Lmod is a muscle-specific actin nucleator that displays structural similarity to the filament pointed-end–capping protein, Tmod. The mechanisms of localizations of Lmod and Tmod in muscle sarcomeres are strikingly different. Lmod contributes to the organization of mature myofibrils through a mechanism that requires interaction with tropomyosin.
Abstract
Leiomodin (Lmod) is a muscle-specific F-actin–nucleating protein that is related to the F-actin pointed-end–capping protein tropomodulin (Tmod). However, Lmod contains a unique ∼150-residue C-terminal extension that is required for its strong nucleating activity. Overexpression or depletion of Lmod compromises sarcomere organization, but the mechanism by which Lmod contributes to myofibril assembly is not well understood. We show that Tmod and Lmod localize through fundamentally different mechanisms to the pointed ends of two distinct subsets of actin filaments in myofibrils. Tmod localizes to two narrow bands immediately adjacent to M-lines, whereas Lmod displays dynamic localization to two broader bands, which are generally more separated from M-lines. Lmod's localization and F-actin nucleation activity are enhanced by interaction with tropomyosin. Unlike Tmod, the myofibril localization of Lmod depends on sustained muscle contraction and actin polymerization. We further show that Lmod expression correlates with the maturation of myofibrils in cultured cardiomyocytes and that it associates with sarcomeres only in differentiated myofibrils. Collectively, the data suggest that Lmod contributes to the final organization and maintenance of sarcomere architecture by promoting tropomyosin-dependent actin filament nucleation.
INTRODUCTION
Actin filaments play a central role in cells by promoting membrane dynamics and by forming contractile structures. Processes involving membrane dynamics rely on the coordinated polymerization/depolymerization of actin filaments under the control of a large number of proteins, including filament nucleation, elongation, and disassembly factors (Chhabra and Higgs, 2007). By contrast, force in contractile actin filament structures, such as the myofibrils of muscle cells, is generated by ATP-dependent myosin movement along actin filaments. Each myofibril consists of a large number of sarcomeres, which is the smallest functional unit of the muscle. Neighboring sarcomeres share a Z-disk, to which the barbed ends of the actin filaments from adjacent sarcomeres are anchored by α-actinin and other F-actin–binding/cross-linking proteins. In the middle of the sarcomere, M-line proteins, such as myomesin, cross-link and anchor the myosin filaments to each other (Agarkova and Perriard, 2005).
The actin filaments in cardiac and striated muscle sarcomeres appear regular in length and spacing and are stabilized by interactions with a number of muscle-specific proteins, such as the troponin complex, tropomyosin (TM), and the barbed- and pointed-end–capping proteins CapZ and tropomodulin (Tmod), respectively. Toward the center of sarcomeres, the actin “thin” filaments overlap with the myosin “thick” filaments, forming a tight hexagonal lattice (Clark et al., 2002; Cooper and Sept, 2008; Littlefield and Fowler, 2008). The appearance is that of a rigid structure, and it is not surprising that it has been traditionally thought that the actin filaments in sarcomeres are less dynamic than in nonmuscle cells. This view is evolving with new evidence suggesting that Z-disk components and actin itself display relatively rapid dynamics in smooth, skeletal and cardiac muscle cells (Wang et al., 2005; Gunst and Zhang, 2008; Sanger and Sanger, 2008; Skwarek-Maruszwska et al., 2009). At least in developing cardiomyocytes, rapid actin dynamics depends on myofibril contractility and appears to play an important role in the organization and maintenance of regular sarcomeric actin filament arrays (Skwarek-Maruszewska et al., 2009). The dynamic remodeling of muscle sarcomeres would be consistent with the need for proteins that could stimulate filament assembly, such as actin filament nucleators. However, the contribution of actin filament nucleators to these processes is poorly understood.
