ZFP36L1 negatively regulates erythroid differentiation of human hematopoietic progenitors by directly binding the 3′ UTR of Stat5b mRNA, thereby triggering its degradation. This study shows that posttranscriptional regulation is involved in the control of hematopoietic differentiation.
Abstract
ZFP36L1 is a member of a family of CCCH tandem zinc finger proteins (TTP family) able to bind to AU-rich elements in the 3′-untranslated region of mRNAs, thereby triggering their degradation. The present study suggests that such mechanism is used during hematopoiesis to regulate differentiation by posttranscriptionally modulating the expression of specific target genes. In particular, it demonstrates that ZFP36L1 negatively regulates erythroid differentiation by directly binding the 3′ untranslated region of Stat5b encoding mRNA. Stat5b down-regulation obtained by ZFP36L1 overexpression results, in human hematopoietic progenitors, in a drastic decrease of erythroid colonies formation. These observations have been confirmed by silencing experiments targeting Stat5b and by treating hematopoietic stem/progenitor cells with drugs able to induce ZFP36L1 expression. Moreover, this study shows that different members of ZFP36L1 family act redundantly, because cooverexpression of ZFP36L1 and family member ZFP36 determines a cumulative effect on Stat5b down-regulation. This work describes a mechanism underlying ZFP36L1 capability to regulate hematopoietic differentiation and suggests a new target for the therapy of hematopoietic diseases involving Stat5b/JAK2 pathway, such as chronic myeloproliferative disorders.
INTRODUCTION
The TTP family of tandem zinc finger proteins includes TTP/ZFP36, TIS11b/ZFP36L1, and TIS11d/ZFP36L2, all of which have been shown to directly bind AU-rich elements (ARE) and promote degradation of the host transcript (Carballo et al., 2000; Ogilvie et al., 2005). Their central RNA-binding domain interacts with AU-rich elements (UUAUUUAUU), whereas the N- and C-terminal domains recruit enzymes involved in the mRNA degradation pathway. Expression of all TTP family genes can be induced by phorbol esters (TPA) and various other mitogenic stimuli, such as growth factors, in a broad variety of cell types (Gomperts et al., 1992; Corps and Brown, 1995). For ZFP36 and in part also for other members, binding to and destabilization of TNF-α, granulocyte macrophage colony-stimulating factor (GM-CSF), and Vegf mRNAs has been demonstrated (Carballo et al., 1998, 2000; Lai et al., 1999; Ciais et al., 2004). Furthermore, ZFP36 is a transcriptional regulator (Murata et al., 2000, 2002) and mitogens induce its rapid nuclear to cytosolic translocation (Taylor et al., 1996). Recent publications suggest that TTP family proteins are phosphorylated (Taylor et al., 1995), might be targets of the p38 MAPK pathway (Carballo et al., 2001; Mahtani et al., 2001), and might be involved in the regulation of apoptosis (Johnson et al., 2000; Johnson and Blackwell, 2002). Finally, TTP family members have recently been involved in cell differentiation and cancer. In particular, ZFP36 has been indicated as a critical regulator of dendritic cells maturation (Emmons et al., 2008), and it has been demonstrated that targeted disruption of ZFP36L2 results in defective hematopoiesis (Stumpo et al., 2009) and that ZFP36 is suppressed in many cancers, altering tumorigenic phenotypes and patient prognosis (Brennan et al., 2009).
The Stat5/JAK2 pathway is widely studied. Its aberrant activation is involved in several diseases (Benekli et al., 2003; Choudhary et al., 2007; Kotecha et al., 2008; Wagner and Rui, 2008; Funakoshi-Tago et al., 2009), and particularly its constitutive activation dependent on JAK2 V617F mutation underlies chronic myeloproliferative disorders (Kralovics et al., 2005; Heller et al., 2006; Guglielmelli et al., 2007; Levine and Gilliland, 2008; Grimwade et al., 2009). Conversely, the same pathway is also involved in the control of normal hematopoiesis, because it plays a fundamental role during erythroid maturation (Grebien et al., 2008; Olthof et al., 2008). The Stat5a and Stat5b genes belong to the signal transducer and activator of transcription (Stat) family of transcription factors. The family of Stat genes is believed to originate from successive genome duplication and functional divergence of a single ancestral Stat gene early in vertebrate evolution. A recent duplication of Stat5 is believed to have given rise to the closely related Stat5a and Stat5b homologous genes that encode proteins that in humans are ∼91% identical, the main difference between the two proteins residing in a five-amino acid insertion at the COOH-terminus of Stat5b (Crispi et al., 2004). Stat5a and Stat5b exert redundant yet distinct functions (Basham et al., 2008) both in physiological and pathological conditions; for instance Stat5b and not Stat5a is responsible for inducing an increase of cell motility and invasiveness in hepatocellular carcinoma (Lee et al., 2006). The common aspects and the differences between Stat5a and Stat5b's behavior depend on multiple factors such as differences in mRNA levels (Ambrosio et al., 2002), activation by phosphorylation (Moucadel and Constantinescu, 2005), or as we point out in the present work, a different pattern of tissue specific expression.
The present study has its starting point in the analysis of a gene expression profile of human hematopoietic cell populations (data not shown) that consistently shows that ZFP36L1 expression is substantially low in erythroid progenitors compared with the other hematopoietic cell types. On these premises, the role of ZFP36L1 was evaluated by ectopic expression and gene silencing experiments in human hematopoietic cord blood–derived CD34+ stem/progenitor cells and on the erythroleukemia cell line carrying the JAK2 V617F mutation, Hel. Such experiments confirmed that ZFP36L1 expression negatively regulates erythroid maturation, and the results were further validated by monitoring the effect of cinnamon-derived polyphenols, a drug able to induce the expression of TTP gene family (Schoene et al., 2005; Cao et al., 2007), on the same cell populations. To investigate the mechanisms underlying such biological behavior, we hypothesized that ZFP36L1 might interfere with Stat5/JAK2 pathway, whose activity is pivotal for erythroid differentiation and survival. We demonstrated that ZFP36L1 and also ZFP36 are able to directly bind the 3′-untranslated region of Stat5b mRNA, thereby triggering its degradation. As confirmed by gene silencing experiments targeting Stat5b, its down-regulation is sufficient to inhibit the formation of erythroid precursors from CD34+ human stem/progenitor cells. Moreover, the current study demonstrates that at least two members of the TTP family, ZFP36L1 and ZFP36, are capable of acting redundantly, because their common overexpression results in a cumulative effect on Stat5b down-regulation.
MATERIALS AND METHODS
Cell Cultures and Treatments
The U937 cell line was obtained from ATCC (Rockville, MD) and cultured in RPMI1640 medium (Euroclone, Devon, United Kingdom), supplemented with 10% heat-inactivated fetal bovine serum (Biowhittaker, Walkersville, MD) and 1 mM l-glutamine (Euroclone). The Hel cell line was obtained from ATCC and was cultured in IMDM medium (Euroclone) supplemented with 10% heat-inactivated fetal bovine serum and 1 mM l-glutamine. GP+envAm12 cells and human embryonic kidney (HEK) 293 cells were cultured in DMEM (Euroclone), supplemented with 10% heat-inactivated fetal bovine serum and 1 mM l-glutamine. Ecotropic Phoenix cells were cultured in IMDM medium supplemented with 10% heat-inactivated fetal bovine serum and 1 mM l-glutamine.
Human CD34+ hematopoietic stem/progenitor cells (HSCs) were purified from umbilical cord blood (CB) samples as already described (Piacibello et al., 1998; Montanari et al., 2005) and seeded (5–10 × 104/ml) in IMDM (Euroclone) containing 20% heat-inactivated fetal bovine serum and early acting human hematopoietic cytokines: 50 ng/ml stem cell factor (SCF) and Flt3-ligand (Flt3-l), and 10 ng/ml interleukin-6 (IL-6) and interleukin-3 (IL-3; R&D Systems, Minneapolis, MN). These experimental conditions normally promote a mixed granulo-monocyte differentiation of CD34+ cells that is generally achieved within 14 d from plating (Piacibello et al., 1998; Montanari et al., 2005).
For CFC assay CD34+ cells were cultured for 48 h before transfection with nontargeting control siRNA/anti-Stat5b siRNAs or before infection with viral supernatants; cells were then plated as already described in a complete methylcellulose medium (MethoCult H4434, Stem Cell Technologies,Vancouver, BC, Canada) between 24 and 48 h after transfection or 24 h after purification of transduced cells. Colonies were scored 14 d after plating as erythroid (burst forming unit-erythroid [BFU-E] + colony forming unit-erythroid [CFU-E]), CFU-GEMM, CFU-GM, CFU-G, or CFU-M. Error bars represent SEM calculated on a set of three to four independent experiments (*p < 0.05).
Human primary myeloblasts, monoblasts, erythroblasts, and megakaryoblasts were generated by in vitro culture of cord blood CD34+ HSCs performed as already described (Gemelli et al., 2008).
Cinnamon extracts were kindly provided by Dr. R. A. Anderson (Human Nutrition Research Center, Beltsville, MD); they were resuspended in water and added once to IMDM culture medium/methylcellulose medium at the concentrations indicated in figure legends, and medium was not changed until cells were lysed or colonies were scored.
Plasmids, Expression Constructs, and Retroviral Vectors
Full-length ZFP36 and ZFP36L1 cDNAs were generated by RT-PCR performed on total RNA extracted from U937 cells using ZFP36-direct primer (DP) (5′-ATGGATCTGACTGCCATCTACG-3′) and ZFP36-reverse primer (RP) (5′-CGGGCAGTCACTTTGTCACT-3′) or ZFP36L1-DP (5′-GAACGCACAGGATGACCACCA-3′) and ZFP36L1-RP (5′-CCTACCCTGGCTTAGTCATCTGA-3′). Amplification was carried out using FastStart High Fidelity PCR System (Roche Applied Science, Penzberg, Germany), and the amplified fragments were inserted into the pCR2.1-TOPO T/A cloning vector (Invitrogen, Carlsbad, CA) where they were fully sequenced to exclude polymerase-induced mutations.
