels of CENP-A in human PSCs and day 7 differentiated progeny are significantly higher than fully differentiated cycling somatic fibroblast cells. This elevated level of CENP-A mRNA is found regardless of whether hPSCs were derived by culture-induced conversion of the inner cell mass, or by induced reprogramming of human fibroblasts. However, despite this higher mRNA load, protein analysis indicates that the amount of CENP-A in hPSC chromatin is identical to fibroblasts. A second novel finding is that depletion of CENP-A mRNA and protein in hPSCs under self-renewing conditions causes no significant phenotype, whereas inducing differentiation of the CENP-A depleted cells causes accumulation of cells in G2/M, and a significant increase in p53-dependent apoptosis. Together these results suggest that the amount of CENP-A protein required to define the centromere may be cell type, and context specific in mammalian cells, with undifferentiated hPSCs requiring less CENP-A protein to faithfully retain the centromeric epigenetic mark relative to fibroblasts (Fig. 6). Finally, given that the threshold for significantly depleting CENP-A protein in hPSCs was lower than fibroblasts, we induced DNA damage in CENP-A-depleted self-renewing hPSCs, and determined that CENP-A-depleted hPSCs undergo significantly more apoptosis 24 h after irradiation, which is 22 h after the dynamic remodeling of CENP-A foci that occur in response to DNA damage.
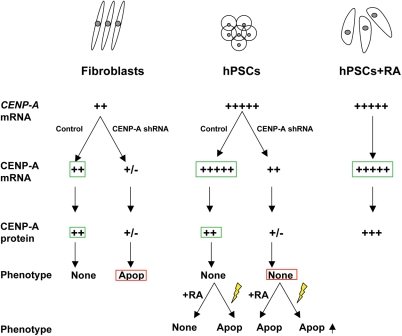
Figure 6.
Summary of differences between CENP-A expression and functional dynamics in fibroblasts and hPSCs. Our data demonstrate that the relative levels of CENP-A mRNA are significantly higher in undifferentiated hPSCs and hPSCs treated with retinoic acid (RA) relative to fibroblasts (green boxes). However, this higher load of CENP-A mRNA in control or unmanipulated hPSCs does not translate into significantly higher levels of CENP-A protein in the undifferentiated state (green boxes). Depletion of CENP-A mRNA and protein in fibroblasts using shRNA causes apoptosis (Apop) and G2/M delay as previously reported (red box). In contrast, depletion of CENP-A mRNA and protein in hPSCS using shRNAs had no effect on the maintenance of a functional epigenetic mark (red box). Treatment of CENP-A-depleted hESCs with either retinoic acid (RA) to induce differentiation or irradiation (yellow lightening rod) to induce DNA damage caused significant apoptosis above control, suggesting that in the CENP-A-depleted state, hPSCs cannot effectively mount a differentiation response, or overcome DNA damage and continue to proliferate (+/−, detection at the lower limit of resolution for the assay; +, low expression; +++++, high expression).
Previous reports using microarray analysis determined that CENP-A mRNA levels were higher in oocytes and hESCs relative to somatic cells (27–31). Our study confirms these findings; however, we further show that despite the elevated CENP-A mRNA in undifferentiated hPSCs, protein levels of CENP-A are equivalent to fibroblasts. This uncoupling of relative RNA to protein levels could be explained by the unique RNA translational controls recently identified in murine PSCs (41,42). For example, in murine ESCs, ‘parsimonious translation’ is hypothesized to define the pluripotent state, with undifferentiated ESCs containing 78% lower ribosome loading of RNA transcripts relative to their differentiated progeny, which establishes an RNA pool of specific transcripts that do not undergo productive protein synthesis (41). Translational regulation is also a critical feature of oocyte growth and the transition to an embryonic developmental program after fertilization. Translational regulation in the oocyte is a complex process involving multi-component RNA-binding complexes, compartmentalization of maternal RNAs and polyadenylation-induced translation (reviewed in 43,44). Whether the elevated levels of CENP-A mRNA are a product of poor polyribosome activity and content in hPSCs and/or the presence of a hPSC-specific CENP-A RNA-binding protein that regulate translation remain to be determined.