Myofibril assembly begins at the edges of muscle cells, with premyofibrils composed of α-actinin and actin-enriched Z-bodies and nonmuscle myosin II filaments. Subsequently, as the premyofibril moves away from the cell periphery, Z-bodies mature into Z-disks, and nonmuscle myosin II is replaced by muscle myosin II. With maturation, the α-actinin/myosin II periodicity becomes more regular, and the Z-bodies arrange into linear Z-disks (Sanger et al., 2005; Sparrow and Shöck, 2009). However, to date, the mechanisms by which actin filament nucleation begins in Z-bodies have not been identified. Furthermore, mature sarcomeres appear to undergo constant remodeling (Skwarek-Maruszewska et al., 2009), but the possible contribution of actin filament nucleating proteins to this process remains to be established. Our recent study revealed leiomodin (Lmod) as a powerful muscle-specific actin filament nucleator, suggesting that it could play a critical role in these processes. Consistent with this idea, overexpression or depletion of Lmod had dramatic effects on sarcomeric structure and organization (Chereau et al., 2008). Moreover, Lmod interacts with TM, and this interaction appears to modulate its nucleation activity and localization.
The first ∼340 amino acids of Lmod are ∼40% identical to Tmod, a pointed-end–capping protein that interacts with tropomyosin to regulate actin filament stability in myofibrils (Conley et al., 2001; Fowler et al., 2003; Fritz-Six et al., 2003; Mudry et al., 2003; Fischer and Fowler, 2003; Kostyukova et al., 2007; Chereau et al., 2008; Yamashiro et al., 2008). The N-terminal portion of Tmod is unstructured, except for three predicted helical segments, one of which binds actin, whereas the other two are thought to mediate the binding of two TM coiled coil dimers. Tmod has a second actin-binding site within the C-terminal Leu-rich repeat (LRR) domain (Krieger et al., 2002; Fowler et al., 2003). Lmod shares this domain organization, except for one important difference: only one of the two TM-binding helices of Tmod is conserved in Lmod. Furthermore, Lmod has a ∼150-amino acid (aa) C-terminal extension featuring a third actin-binding site, consisting of a WASP-homology 2 (WH2) domain. The presence of the WH2 domain brings the total number of actin-binding sites in Lmod to three, allowing it to stabilize a trimeric actin seed for filament nucleation (Chereau et al., 2008). The C-terminal extension also features a basic patch, which constitutes a potential nuclear localization signal (NLS).
Here, we examined the timing of Lmod expression in cardiomyocytes and compared its sarcomeric localization and dynamics to those of Tmod. Our data suggest that Lmod contributes to the organization and/or maintenance of mature myofibrils through a mechanism that requires its interaction with TM. Importantly, despite their apparent similarities, the mechanisms of localizations and functions of Lmod and Tmod in muscle sarcomeres are strikingly different.
MATERIALS AND METHODS
Preparation of Constructs for Cell Biology
The DNA encoding for cardiac Lmod2 constructs LmodFL, Lmod1-342, Lmod162-495, and Lmod44-495 were cloned between the XhoI and EcoRI sites of vectors pEGFP-N1 and pEGFP-C1 (Clontech, Palo Alto, CA), with enhanced green fluorescent protein (EGFP) fused at the C- or N-terminal ends. In addition, Lmod44-495(5xGS) was obtained by substituting residues 426-435 of construct Lmod44-495 (corresponding to a basic patch within the C-terminal extension) with a glycine-serine repeats of equal length.
Isolation of Myocytes and Cell Transfection
Neonatal rat cardiomyocytes were isolated and treated as described (Skwarek-Maruszewska et al., 2009). Briefly, neonatal rat hearts were dissected and enzymatically digested. After plating for ∼60 min to discard fibroblasts, the cardiomyocytes were replated on coated dishes. After culturing for 24 h, the plating medium was exchanged for “maintaining medium” (see Skwarek-Maruszewska et al., 2009). For fluorescence recovery after photobleaching (FRAP) experiments, cells were plated on fibronectin-coated glass-bottom culture dishes (MatTek, Ashland, MA) and treated as described above. Embryonic chick cardiomyocytes were isolated as described (Dabiri et al., 1999). Briefly, hearts of 7-d-old embryos were dissected and digested with trypsin. After preplating on plastic dishes for ∼60 min to remove fibroblasts, cardiomyocytes were transferred to dishes containing laminin-coated glass coverslips. Cells were used for immunostaining 24–48 h after plating. Skeletal muscle myocytes were isolated from the tails of 10-d-old zebra fish larvae using a similar procedure. Animals were anesthetized with tricane, and the tail region of larvae were dissected and digested for 1 h at 37°C with 0.05% trypsin dissolved in 60% strength PBS. Dissociated cells were spun down and resuspended in culture medium consisting of 60% L-15 medium, 34% water, 3% FBS, and 3% horse serum and supplemented with penicillin and streptomycin. Cells were plated on laminin-coated coverslips and cultured at room temperature. Cells were transiently transfected with GFP constructs 1 d after isolation using Escort III (Sigma-Aldrich, St. Louis, MO) or Lipofectamine 2000 (Invitrogen, Carlsbad, CA), according to manufacturer's specifications and cultured in the maintenance medium.