ZFP36/ZFP36L1 cDNAs were then EcoRI excised and cloned into an EcoRI-digested pcDNA3.1 expression vector (Invitrogen) or into an EcoRI-digested LxIΔN retroviral vector, resulting in the construction of LZFP36IΔN and LZFP36L1IΔN retroviral vectors.
FlagZFP36/FlagZFP36L1 fragments were RT-PCR amplified using primers FlagZFP36-DP (5′-ATGGACTACAAAGATGACGACGACAAGGATCTGACTGCCATCTAC-3′) and ZFP36-RP or FlagZFP36L1-DP (5′-ATGGACTACAAAGATGACGACGACAAGACCACCACCCTCGTGTCT-3′) and ZFP36L1-RP and the amplified fragments were inserted into the pCR2.1-TOPO T/A cloning vector, from which they were EcoRI excised and cloned into an EcoRI-digested pcDNA3.1 vector.
To obtain vectors for in vitro transcription and translation, FlagZFP36/ZFP36L1 RT-PCR amplified fragments were inserted into the pEXP5-CT-TOPO T/A cloning vector (Invitrogen) where they were fully sequenced to exclude polymerase-induced mutations.
Vegfa and Stat5b ARE-containing 3′UTR were amplified by RT-PCR on total RNA extracted from U937 cells using Vegfa-3′UTR-DP (5′-ACAGAGAGACAGGGCAGGAT-3′) and Vegfa-3′UTR-RP (5′-GGAATATCTCGAAAAACTGCAC-3′) or Stat5b-3′UTR-DP (5′-GATGGCCCCAAAAACCTTAT-3′) and Stat5b-3′UTR-RP (5′-TCCCTTCAGAGGAAGGCTTTA-3′). The amplified fragments were then inserted into the pCR2.1-TOPO T/A cloning vector and were fully sequenced. The 3′UTR fragments were then EcoRI excised, blunted, and cloned into a SmaI-digested pGL3-Promoter Vector (Promega, Madison, WI) that had previously been modified in order to transfer the multiple cloning region downstream the luciferase reporter gene.
DNA Transfection and Retroviral Infection
Transient transfection of pcDNA3.1-based constructs in Hel cells was obtained using the Amaxa Nucleofector Technology (Lonza Cologne, Cologne, Germany). Briefly, 1 × 106 Hel cells were electroporated in 100 μl Ingenio Electroporation Solution (Mirus Bio, Madison, WI) containing 5 μg of the desired DNA and using T016 electroporation program. Cells were lysed for RNA or protein extraction 72 h after electroporation.
The U937 cell line and CD34+ HSCs were transduced using LxIΔN empty control vector or LZFP36IΔN/LZFP36L1IΔN retroviral vectors. Packaging lines for the LxIΔN-based constructs were generated by transinfection in the ecotropic Phoenix and amphotropic GP+envAm12 cells, as previously described. Viral titers were assessed by flow cytometry analysis of the expression percentage of the reporter gene ΔLNGFR upon infection of U937 or CD34+ hematopoietic progenitors. Incubation of cells with viral supernatants and purification of transduced cells were performed as previously described (Zanocco-Marani et al., 2006). Transduced U937/CD34+ HSCs were lysed for RNA extraction between 7 and 10 d after transduction, as indicated in figure legends.
Gene Silencing
Stat5b silencing in Hel cells and in CD34+ cells was obtained by using a mix of two predesigned small interfering RNA (siRNA; Cat. no.: AM16708; siRNA ID no.: 115945 and 3618; Applied Biosystems, Foster City, CA). Electroporation was performed using the Amaxa Nucleofector Technology (Lonza Cologne) according to the manufacturer's instructions. For each cycle, 5–10 × 105 cells were electroporated in 100 μl Ingenio Electroporation Solution (Mirus Bio) containing 3 μg of each siRNA duplex. Every experiment included a negative control represented by 6 μg of a nontargeting siRNA (Dharmacon, Lafayette, CO) in order to exclude nonspecific effects of siRNA nucleofection. Hel cells were lysed for protein or RNA extraction 48 h after electroporation; CD34+ HSCs were lysed between 4 and 7 d after electroporation, as indicated in figure legends.
Western Blot
Twenty micrograms of total protein extracts obtained by cell lysis in RIPA buffer or 2 μl of in vitro–translated proteins was loaded onto 10% SDS-polyacrylamide gel and blotted. Membranes were preblocked in blocking solution, 5% milk in 0.1% TBST, for 1 h at room temperature and then incubated with a 1:200 dilution of mouse anti-Stat5b (G-2) monoclonal Ab (MoAb; Santa Cruz Biotechnology, Santa Cruz, CA), followed by a 1-h incubation at room temperature with goat anti-mouse IgG conjugated to horseradish peroxidase 1:10,000 (sc-2005, Santa Cruz Biotechnology). Expression of actin was analyzed with a mouse anti-human pan-actin MoAb (Sigma-Aldrich, St. Louis, MO) to normalize protein samples. Detection of in vitro–translated proteins was carried out with anti-ZFP36 polyclonal antibody (Abcam, Cambridge, MA; ab36558), anti-ZFP36L1 polyclonal antibody (Abcam; ab35801), or anti-Flag M2 mouse MoAb (Sigma-Aldrich). Detection was carried out by using the BM Chemiluminescence Blotting Substrate (Roche Applied Science).
Microarray Data Analysis
Expression profiles of CD34+ HSCs and CD34-derived erythroblasts, megakaryoblasts, monoblasts, and myeloblasts were obtained and analyzed as previously described (Gemelli et al., 2008).
Stat5a, Stat5b, and ZFP36L1 expression levels during in vitro erythroid differentiation of CD34+ hematopoietic progenitor cells were obtained from the GEO-Profiles database (http://www.ncbi.nlm.nih.gov/geo/; GDS2431).
RT-PCR and Real-Time PCR
Total cellular RNA was extracted using the EuroGOLD Total RNA kit (Euroclone) or RNeasy MicroKit (Qiagen, Hilden, Germany). RNA integrity and concentration was then assessed by the Bio-Analyzer technique (Applied Biosystems).
For RT-PCR 3 μg total RNA were reverse transcribed using M-MLV reverse transcriptase (Invitrogen), and then expression of specific genes was assessed by PCR amplification performed using specific primers: GAPDH-DP (5′-GAAGGTGAAGGTCGGAGTC-3′), GAPDH-RP (5′-GAAGGCCATGCCAGTGAGCT-3′); ZFP36L1-DP (5′-GAACGCACAGGATGACCACCA-3′); ZFP36L1-RP (5′-CCTACCCTGGCTTAGTCATCTGA-3′); Stat5b-DP (5′-GATGGCCCCAAAAACCTTAT-3′); Stat5b-RP (5′-TCCCTTCAGAGGAAGGCTTTA-3′); Jak2-DP (5′-GAGCCTATCGGCATGGAATA-3′); Jak2-RP (5′-TCACCTGAAGGACCACTTCC-3′); IRF8-DP (5′-ATGTGTGACCGGAATGGTG-3′); and IRF8-RP (5′-CCAGACAGAGGGATCCACAT-3′). Normalization of the amplified samples was obtained by the glyceraldehyde 3-phosphate dehydrogenase (GAPDH) housekeeping gene.
For quantitative real-time PCR (QRT-PCR) 100 ng total RNA were reverse-transcribed using the High-Capacity cDNA Archive Kit (Applied Biosystems) according to the manufacturer's instructions; QRT-PCR was then performed with an ABI PRISM 7900 sequence detection system (Applied Biosystems) as already described (Zanocco-Marani et al., 2006). Primers and probes for the different genes' amplification were provided by Applied Biosystems; quantitation was performed by amplifying GAPDH mRNA as endogenous control. Error bars represent SEM calculated on a set of three to four independent experiments (*p < 0.05; **p < 0.01).
RNA Electrophoretic Mobility Shift Assays
RNA electrophoretic mobility shift assays (REMSAs) were performed on in vitro–transcribed/translated FlagZFP36 and FlagZFP36L1 proteins. Such samples were produced by using the Expressway Cell-Free E. coli Expression System (Invitrogen) following the manufacturer's guidelines. Mobility shift reactions were carried out in gel shift buffer (10 mM HEPES, pH 7.9, 10 mM KCl, 2 mM MgCl2, 0.5 mM DTT, 10% glycerol, 0.15% NP40) in a total volume of 20 μl. RNA probes were synthesized by Eurofins MWG Operon (Ebersberg, Germany); their sequences are as follows: GMCSF-ARE: 5′-UAUUUAUUUAUUUAUUUAUUUA-3′ (NM_000758, nt from 695 to 716); Stat5b-ARE: 5′-AUAGUAAAUUAUUUAUUGGAAGAU-3′(NM_012448, nt from 5005 to 5028). Single-stranded RNA probes were 5′-end labeled using γ32P-ATP (6000Ci/mmol; PerkinElmer, Waltham, MA) and T4 polynucleotide kinase (New England Biolabs, Ipswich, MA). The binding reactions contained 1.0 × 105 cpm of the indicated labeled probe and 2 μl of in vitro–transcribed/translated protein. Supershifts were determined by an extra 20-minute incubation in the presence of 1 μg Flag M2 mouse MoAb (Sigma-Aldrich). Reactions were resolved on a nondenaturing 5% polyacrylamide gel. Electrophoresis was performed at 4°C at 160 V for 3 h.
Luciferase Assays
HEK293 cells were plated at a density of 50,000 cells/well 12 h before transfection in 24-well plates. Transfections were carried using Lipofectamine 2000 reagent (Invitrogen). In a typical assay, each well received 200 ng of pGL3-based reporter construct, 200 ng of CMV-β-galactosidase plasmid (Clontech Laboratories, Mountain View, CA), and 10–20 ng of pcDNA3.1 FlagZfp36 or pcDNA3.1 FlagZfp36L1, as indicated in figure legends; the difference in total DNA transfected was scaled up with pcDNA3.1 empty vector. After 24–48 h cells were harvested, and cell lysates were assayed for luciferase and β-galactosidase activity (Zappavigna et al., 1994). Each transfection was done in duplicate in the same experiment, and the plotted luciferase activities represent the average of five different experiments; error bars represent SEM (*p < 0.05; **p < 0.01).