The depletion experiments in the current study suggest that under circumstances where CENP-A mRNA and protein levels are reduced, the threshold of CENP-A required to maintain a functional centromeric mark must be lower in hPSCs relative to fibroblasts because proliferation and self-renewal were unaffected. Depletion of CENP-A was achieved using shRNAs which are known to mostly act through RNA degradation (45). This would suggest that the high mRNA reserves of CENP-A in hPSCs are required to sustain wild-type levels of CENP-A protein. The paradox in this analysis is that reducing CENP-A mRNA and protein levels in undifferentiated hPSCs using shRNAs did not affect proliferation or self-renewal, whereas reducing CENP-A by the same technique in fibroblasts was lethal. One possible explanation is the unique chromatin configuration of PSCs compared with fibroblasts. For example, hPSCs have what is termed an ‘open’ chromatin configuration largely devoid of heterochromatin (37,46). In support of this, we show that the heterochromatic mark H3K27me3 is low in the undifferentiated hPSCs used in the current study, and that this epigenetic mark is significantly increased upon hPSC differentiation. We also show that as H3K27me3 levels increase during differentiation so do the levels of CENP-A in chromatin. The purpose of an open chromatin state is hypothesized to create the plasticity for mounting a rapid response to differentiation signals (reviewed in 47). Using human artificial chromosomes, it has been shown that heterochromatin can act as one of the major determinants of minimal centromeric length (48). Therefore, if reduced heterochromatin is also found at the centromeres of undifferentiated hPSCs, this unique chromatin architecture may create the opportunity for sustaining a functional centromere under circumstances where CENP-A protein levels become significantly reduced. This hypothesis would only be plausible if heterochromatin were not necessary to sustain a functional centromere, and indeed this was recently shown using neocentromeres as a model in lymphoblast and fibroblast BBB lines (49).
Our data indicate that once CENP-A-depleted hPSCs exit the self-renewing state, lineage-committed CENP-A-depleted cells undergo P53-dependent apoptosis. In support of this, a recent study using human primary fibroblasts revealed that entry into senescence in CENP-A-depleted fibroblasts is also P53 dependent (50). Therefore, our data support the hypothesis proposed by Maehara et al. (50) that p53 is a major surveillance factor for centromeric defects, and our study extends these original findings to indicate that reduced levels of CENP-A during early lineage commitment in the embryo would also act to prevent aneupoidy as a consequence of compromised centromere function.
A non-centromeric role for CENP-A in DNA damage repair has previously been proposed using Xenopus sperm as well as mouse and human somatic cells (38,39). In order to evaluate the dynamics of CENP-A in hPSCs following DNA damage, we used irradiation to induce double-strand breaks, and in agreement with previous studies, we identified an increase in the number of CENP-A foci shortly after inducing damage (38,39). However, our data stand in contrast to previous reports, as we did not observe an increase in foci size, or direct correlation with γH2AX staining. These differences may be explained by species differences, or an inherent difference between diploid hPSCs and haploid sperm. Given that in our study, CENP-A foci did not correlate with γH2AX after DNA damage, this would indicate that CENP-A is not localizing stably to induced break points in hPSCs (38). Furthermore, at 60 min after DNA damage, CENP-A foci also do not co-localize with CENP-C. This would indicate that the centromere is no longer functional during this period and therefore, before the cells can resume mitosis after DNA damage and repair, functional centromeres will need to be rebuilt. Inducing DNA damage of CENP-A-depleted hPSCs revealed that 24 h after DNA damage, CENP-A depleted cells are undergoing significantly higher levels of apoptosis relative to control. Combined with the result that CENP-C and CENP-A foci do not correlate following DNA damage, our interpretation is that increased apoptosis at 24 h is not due to defects in DNA repair associated with γH2AX, because CENP-A and γH2AX show no major overlap. Instead, we favor the hypothesis that increased apoptosis in CENP-A-depleted cells following DNA damage is due to reduced availability of new CENP-A protein to rebuild the CENP-A/CENP-C uncoupled centromeres following DNA damage.
In conclusion, cultured hPSCs are unique cell types that closely resemble the inner cell mass cells in blastocysts that normally persist for only a few days before changing fate to create every cell type in the body including the germline. In the first week post-fertilization, the human embryo is dividing on a rigid time scale governed by female reproductive physiology that creates a small window for successful implantation. If cell division occurs too slowly, then the embryo is lost because the window for receptive implantation is over. Our results suggest that hPSCs create both a reserve of CENP-A mRNA as well as the flexibility to reduce the amount of CENP-A necessary to create a functional centromere. This mechanism functions in favor of cell division, while minimizing the potential for post-fertilization aneuploidies due to defective centromeres. The fact that hIPS cells maintain this same phenomenon indicates that establishing a dynamic range for CENP-A is not merely a relic from the pre-implantation embryo, but an integral aspect of hPSC self-renewal. Finally, our data indicate that hPSCs provide a new and unique tool with which to evaluate the biology of CENP-A (Fig. 6) and the pathways by which centromeric nucleosomes in primary human cells of the same genotype are dynamically remodeled under conditions of self-renewal, differentiation and DNA damage.
MATERIALS AND METHODS
Cell culture and treatments
Information regarding the hESCs, HSF-6 (UC06, 46XX) and HSF-1 (46XY) can be obtained at http://stemcells.nih.gov/stemcells. Undifferentiated hESC colonies were maintained as previously described (51). For all experiments, hESCs were used between passages 40 and 60. The hIPS cell line 2 (hIPS2, 46XY) was cultured as previously described (40). All hESC and hIPS experiments were conducted with prior approval from the UCLA Embryonic Stem Cell Research Oversight Committee.