Immunofluorescence Microscopy and Western Blotting
Rat cardiomyocytes were fixed for 20 min in 4% paraformaldehyde in PBS and permeabilized with 0.2% Triton X-100 in PBS for 7 min. Chick cardiomyocytes and zebrafish skeletal muscle myocytes were extracted with 1% Triton X-100 in PEM buffer (100 mM PIPES, pH 6.9, 1 mM EGTA. 1 mM MgCl2) supplemented with 2 μM phalloidin for 5 min before fixation in 4% paraformaldehyde in PBS for 20 min. Indirect immunofluorescence was carried out as described (Vartiainen et al., 2000), using the following antibodies and dilutions: anti-sarcomeric-α-actinin (clone EA 53, Sigma-Aldrich) 1:500, anti-tropomodulin 1:750 (gifts from Carol C. Gregorio, University of Arizona, Tucson, AZ, and Velia Fowler, the Scripps Research Institute, La Jolla, CA), anti-Lmod 1:20 (Chereau et al., 2008) monoclonal anti-myomesin (mMaC) 1:100 and muscle myosin (MF20) 1:100 (both from Developmental Studies Hybridoma Bank). FITC or Cy5-conjugated secondary antibodies (Invitrogen) were used at a 1:250 dilution. Actin filaments were visualized with Alexa Fluor 488– (or 647) or Rhodamine-conjugated phalloidin diluted 1:100–1:300 (Molecular Probes, Invitrogen, Eugene, OR). Images were acquired as described (Hotulainen and Lappalainen, 2006; Korobova and Svitkina, 2010), and cells for Western blotting were treated as described (Hotulainen et al., 2005). For Western blotting 1:10 dilutions of anti-Lmod (Chereau et al., 2008) was used, and anti-actin (clone AC-40, Sigma-Aldrich) was used as a loading control at a dilution of 1:10,000.
FRAP Assay
Cells expressing various GFP constructs were cultured for 24–36 h on glass-bottomed dishes in maintaining medium. Imaging was performed on a confocal microscope (TCS, SP2AOBS, Leica, Deerfield, IL) with heating (37°C) and CO2 controlling. After three prebleached scans of an entire image, three bleaching scans (1 s each), with 100% intensity of a 476-, 488, and 496-nm laser, were performed over the regions of interest. After photobleaching, the fluorescence recovery was monitored 15 times every 1 s and 10 times every 10 s. The intensity of the GFP fluorescence was measured using the Leica confocal software. The intensity of the bleached area was normalized to a neighboring nonbleached area to diminish error caused by normal photobleaching during the monitoring period. The data were fitted to curves using the SigmaPlot graphical analysis software. The fluorescence recovery curves from FRAP experiments were fitted to a single exponential equation: y(t) = y0+R*(1 − exp[−τ*x]), where y0 is the offset and R the mobile fraction. Half-time (t/2) was calculated as t/2 = ln0.5/τ. Only cells with low-to-moderate levels of protein expression were analyzed. During and after the experiments, cells were contracting. Each experiment was repeated four times.