Ribonucleoprotein Complex Immunoprecipitation and Analysis by RT-PCR
HEK293 cells were cotransfected with pGL3Stat5b-3′UTR and pcDNA3.1-ZFP36L1 plasmids using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's guidelines. Twenty-four hours after transfection ribonucleoprotein complexes were immunoprecipitated following a protocol described in literature (Niranjanakumari et al., 2002). Lysates were immunoprecipitated in the presence of protein A Sepharose (CL-4B; GE Healthcare, Waukesha, WI) previously incubated with anti-ZFP36L1 polyclonal antibody (Abcam; ab35801), anti-glutathione S-transferase (GST) polyclonal antibody (Santa Cruz Biotechnology; sc-459), or with no antibody. Immunoprecipitated mRNAs were retro-transcribed using random hexamers and M-MLV (Moloney murine leukemia virus) reverse transcriptase (Invitrogen). Primers used for subsequent PCR analysis were DP (5′-TCAAAGAGGCGAACTGTGTG-3′) and RP (5′-GCTCCTCAGAGGCAGAGAGA-3′).
RESULTS
ZFP36L1 Ectopic Expression Down-Regulates Stat5b Expression and Interferes with Erythroid Differentiation of Human CD34+ Cord Blood–derived Stem/Progenitor Cells
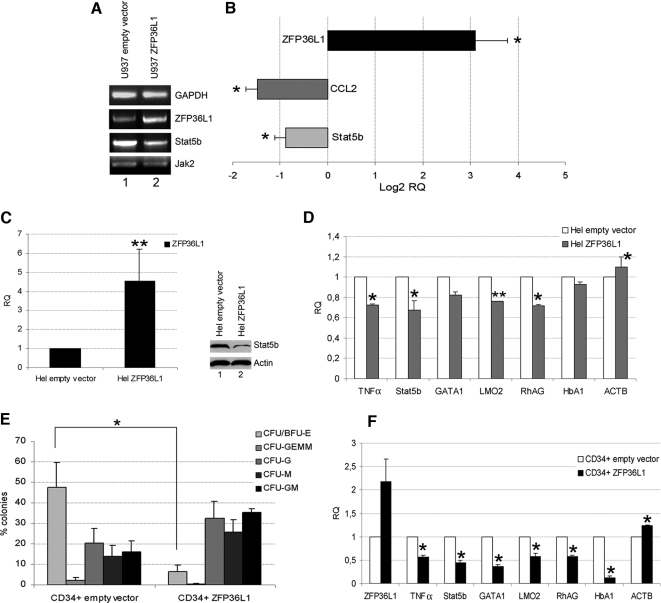
On the basis of the main hypothesis that ZFP36L1 could be a negative regulator of erythroid differentiation, we searched the 3′UTR regions of mRNAs encoding proteins involved in such maturation process, in order to find AREII sequences. Among several others not included in the current study, Stat5b appeared to be a suitable candidate, because its 3′UTR carries several canonical and noncanonical AREII sequences. To preliminarily evaluate a possible relation between ZFP36L1 and Stat5b expression, we overexpressed ZFP36L1 in U937 monoblastic and Hel erythroleukemia cell lines and monitored changes in Stat5b expression. The results are shown in Figure 1; in particular, panels A and B show the results of overexpression in U937 cells, whereas panels C and D show the results of the same experiment performed on Hel cells. Figure 1A shows an RT-PCR, showing at the mRNA level that ZFP36L1 overexpression is coupled to a decrease of Stat5b mRNA. Figure 1B shows the same results analyzed by QRT-PCR, showing that ZFP36L1 overexpression is also coupled to the down-regulation of CCL2, a gene previously described as a target of TTP family proteins (Sauer et al., 2006). Figure 1C shows a QRT-PCR showing the extent of ZFP36L1 overexpression obtained in Hel cells. The same panel includes a Western blot showing Stat5b protein down-regulation after ZFP36L1 overexpression. To gather further information on the effect of ZFP36L1 ectopic expression in Hel cells, we also analyzed by QRT-PCR whether transgene expression is capable of perturbing the expression of a set of erythroid differentiation genes. The results are shown in Figure 1D and show that indeed such perturbation occurs. In the same panel tumor necrosis factor (TNF)-α expression, previously described as a TTP family target (Carballo et al., 1997; Johnson and Blackwell, 2002), and β-actin were monitored as positive and negative controls.
Figure 1.
Overexpression of ZFP36L1 negatively regulates Stat5b and impairs erythroid differentiation of CD34+ HSCs. (A) RT-PCR showing the expression levels of ZFP36L1, Stat5b, and Jak2 in U937 cells transduced with empty vector (lane 1) or with ZFP36L1-overexpressing vector (lane 2); RNAs were extracted 7 d after transduction. (B) Real-time PCR performed on U937 cells transduced with ZFP36L1-overexpressing vector showing that overexpression of ZFP36L1 down-regulates the expression levels of Stat5b; CCL2 was used as a positive control. Data are provided in terms of Log2 of relative quantity compared with the expression levels of the same genes in U937 cells transduced with empty vector; error bars, SEM calculated on a set of three independent experiments (*p < 0.05). (C) Left, real-time PCR demonstrating the expression levels obtained in Hel cells via transduction with ZFP36L1-overexpressing vector compared with cells transduced with empty vector; error bar, SEM calculated on three independent experiments (**p < 0.01); RNAs were extracted 72 h after transduction. Right, Western blot analysis of Stat5b expression in Hel cells transduced with empty vector (lane 1) or with ZFP36L1-expressing vector (lane 2); cells were lysed 72 h after transduction. (D) Real-time PCR showing that Hel cells overexpressing ZFP36L1 (▩) have reduced levels of Stat5b and of the erythroid markers GATA1, LMO2, RhAG, and HbA1 compared with cells transduced with empty vector (□); TNF-α was used as a positive control, and ACTB was used as control housekeeping gene; error bars, SEM calculated on three independent experiments (*p < 0.05; **p < 0.01). RNAs were extracted 72 h after transduction. (E) Clonogenic assay performed on CD34+ HSCs transduced with empty vector (left) or overexpressing ZFP36L1 (right); error bars, SEM calculated on four independent experiments (*p < 0.05). (F) Real-time PCR showing that CD34+ HSCs overexpressing ZFP36L1 (■) have reduced levels of Stat5b and of the erythroid markers GATA1, LMO2, RhAG, HbA1 compared with cells transduced with empty vector (□); TNF-α was used as a positive control and ACTB as control housekeeping gene (SEM calculated on three experiments; *p < 0.05); RNAs were extracted 10 d after transduction.
Consequently to these preliminary experiments, the biological effect of ZFP36L1 overexpression was assessed in CD34+ human cord blood–derived stem/progenitor cells. Therefore, CD34+ cells were infected with a recombinant retrovirus allowing ZFP36L1 ectopic expression and then seeded in methylcellulose to evaluate a possible interference with differentiation. The results are shown in Figure 1E and show that, compared with the control (empty vector), ZFP36L1 ectopic expression determines a drastic decrease in the formation of erythroid colonies (CFU/BFU-E). In the same primary cell populations we monitored by QRT-PCR the expression of several erythroid differentiation markers (Figure 1F) and observed that expression of several among them (GATA1, LMO2, HbA1, and RhAG) decreases accordingly to the observed suppression of erythroid differentiation.
ZFP36L1 Directly Binds Stat5b 3′-UTR and Thereby Affects Its Stability.
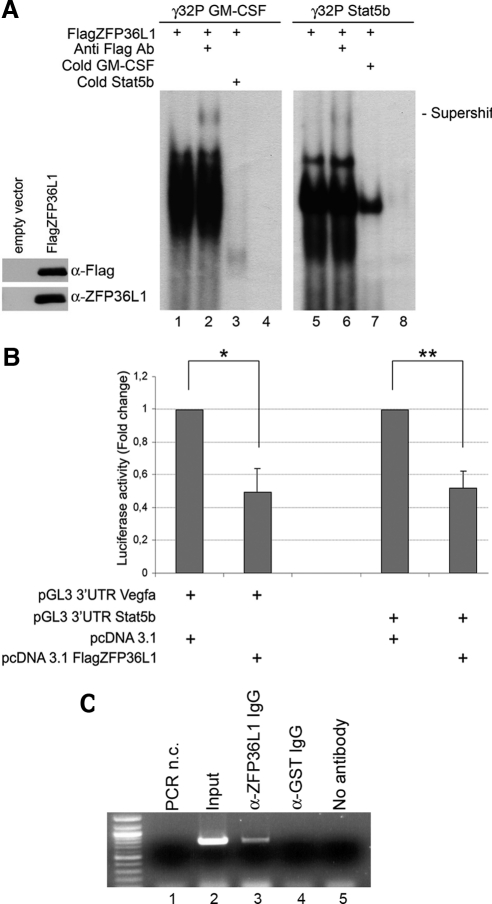
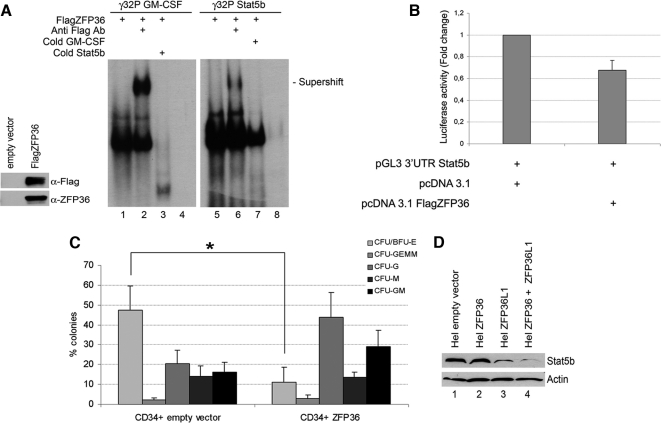
Figure 2A is a REMSA assay showing that an in vitro–translated, Flag-carrying ZFP36L1 protein is able to directly bind to radioactively labeled RNA probes corresponding to the putative AREII regions of Stat5b mRNA (lanes 5–8) and to the AREII regions belonging to GM-CSF mRNA described in literature (lanes 1–4; Carballo et al., 2000). The REMSA assay also shows Flag antibody dependent supershift formation (lanes 2 and 6) and cold probe competition (lanes 3 and 7) for both Stat5b and GM-CSF probes. Figure 2A also includes a Western blot performed on the in vitro–translated proteins used in the REMSA assay showing that both α-Flag and α-ZFP36L1 antibodies are capable of binding such proteins. The sequences of the probes are described in Materials and Methods.
