This study demonstrates that the SPG20 hereditary spastic paraplegia protein spartin interacts with the ESCRT-III protein Ist1. This interaction is required for completion of the abscission phase of cytokinesis.
Abstract
Hereditary spastic paraplegias (HSPs, SPG1-46) are inherited neurological disorders characterized by lower extremity spastic weakness. Loss-of-function SPG20 gene mutations cause an autosomal recessive HSP known as Troyer syndrome. The SPG20 protein spartin localizes to lipid droplets and endosomes, and it interacts with tail interacting protein 47 (TIP47) as well as the ubiquitin E3 ligases atrophin-1-interacting protein (AIP)4 and AIP5. Spartin harbors a domain contained within microtubule-interacting and trafficking molecules (MIT) at its N-terminus, and most proteins with MIT domains interact with specific ESCRT-III proteins. Using yeast two-hybrid and in vitro surface plasmon resonance assays, we demonstrate that the spartin MIT domain binds with micromolar affinity to the endosomal sorting complex required for transport (ESCRT)-III protein increased sodium tolerance (Ist)1 but not to ESCRT-III proteins charged multivesicular body proteins 1–7. Spartin colocalizes with Ist1 at the midbody, and depletion of Ist1 in cells by small interfering RNA significantly decreases the number of cells where spartin is present at midbodies. Depletion of spartin does not affect Ist1 localization to midbodies but markedly impairs cytokinesis. A structure-based amino acid substitution in the spartin MIT domain (F24D) blocks the spartin–Ist1 interaction. Spartin F24D does not localize to the midbody and acts in a dominant-negative manner to impair cytokinesis. These data suggest that Ist1 interaction is important for spartin recruitment to the midbody and that spartin participates in cytokinesis.
INTRODUCTION
The hereditary spastic paraplegias (HSPs) are a group of inherited neurological disorders characterized by lower extremity spastic weakness (Soderblom and Blackstone, 2006; Dürr, 2008; Salinas et al., 2008). Classically, the HSPs have been divided into two forms: “pure” when lower extremity spasticity and weakness are the only features and “complicated” when additional neurological symptoms are present (Harding, 1983). More recently, a genetic classification scheme has come into wide use, with >40 distinct genetic loci reported (SPG1-46) and 20 genes identified (Dürr, 2008). The involved proteins have been subdivided into several functional groups that may be relevant for cellular pathogenesis. These groups include neuronal cell recognition and pathfinding, myelination, mitochondrial function, and intracellular trafficking and transport. Of these, the majority of proteins mutated in the HSPs seem to be involved in intracellular membrane and protein trafficking and distribution (Soderblom and Blackstone, 2006).
Troyer syndrome (SPG20; OMIM 275900) is an autosomal recessive, complicated HSP that presents in early childhood and is characterized by spastic dysarthria, cognitive impairment, short stature, and distal muscle wasting in addition to lower extremity spastic weakness (Cross and McKusick, 1967; Proukakis et al., 2004; Bakowska et al., 2008; Manzini et al., 2010). To date, there are only two known homozygous mutations associated with Troyer syndrome. The first mutation is a single base-pair deletion in the Old Order Amish resulting in a 29-amino acid substitution at the C terminus and premature truncation of the 666-amino acid protein by 268 residues (Patel et al., 2002). The second is a two base-pair deletion in an Omani kindred that results in an amino acid substitution followed by a stop codon in the first coding exon (p.M122VfsX1; Manzini et al., 2010). Because cell lines derived from Troyer syndrome patients have no detectable truncated spartin protein, loss of function represents the likely pathogenesis (Bakowska et al., 2008; Manzini et al., 2010).
Spartin harbors several distinct domains including an N-terminal MIT (contained within microtubule-interacting and transport molecules) domain, a –P-P-x-Y– motif, and a less well characterized plant-related region at the C terminus (Ciccarelli et al., 2003). Several studies investigating the distribution of both overexpressed and endogenous spartin have reported localizations to a variety of subcellular structures, including endosomes, midbodies, lipid droplets, and mitochondria (Lu et al., 2006; Robay et al., 2006; Bakowska et al., 2007; Eastman et al., 2009; Edwards et al., 2009). Spartin is monoubiquitinated (Bakowska et al., 2007) and interacts with the homologous to the E6-AP carboxy terminus domain (HECT) ubiquitin E3 ligases atrophin-1-interacting protein (AIP)4 and AIP5 as well as the lipid droplet protein tail interacting protein (TIP)47 (Eastman et al., 2009; Edwards et al., 2009; Hooper et al., 2010).
MIT domains of many other proteins interact with components of the endosomal sorting complexes required for transport (ESCRT) machinery, specifically C-terminal MIT-interacting motifs (MIMs) of ESCRT-III proteins (reviewed in Hurley and Yang, 2008). The ESCRT machinery is a series of multiprotein complexes (ESCRT-0, -I,-II, and -III) that assemble at target membranes. It has been implicated in several physiological and pathological processes in eukaryotes, including multivesicular body (MVB) formation, membrane abscission during cytokinesis, and human immunodeficiency virus budding (McDonald and Martin-Serrano, 2009; Hurley and Hanson, 2010), and defects in the ESCRTs are associated with neurodegeneration (Lee et al., 2007). ESCRT-0, -I, and -II contain ubiquitin-binding domains, sort ubiquitinated cargo into MVBs, and also may interact with ubiquitinated proteins in the course of their other functions. ESCRT-III seems to comprise the core machinery mediating membrane scission events in all ESCRT-dependent pathways (Wollert et al., 2009). C-Terminal MIM1 (Obita et al., 2007; Stuchell-Brereton et al., 2007) and MIM2 (Kieffer et al., 2008) motifs in ESCRT-III proteins mediate the recruitment of Vps4, which is responsible for the ATP-dependent disassembly of ESCRT-III after membrane scission. These MIMs also coordinate the interaction of ESCRT-III proteins with the microtubule-severing enzyme and SPG4 gene product spastin (Yang et al., 2008; Connell et al., 2009) as well as deubiquitinating enzymes and other proteins (Rigden et al., 2009, and references therein; Hurley and Hanson, 2010).