BJ fibroblast somatic cells were cultured in 90% minimum essential medium (MEM) with Earle's salt and l-glutamine, 10% fetal bovine serum, 1% non-essential amino acids and 1 mm sodium pyruvate. Cells were passaged using 0.25% trypsin every 7 days.
Differentiation from hESCs and hIPS
Differentiation was performed on matrigel (BD Biosciences) treated six-well plates. Differentiation media included DMEM/F12 (Gibco BRL) supplemented with Lot-tested 20% FBS (Hyclone), 0.1 mm non-essential amino acids (Gibco BRL), 0.1 mm β-mercaptoethanol (Gibco BRL), 1 mm l-glutamine (Gibco BRL) and 10 µm retinoic Acid (RA, Sigma, St Louis, MO, USA, http://www.sigmaaldrich.com). Media were changed every 2 days during differentiation with the addition of fresh 10 µM RA at each medium change.
Irradiation
Human ES cells were irradiated 2 days after passaging with 5 Gy of γ-radiation using a Mark-1 cesium137 irradiator. Immediately after irradiation, cells were returned to the incubator for recovery until the appropriate time point.
Vectors
Sense and antisense oligonucleotides of the CENP-A shRNA duplex were as follows: sense ttc gga tct cct tta gcc a, antisense ttc gga tct cct tta gcc a. The oligonucleotides were synthesized and HPLC purified by Invitrogen. After annealing, the duplexes were cloned into the H1P lentiviral vector as already described (52).
Lentivirus production and ES cell transduction
Lentiviral vectors were packaged, concentrated by ultracentrifugation at 50 000g for 1.5 h, resuspended in hESC media and stored at −80°C as previously described (53). Titering was performed on 293 FT cells. For transduction, ES cells were treated with 1 mg/ml collagenase, resuspended in conditioned media and 2.5 × 106 IU of virus was added to 1 × 105 cells and rocked in the incubator for 2.5 h as already described (54). Polybrene (Sigma) was added at a concentration of 10 µg/ml. Subsequently, cells were pelleted, virus washed off and plated on matrigel coated plates in conditioned media. Selection with hygromycin at 200 µg/ml was started at 24–48 h post-transduction. Majority of the experiments were performed after having cells under selection for 5 days. Knockdown efficiency was confirmed using quantitative PCR, western blots and immunohistochemistry.
Competition assay
shRNA-transduced BJ cells were mixed with non-transduced cells at a ratio of 4:1 and plated into gelatinized 12-well plates. Every 48 h (one passage), the cells were trypsinized and replated. At each passage, the proportion of GFP+/GFP− cells was measured by flow cytometry on a LSRII (BD Biosciences). Analyses were carried out for six consecutive passages. Similar analysis was performed in shRNA-transduced hESCs, but cells were plated on matrigel plates in conditioned media and analyzed at each passage (approximately every 5–6 days) for two passages.
RNA extraction and polymerase chain reaction
Total RNA was extracted using the RNeasy kit (Qiagen, Hilden, Germany, http://www.qiagen.com) according to manufacturer's instructions. Total RNA was quantified, and 1 µg was used for cDNA synthesis using random primers (Invitrogen, Carlsbad, CA, USA) under standard conditions. Reverse transcriptase–polymerase chain reaction (RT–PCR) amplifications were conducted for 35 cycles of 95°C, 30 s; 60°C, 30 s and 72°C, 30 s. Primer sequences are as follows: glyceraldehydes-3-phosphate dehydrogenase (GAPDH) F 5′-acc aca gtc cat gcc atc ac-3′, GAPDH R 5′-tcc acc acc ctg ttg ctg ta-3′, centromere protein A (CENP-A) F 5′-ata tgt gtt aaa ttc act cgt ggt gt-3′, CENP-A R 5′-cag gaa aga ctg aca gaa aca ctg-3′, OCT-4 F 5′-aca tca aag ctc tgc aga aag aac t-3′, OCT-4 R 5′-ctg aat acc ttc cca aat aga acc c-3′, NANOG F 5′-cag ctg tgt gta ctc aat gat aga ttt-3′, NANOG R 5′-aca cca ttg cta ttc ttc ggc cag ttg-3′, SOX2 F 5′-agt ctc caa gcg acg aaa aa-3′, SOX2 R 5′-gca aga agc ctc tcc ttg aa-3′.
Quantitative real-time PCR was performed in duplicate for each sample from at least three separate experiments using Sybr Green master mix (Roche) according to manufacturer's instructions. Sybr green PCR was initiated at 95°C for 3 min followed by cycles of 95°C, 30 s; 60°C, 30 s and 72°C, 30 s. All primers were used as stated above. All results were normalized against GAPDH.