Figure 2.
ZFP36L1 binding to AU-rich-elements (ARE) in the 3′UTR of Stat5b mRNA confers instability to mRNAs containing such elements. (A) Left, Western blot demonstrating identity and integrity of in vitro–translated FlagZFP36L1 protein: immunoblotting was performed with anti-Flag or anti-ZFP36L1 antibody as indicated. Right, RNA mobility shift assay performed by incubating in vitro–translated FlagZFP36L1 protein with labeled RNA probes corresponding to ARE in the 3′UTR of GM-CSF mRNA (used as a positive control, lanes 1–4) or to ARE in the 3′UTR of Stat5b mRNA (lanes 5–8). Probe sequences are: GM-CSF: 5′-UAUUUAUUUAUUUAUUUAUUUA-3′; Stat5b: 5′-AUAGUAAAUUAUUUAUUGGAAGAU-3′. Supershifts were obtained by an additional incubation with anti-Flag Ab (lanes 2 and 6). Competition experiments were performed with cold Stat5b probe (lane 3) or with cold GM-CSF probe (lane 7). Lanes 4 and 8 represent labeled probes incubated with an in vitro translation reaction mix performed on empty vector. (B) Luciferase activity assay performed in HEK293 cells transfected with pcDNA3.1 empty expression vector or with pcDNA3.1 overexpressing FlagZFP36L1 (20 ng) together with pGL3 reporter construct encoding for a luciferase gene fused to the 3′UTR of Vegfa (positive control) or to the 3′UTR of Stat5b. Luciferase activity is represented in terms of fold change; error bars, SEM calculated on a set of five independent experiments. (C) Ribonucleoprotein complexes immunoprecipitation assay demonstrating that binding of ZFP36L1 protein to the ARE in the 3′UTR of Stat5b mRNA occurs in vivo: ribonucleoprotein complexes were immunoprecipitated from lysates of HEK293 cells transfected with pcDNA3.1 ZFP36L1 and pGL3 3′UTR Stat5b vectors, RNA was extracted, reverse-transcribed, and amplified by PCR using primers specific for Stat5b 3′UTR fused to the luciferase gene. Lanes 1 and 2, negative and positive control respectively, i.e., PCR amplification performed on no template or on RNA extracted from cell lysates before immunoprecipitation; lane 3, specific immunoprecipitation obtained with anti-ZFP36L1 antibody; lanes 4 and 5, control immunoprecipitations performed with nonspecific antibody or with no antibody.
To complete the previous observations, data had to be gathered on the functional meaning of the bond between ZFP36L1 and Stat5b mRNA. Therefore, we performed a luciferase reporter assay, generating luciferase reporter constructs (pGL3 based) allowing transcription of a luciferase mRNA carrying the 3′-UTR of Stat5b or, as a control, the 3′-UTR of Vegfa, a target gene of TTP family (Ciais et al., 2004; Bell et al., 2006; Suswam et al., 2008). The results are shown in Figure 2B and show that coexpression of the reporter vectors encoding Stat5b or Vegfa 3′UTRs with a vector expressing ZFP36L1 significantly decreases the basal reporter activity, demonstrating that by directly binding the described mRNAs, ZFP36L1 impairs protein production by either promoting mRNA degradation or by inhibiting mRNA translation.
After characterizing the interaction between ZFP36L1 and Stat5b 3′UTR in vitro, next step was to demonstrate that this interaction could also occur in live cells. For this purpose, we transfected HEK293 cells with pcDNA3.1-ZFP36L1 and pGL3-Stat5b-3′UTR plasmids and subsequently subjected them to in vivo cross-linking. ZFP36L1-containing ribonucleoprotein complexes were then immunoprecipitated from cell lysates using a specific anti-ZFP36L1 antibody and the immunoprecipitates were subjected to RT–PCR amplification of the firefly luciferase mRNA. As shown in Figure 2 panel C, luciferase mRNA was specifically detected in anti-ZFP36L1 immunoprecipitates and in the input sample, whereas it was not detectable in the control immunoprecipitations (α-GST Ab, no antibody). These results demonstrate that the ZFP36L1–Stat5b 3′UTR interaction occurs not only in reconstituted in vitro systems but also in living cells.
Comparison between the Effects of Cinnamon Derived Polyphenols and ZFP36L1 Expression on Erythroid Differentiation.
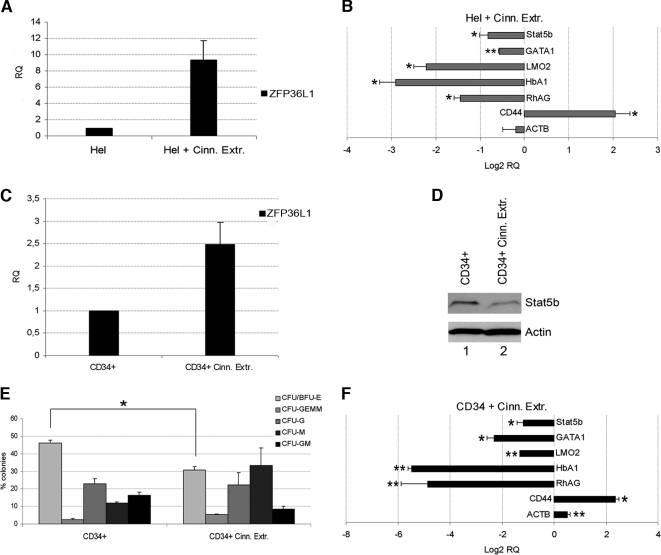
To further validate the data collected so far, the effects of ZFP36L1 ectopic expression were compared with the effects observed on Hel cells and on CD34+ cells after treatment with cinnamon derived polyphenols, that have been described in literature as potent inducers of the genes belonging to the TTP family (Schoene et al., 2005; Cao et al., 2007). The results are shown in Figure 3. Figure 3A shows by QRT-PCR the induction of ZFP36L1 obtained after administration of cinnamon extracts in Hel cells (24-h treatment with 0.5 μg/μl cinnamon extracts). Figure 3B shows the subsequent changes in the expression of Stat5b and other erythroid related genes, which are quite comparable to those observed after ZFP36L1 ectopic expression. Figure 3B also includes, as controls, the expression analysis of the housekeeping gene β-actin, whose expression does not change, and of CD44, a marker of monocyte differentiation whose expression increases after cinnamon extract administration. The up-regulation of CD44 is an event we didn't further investigate on, but could be related to the fact that ZFP family genes are highly expressed in monocytes (ZFP36L1 expression in monoblasts is shown in Figure 4A) and are induced by phorbol esters, that are capable of triggering monocyte differentiation. Figure 3C shows cinnamon extracts -driven ZFP36L1 induction in CD34+ cells 24 h after drug administration (0.25 μg/μl cinnamon extracts). Figure 3D is a Western blot of the same cells showing the decrease in Stat5b expression. In parallel to these experiments, CD34+ cells treated with cinnamon extracts were seeded in methylcellulose. Colony formation was monitored 2 wk after plating and, as well as in CD34+ cells ectopically expressing ZFP36L1, a statistically significant decrease in erythroid colony formation was observed (Figure 3E). Figure 3F depicts the QRT-PCR analysis of the expression of Stat5b and erythroid markers in CD34+ cells treated with cinnamon extracts. Again, the results are in accord with those observed during ZFP36L1 overexpression. Altogether, these experiments show that the biological behavior of cells ectopically expressing ZFP36L1 is essentially superimposable to that of the same cells treated with a drug capable of inducing the expression of the genes belonging to the same family.
Figure 3.
Treatment with cinnamon extracts up-regulates endogenous expression of ZFP36L1, down-regulates Stat5b levels, and inhibits erythroid differentiation of CD34+ HSCs. (A) Real-time PCR showing up-regulation of ZFP36L1 after treatment of Hel cells with 0.5 μg/μl cinnamon extracts for 24 h (SEM calculated on three experiments). (B) Real-time PCR performed on Hel cells treated with cinnamon extracts showing decreased expression of Stat5b and of the erythroid markers GATA1, LMO2, HbA1, and RhAG; CD44 monocyte differentiation marker expression is increased after treatment with cinnamon extracts; ACTB was used as control housekeeping gene. Results are provided in terms of Log2 of relative quantity compared with the expression levels of the same genes in untreated Hel cells; error bars, SEM calculated on a set of three independent experiments (*p < 0.05; **p < 0.01). (C) Real-time PCR showing up-regulation of ZFP36L1 after treatment of CD34+ HSCs in liquid culture with 0.25 μg/μl cinnamon extracts for 24 h (SEM calculated on three experiments). (D) Western blot analysis of Stat5b levels in untreated CD34+ HSCs (lane 1) and in the same cells treated with 0.25 μg/μl for 48 h (lane 2). (E) Clonogenic assay performed on CD34+ HSCs in the absence (left) on in the presence (right) of cinnamon extracts. Cinnamon extracts were added directly to methylcellulose medium at a concentration of 0.25 μg/μl (SEM calculated on three independent experiments; *p < 0.05). (F) Real-time PCR showing down-regulation of Stat5b and erythroid markers in CD34+ HSCs treated with 0.25 μg/μl cinnamon extracts in liquid culture medium for 48 h; CD44 expression is up-regulated after treatment with cinnamon extracts; ACTB was used as control housekeeping gene. Data are provided in terms of Log2 of relative quantity compared with the expression levels of the same genes in untreated CD34+ HSCs; error bars, SEM calculated on a set of three independent experiments (*p < 0.05; **p < 0.01).