Spartin and spastin are the only known HSP proteins with an MIT domain, and both inhibit bone morphogenic protein signaling (Tsang et al., 2009), indicating that they may have common interactions. To gain insight into the cellular function of spartin, we explored whether it interacted with the ESCRT machinery via its N-terminal MIT domain. Because MIT domains interact with ESCRT-III subunits that are present at both endosomes and midbodies, localization of spartin to these structures might be predicted. The SPG4 protein spastin harbors an MIT domain with moderate sequence similarity to that of spartin and binds with high affinity to the ESCRT-III subunit charged multivesicular body protein (CHMP)1B (Reid et al., 2005; Yang et al., 2008; Connell et al., 2009). The spartin MIT domain, however, binds neither to CHMP1B nor to any of the other “traditional” human ESCRT-III proteins CHMP1-7. Recently, the human orthologue of yeast increased sodium tolerance (Ist)1 was characterized as a novel ESCRT-III–related protein containing MIM1 and MIM2 motifs (Bajorek et al., 2009a; Agromayor et al., 2009) and an ESCRT-III core (Xiao et al., 2009; Bajorek et al., 2009b). After a systematic analysis of all known traditional ESCRT-III proteins and Ist1, we report a highly selective, high-affinity interaction of the spartin MIT domain with Ist1. We also demonstrate that Ist1 recruits spartin to midbodies. Ist1 plays a role in cytokinesis (Bajorek et al., 2009a, Agromayor et al., 2009), and the colocalization of Ist1 and spartin at midbodies suggested that spartin also might play a role in cytokinesis, which we describe in this study.
MATERIALS AND METHODS
DNA Constructs
The human Ist1 (also known as KIAA0174) coding sequence (GenBank accession NM_014761) was cloned into the EcoRI site of the mammalian expression vector pGW1-HA. Amino acid numbering for Ist1 throughout the text refers to isoform 2 (UniProtKB/Swiss-Prot P53990-2), which has an MIM1 sequence identical to that of the canonical isoform 1 (P53990-1). The eukaryotic expression construct for HA-tagged spartin was described in Bakowska et al. (2007). The pBHA-spartin (1-107) and pBHA-spastin (110-195) yeast two-hybrid bait vectors and the pGAD10 and pGADT7 prey vectors for CHMP1-7 also were described previously (Bakowska et al., 2005; Yang et al., 2008). The full coding sequence and indicated deletion constructs for Ist1 were cloned into the EcoRI or EcoRI/XhoI sites of pGAD10 (Clontech, Mountain View, CA). Site-directed mutagenesis was performed using QuikChange (Stratagene, La Jolla, CA).
Yeast Two-Hybrid Assays
Yeast two-hybrid assays using the L40 yeast strain were performed as described previously (Yang et al., 2008).
Antibodies
Mouse monoclonal antibodies were used against β-tubulin (IgG1, clone D66; Sigma-Aldrich, St. Louis, MO), actin (clone AC-40; Sigma-Aldrich), phospholipase C (PLC)γ-1 (clone B-6-4; Millipore, Billerica, MA), and hemagglutinin (HA)-epitope (ab9110; Abcam, Cambridge, MA). Rabbit polyclonal antibodies were used against spartin (Bakowska et al., 2007) and Ist1 (KIAA0174; GeneTex, Irvine, CA). Mouse polyclonal anti-CEP55 antibodies were obtained from Abnova (Taipei City, Taiwan). The goat polyclonal anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibody was from Abcam.
Cell Culture and Transfection
Human HeLa and U-2 OS osteosarcoma (HTB-96; American Type Culture Collection, Manassas, VA) cells were maintained in DMEM supplemented with 10% (vol/vol) fetal bovine serum at 37°C in 5% CO2-humidified incubators. For confocal microscopy and biochemical studies, cells were transiently transfected with DNA plasmids by using calcium phosphate precipitation for 48 h or with predesigned small interfering RNAs (siRNAs) by using Oligofectamine (Invitrogen, Carlsbad, CA) for 72 h (Yang et al., 2008). The siRNA sequences (Invitrogen) used were as follows: Ist1 #1, 5′-GGGUAGAGACAGAUCUUAUUGAUGU-3′; Ist1 #2, 5′-CCCAUCGUAUGAAUCUAUGACAUUA-3′; Ist 1 #3, 5′-CCAGUCAGAAGUGGCUGAGUUGAAA-3′; and spartin, 5′-GGCAAGGAUUGGAAUGUGCAGCUAA-3′ (Bakowska et al., 2007). Control siRNAs were from Applied Biosystems/Ambion (Austin, TX).
Confocal Immunofluorescence Microscopy
To observe the midbody, cells plated on coverslips were fixed for 4 min with ice-cold methanol and blocked for 45 min with 10% goat serum, 0.5% Triton X-100 in phosphate-buffered saline (PBS; pH 7.4) at room temperature. After three washes, coverslips were incubated with primary antibodies diluted in 1% goat serum. Alexa Fluor anti-rabbit and anti-mouse secondary antibodies (Invitrogen) were used at 1:300 dilution. Cells were counterstained with 4′,6-diamidino-2-phenylindole (DAPI; 0.1 mg/ml) where indicated and mounted using Fluoromount-G (Southern Biotechnology, Birmingham, AL). Cells were imaged using an LSM510 confocal microscope (Carl Zeiss Microimaging, Thornwood, NY) with a 63× 1.4 numerical aperture (NA) Plan-Apochromat oil differential interference contrast (DIC) objective, and image acquisition was performed using LSM510 version 3.2 SP2 software (Carl Zeiss Microimaging). Images were processed with ImageJ (National Institutes of Health, Bethesda, MD), Photoshop 7.0 (Adobe Systems, Mountain View, CA), and Illustrator CS2 software (Adobe Systems).
For live-cell imaging experiments, HeLa cells were cotransfected with spartin siRNA and pmaxGFP (Lonza Walkersville, Walkersville, MD) to label the transfected cells, and cultured on two-well glass-bottomed chambers (Lab-Tek II chamber slides; Nalge Nunc International, Rochester, NY). Time-lapse recordings were made using an LSM 710 confocal microscope (Zeiss/PeCon XL LSM 710S1 live-cell incubator system with TempModule S, CO2 Module S1 and Heating Unit XL S; Carl Zeiss Microimaging) fitted under thermostat conditions (37°C in 5% CO2-humidified chamber). Images were collected with a 40× 1.4 NA Plan-Apochromat oil DIC objective as one picture of 1-μm Z-stacks at 20-min intervals, together with a single DIC reference image. Images were exported in 8-bit TIFF format by using ImageJ software. For quantification studies, DIC images were used to determine the timing of mitosis.
Gel Electrophoresis and Immunoblotting
Preparation of cell extracts, gel electrophoresis, and immunoblotting were performed as described previously (Zhu et al., 2003).