Flow cytometry
Cells were dissociated with 0.05% trypsin and 0.5 mm EDTA (Gibco BRL) at 37°C for 5 min and collected by centrifugation at 156.5g for 5 min. Cells were incubated in 1% BSA in PBS containing primary antibodies on ice for 20 min. Cells were then washed and incubated in FITC- or Cy5-conjugated secondary antibodies on ice for another 20 min. For the Annexin assays, cells were washed and incubated with 5 µl PE-conjugated annexin per 1 × 105 cells at room temperature in the dark for 15 min according to manufacturer's instructions (BD Pharmingen). Analysis was performed using LSR II (Becton Dickinson, Franklin Lakes, NJ, USA, http://www.bd.com) and FlowJo software (Tree Star Inc., Ashland, OR, USA, http://www.treestar.com).
Cell cycle analysis
Cells were dissociated with 0.05% trypsin and 0.5 mm EDTA (Gibco BRL) at 37°C for 5 min and collected by centrifugation at 156.5g for 5 min. Cells were then washed once in PBS and subsequently resuspended in hypotonic propidium iodide buffer as described previously (55) at a concentration of 1 × 106 cells/ml. Cells were then immediately analysed using FacScan I (Becton Dickinson) and ModFit software (http://www.vsh.com) subsequently.
Immunofluorescence
Immunofluorescence was performed on cells grown on slides coated with matrigel fixed with 4% paraformaldehyde for 15 min. Blocking was performed in 10% heat-inactivated FBS (Hyclone) with 0.05% Tween 20 (Sigma) in phosphate-buffered saline (PBS) for 1 h prior to incubation with primary antibody [CENP-A (Millipore), γH2AX (Millipore) and Tubulin (Sigma)]. The primary antibody was detected with fluorescent dye-conjugated secondary antibody. TO-PRO-3 iodide (Invitrogen, http://www.invitrogen.como) was used as a marker for nucleus.
Western blot
Protein was harvested using M-PER cell lysis reagent (Thermo Scientific, Rockford, IL, USA, http://www.thermo.com). Protein concentration was measured using BCA Protein assay (Thermo Scientific). One hundred micrograms of total protein were electrophoresed through 12% NuPAGE Novex Bis-Tris gels (Invitrogen) and transferred according to standard procedures. For immunoblotting, primary antibody was rabbit anti-human CENP-A protein (Millipore) and rabbit anti-human β-actin (Abcam). Secondary HRP-conjugate was used at a 1:10 000 dilution. Blots were developed using SuperSignal West Pico Chemiluminescent Substrate (Pierce). Soluble proteins were separated from chromatin-hydroxyapatite using 0.35 m NaCl-buffer washes, and concentrated by TCA precipitation, followed by acetone rinse. Air-dried pellets were dissolved directly in 1× SDS–PAGE loading buffer. The H3K27Me3 antibody (Millipore) was used at 1:5000 dilution o/n, with secondary at 1:50 000 for 2 h.
Chromatin purification and CENP-A detection
Bulk chromatin was purified from HSF-6 and BJ cells following published protocol with slight modification (56). Briefly, cells were pelleted at low speed, washed three times with PBS containing 0.1% Tween 20. Nuclei were releases by resuspending cells in TM2 (20 mm Tris–HCl, pH 8.0, 2 mm MgCl2) containing 0.5% NP40 for 2 min on ice. Released nuclei were collected by centrifugation at 800 rpm for 10 min at 4°C. Nuclei were washed once with TM2 and 0.1 m TE. Nuclei were resuspended ice-cold nuclear extraction buffer (1× PBS containing 0.35 m NaCl2, 2 mm EDTA and 0.5 mm PMSF) containing 10% hydroxylapatite (HAP) to purify the DNA-bound histones, and left in an end-over-end rotary shaker in a cold box over night. HAP chromatin was collected by centrifugation 1000 rpm for 10 min. HAP chromatin was washed four times with ice-cold nuclear extraction buffer. HAP-/DNA-bound histones were eluted with high salt buffer (1× PBS containing 2 m NaCl2, 2 mm EDTA and 0.5 mm PMSF) for 16 h. Eluted proteins were collected by centrifugation and tri-chloroacetic acid (TCA, Sigma) precipitated for 10 min, rinsed with 70% acetone and then gently dried. Pellet was resuspended 1× SDS–PAGE sample buffer containing β-mercaptoethanol (Invitrogen), boiled for 10 min and electrophoretically resolved on 4–20% gradient TGX gels in Tris/Glycin/SDS buffer (Biorad). The gels were either fixed and stained with Coomassie Brilliant Blue or transferred to Optitran membrane for western blot with anti-CENP-A antibody according to manufacturer's protocol (Millipore).
Statistics
Two-sided Student's t-tests were performed to evaluate significance between two groups. Significance was accepted at P < 0.05.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
Conflict of Interest statement. None declared.