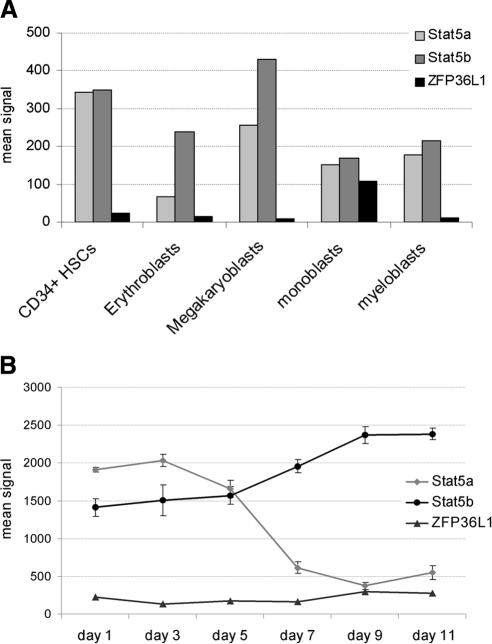
Figure 4.
Stat5a, Stat5b, and ZFP36L1 expression in hemopoietic progenitors/precursors and during erythroid differentiation. (A) Microarray analysis results showing Stat5a, Stat5b, and ZFP36L1 expression levels in primary hemopoietic cell contexts: normal cord blood CD34+ HSCs, CD34-derived erythroblasts, megakaryoblasts, monoblasts, and myeloblasts. (B) Microarray analysis showing the expression levels of Stat5a, Stat5b, and ZFP36L1 during in vitro erythroid differentiation of adult CD34+ hematopoietic progenitor cells at various time points in serum-free medium containing erythropoietin, IL-3, and stem cell factor (data from GEO Profiles database: GDS2431).
Stat5b Gene Silencing Is Sufficient to Impair Erythroid Differentiation of Human CD34+ Cord Blood–derived Stem/Progenitor Cells.
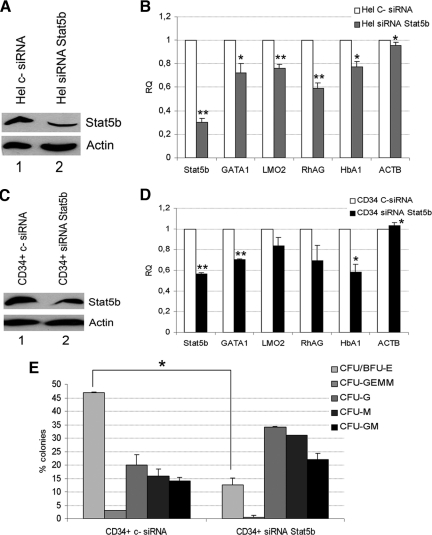
Because in several contexts Stat5a and Stat5b act redundantly, we evaluated the expression levels of the two genes in the different human hematopoietic cell contexts. Figure 4A is an extract of a microarray analysis previously performed in our laboratory showing the expression levels of Stat5a, Stat5b, and ZFP36L1 in human CD34+ stem/progenitor cells and in the various myeloid precursors. Figure 4A shows that Stat5a and b are expressed at comparable levels in all the contexts analyzed with the exception of erythroblasts, where Stat5a is scarcely represented compared with Stat5b, and also shows that ZFP36L1 is scarcely expressed among the different lineages with the exception of monoblasts. To consolidate this observation, we monitored the expression of Stat5a, Stat5b, and ZFP36L1 in a gene expression profile deriving from adult CD34+ hematopoietic progenitor cells differentiating toward the erythroid lineage at various time points up to 11 d, published on the GEO-Profile database (GDS2431). The results are shown in Figure 4B and show that during maturation of human CD34+ progenitors toward the erythroid lineage a down-regulation of Stat5a occurs, whereas high levels of Stat5b are maintained throughout the process. Again, ZFP36L1 remains absent along the erythroid differentiation process. On these grounds, we hypothesized that erythroid maturation of human CD34+ stem/progenitor cells is sustained mainly by Stat5b, rather than Stat5a. Consequently, in order to evaluate whether the decrease of erythroid differentiation that follows ectopic expression of ZFP36L1 relies specifically on the down-regulation of Stat5b, we performed gene silencing experiments targeting Stat5b in Hel and CD34+ cells. The results of such experiment are described in Figure 5. Figure 5, A and C, are Western blots showing that Stat5b expression decreases after siRNA treatment both in Hel and CD34+ cells. Figure 5, B and D, show by QRT-PCR the decrease of the expression of erythroid markers and the expression of β-actin as a control in Hel and CD34+ cells, respectively, after Stat5b gene silencing. Figure 5E shows the results of a colony assay performed on CD34+ cells treated with siRNA oligonucleotides targeting Stat5b. The assay shows a strong decrease in the formation of erythroid colonies (CFU/BFU-E) in the cell population where Stat5b has been down-regulated. This result in particular, mimics the effect of ZFP36L1 overexpression in the same cell population.
Figure 5.
Stat5b silencing is sufficient to down-regulate the expression of erythroid markers and to impair erythroid differentiation of CD34+ HSCs. (A) Western blot analysis showing the down-regulation of Stat5b obtained in Hel cells transfected with anti-Stat5b siRNAs (lane 2) compared with cells transfected with a nontargeting control siRNA (lane 1); cells were lysed 48 h after transfection. (B) Real-time PCR performed on Hel cells transfected with control siRNA (□) or with anti-Stat5b siRNAs (▩) demonstrating the efficacy of Stat5b silencing and the down-regulation of erythroid markers; ACTB was used as control housekeeping gene (error bars, SEM calculated on a set of three experiments; *p < 0.05; **p < 0.01). RNAs were extracted 48 h after transfection with siRNAs. (C) Western blot analysis showing the down-regulation of Stat5b obtained in CD34+ HSCs transfected with anti-Stat5b siRNAs (lane 2) compared with cells transfected with a nontargeting control siRNA (lane 1); cells were lysed 7 d after transfection. (D) Real-time PCR performed on CD34+ HSCs transfected with control siRNA (□) or with anti-Stat5b siRNAs (■) demonstrating the efficacy of Stat5b silencing and the down-regulation of erythroid markers; ACTB was used as control housekeeping gene (error bars, SEM calculated on a set of three experiments; *p < 0.05; **p < 0.01). RNAs were extracted 96 h after transfection with siRNAs. (E) Clonogenic assay performed on CD34+ HSCs transfected with a nontargeting control siRNA (left) or with anti-Stat5b siRNAs (right); SEM calculated on three independent experiments; *p < 0.05.
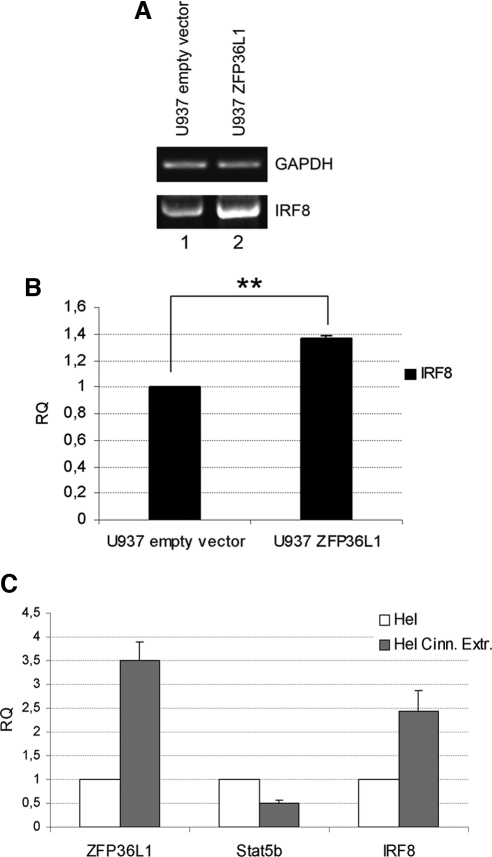
Transcription Factor IRF8 Is Up-Regulated after ZFP36L1 Ectopic Expression.
Because published data suggest that Stat5 is able to inhibit dendritic development by directly suppressing transcription factor IRF8 (Esashi et al., 2008), we evaluated, to further validate our data, whether ZFP36L1-mediated Stat5b down-regulation indirectly determines a change in the expression of transcription factor IRF8. The results of such analysis are shown in Figure 6. Figure 6A depicts an RT-PCR showing that indeed IRF8 mRNA increases in U937 cells after ZFP36L1 ectopic expression. Figure 6B shows the same data analyzed by QRT-PCR, whereas panel C exhibits the changes in IRF8 expression in Hel cells after treatment with cinnamon extracts. As in the experiments described in Figure 3, cinnamon extracts induce ZFP36L1 expression; Figure 6C shows that an increase in IRF8 expression is associated with Stat5b down-regulation. Because the increase of IRF8 expression is observed both after ZFP36L1 ectopic expression and cinnamon extracts administration, it is plausible that, in both cases, this event depends on ZFP36L1-mediated Stat5b down-regulation.
Figure 6.
Increased levels of ZFP36L1 result in up-regulation of IRF8 gene. (A) RT-PCR showing increased expression of IRF8 in U937 cells transduced with ZFP36L1-overexpressing vector (lane 2) compared with cells transduced with empty vector (lane 1); RNAs were extracted 7 d after transfection. (B) Real-time PCR showing IRF8 up-regulation after transduction of U937 cells with ZFP36L1-expressing vector (right); data are provided in terms of relative quantity compared with cells transduced with empty vector (left); error bars, SEM calculated on a set of four independent experiments; **p < 0.01. (C) Real-time PCR showing that treatment of Hel cells with 0.5 μg/μl cinnamon extracts for 48 h results in up-regulation of ZFP36L1, down-regulation of Stat5b, and up-regulation of IRF8 (SEM calculated on three different experiments).