Epidermal Growth Factor Receptor (EGFR) Degradation Assay
EGFR degradation assays were performed essentially as described by Bakowska et al. (2007). In brief, HeLa cells were transfected with HA-spartin, HA-spartin F24D, or empty pGW1-HA vector; serum starved for 16 h; and then treated with EGF (100 ng/ml) and cycloheximide (10 μg/ml) for the indicated times. After washing with ice-cold PBS, cells were rapidly lysed with Laemmli sample buffer; resolved on SDS-polyacrylamide gel electrophoresis gels; and immunoblotted for EGFR, HA-epitope, and actin. EGFR immunoreactive bands were quantified using ImageJ software.
Fusion Protein Production in Bacteria
The expression and purification of CHMP1B C-terminal region (CTR) and spastin MIT was performed as reported previously (Yang et al., 2008). The CTRs of human Ist1 (residues 296-335) and the spartin MIT domain (residues 1-107) were cloned into pGST2 and pGEX6P-1 vectors, respectively, and expressed in Escherichia coli Rosetta (DE3) cells. Expression was induced by 1 mM isopropyl-β-d-thiogalactopyranoside at 18°C for 20 h. Cells were then lysed in PBS with 7 mM β-mercaptoethanol (β-ME) by using sonication. Lysates were applied to glutathione-Sepharose resin (GE Healthcare, Piscataway, NJ) and then washed with PBS with 7 mM β-ME for 50 column volumes. Glutathione transferase (GST)-Ist1-CTR was eluted from the glutathione-Sepharose resin by using 20 mM glutathione (reduced form) in PBS with 7 mM β-ME (pH 7.1). The protein was dialyzed in 10 mM HEPES, pH 7.0, with 150 mM NaCl. For the spartin MIT domain, the GST tag was cleaved on the column by incubating with tobacco etch virus (TEV) protease at room temperature overnight. The cleaved protein was eluted by PBS with 7 mM β-ME and then passed through a HisTrap HP column to remove His-tagged TEV protease. The protein was dialyzed in 20 mM Tris, pH 7.6, with 100 mM NaCl and 7 mM β-ME.
Surface Plasmon Resonance (SPR)
Binding of MIT domains to CHMP1B-CTR or Ist1-CTR constructs was analyzed using a Biacore T100 instrument at 25°C with a flow rate of 10 μl/min (Biacore Life Sciences, Piscataway, NJ). Hexahistidine-tagged CHMP1B-CTR, GST-tagged Ist1-CTR, and GST samples were immobilized by first being diluted in 10 mM acetate buffer, pH 4.0 and then passed over a CM5 chip that had been activated with a 1:1 mixture of N-hydroxysuccinimide and 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide at a flow rate of 10 μl/min. After the immobilization step, the CM5 surface was blocked using 1 M ethanolamine, pH 8.5. Binding studies were performed by passing the spartin or spastin MIT domains over the chip surface at a flow rate of 10 μl/min. The surface was regenerated with an injection of 10 mM glycine-HCl, pH 2.2, at a flow rate of 10 μl/min for 30 s. The reference cell contained immobilized recombinant GST. The data were fit with the following equation:
where [MIT] is the protein concentration of the flowing analyte, Kd is the dissociation constant, Rmax is the maximal response, and “offset” is the background signal. Data were processed using BiaEvaluation (Biacore Life Sciences) and SigmaPlot software (Systat Software, San Jose, CA).
Protein Content Determination
Protein content was assessed using the bicinchoninic acid assay (Pierce Chemical, Rockford, IL), with bovine serum albumin as the standard.
Statistical Analysis
Statistical significance was assessed using two-tailed, unpaired Student's t tests, assuming unequal variance.
RESULTS
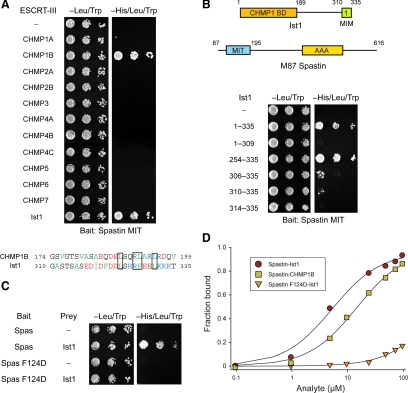
Spastin has been recently shown to interact with Ist1 as well as with CHMP1B. We investigated this interaction by using yeast two-hybrid tests, and we confirmed the specificity of these interactions (Figure 1A). Similar results were observed using just C-terminal domains of the ESCRT-III proteins (Supplemental Figure S1A). This may be because residues important for the spastin–CHMP1B interaction are conserved in Ist1 (Figure 1A). We were able to narrow down this interaction to the C-terminal 26 amino acid residues of Ist1, making up the region known as MIM1 (Figure 1B). A structure-based mutation that interfered markedly with spastin–CHMP1B binding (Yang et al., 2008), F124D, also substantially inhibited interaction with Ist1, as shown by yeast two-hybrid analysis (Figure 1C). Analogous results were obtained for the interaction of spastin with Ist1 by SPR; wild-type and F124D mutant spastin MITs interacted with Kd = 4.6 ± 0.1 and 471 ± 16 μM, respectively (Figure 1D).
Figure 1.
Spastin binds to MIM1 of the ESCRT-III protein Ist1. (A) Top, yeast two-hybrid interactions between the spastin MIT domain bait (residues 110-195) and the indicated ESCRT-III prey constructs were assayed using the HIS3 reporter (sequential 10-fold yeast dilutions are shown). Bottom, amino acid sequence alignment of the C-terminal MIM domains of CHMP1B and Ist1. Hydrophobic residues are in green, positively charged residues are in blue, and negatively charged residues are in red. Conserved residues important for MIM–MIT domain interactions in several other studies are boxed. (B) Spastin MIT bait was tested for yeast two-hybrid interactions as described in A with the indicated Ist1 prey constructs. Boundary amino acid residues are indicated. BD, binding domain. (C) Wild-type spastin and mutant spastin F124D MIT baits were tested for yeast two-hybrid interactions with the Ist1 prey. Schematic diagrams of the Ist1 and spastin (M87 isoform) protein structures are at the top, with amino acid numbers shown. (D) SPR analysis of wild-type or mutant spastin F124D MIT domain (analyte) binding to immobilized Ist1-CTR or CHMP1B-CTR, as shown.