FUNDING
This work was supported by an R01 grant from the NIH/NICHD (R01 1R01HD058047-01), P01 funds from the NIH/NIGMS (P01 P01GM081621), funds from the Fuller Foundation (A.T.C.) and intramural research funding NIH/NCI (Y.D., R.K.G.). G.A. was supported by a training grant from the NIH/NICHHD UCLA Women's Reproductive Health Research Center (5 K12 HD001281) and a UCLA Clinical Fellows California Institute for Regenerative Medicine (CIRM) training grant. J.V. is supported by a graduate student training grant from CIRM. Fluorescence microscopy for some experiments was performed at the California NanoSystems Institute Advanced Light Microscopy/Spectroscopy Shared Facility at UCLA, directed by Dr Laurent Bentolila and supported with funding from NIH-NCRR shared resources grant (CJX1-443835-WS-29646) and NSF Major Research Instrumentation grant (CHE-0722519). Funding to pay the Open Access Charge was provided by UCLA Eli and Edythe Broad Center of Regenerative Medicine and Stem Cell Research.
Supplementary Material
REFERENCES
- 1.Macklon N.S., Geraedts J.P., Fauser B.C. Conception to ongoing pregnancy: the 'black box' of early pregnancy loss. Hum. Reprod. Update. 2002;8:333–343. doi: 10.1093/humupd/8.4.333. doi:10.1093/humupd/8.4.333. [DOI] [PubMed] [Google Scholar]
- 2.Martin R.H. Meiotic errors in human oogenesis and spermatogenesis. Reprod. Biomed. Online. 2008;16:523–531. doi: 10.1016/s1472-6483(10)60459-2. doi:10.1016/S1472-6483(10)60459-2. [DOI] [PubMed] [Google Scholar]
- 3.Humpherys D., Eggan K., Akutsu H., Hochedlinger K., Rideout W.M., 3rd, Biniszkiewicz D., Yanagimachi R., Jaenisch R. Epigenetic instability in ES cells and cloned mice. Science. 2001;293:95–97. doi: 10.1126/science.1061402. [DOI] [PubMed] [Google Scholar]
- 4.Pera M.F. Unnatural selection of cultured human ES cells? Nat. Biotechnol. 2004;22:42–43. doi: 10.1038/nbt0104-42. doi:10.1038/nbt0104-42. [DOI] [PubMed] [Google Scholar]
- 5.Draper J.S., Moore H.D., Ruban L.N., Gokhale P.J., Andrews P.W. Culture and characterization of human embryonic stem cells. Stem Cells Dev. 2004;13:325–336. doi: 10.1089/scd.2004.13.325. doi:10.1089/scd.2004.13.325. [DOI] [PubMed] [Google Scholar]
- 6.Draper J.S., Smith K., Gokhale P., Moore H.D., Maltby E., Johnson J., Meisner L., Zwaka T.P., Thomson J.A., Andrews P.W. Recurrent gain of chromosomes 17q and 12 in cultured human embryonic stem cells. Nat. Biotechnol. 2004;22:53–54. doi: 10.1038/nbt922. doi:10.1038/nbt922. [DOI] [PubMed] [Google Scholar]
- 7.Pyle A.D., Lock L.F., Donovan P.J. Neurotrophins mediate human embryonic stem cell survival. Nat. Biotechnol. 2006;24:344–350. doi: 10.1038/nbt1189. doi:10.1038/nbt1189. [DOI] [PubMed] [Google Scholar]
- 8.Watanabe K., Ueno M., Kamiya D., Nishiyama A., Matsumura M., Wataya T., Takahashi J.B., Nishikawa S., Nishikawa S., Muguruma K., et al. A ROCK inhibitor permits survival of dissociated human embryonic stem cells. Nat. Biotechnol. 2007;25:681–686. doi: 10.1038/nbt1310. doi:10.1038/nbt1310. [DOI] [PubMed] [Google Scholar]
- 9.Lavon N., Narwani K., Golan-Lev T., Buehler N., Hill D., Benvenisty N. Derivation of euploid human embryonic stem cells from aneuploid embryos. Stem Cells. 2008;26:1874–1882. doi: 10.1634/stemcells.2008-0156. doi:10.1634/stemcells.2008-0156. [DOI] [PubMed] [Google Scholar]
- 10.Heun P., Erhardt S., Blower M.D., Weiss S., Skora A.D., Karpen G.H. Mislocalization of the Drosophila centromere-specific histone CID promotes formation of functional ectopic kinetochores. Dev. Cell. 2006;10:303–315. doi: 10.1016/j.devcel.2006.01.014. doi:10.1016/j.devcel.2006.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Howman E.V., Fowler K.J., Newson A.J., Redward S., MacDonald A.C., Kalitsis P., Choo K.H. Early disruption of centromeric chromatin organization in centromere protein A (Cenpa) null mice. Proc. Natl Acad. Sci. USA. 2000;97:1148–1153. doi: 10.1073/pnas.97.3.1148. doi:10.1073/pnas.97.3.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sugimoto K., Fukuda R., Himeno M. Centromere/kinetochore localization of human centromere protein A (CENP-A) exogenously expressed as a fusion to green fluorescent protein. Cell Struct. Funct. 2000;25:253–261. doi: 10.1247/csf.25.253. doi:10.1247/csf.25.253. [DOI] [PubMed] [Google Scholar]