ZFP36 (TTP) Behaves Comparably to ZFP36L1 and Their Effect on Stat5b Expression is Cumulative.
Because ZFP36 and ZFP36L1 share several targets, we evaluated if ZFP36 also is capable of down-regulating Stat5b and interfering with hematopoietic differentiation. Figure 7A is a REMSA assay showing that ZFP36 is able to bind the same AREII regions of GM-CSF and Stat5b mRNA described in Figure 2A as targets of ZFP36L1. Figure 7A also includes a Western blot showing that both α-Flag and α-ZFP36 antibodies are capable of binding in vitro–translated ZFP36, suggesting that such protein is folded correctly. Figure 7B is a luciferase reporter assay performed by cotransfecting a plasmid expressing ZFP36 and a reporter vector encoding a luciferase mRNA carrying Stat5b 3′-UTR. The results show that ZFP36, as well as ZFP36L1, determines a decrease of luciferase activity by binding Stat5b 3′-UTR. Next we assessed whether ZFP36 is also able to interfere with erythroid differentiation. Therefore, ZFP36 was ectopically expressed in CD34+ human stem/progenitor cells that successively underwent a methylcellulose colony assay. The results are described in Figure 7C and show that, compared with the control population, after 2 wk a decrease of erythroid colonies (CFU/BFU-E) is visible, and it is super imposable to that observed in CD34+ cells overexpressing ZFP36L1. On these grounds, the hypothesis that the two proteins could act redundantly was tested. To do so, Hel cells were cotransfected with expression plasmids encoding ZFP36 and ZFP36L1, and the effect on Stat5b expression was monitored by Western blot. The results are shown in Figure 7D and demonstrate that although Stat5b expression decreases after transfection of one or the other mRNA binding protein (lanes 2 and 3), their cotransfection (lane 4) is capable of completely abrogating it. To assess whether the effect on Stat5b depends on a synergistic or redundant behavior, we performed another REMSA that is included in the Supplementary Material, showing that ZFP36 and ZFP36L1 bind to Stat5b 3′UTR with equal affinity. Such observation, together with published data demonstrating that ZFP36 and ZFP36L1 share the domains that recruit deadenylases and therefore trigger mRNA decay (Lykke-Andersen and Wagner, 2005), suggests that the two proteins are redundant when coexpressed, rather than synergic.
Figure 7.
ZFP36 behaves similarly to ZFP36L1 in binding to and destabilizing mRNAs spanning Stat5b 3′UTR and in inhibiting erythroid differentiation of CD34+ HSCs. (A) Left, Western blot demonstrating identity and integrity of in vitro–translated FlagZFP36 protein: immunoblotting was performed with anti-Flag or anti-ZFP36 antibody as indicated. Right, RNA mobility shift assay performed by incubating in vitro–translated FlagZFP36 protein with labeled RNA probes spanning the ARE in the 3′UTR of GM-CSF mRNA (used as a positive control, lanes 1–4) or the ARE in the 3′UTR of Stat5b mRNA (lanes 5–8). Supershifts were obtained by an additional incubation with anti-Flag Ab (lanes 2 and 6). Competition experiments were performed with cold Stat5b probe (lane 3) or with cold GM-CSF probe (lane 7). Lanes 4 and 8 represent labeled probes incubated with an in vitro translation reaction mix performed on empty vector. (B) Luciferase activity assay performed in HEK293 cells transfected with pcDNA3.1 empty expression vector (left) or with pcDNA3.1 overexpressing FlagZFP36 (10 ng; right) together with pGL3 reporter construct encoding for a luciferase gene fused to the 3′UTR of Stat5b. Luciferase activity is represented in terms of fold change; error bars, SEM calculated on a set of five independent experiments. (C) Clonogenic assay performed on CD34+ HSCs transduced with empty vector (left) or overexpressing ZFP36 (right); error bars, SEM calculated on four independent experiments (*p < 0.05). (D) Western blot analysis showing Stat5b levels in Hel cells transfected with empty vector (lane 1), with ZFP36-overexpressing vector (lane 2), with ZFP36L1-overexpressing vector (lane 3), or transfected with both overexpressing vectors (lane 4). Cells were lysed 72 h after transfection.
DISCUSSION
Human hematopoietic differentiation is a strictly controlled process in which regulatory events take place at all the levels of gene expression. Starting from chromatin conformation moving downstream toward the role of transcription factors and microRNAs, a wide amount of data has been gathered so far. With the current study we suggest a novel mechanism to control hematopoietic differentiation based on the activity of mRNA-binding proteins whose role, to date, was mostly related to the control of the inflammatory response (Sandler and Stoecklin, 2008; Schaljo et al., 2009). In our hypothesis, such proteins act by repressing the development of a specific blood lineage, namely the erythroid, rather than by promoting the differentiation of specific progenitors and precursors as classically described in many studies that focus on the role of transcription factors.
Starting from the analysis of a gene expression database showing that ZFP36L1 is differentially expressed in the different human blood cell contexts and, particularly, that its expression is consistently low in the erythroid compared with the other lineages (data not shown), we hypothesized that this protein and other members of the same family could play a role during hematopoiesis. By this study we demonstrated that ZFP36L1 and ZFP36 inhibit erythroid development of CD34+ human cord blood–derived stem/progenitor cells by inhibiting proliferation of erythroid precursors and suggest that this behavior depends on ZFP36L1-mediated Stat5b mRNA degradation. Moreover, we demonstrated that ZFP36L1 and ZFP36 share Stat5b as a common target and are able to directly bind its mRNA on a canonical AREII sequence located in the 3′-UTR. The inhibition of erythroid colonies formation observed after ZFP36L1 and ZFP36 ectopic expression is statistically significant, and it is coupled to a decrease of precursors' proliferation rather than to apoptosis or differentiation arrest (see Supplementary Material). To associate such observation with the sole down-regulation of Stat5b was not an obvious step, because this gene is highly homologous to Stat5a, and in murine erythropoiesis the proteins Stat5a and Stat5b are capable to some degree of substituting each other (Socolovsky et al., 2001). Therefore, we monitored the expression of Stat5a and Stat5b in human myeloid cell contexts, at different time points during human erythroid differentiation and then performed gene-silencing experiments targeting Stat5b. The expression analysis revealed that, unlike in murine erythropoiesis, during erythroid differentiation of human CD34+ stem/progenitor cells, a down-regulation of Stat5a expression occurs, whereas Stat5b expression persists at high levels (Figure 4). Accordingly, the gene silencing approach showed that Stat5b down-regulation is sufficient to impair erythroid maturation. It remains to be elucidated whether the different expression pattern of Stat5a and Stat5b in human erythropoiesis depends on transcriptional or posttranscriptional events. The presented results show that the idea underneath is correct, although a more complete hypothesis that rises from the present study suggests that ZFP36L1, ZFP36, and possibly other members of the same family could have among their targets other members of the Stat5b/JAK2 signal transduction pathway that have not been described in the experiments presented here.
The relation between ZFP36L1 and Stat5b is also indirectly described in the experiment (Figure 6) showing that ZFP36L1 ectopic expression results in the increase of transcription factor IRF8. Such a gene is involved at different levels in hematopoietic differentiation as a promoter of specific myeloid and lymphoid lineages (Qi et al., 2009; Saberwal et al., 2009; Wang and Morse, 2009) but not the erythroid one, it has been described for being negatively regulated by Stat5 (Esashi et al., 2008) and is capable of acting as a proapoptotic factor (Yang et al., 2007, 2009). The ability of ZFP36L1 to down-regulate Stat5b, thereby promoting the expression of the proapoptotic factor IRF8, suggests that this gene family encodes proteins that in specific contexts are able to function as tumor suppressors. Such an observation is strengthened by a recently published report that shows that TTP family members are down-regulated in different tumors (Brennan et al., 2009).
Another relevant aspect of the present work resides in the fact that drugs capable of inducing the expression of TTP family genes, such as cinnamon-derived polyphenols, do in fact down-regulate Stat5b expression and exhibit a very strong influence on erythroid maturation of human CD34+ stem/progenitor cells. The effect of cinnamon polyphenols is observed even at low concentrations of drug (0.25 μg/μl), and this efficiency could depend on the fact that they induce expression of both ZFP36 and ZFP36L1 that, as demonstrated by the experiment shown in Figure 7D, share the ability to bind Stat5b mRNA and therefore cooperate in promoting its degradation.
The present study establishes a relation between the TTP family of genes and the Stat5b/JAK2 pathway. Alterations in the genes belonging to such pathway, and precisely the JAK2 V617F mutation that leads to constitutive signal transduction, underlie chronic myeloproliferative disorders (Baxter et al., 2005; Kralovics et al., 2005; Levine and Gilliland, 2008), such as polycythemia vera (PV), essential thrombocythemia (ET), and primary myelofibrosis (PMF). Today there is considerable optimism that these diseases can be successfully treated with tyrosine kinase inhibitors. In fact specific inhibitors of JAK2 kinase activity have been designed (Gozgit et al., 2008; Hexner et al., 2008; Wernig et al., 2008) and have entered the clinic in Phase I/II trials in PMF and post-PV/ET myelofibrosis. Given the central role of JAK2 signaling to a myriad of cellular processes, there may be significant toxicities associated with JAK2 inhibition, and “off-target” inhibition of JAK1, JAK3, or TYK2 might lead to hematologic, immunologic, and endocrine side effects. Such toxicities could likely preclude their prolonged use; consequently, it is possible that alternative or synergic therapies will have to be developed. On these premises and on the ground of the results presented in this work, we hypothesize an approach for PV, characterized by an increase in absolute quantity of erythroid cells, based on the utilization of drugs that induce TTP gene family expression and that do not present the toxicity problems suggested above, determined by JAK2 inhibitors. TTP-inducing drugs would determine Stat5b down-regulation and would thereby allow decreasing the activity of JAK2-Stat5 pathway without interfering with the activity of other necessary, unmutated tyrosine kinases such as JAK1, JAK3, or TYK2 and others. Such an approach would not likely replace JAK2 inhibitors, but a combined strategy could possibly be useful to lower the dosage of tyrosine kinase inhibitors and therefore limit their toxicity.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Dr. Richard A. Anderson for kindly providing cinnamon extracts and the cinnamon-derived polyphenols used in this study.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-01-0040) on August 11, 2010.