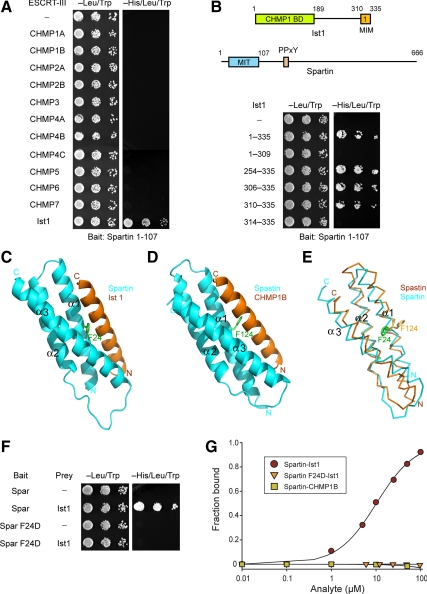
The spartin MIT domain has no known interactions with ESCRT-III proteins, but it is most similar to the MIT domain of the SPG4 protein spastin. We investigated the interactions of spartin MIT with the 12 known human ESCRT-III subunits, making up CHMP1-7 and Ist1, by using yeast two-hybrid tests. The spartin MIT domain interacted robustly with Ist1 but not with any other ESCRT-III proteins, including CHMP1B (Figure 2A). Because C-terminal regions (CTRs) of ESCRT-III proteins act as autoinhibitory switches (Bajorek et al., 2009b), we considered the possibility that using isolated C-termini of other ESCRT-IIIs might uncover weaker interactions. However, only the Ist1 CTR interacted with the spartin MIT domain (Supplemental Figure S1B), as for the full-length proteins shown in Figure 2A. The C-terminal MIM1 domain (residues 310-335 in isoform 2) was both necessary and sufficient for this interaction (Figure 2B), similar to the findings for interaction with the spastin MIT (Figure 1B).
Figure 2.
Spartin MIT domain binds selectively to Ist1. (A) Yeast two-hybrid interactions between the spartin MIT domain bait and the indicated ESCRT-III prey constructs were assayed using the HIS3 reporter (sequential 10-fold yeast dilutions are shown). (B) Spartin MIT bait was tested for yeast two-hybrid interactions as described in A with the indicated Ist1 prey constructs. Boundary amino acid residues are indicated. Schematic diagrams for Ist1 and spartin are shown at the top, with amino acid residues numbered. BD, binding domain. (C) Structural model of spartin MIT domain with Ist1 MIM. Spartin is colored in cyan and Ist1 in orange. Residue Phe24 (F24) in the spartin MIT domain is shown in green. (D) Crystal structure of spastin MIT in complex with CHMP1B, from Yang et al. (2008). Spastin is colored in cyan and CHMP1B in orange. Residue Phe124 (F124) in the spastin MIT domain is colored in green. (E) Structural alignment model for the spartin (cyan) and spastin (orange) MIT domains. Spartin residue F24 is colored in green and spastin residue F124 is in yellow. (F) Wild-type and mutant spartin F24D MIT baits were tested for yeast two-hybrid interactions with Ist1. BD, binding domain. (G) SPR analysis of wild-type or mutant spartin F24D MIT domain (analyte) binding to immobilized Ist1-CTR or CHMP1B-CTR.
In the solution structure of the human spartin MIT domain (Protein Data Bank accession code 2DL1), Phe24 (Phe23 in 2DL1 numbering) is in the same three-dimensional position as Phe124 in spastin, with the spartin Phe24 side chain rotated to fill the void left by the absence of the CHMP1B MIM helix in the spastin MIT-CHMP1B MIM structure (Yang et al., 2008; Figure 2, C–E). This conserved Phe residue is critical for interactions of the spastin MIT with ESCRT-III proteins, because its replacement with Glu (F124D) abolishes spastin MIT interactions with both CHMP1B (Yang et al., 2008) and Ist1 (Figure 1C). Similarly, replacement of the equivalent Phe24 in spartin with Glu (F24D) abolished the interaction between spartin MIT and Ist1 in yeast two-hybrid tests (Figure 2F). We measured the affinity of the wild-type spartin MIT domain–Ist1 interaction by using SPR and obtained a Kd = 10.4 ± 0.3 μM. Consistent with the yeast two-hybrid results, no interaction between spartin F24D MIT and Ist1 was detected using SPR (Figure 2G). Thus, spartin F24D is unable to interact with Ist1. Although CHMP1B interacts robustly with the spastin MIT domain (Yang et al., 2008), the spartin MIT domain shows no interaction with CHMP1B in yeast two-hybrid tests or in vitro (Figure 2, A and G).
Because spartin is modified by monoubiquitination (Bakowska et al., 2007), we examined whether interaction with Ist1 is required for this modification in HeLa cells. HA-tagged spartin F24D was monoubiquitinated to a similar extent as HA-tagged, wild-type spartin (Supplemental Figure S2A), indicating that interaction with Ist1 is not required for spartin monoubiquitination. Both spartin overexpression and depletion by siRNA have been shown to decrease EGFR degradation (Bakowska et al., 2007; Edwards et al., 2009), indicating that overexpressed spartin might act in a dominant-negative manner by binding to proteins outside of the proper cellular context, consistent with its proposed role as an adaptor protein (Bakowska et al., 2007; Edwards et al., 2009). Overexpression of HA-spartin F24D had a significant effect on EGFR degradation compared with control cells or those overexpressing wild-type spartin (Supplemental Figure S2, B and C; Bakowska et al., 2007). However, similar to a recent study, we found no significant difference in EGFR degradation upon suppression of Ist1 expression by using siRNA (Supplemental Figure S2, D–F; Agromayor et al., 2009), suggesting that the observed effects of spartin on EGFR degradation do not require interaction with Ist1.
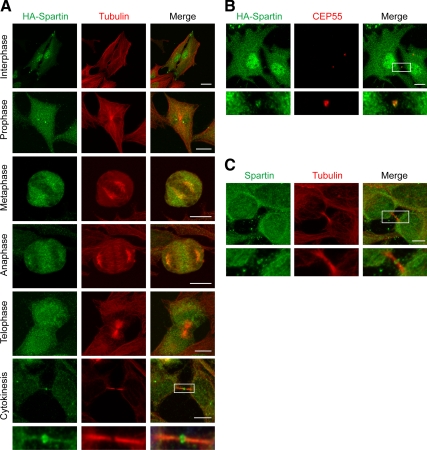
Given the lack of significant effect of the spartin–Ist1 interaction in EGFR degradation, we sought to establish further the functional importance for the interaction of Ist1 and spartin in cells. Thus, we examined the distribution of Ist1 and spartin in HeLa cells during different stages of the cell cycle. Consistent with previous reports (Bajorek et al., 2009a; Agromayor et al., 2009), HA-tagged Ist1 was found within the microtubule-organizing center/centrosome during the cell cycle, with this distribution particularly evident from prophase through anaphase (Figure 3A). In addition, Ist1 appeared at the midbody during late cytokinesis, before abscission (Figure 3, B and C). Ist1 immunostaining was usually concentrated at both ends of the Flemming body, which was most evident when immunostaining was performed for endogenous Ist1 (Figure 3D). Similar localization studies investigating HA-spartin showed more diffuse distributions of immunoreactive puncta throughout the cell cycle, although there was a clear localization to the midbody during late cytokinesis before abscission in HeLa cells (Figure 4A). Specifically, immunoreactivity for both HA-spartin and endogenous spartin typically concentrated at the ends of the Flemming body (Figure 4, B and C). The localization of endogenous Ist1 and spartin at the ends of the Flemming body also was clearly observed in another human cell type, U-2 OS cells (Supplemental Figure S3).