- 13.Sullivan B.A., Blower M.D., Karpen G.H. Determining centromere identity: cyclical stories and forking paths. Nat. Rev. Genet. 2001;2:584–596. doi: 10.1038/35084512. doi:10.1038/35084512. [DOI] [PubMed] [Google Scholar]
- 14.Earnshaw W.C., Rothfield N. Identification of a family of human centromere proteins using autoimmune sera from patients with scleroderma. Chromosoma. 1985;91:313–321. doi: 10.1007/BF00328227. doi:10.1007/BF00328227. [DOI] [PubMed] [Google Scholar]
- 15.Palmer D.K., Margolis R.L. Kinetochore components recognized by human autoantibodies are present on mononucleosomes. Mol. Cell Biol. 1985;5:173–186. doi: 10.1128/mcb.5.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sullivan K.F., Hechenberger M., Masri K. Human CENP-A contains a histone H3 related histone fold domain that is required for targeting to the centromere. J. Cell Biol. 1994;127:581–592. doi: 10.1083/jcb.127.3.581. doi:10.1083/jcb.127.3.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bernad R., Sanchez P., Losada A. Epigenetic specification of centromeres by CENP-A. Exp. Cell Res. 2009;315:3233–3241. doi: 10.1016/j.yexcr.2009.07.023. doi:10.1016/j.yexcr.2009.07.023. [DOI] [PubMed] [Google Scholar]
- 18.Dalal Y., Bui M. Down the rabbit hole of centromere dynamics and assembly. Curr. Opinion Cell Biol. 2010 doi: 10.1016/j.ceb.2010.02.005. Epub ahead of print March 19 doi:10.1139/O08-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Talbert P.B., Henikoff S. Histone variants—ancient wrap artists of the epigenome. Nat. Rev. Mol. Cell. Biol. 2010;11:264–275. doi: 10.1038/nrm2861. doi:10.1038/nrm2861. [DOI] [PubMed] [Google Scholar]
- 20.Bloom K., Joglekar A. Towards building a chromosome segregation machine. Nature. 2010;463:446–456. doi: 10.1038/nature08912. doi:10.1038/nature08912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Okada M., Okawa K., Isobe T., Fukagawa T. CENP-H-containing complex facilitates centromere deposition of CENP-A in cooperation with FACT and CHD1. Mol. Biol. Cell. 2009;20:3986–3995. doi: 10.1091/mbc.E09-01-0065. doi:10.1091/mbc.E09-01-0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jansen L.E., Black B.E., Foltz D.R., Cleveland D.W. Propagation of centromeric chromatin requires exit from mitosis. J. Cell Biol. 2007;176:795–805. doi: 10.1083/jcb.200701066. doi:10.1083/jcb.200701066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Regnier V., Vagnarelli P., Fukagawa T., Zerjal T., Burns E., Trouche D., Earnshaw W., Brown W. CENP-A is required for accurate chromosome segregation and sustained kinetochore association of BubR1. Mol. Cell. Biol. 2005;25:3967–3981. doi: 10.1128/MCB.25.10.3967-3981.2005. doi:10.1128/MCB.25.10.3967-3981.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goshima G., Kiyomitsu T., Yoda K., Yanagida M. Human centromere chromatin protein hMis12, essential for equal segregation, is independent of CENP-A loading pathway. J. Cell Biol. 2003;160:25–39. doi: 10.1083/jcb.200210005. doi:10.1083/jcb.200210005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Black B.E., Jansen L.E., Maddox P.S., Foltz D.R., Desai A.B., Shah J.V., Cleveland D.W. Centromere identity maintained by nucleosomes assembled with histone H3 containing the CENP-A targeting domain. Mol. Cell. 2007;25:309–322. doi: 10.1016/j.molcel.2006.12.018. doi:10.1016/j.molcel.2006.12.018. [DOI] [PubMed] [Google Scholar]
- 26.Tomonaga T., Matsushita K., Ishibashi M., Nezu M., Shimada H., Ochiai T., Yoda K., Nomura F. Centromere protein H is up-regulated in primary human colorectal cancer and its overexpression induces aneuploidy. Cancer Res. 2005;65:4683–4689. doi: 10.1158/0008-5472.CAN-04-3613. doi:10.1158/0008-5472.CAN-04-3613. [DOI] [PubMed] [Google Scholar]