REFERENCES
- Ambrosio R., Fimiani G., Monfregola J., Sanzari E., De Felice N., Salerno M. C., Pignata C., D'Urso M., Ursini M. V. The structure of human STAT5A and B genes reveals two regions of nearly identical sequence and an alternative tissue specific STAT5B promoter. Gene. 2002;285:311–318. doi: 10.1016/s0378-1119(02)00421-3. [DOI] [PubMed] [Google Scholar]
- Basham B., Sathe M., Grein J., McClanahan T., D'Andrea A., Lees E., Rascle A. In vivo identification of novel STAT5 target genes. Nucleic Acids Res. 2008;36:3802–3818. doi: 10.1093/nar/gkn271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter E. J., et al. Acquired mutation of the tyrosine kinase JAK2 in human myeloproliferative disorders. Lancet. 2005;365:1054–1061. doi: 10.1016/S0140-6736(05)71142-9. [DOI] [PubMed] [Google Scholar]
- Bell S. E., Sanchez M. J., Spasic-Boskovic O., Santalucia T., Gambardella L., Burton G. J., Murphy J. J., Norton J. D., Clark A. R., Turner M. The RNA binding protein Zfp36l1 is required for normal vascularisation and post-transcriptionally regulates VEGF expression. Dev. Dyn. 2006;235:3144–3155. doi: 10.1002/dvdy.20949. [DOI] [PubMed] [Google Scholar]
- Benekli M., Baer M. R., Baumann H., Wetzler M. Signal transducer and activator of transcription proteins in leukemias. Blood. 2003;101:2940–2954. doi: 10.1182/blood-2002-04-1204. [DOI] [PubMed] [Google Scholar]
- Brennan S. E., Kuwano Y., Alkharouf N., Blackshear P. J., Gorospe M., Wilson G. M. The mRNA-destabilizing protein tristetraprolin is suppressed in many cancers, altering tumorigenic phenotypes and patient prognosis. Cancer Res. 2009;69:5168–5176. doi: 10.1158/0008-5472.CAN-08-4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H., Polansky M. M., Anderson R. A. Cinnamon extract and polyphenols affect the expression of tristetraprolin, insulin receptor, and glucose transporter 4 in mouse 3T3–L1 adipocytes. Arch. Biochem. Biophys. 2007;459:214–222. doi: 10.1016/j.abb.2006.12.034. [DOI] [PubMed] [Google Scholar]
- Carballo E., Cao H., Lai W. S., Kennington E. A., Campbell D., Blackshear P. J. Decreased sensitivity of tristetraprolin-deficient cells to p38 inhibitors suggests the involvement of tristetraprolin in the p38 signaling pathway. J. Biol. Chem. 2001;276:42580–42587. doi: 10.1074/jbc.M104953200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carballo E., Gilkeson G. S., Blackshear P. J. Bone marrow transplantation reproduces the tristetraprolin-deficiency syndrome in recombination activating gene-2 (−/−) mice. Evidence that monocyte/macrophage progenitors may be responsible for TNFalpha overproduction. J. Clin. Invest. 1997;100:986–995. doi: 10.1172/JCI119649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carballo E., Lai W. S., Blackshear P. J. Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science. 1998;281:1001–1005. doi: 10.1126/science.281.5379.1001. [DOI] [PubMed] [Google Scholar]
- Carballo E., Lai W. S., Blackshear P. J. Evidence that tristetraprolin is a physiological regulator of granulocyte-macrophage colony-stimulating factor messenger RNA deadenylation and stability. Blood. 2000;95:1891–1899. [PubMed] [Google Scholar]
- Choudhary C., Brandts C., Schwable J., Tickenbrock L., Sargin B., Ueker A., Bohmer F. D., Berdel W. E., Muller-Tidow C., Serve H. Activation mechanisms of STAT5 by oncogenic Flt3-ITD. Blood. 2007;110:370–374. doi: 10.1182/blood-2006-05-024018. [DOI] [PubMed] [Google Scholar]
- Ciais D., Cherradi N., Bailly S., Grenier E., Berra E., Pouyssegur J., Lamarre J., Feige J. J. Destabilization of vascular endothelial growth factor mRNA by the zinc-finger protein TIS11b. Oncogene. 2004;23:8673–8680. doi: 10.1038/sj.onc.1207939. [DOI] [PubMed] [Google Scholar]
- Corps A. N., Brown K. D. Insulin and insulin-like growth factor I stimulate expression of the primary response gene cMG1/TIS11b by a wortmannin-sensitive pathway in RIE-1 cells. FEBS Lett. 1995;368:160–164. doi: 10.1016/0014-5793(95)00635-m. [DOI] [PubMed] [Google Scholar]
- Crispi S., Sanzari E., Monfregola J., De Felice N., Fimiani G., Ambrosio R., D'Urso M., Ursini M. V. Characterization of the human STAT5A and STAT5B promoters: evidence of a positive and negative mechanism of transcriptional regulation. FEBS Lett. 2004;562:27–34. doi: 10.1016/S0014-5793(04)00166-8. [DOI] [PubMed] [Google Scholar]
- Emmons J., Townley-Tilson W. H., Deleault K. M., Skinner S. J., Gross R. H., Whitfield M. L., Brooks S. A. Identification of TTP mRNA targets in human dendritic cells reveals TTP as a critical regulator of dendritic cell maturation. RNA. 2008;14:888–902. doi: 10.1261/rna.748408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esashi E., Wang Y. H., Perng O., Qin X. F., Liu Y. J., Watowich S. S. The signal transducer STAT5 inhibits plasmacytoid dendritic cell development by suppressing transcription factor IRF8. Immunity. 2008;28:509–520. doi: 10.1016/j.immuni.2008.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funakoshi-Tago M., Tago K., Sumi K., Abe M., Aizu-Yokota E., Oshio T., Sonoda Y., Kasahara T. The acute lymphoblastic leukemia-associated JAK2 L611S mutant induces tumorigenesis in nude mice. J. Biol. Chem. 2009;284:12680–12690. doi: 10.1074/jbc.M808879200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemelli C., et al. The vitamin D3/Hox-A10 pathway supports MafB function during the monocyte differentiation of human CD34+ hemopoietic progenitors. J. Immunol. 2008;181:5660–5672. doi: 10.4049/jimmunol.181.8.5660. [DOI] [PubMed] [Google Scholar]
- Gomperts M., Corps A. N., Pascall J. C., Brown K. D. Mitogen-induced expression of the primary response gene cMG1 in a rat intestinal epithelial cell-line (RIE-1) FEBS Lett. 1992;306:1–4. doi: 10.1016/0014-5793(92)80825-2. [DOI] [PubMed] [Google Scholar]
- Gozgit J. M., et al. Effects of the JAK2 inhibitor, AZ960, on Pim/BAD/BCL-xL survival signaling in the human JAK2 V617F cell line SET-2. J. Biol. Chem. 2008;283:32334–32343. doi: 10.1074/jbc.M803813200. [DOI] [PubMed] [Google Scholar]
- Grebien F., et al. Stat5 activation enables erythropoiesis in the absence of EpoR and Jak2. Blood. 2008;111:4511–4522. doi: 10.1182/blood-2007-07-102848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimwade L. F., et al. Phospho-STAT5 and phospho-Akt expression in chronic myeloproliferative neoplasms. Br. J. Haematol. 2009;147:495–506. doi: 10.1111/j.1365-2141.2009.07870.x. [DOI] [PubMed] [Google Scholar]
- Guglielmelli P., et al. Molecular profiling of CD34+ cells in idiopathic myelofibrosis identifies a set of disease-associated genes and reveals the clinical significance of Wilms' tumor gene 1 (WT1) Stem Cells. 2007;25:165–173. doi: 10.1634/stemcells.2006-0351. [DOI] [PubMed] [Google Scholar]
- Heller P. G., Lev P. R., Salim J. P., Kornblihtt L. I., Goette N. P., Chazarreta C. D., Glembotsky A. C., Vassallu P. S., Marta R. F., Molinas F. C. JAK2V617F mutation in platelets from essential thrombocythemia patients: correlation with clinical features and analysis of STAT5 phosphorylation status. Eur. J. Haematol. 2006;77:210–216. doi: 10.1111/j.1600-0609.2006.00688.x. [DOI] [PubMed] [Google Scholar]
- Hexner E. O., et al. Lestaurtinib (CEP701) is a JAK2 inhibitor that suppresses JAK2/STAT5 signaling and the proliferation of primary erythroid cells from patients with myeloproliferative disorders. Blood. 2008;111:5663–5671. doi: 10.1182/blood-2007-04-083402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson B. A., Blackwell T. K. Multiple tristetraprolin sequence domains required to induce apoptosis and modulate responses to TNFalpha through distinct pathways. Oncogene. 2002;21:4237–4246. doi: 10.1038/sj.onc.1205526. [DOI] [PubMed] [Google Scholar]
- Johnson B. A., Geha M., Blackwell T. K. Similar but distinct effects of the tristetraprolin/TIS11 immediate-early proteins on cell survival. Oncogene. 2000;19:1657–1664. doi: 10.1038/sj.onc.1203474. [DOI] [PubMed] [Google Scholar]
- Kotecha N., et al. Single-cell profiling identifies aberrant STAT5 activation in myeloid malignancies with specific clinical and biologic correlates. Cancer Cell. 2008;14:335–343. doi: 10.1016/j.ccr.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kralovics R., Passamonti F., Buser A. S., Teo S. S., Tiedt R., Passweg J. R., Tichelli A., Cazzola M., Skoda R. C. A gain-of-function mutation of JAK2 in myeloproliferative disorders. N. Engl. J. Med. 2005;352:1779–1790. doi: 10.1056/NEJMoa051113. [DOI] [PubMed] [Google Scholar]