Figure 3.
Ist1 localizes to centrosome and midbody during mitosis. (A–C) HA-Ist1 (green) accumulates at the microtubule-organizing center/centrosome during prophase, metaphase (arrowheads), and early anaphase (A), and at the midbody during late cytokinesis (A–C). Corresponding images show the localizations of β-tubulin (red; B) and CEP55 (red; C), and merged images are at the right. A DIC image centered on the midbody is shown in an inset in B. (D) HeLa cells were costained for endogenous Ist1 (green) and β-tubulin (red), with the merged image at the right. Boxed areas are enlarged in the panels below. Bars, 10 μm.
Figure 4.
Spartin localizes to midbodies during cytokinesis. (A) HA-spartin (green) accumulates at the midbody during late cytokinesis. Corresponding images show the localization of β-tubulin (red), and merged images are at the right. The boxed area is enlarged in the panels below. (B) HA-spartin (green) localizes to the midbody, as shown with costaining for CEP55 (red). The merged image is at the right. Boxed area is enlarged in the panels below. (C) HeLa cells were stained for endogenous spartin (green) and β-tubulin (red), with the merged image at the right. Boxed area is enlarged in the bottom panels. Bars, 10 μm.
Given the high-affinity interaction of spartin and Ist1 and their similar distribution at the ends of the Flemming body during cytokinesis, we examined their roles in recruitment to midbody. In cells depleted of Ist1 by using siRNA, there was a significant loss of both spartin and Ist1 at midbodies (Figure 5, A–C). Consistent with previous reports (Bajorek et al., 2009a; Agromayor et al., 2009), Ist1 depletion caused a significant increase in the number of multinucleated cells, indicating a clear impairment in cytokinesis (Figure 5, D and E). By contrast, depletion of spartin by using siRNA had no effect on Ist1 localization to the midbody even when the proportion of cells with spartin at the midbodies was significantly down-regulated (Figure 6, A–C). Similarly, depletion of spartin had no effects on the midbody localization of CHMP1B (Supplemental Figure S4A). Although depletion of spartin resulted in no significant differences in number of mitotic cells or interphase cells (Figure 6, D and E), the number of cells interconnected by a midbody and the number of multinucleated cells were both increased (Figure 6, F and G). Together, these studies indicate that although Ist1 is required for spartin localization to the midbody, spartin is not required for the midbody localization of Ist1. In cells depleted of either Ist1 or spartin, the number of multinucleated cells was substantially increased, consistent with a defect in cytokinesis. The Flemming body itself was consistently visible on DIC imaging (Figure 6B), indicating that any localization changes are not secondary to disruption of midbody structure.
Figure 5.
Ist1 depletion decreases spartin recruitment to midbodies and impairs cell division. (A) HeLa cells were transfected with control siRNA or else Ist1 siRNA and immunostained for Ist1 (green) or β-tubulin (red). Boxed areas are enlarged in the panels below. Merged images are shown at the right. (B) Ist siRNA cells were imunostained for spartin (green) and β-tubulin (red). There is no spartin at the midbody, as shown in the boxed area enlarged in the panels below. (C) Cell extracts from HeLa cells treated with control or Ist1-specific siRNA were immunoblotted for Ist1. PLCγ levels were monitored to control for protein loading (left). Percentages of control and Ist1 siRNA cells with spartin (middle) or Ist1 (right) present at midbodies are shown (means ± SD of at least three trials, with 100 cells/experiment). (D and E) Multinucleated cells were evident upon Ist1 siRNA treatment of HeLa cells (D). Nuclei were identified by costaining with DAPI. (E) Quantification of the percentage of multinucleated cells after control versus Ist1 siRNA treatment (means ± SD of at least three trials, with 100 cells per experiment). *p < 0.001. Bars, 10 μm.
Figure 6.
Spartin is not required for Ist1 localization to midbodies. (A) HeLa cells were transfected with control siRNA or else spartin-specific siRNA and immunostained for spartin (green) or β-tubulin (red). Boxed areas are enlarged in the panels below. Merged images are shown at the right. (B) Cells treated with spartin siRNA were immunostained for Ist1 (green) and β-tubulin (red). Ist1 is present at the midbody, as shown in the boxed area enlarged in the panels below. A DIC image centered on the midbody is shown in the inset. (C) Cell extracts from HeLa cells treated with control or spartin-specific siRNA were immunoblotted for spartin. GAPDH levels were monitored to control for protein loading (left). Quantification of the percentage of control and Ist1 siRNA cells with spartin (middle) or Ist1 (right) present at midbodies (means ± SD of at least three trials, with 100 cells per experiment). (D–G) Quantifications of cells prepared as described in C are shown for percentages of mitotic cells (D), cells in interphase (E), cells interconnected with a visible midbody (F), and multinucleated cells (G). Means ± SD of at least three trials are shown, with 100 cells per experiment. **p < 0.001. Bars, 10 μm.
To examine in more detail the effects of spartin depletion on cytokinesis, we conducted live imaging experiments. Although cytokinesis occurred normally in control siRNA cells (Figure 7A, top; and Supplemental Video 1), spartin-depleted cells showed failure of abscission (Figure 7A, middle; and Supplemental Video 2) as well as extension of the abscission phase (Figure 7A, bottom; and Supplemental Video 3). Overall, time to complete cytokinesis was substantially increased in cells lacking spartin (Figure 7B), but there was no significant difference in formation or ingression of the cleavage furrow in these cells, as assessed by changes in furrow diameter over time after the onset of anaphase (Figure 7C).
Figure 7.
Spartin depletion inhibits the abscission phase of cytokinesis. (A) HeLa cells transfected with control or spartin-specific siRNA were imaged live at the indicated times (in minutes). Bars, 10 μm. (B) Quantification of the time required to complete cytokinesis (12 cells/condition). *p < 0.001. (C) Plot of cleavage furrow diameter over time after anaphase onset (10 cells/condition, means ± SD).