- 27.Dobson A.T., Raja R., Abeyta M.J., Taylor T., Shen S., Haqq C., Pera R.A. The unique transcriptome through day 3 of human preimplantation development. Hum. Mol. Genet. 2004;13:1461–1470. doi: 10.1093/hmg/ddh157. doi:10.1093/hmg/ddh157. [DOI] [PubMed] [Google Scholar]
- 28.Kocabas A.M., Crosby J., Ross P.J., Otu H.H., Beyhan Z., Can H., Tam W.L., Rosa G.J., Halgren R.G., Lim B., et al. The transcriptome of human oocytes. Proc. Natl Acad. Sci. USA. 2006;103:14027–14032. doi: 10.1073/pnas.0603227103. doi:10.1073/pnas.0603227103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sudheer S., Adjaye J. Functional genomics of human pre-implantation development. Brief. Funct. Genomic Proteomic. 2007;6:120–132. doi: 10.1093/bfgp/elm012. doi:10.1093/bfgp/elm012. [DOI] [PubMed] [Google Scholar]
- 30.Wells D., Patrizio P. Gene expression profiling of human oocytes at different maturational stages and after in vitro maturation. Am. J. Obstet. Gynecol. 2008;198:455 e1–455 e1. doi: 10.1016/j.ajog.2007.12.030. (discussion 455 e9–455 11) [DOI] [PubMed] [Google Scholar]
- 31.Zhang P., Kerkela E., Skottman H., Levkov L., Kivinen K., Lahesmaa R., Hovatta O., Kere J. Distinct sets of developmentally regulated genes that are expressed by human oocytes and human embryonic stem cells. Fertil. Steril. 2007;87:677–690. doi: 10.1016/j.fertnstert.2006.07.1509. doi:10.1016/j.fertnstert.2006.07.1509. [DOI] [PubMed] [Google Scholar]
- 32.Shaw M.L., Williams E.J., Hawes S., Saffery R. Characterisation of histone variant distribution in human embryonic stem cells by transfection of in vitro transcribed mRNA. Mol. Reprod. Dev. 2009;76:1128–1142. doi: 10.1002/mrd.21077. doi:10.1002/mrd.21077. [DOI] [PubMed] [Google Scholar]
- 33.Bartova E., Galiova G., Krejci J., Harnicarova A., Strasak L., Kozubek S. Epigenome and chromatin structure in human embryonic stem cells undergoing differentiation. Dev. Dyn. 2008;237:3690–3702. doi: 10.1002/dvdy.21773. doi:10.1002/dvdy.21773. [DOI] [PubMed] [Google Scholar]
- 34.Mattout A., Meshorer E. Chromatin plasticity and genome organization in pluripotent embryonic stem cells. Curr. Opin. Cell Biol. 2010 doi: 10.1016/j.ceb.2010.02.001. doi:10.1016/j.ceb.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 35.Meshorer E., Misteli T. Chromatin in pluripotent embryonic stem cells and differentiation. Nat. Rev. Mol. Cell. Biol. 2006;7:540–546. doi: 10.1038/nrm1938. doi:10.1038/nrm1938. [DOI] [PubMed] [Google Scholar]
- 36.Wiblin A.E., Cui W., Clark A.J., Bickmore W.A. Distinctive nuclear organisation of centromeres and regions involved in pluripotency in human embryonic stem cells. J. Cell Sci. 2005;118:3861–3868. doi: 10.1242/jcs.02500. doi:10.1242/jcs.02500. [DOI] [PubMed] [Google Scholar]
- 37.Meshorer E., Yellajoshula D., George E., Scambler P.J., Brown D.T., Misteli T. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev. Cell. 2006;10:105–116. doi: 10.1016/j.devcel.2005.10.017. doi:10.1016/j.devcel.2005.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zeitlin S.G., Baker N.M., Chapados B.R., Soutoglou E., Wang J.Y., Berns M.W., Cleveland D.W. Double-strand DNA breaks recruit the centromeric histone CENP-A. Proc. Natl Acad. Sci. USA. 2009;106:15762–15767. doi: 10.1073/pnas.0908233106. doi:10.1073/pnas.0908233106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zeitlin S.G., Patel S., Kavli B., Slupphaug G. Xenopus CENP-A assembly into chromatin requires base excision repair proteins. DNA Repair. 2005;4:760–772. doi: 10.1016/j.dnarep.2005.02.007. doi:10.1016/j.dnarep.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 40.Lowry W.E., Richter L., Yachechko R., Pyle A.D., Tchieu J., Sridharan R., Clark A.T., Plath K. Generation of human induced pluripotent stem cells from dermal fibroblasts. Proc. Natl Acad. Sci. USA. 2008;105:2883–2888. doi: 10.1073/pnas.0711983105. doi:10.1073/pnas.0711983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sampath P., Pritchard D.K., Pabon L., Reinecke H., Schwartz S.M., Morris D.R., Murry C.E. A hierarchical network controls protein translation during murine embryonic stem cell self-renewal and differentiation. Cell Stem Cell. 2008;2:448–460. doi: 10.1016/j.stem.2008.03.013. doi:10.1016/j.stem.2008.03.013. [DOI] [PubMed] [Google Scholar]