- Lai W. S., Carballo E., Strum J. R., Kennington E. A., Phillips R. S., Blackshear P. J. Evidence that tristetraprolin binds to AU-rich elements and promotes the deadenylation and destabilization of tumor necrosis factor alpha mRNA. Mol. Cell. Biol. 1999;19:4311–4323. doi: 10.1128/mcb.19.6.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T. K., Man K., Poon R. T., Lo C. M., Yuen A. P., Ng I. O., Ng K. T., Leonard W., Fan S. T. Signal transducers and activators of transcription 5b activation enhances hepatocellular carcinoma aggressiveness through induction of epithelial-mesenchymal transition. Cancer Res. 2006;66:9948–9956. doi: 10.1158/0008-5472.CAN-06-1092. [DOI] [PubMed] [Google Scholar]
- Levine R. L., Gilliland D. G. Myeloproliferative disorders. Blood. 2008;112:2190–2198. doi: 10.1182/blood-2008-03-077966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen J., Wagner E. Recruitment and activation of mRNA decay enzymes by two ARE-mediated decay activation domains in the proteins TTP and BRF-1. Genes Dev. 2005;19((3)):351–361. doi: 10.1101/gad.1282305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahtani K. R., Brook M., Dean J. L., Sully G., Saklatvala J., Clark A. R. Mitogen-activated protein kinase p38 controls the expression and posttranslational modification of tristetraprolin, a regulator of tumor necrosis factor alpha mRNA stability. Mol. Cell. Biol. 2001;21:6461–6469. doi: 10.1128/MCB.21.9.6461-6469.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montanari M., et al. Correlation between differentiation plasticity and mRNA expression profiling of CD34+-derived CD14- and CD14+ human normal myeloid precursors. Cell Death Differ. 2005;12:1588–1600. doi: 10.1038/sj.cdd.4401679. [DOI] [PubMed] [Google Scholar]
- Moucadel V., Constantinescu S. N. Differential STAT5 signaling by ligand-dependent and constitutively active cytokine receptors. J. Biol. Chem. 2005;280:13364–13373. doi: 10.1074/jbc.M407326200. [DOI] [PubMed] [Google Scholar]
- Murata T., Hikita K., Kaneda N. Transcriptional activation function of zinc finger protein TIS11 and its negative regulation by phorbol ester. Biochem. Biophys. Res. Commun. 2000;274:526–532. doi: 10.1006/bbrc.2000.3182. [DOI] [PubMed] [Google Scholar]
- Murata T., Yoshino Y., Morita N., Kaneda N. Identification of nuclear import and export signals within the structure of the zinc finger protein TIS11. Biochem. Biophys. Res. Commun. 2002;293:1242–1247. doi: 10.1016/S0006-291X(02)00363-7. [DOI] [PubMed] [Google Scholar]
- Niranjanakumari S., Lasda E., Brazas R., Garcia-Blanco M. Reversible cross-linking combined with immunoprecipitation to study RNA-protein interactions in vivo. Methods. 2002;26((2)):182–190. doi: 10.1016/S1046-2023(02)00021-X. [DOI] [PubMed] [Google Scholar]
- Ogilvie R. L., Abelson M., Hau H. H., Vlasova I., Blackshear P. J., Bohjanen P. R. Tristetraprolin down-regulates IL-2 gene expression through AU-rich element-mediated mRNA decay. J. Immunol. 2005;174:953–961. doi: 10.4049/jimmunol.174.2.953. [DOI] [PubMed] [Google Scholar]
- Olthof S. G., Fatrai S., Drayer A. L., Tyl M. R., Vellenga E., Schuringa J. J. Downregulation of signal transducer and activator of transcription 5 (STAT5) in CD34+ cells promotes megakaryocytic development, whereas activation of STAT5 drives erythropoiesis. Stem Cells. 2008;26:1732–1742. doi: 10.1634/stemcells.2007-0899. [DOI] [PubMed] [Google Scholar]
- Piacibello W., Sanavio F., Garetto L., Severino A., Dane A., Gammaitoni L., Aglietta M. Differential growth factor requirement of primitive cord blood hematopoietic stem cell for self-renewal and amplification vs proliferation and differentiation. Leukemia. 1998;12:718–727. doi: 10.1038/sj.leu.2401003. [DOI] [PubMed] [Google Scholar]
- Qi C. F., Li Z., Raffeld M., Wang H., Kovalchuk A. L., Morse H. C., 3rd Differential expression of IRF8 in subsets of macrophages and dendritic cells and effects of IRF8 deficiency on splenic B cell and macrophage compartments. Immunol. Res. 2009;45:62–74. doi: 10.1007/s12026-008-8032-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saberwal G., Horvath E., Hu L., Zhu C., Hjort E., Eklund E. A. The interferon consensus sequence binding protein (ICSBP/IRF8) activates transcription of the FANCF gene during myeloid differentiation. J. Biol. Chem. 2009;284:33242–33254. doi: 10.1074/jbc.M109.010231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandler H., Stoecklin G. Control of mRNA decay by phosphorylation of tristetraprolin. Biochem. Soc. Trans. 2008;36:491–496. doi: 10.1042/BST0360491. [DOI] [PubMed] [Google Scholar]
- Sauer I., Schaljo B., Vogl C., Gattermeier I., Kolbe T., Muller M., Blackshear P. J., Kovarik P. Interferons limit inflammatory responses by induction of tristetraprolin. Blood. 2006;107:4790–4797. doi: 10.1182/blood-2005-07-3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaljo B., et al. Tristetraprolin is required for full anti-inflammatory response of murine macrophages to IL-10. J. Immunol. 2009;183:1197–1206. doi: 10.4049/jimmunol.0803883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoene N. W., Kelly M. A., Polansky M. M., Anderson R. A. Water-soluble polymeric polyphenols from cinnamon inhibit proliferation and alter cell cycle distribution patterns of hematologic tumor cell lines. Cancer Lett. 2005;230:134–140. doi: 10.1016/j.canlet.2004.12.039. [DOI] [PubMed] [Google Scholar]
- Socolovsky M., Nam H., Fleming M. D., Haase V. H., Brugnara C., Lodish H. F. Ineffective erythropoiesis in Stat5a(−/−)5b(−/−) mice due to decreased survival of early erythroblasts. Blood. 2001;98:3261–3273. doi: 10.1182/blood.v98.12.3261. [DOI] [PubMed] [Google Scholar]
- Stumpo D. J., et al. Targeted disruption of Zfp36l2, encoding a CCCH tandem zinc finger RNA-binding protein, results in defective hematopoiesis. Blood. 2009;114:2401–2410. doi: 10.1182/blood-2009-04-214619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suswam E., Li Y., Zhang X., Gillespie G. Y., Li X., Shacka J. J., Lu L., Zheng L., King P. H. Tristetraprolin down-regulates interleukin-8 and vascular endothelial growth factor in malignant glioma cells. Cancer Res. 2008;68:674–682. doi: 10.1158/0008-5472.CAN-07-2751. [DOI] [PubMed] [Google Scholar]
- Taylor G. A., Thompson M. J., Lai W. S., Blackshear P. J. Phosphorylation of tristetraprolin, a potential zinc finger transcription factor, by mitogen stimulation in intact cells and by mitogen-activated protein kinase in vitro. J. Biol. Chem. 1995;270:13341–13347. doi: 10.1074/jbc.270.22.13341. [DOI] [PubMed] [Google Scholar]
- Taylor G. A., Thompson M. J., Lai W. S., Blackshear P. J. Mitogens stimulate the rapid nuclear to cytosolic translocation of tristetraprolin, a potential zinc-finger transcription factor. Mol. Endocrinol. 1996;10:140–146. doi: 10.1210/mend.10.2.8825554. [DOI] [PubMed] [Google Scholar]
- Wagner K. U., Rui H. Jak2/Stat5 signaling in mammogenesis, breast cancer initiation and progression. J. Mammary Gland Biol. Neoplasia. 2008;13:93–103. doi: 10.1007/s10911-008-9062-z. [DOI] [PubMed] [Google Scholar]
- Wang H., Morse H. C., 3rd IRF8 regulates myeloid and B lymphoid lineage diversification. Immunol. Res. 2009;43:109–117. doi: 10.1007/s12026-008-8055-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernig G., et al. Efficacy of TG101348, a selective JAK2 inhibitor, in treatment of a murine model of JAK2V617F-induced polycythemia vera. Cancer Cell. 2008;13:311–320. doi: 10.1016/j.ccr.2008.02.009. [DOI] [PubMed] [Google Scholar]
- Yang D., Thangaraju M., Browning D. D., Dong Z., Korchin B., Lev D. C., Ganapathy V., Liu K. IFN regulatory factor 8 mediates apoptosis in nonhemopoietic tumor cells via regulation of Fas expression. J. Immunol. 2007;179:4775–4782. doi: 10.4049/jimmunol.179.7.4775. [DOI] [PubMed] [Google Scholar]
- Yang D., et al. IFN regulatory factor 8 sensitizes soft tissue sarcoma cells to death receptor-initiated apoptosis via repression of FLICE-like protein expression. Cancer research. 2009;69:1080–1088. doi: 10.1158/0008-5472.CAN-08-2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanocco-Marani T., Vignudelli T., Gemelli C., Pirondi S., Testa A., Montanari M., Parenti S., Tenedini E., Grande A., Ferrari S. Tfe3 expression is closely associated to macrophage terminal differentiation of human hematopoietic myeloid precursors. Exp. Cell Res. 2006;312:4079–4089. doi: 10.1016/j.yexcr.2006.09.015. [DOI] [PubMed] [Google Scholar]
- Zappavigna V., Sartori D., Mavilio F. Specificity of HOX protein function depends on DNA–protein and protein–protein interactions, both mediated by the homeo domain. Genes Dev. 1994;8:732–744. doi: 10.1101/gad.8.6.732. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.