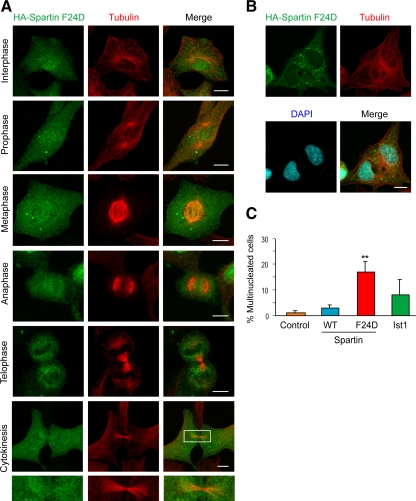
Because spartin F24D is unable to bind Ist1 or localize to midbodies (Figures 2, F and G, and 8A), we investigated the effects of spartin F24D overexpression on cytokinesis. There was a dramatic increase in the number of multinucleated cells in HA-spartin F24D expressing cells, compared with control cells and cells overexpressing either wild-type HA-spartin or HA-Ist1 (Figure 8, B and C). Overexpression of spartin F24D had no effect on Ist1 or CEP55 localization to the Flemming body (Supplemental Figure S4B). Spartin F24D probably exerts a strong dominant-negative effect on cytokinesis by interacting with other proteins and sequestering them from Ist1 interactions at the midbody or elsewhere.
Figure 8.
Spartin F24D does not localize to midbodies and causes dominant-negative impairment in cell division. (A) HA-spartin F24D (green) does not accumulate at the midbody during cytokinesis in HeLa cells. Corresponding images show the localization of β-tubulin (red), and merged images are at the right. The boxed area is enlarged in the panels below. (B) Multinucleated cells, with nuclei identified using DAPI staining (blue), were frequently seen upon HA-spartin F24D expression (green). β-Tubulin staining is shown in red. (C) Quantification of the percentage of multinucleated cells upon expression of empty vector, HA-spartin, HA-spartin F24D, and HA-Ist1 (means ± SD of at least three trials, with 100 cells/experiment). **p < 0.001 for wild type (WT) versus F24D. Bars, 10 μm.
DISCUSSION
In this study, we have demonstrated the high-affinity, selective interaction of the MIT domain of the spartin protein that is mutated in Troyer syndrome (SPG20) with the MIM1 of the ESCRT-III protein Ist1. The in vitro affinity of the spartin MIT domain with the Ist1 MIM1, with a Kd of ∼10 μM, is among the highest known for MIT–MIM1 interactions, which are typically in the range of Kd of ∼30–35 μM (Obita et al., 2007, Stuchell-Brereton et al., 2007) and similar to the affinity of the extended spastin MIM1 for CHMP1B (Yang et al., 2008). Given the importance of the Phe124 residue in the spastin MIT–CHMP1B complex (Yang et al., 2008) and that this residue is conserved in the spartin MIT domain, we investigated the role of this residue in the spartin–Ist1 interaction and found that mutation of this residue in spartin (F24D) abolished the interaction with Ist1. Despite that this conserved Phe is critical for interactions of both spastin and spartin MIT domains with Ist1 as well as the spastin MIT interaction with CHMP1B (this study and Yang et al., 2008), the two MIT domains show exquisite discrimination in terms of CHMP1B MIM interaction. The spastin MIT binds CHMP1B robustly, with a Kd of ∼12 μM (Yang et al., 2008), but the spartin MIT does not interact at all with CHMP1B (Figure 2, A and F), indicating that other residues are critical for this discrimination.
The interaction with Ist1 provides support for previous studies localizing spartin to endosomes, because Ist1 in Saccharomyces cerevisiae interacts with Did2 (analogous to human CHMP1A,B) and is a positive component of the MVB-sorting pathway (Dimaano et al., 2008; Rue et al., 2008). Thus, we suggest that spartin also may have a modulatory effect on MVB sorting in mammalian cells through interaction with Ist1 but that interaction with Ist1 is not required for this effect (Bakowska et al., 2007; Edwards et al., 2009).
A recurring theme in several studies investigating spartin localization is its recruitment to various cellular compartments. For example, spartin is recruited to lipid droplets upon treatment with oleic acid (Eastman et al., 2009; Edwards et al., 2009) and to endosomes upon treatment with EGF (Bakowska et al., 2007; Edwards et al., 2009). The specific recruitment of spartin to midbodies by Ist1 discovered here fits this scheme well, because like other ESCRT-III proteins Ist1 is probably present in an autoinhibited conformation in cells (Bajorek et al., 2009b), and the autoinhibition may be released as ESCRT-III forms multimeric circular arrays during cytokinesis (Hanson et al., 2008), permitting interactions of ESCRT-III MIMs with MIT domain-containing proteins such as Vps4 and spartin. Based on the prominent impairment of cytokinesis in cells depleted of spartin as well as in cells overexpressing spartin F24D, we suggest that spartin may serve as an adaptor platform for recruitment of other proteins important for the abscission phase of cytokinesis. For example, overexpression of a dominant-negative form of the deubiquitinating protease UBPY, which also binds Ist1, similarly leads to the appearance of multinucleated cells (Agromayor et al., 2009). This is consistent with a requirement for ubiquitin modification at the midbody for efficient cytokinesis (Pohl and Jentsch, 2008). The spartin interaction with Ist1 we report here, in conjunction with previous reports that spartin interacts with E3 ubiquitin ligases (AIP4 and AIP5) and is both monoubiquitinated and binds ubiquitin (Bakowska et al., 2007; Eastman et al., 2009; Edwards et al., 2009), provides additional support for the participation of spartin in an ubiquitin-based regulatory network during cytokinesis.
Importantly, this study provides pathogenic insights into mechanisms underlying the hereditary spastic paraplegias. Both the SPG4 protein spastin and SPG20 protein spartin interact with the ESCRT-III protein Ist1. In addition, recent studies of the SPG15 protein FYVE-CENT have identified an interaction of FYVE-CENT with ESCRT complex via FYVE-CENT interactions with the TTC19 protein that in turn interacts with the ESCRT-III protein CHMP4B (Sagona et al., 2010). Together, these findings suggest a converging pathway implicating the ESCRT complex in many different forms of hereditary spastic paraplegia. It will be important to determine whether this has specific functions in development or maintenance of axons in neurons.