- 42.Chang W.Y., Stanford W.L. Translational control: a new dimension in embryonic stem cell network analysis. Cell Stem Cell. 2008;2:410–412. doi: 10.1016/j.stem.2008.04.009. doi:10.1016/j.stem.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 43.Li L., Zheng P., Dean J. Maternal control of early mouse development. Development. 2010;137:859–870. doi: 10.1242/dev.039487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pique M., Lopez J.M., Mendez R. Cytoplasmic mRNA polyadenylation and translation assays. Methods Mol. Biol. 2006;322:183–198. doi: 10.1007/978-1-59745-000-3_13. doi:10.1007/978-1-59745-000-3_13. [DOI] [PubMed] [Google Scholar]
- 45.Martinez J., Patkaniowska A., Urlaub H., Luhrmann R., Tuschl T. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell. 2002;110:563–574. doi: 10.1016/s0092-8674(02)00908-x. doi:10.1016/S0092-8674(02)00908-X. [DOI] [PubMed] [Google Scholar]
- 46.Gaspar-Maia A., Alajem A., Polesso F., Sridharan R., Mason M.J., Heidersbach A., Ramalho-Santos J., McManus M.T., Plath K., Meshorer E., et al. Chd1 regulates open chromatin and pluripotency of embryonic stem cells. Nature. 2009;460:863–868. doi: 10.1038/nature08212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chi A.S., Bernstein B.E. Developmental biology. Pluripotent chromatin state. Science. 2009;323:220–221. doi: 10.1126/science.1166261. doi:10.1126/science.1166261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Okamoto Y., Nakano M., Ohzeki J., Larionov V., Masumoto H. A minimal CENP-A core is required for nucleation and maintenance of a functional human centromere. EMBO J. 2007;26:1279–1291. doi: 10.1038/sj.emboj.7601584. doi:10.1038/sj.emboj.7601584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alonso A., Hasson D., Cheung F., Warburton P.E. A paucity of heterochromatin at functional human neocentromeres. Epigenetics Chromatin. 2010;3:6. doi: 10.1186/1756-8935-3-6. doi:10.1186/1756-8935-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maehara K., Takahashi K., Saitoh S. CENP-A reduction induces a p53-dependent cellular senescence response to protect cells from executing defective mitoses. Mol. Cell. Biol. 2010;30:2090–2104. doi: 10.1128/MCB.01318-09. doi:10.1128/MCB.01318-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Clark A.T., Bodnar M.S., Fox M., Rodriquez R.T., Abeyta M.J., Firpo M.T., Pera R.A. Spontaneous differentiation of germ cells from human embryonic stem cells in vitro. Hum. Mol. Genet. 2004;13:727–739. doi: 10.1093/hmg/ddh088. doi:10.1093/hmg/ddh088. [DOI] [PubMed] [Google Scholar]
- 52.Ivanova N., Dobrin R., Lu R., Kotenko I., Levorse J., DeCoste C., Schafer X., Lun Y., Lemischka I.R. Dissecting self-renewal in stem cells with RNA interference. Nature. 2006;442:533–538. doi: 10.1038/nature04915. doi:10.1038/nature04915. [DOI] [PubMed] [Google Scholar]
- 53.Coleman J.E., Huentelman M.J., Kasparov S., Metcalfe B.L., Paton J.F., Katovich M.J., Semple-Rowland S.L., Raizada M.K. Efficient large-scale production and concentration of HIV-1-based lentiviral vectors for use in vivo. Physiol. Genomics. 2003;12:221–228. doi: 10.1152/physiolgenomics.00135.2002. [DOI] [PubMed] [Google Scholar]
- 54.Galic Z., Kitchen S.G., Kacena A., Subramanian A., Burke B., Cortado R., Zack J.A. T lineage differentiation from human embryonic stem cells. Proc. Natl Acad. Sci. USA. 2006;103:11742–11747. doi: 10.1073/pnas.0604244103. doi:10.1073/pnas.0604244103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Krishan A. Rapid flow cytofluorometric analysis of mammalian cell cycle by propidium iodide staining. J. Cell Biol. 1975;66:188–193. doi: 10.1083/jcb.66.1.188. doi:10.1083/jcb.66.1.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dalal Y., Wang H., Lindsay S., Henikoff S. Tetrameric structure of centromeric nucleosomes in interphase Drosophila cells. PLoS Biol. 2007;5:e218. doi: 10.1371/journal.pbio.0050218. doi:10.1371/journal.pbio.0050218. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.