Supplementary Material
ACKNOWLEDGMENTS
We thank J. Nagle and D. Kauffman (DNA Sequencing Facility, National Institute of Neurological Disorders and Stroke, National Institutes of Health, Bethesda, MD) for DNA sequencing. This research was supported by the Intramural Research programs of the National Institute of Neurological Disorders and Stroke (B. R., C. B.) and the National Institute of Diabetes and Digestive Diseases (D. Y., J.H.H.), National Institutes of Health; the National Institutes of Health Bench-to-Bedside Program (J.H.H., C. B.); the Howard Hughes Medical Institute–National Institutes of Health Research Scholars Program (R. P.); and National Institutes of Health K22 training grant NS-050137 (to J.C.B.).
Abbreviations used:
- AIP
atrophin-1-interacting protein
- β-ME
β-mercaptoethanol
- CHMP
charged multivesicular body protein
- CTR
C-terminal region
- DAPI
4′,6-diamidino-2-phenylindole
- DIC
differential interference contrast
- EGFR
epidermal growth factor receptor
- ESCRT
endosomal sorting complex required for transport
- GST
glutathione transferase
- HA
hemagglutinin
- HSP
hereditary spastic paraplegia
- Ist1
increased sodium tolerance-1
- MIM
MIT-interacting motif
- MIT
contained within microtubule-interacting and transport molecules
- MVB
multivesicular body
- SPR
surface plasmon resonance
- TIP47
tail interacting protein 47.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-10-0879) on August 4, 2010.
REFERENCES
- Agromayor M., Carlton J. G., Phelan J. P., Matthews D. R., Carlin L. M., Ameer-Beg S., Bowers K., Martin-Serrano J. Essential role of hIST1 in cytokinesis. Mol. Biol. Cell. 2009;20:1374–1387. doi: 10.1091/mbc.E08-05-0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajorek M., Morita E., Skalicky J. J., Morham S. G., Babst M., Sundquist W. I. Biochemical analyses of human IST1 and its function in cytokinesis. Mol. Biol. Cell. 2009a;20:1360–1373. doi: 10.1091/mbc.E08-05-0475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajorek M., Schubert H. L., McCullough J., Langelier C., Eckert D. M., Stubblefield W.-M. B., Uter N. T., Myszka D. G., Hill C. P., Sundquist W. I. Structural basis for ESCRT-III protein autoinhibition. Nat. Struct. Mol. Biol. 2009b;16:754–762. doi: 10.1038/nsmb.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakowska J. C., Jenkins R., Pendleton J., Blackstone C. The Troyer syndrome (SPG20) protein spartin interacts with Eps15. Biochem. Biophys. Res. Commun. 2005;334:1042–1048. doi: 10.1016/j.bbrc.2005.06.201. [DOI] [PubMed] [Google Scholar]
- Bakowska J. C., Jupille H., Fatheddin P., Puertollano R., Blackstone C. Troyer syndrome protein spartin is mono-ubiquitinated and functions in EGF receptor trafficking. Mol. Biol. Cell. 2007;18:1683–1692. doi: 10.1091/mbc.E06-09-0833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakowska J. C., Wang H., Xin B., Sumner C. J., Blackstone C. Lack of spartin protein in Troyer syndrome: a loss-of-function disease mechanism? Arch. Neurol. 2008;65:520–524. doi: 10.1001/archneur.65.4.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccarelli F. D., Proukakis C., Patel H., Cross H., Azam S., Patton M. A., Bork P., Crosby A. H. The identification of a conserved domain in both spartin and spastin, mutated in hereditary spastic paraplegia. Genomics. 2003;81:437–441. doi: 10.1016/s0888-7543(03)00011-9. [DOI] [PubMed] [Google Scholar]
- Connell J. W., Lindon C., Luzio J. P., Reid E. Spastin couples microtubule severing to membrane traffic in completion of cytokinesis and secretion. Traffic. 2009;10:42–56. doi: 10.1111/j.1600-0854.2008.00847.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross H. E., McKusick V. A. The Troyer syndrome. A recessive form of spastic paraplegia with distal muscle wasting. Arch. Neurol. 1967;16:473–485. doi: 10.1001/archneur.1967.00470230025003. [DOI] [PubMed] [Google Scholar]
- Dimaano C., Jones C. B., Hanono A., Curtiss M., Babst M. Ist1 regulates Vps4 localization and assembly. Mol. Biol. Cell. 2008;19:465–474. doi: 10.1091/mbc.E07-08-0747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dürr A. Genetic testing for the spastic paraplegias: drowning by numbers. Neurology. 2008;71:236–238. doi: 10.1212/01.wnl.0000320131.36091.a5. [DOI] [PubMed] [Google Scholar]
- Eastman S. W., Yassaee M., Bieniasz P. D. A role for ubiquitin ligases and Spartin/SPG20 in lipid droplet turnover. J. Cell Biol. 2009;184:881–894. doi: 10.1083/jcb.200808041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards T. L., Clowes V. E., Tsang H. T. H., Connell J. W., Sanderson C. M., Luzio J. P., Reid E. Endogenous spartin (SPG20) is recruited to endosomes and lipid droplets and interacts with the ubiquitin E3 ligases AIP4 and AIP5. Biochem. J. 2009;423:31–39. doi: 10.1042/BJ20082398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson P. I., Roth R., Lin Y., Heuser J. E. Plasma membrane deformation by circular arrays of ESCRT-III protein filaments. J. Cell Biol. 2008;180:389–402. doi: 10.1083/jcb.200707031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding A. E. Classification of the hereditary ataxias and paraplegias. Lancet. 1983;1:1151–1155. doi: 10.1016/s0140-6736(83)92879-9. [DOI] [PubMed] [Google Scholar]
- Hooper C., Puttamadappa S. S., Loring Z., Shekhtman A., Bakowska J. C. Spartin activates atrophin-1-interacting protein 4 (AIP4) E3 ubiquitin ligase and promotes ubiquitination of adipophilin on lipid droplets. BMC Biol. 2010;8:72. doi: 10.1186/1741-7007-8-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley J. H., Hanson P. I. Membrane budding and scission by the ESCRT machinery: it's all in the neck. Nat. Rev. Mol. Cell Biol. 2010;11:556–566. doi: 10.1038/nrm2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley J. H., Yang D. MIT domainia. Dev. Cell. 2008;14:6–8. doi: 10.1016/j.devcel.2007.12.013. [DOI] [PubMed] [Google Scholar]
- Kieffer C., Skalicky J. J., Morita E., De Domenico I., Ward D. M., Kaplan J., Sundquist W. I. Two distinct modes of ESCRT-III recognition are required for VPS4 functions in lysosomal protein targeting and HIV-1 budding. Dev. Cell. 2008;15:62–73. doi: 10.1016/j.devcel.2008.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- Lee J.-A., Beigneux A., Ahmad S. T., Young S. G., Gao F.-B. ESCRT-III dysfunction causes autophagosome accumulation and neurodegeneration. Curr. Biol. 2007;17:1561–1567. doi: 10.1016/j.cub.2007.07.029. [DOI] [PubMed] [Google Scholar]
- Lu J., Rashid F., Byrne P. C. The hereditary spastic paraplegia protein spartin localises to mitochondria. J. Neurochem. 2006;98:1908–1919. doi: 10.1111/j.1471-4159.2006.04008.x. [DOI] [PubMed] [Google Scholar]
- Manzini M. C., et al. Developmental and degenerative features in a complicated spastic paraplegia. Ann. Neurol. 2010;67:516–525. doi: 10.1002/ana.21923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald B., Martin-Serrano J. No strings attached: the ESCRT machinery in viral budding and cytokinesis. J. Cell Sci. 2009;122:2167–2177. doi: 10.1242/jcs.028308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obita T., Saksena S., Ghazi-Tabatabai S., Gill D. J., Perisic O., Emr S. D., Williams R. L. Structural basis for selective recognition of ESCRT-III by the AAA ATPase Vps4. Nature. 2007;449:735–739. doi: 10.1038/nature06171. [DOI] [PubMed] [Google Scholar]
- Proukakis C., Cross H., Patel H., Patton M. A., Valentine A., Crosby A. H. Troyer syndrome revisited. A clinical and radiological study of a complicated hereditary spastic paraplegia. J. Neurol. 2004;251:1105–1110. doi: 10.1007/s00415-004-0491-3. [DOI] [PubMed] [Google Scholar]
- Patel H., Cross H., Proukakis C., Hershberger R., Bork P., Ciccarelli F. D., Patton M. A., McKusick V. A., Crosby A. H. SPG20 is mutated in Troyer syndrome, an hereditary spastic paraplegia. Nat. Genet. 2002;31:347–348. doi: 10.1038/ng937. [DOI] [PubMed] [Google Scholar]
- Pohl C., Jentsch S. Final stages of cytokinesis and midbody ring formation are controlled by BRUCE. Cell. 2008;132:832–845. doi: 10.1016/j.cell.2008.01.012. [DOI] [PubMed] [Google Scholar]
- Reid E., Connell J., Edwards T. L., Duley S., Brown S. E., Sanderson C. M. The hereditary spastic paraplegia protein spastin interacts with the ESCRT-III complex-associated endosomal protein CHMP1B. Hum. Mol. Genet. 2005;14:19–38. doi: 10.1093/hmg/ddi003. [DOI] [PubMed] [Google Scholar]
- Rigden D. J., Liu H., Hayes S. D., Urbé S., Clague M. J. Ab initio protein modelling reveals novel human MIT domains. FEBS Lett. 2009;583:872–878. doi: 10.1016/j.febslet.2009.02.012. [DOI] [PubMed] [Google Scholar]
- Robay D., Patel H., Simpson M. A., Brown N. A., Crosby A. H. Endogenous spartin, mutated in hereditary spastic paraplegia, has a complex subcellular localization suggesting diverse roles in neurons. Exp. Cell Res. 2006;312:2764–2777. doi: 10.1016/j.yexcr.2006.05.003. [DOI] [PubMed] [Google Scholar]
- Rue S. M., Mattei S., Saksena S., Emr S. D. Novel Ist1-Did2 complex functions at a late step in multivesicular body sorting. Mol. Biol. Cell. 2008;19:475–484. doi: 10.1091/mbc.E07-07-0694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagona A. P., Nezis I. P., Pedersen N. M., Liestøl K., Poulton J., Rusten T. E., Skotheim R. I., Raiborg C., Stenmark H. PtdIns(3)P controls cytokinesis through KIF13A-mediated recruitment of FYVE-CENT to the midbody. Nat. Cell Biol. 2010;12:362–371. doi: 10.1038/ncb2036. [DOI] [PubMed] [Google Scholar]
- Salinas S., Proukakis C., Crosby A., Warner T. T. Hereditary spastic paraplegia: clinical features and pathogenetic mechanisms. Lancet Neurol. 2008;7:1127–1138. doi: 10.1016/S1474-4422(08)70258-8. [DOI] [PubMed] [Google Scholar]
- Soderblom C., Blackstone C. Traffic accidents: molecular genetic insights into the pathogenesis of the hereditary spastic paraplegias. Pharmacol. Ther. 2006;109:42–56. doi: 10.1016/j.pharmthera.2005.06.001. [DOI] [PubMed] [Google Scholar]
- Stuchell-Brereton M. D., Skalicky J. J., Kieffer C., Karren M. A., Ghaffarian S., Sundquist W. I. ESCRT-III recognition by VPS4 ATPases. Nature. 2007;449:740–744. doi: 10.1038/nature06172. [DOI] [PubMed] [Google Scholar]
- Tsang H. T. H., Edwards T. L., Wang X., Connell J. W., Davies R. J., Durrington H. J., O'Kane C. J., Luzio J. P., Reid E. The hereditary spastic paraplegia proteins NIPA1, spastin and spartin are inhibitors of mammalian BMP signaling. Hum. Mol. Genet. 2009;18:3805–3821. doi: 10.1093/hmg/ddp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollert T., Wunder C., Lippincott-Schwartz J., Hurley J. H. Membrane scission by the ESCRT-III complex. Nature. 2009;458:172–177. doi: 10.1038/nature07836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J., Chen X.-W., Davies B. A., Saltiel A. R., Katzmann D. J., Xu Z. Structural basis of Ist1 function and Ist1-Did2 interaction in the multivesicular body pathway and cytokinesis. Mol. Biol. Cell. 2009;20:3514–3524. doi: 10.1091/mbc.E09-05-0403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D., Rismanchi N., Renvoisé B., Lippincott-Schwartz J., Blackstone C., Hurley J. H. Structural basis for midbody targeting of spastin by the ESCRT-III protein CHMP1B. Nat. Struct. Mol. Biol. 2008;15:1278–1286. doi: 10.1038/nsmb.1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu P.-P., Patterson A., Lavoie B., Stadler J., Shoeb M., Patel R., Blackstone C. Cellular localization, oligomerization, and membrane association of the hereditary spastic paraplegia 3A (SPG3A) protein atlastin. J. Biol. Chem. 2003;278:49063–49071. doi: 10.1074/jbc.M306702200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.