Inhibition of Contraction by Blebbistatin and Induction of Actin Monomer Sequestration by Latrunculin B treatment
The cells used in these experiments were 2–4 d old and contained mature sarcomere structures. For examining the localizations of GFP-Lmod constructs after latrunculin B (LatB) (Sigma-Aldrich) treatment, plasmids expressing full-length GFP-Lmod and GFP-Lmod1–342 were transfected to 1-d-old cardiomyocytes using Escort III reagent (Sigma-Aldrich) as described above. The cells were plated on 10 μg/ml fibronectin-coated dishes. For inhibition of contraction, cells were treated with 100 μM blebbistatin in DMSO or DMSO alone (control) and further incubated at 37°C and 5% CO2. Actin monomer sequestration was carried out with addition of 20 μM LatB for the indicated time. In both assays, cells were fixed and immunostained after the indicated times.
Protein Preparation.
Constructs LmodFL (full-length Lmod), Lmod44-495, and Lmod162-495 were cloned between the NdeI and SapI sites of vector pTYB1 (New England Biolabs, Beverly, MA) for protein expression in BL21(DE3) cells (Invitrogen). Protein purification on a chitin affinity column (New England Biolabs) was followed by additional purification on a MonoS column (GE Healthcare, Waukesha, WI). Actin was prepared and pyrene labeled as described (Cooper et al., 1983). Rabbit skeletal muscle tropomyosin was a generous gift of Sherwin Lehrer (Boston Biomedical Research Institute).
Actin Polymerization Assay.
Pyrene-actin polymerization assays were carried out using a Cary Eclipse fluorescence spectrophotometer (Varian, Sunnyvale, CA) and analyzed as described (Harris and Higgs, 2006). Before data acquisition, 2 μM Mg-ATP-actin (6% pyrene-labeled) was mixed with 0.5 μM F-actin seeds or varying amounts of Lmod constructs in F-buffer (10 mM Tris, pH 7.5, 1 mM MgCl2, 50 mM KCl, 1 mM EGTA, 0.1 mM NaN3, 0.02 mg/ml BSA, 0.2 mM ATP). The fluorescence was recorded 10 s after mixing. Control experiments were carried out with addition of F-buffer alone. Polymerization experiments were also performed with addition of varying amounts of TM. F-actin seeds were obtained by incubating 20 μM actin in F-buffer for 1 h at 25°C.
F-Actin Cosedimentation Assay
Actin (25 μM) in G-buffer (2 mM Tris, pH 7.4, 0.2 mM CaCl2, 0.2 mM ATP, 1 mM DTT, 1 mM NaN3) was polymerized by addition of 100 mM KCl, 2 mM MgCl2, and 2 mM EGTA for 8 min at room temperature. Lmod samples (LmodFL or Lmod1-342) were first centrifuged at 400,000 × g for 30 min to remove potential aggregates. F-actin (15 μM) was then incubated with 15 μM Lmod constructs for 30 min at room temperature. Samples were subsequently diluted to a concentration of 2 μM (using the same buffer) and centrifuged at 400,000 × for 30 min. Equal volumes of supernatants and pellets were analyzed on a 4–15% SDS gradient gel (Bio-Rad, Richmond, CA). In the case of experiments carried out in the presence of TM, prepolymerized actin (25 μM) was mixed with 7.1 μM TM and incubated for 20 min at room temperature.
RESULTS
The Expression and Sarcomeric Localization of Lmod Correlate with the Maturation of Myofibrils
We had previously shown that after isolation and plating the myofibrils of rat neonatal cardiomyocytes appear disrupted (Skwarek-Maruszewska et al., 2009). At early time points (1 d after plating) the actin bundles lack the characteristic periodic F-actin pattern, resembling stress fibers or premyofibrils. Subsequently, the cardiomyocytes begin to contract and the stress fiber-like structures mature into myofibrils, displaying the typical periodic F-actin organization. Many sarcomere components, such as Tmod, do not localize to premyofibrils but are only found associated with mature myofibrils (see e.g., Skwarek-Maruszewska et al., 2009). To elucidate whether Lmod functions as an early-stage actin filament nucleator during premyofibril formation or whether it contributes to actin dynamics in mature myofibrils, we examined the expression and subcellular localization of Lmod in cardiomyocytes at different time points after plating.
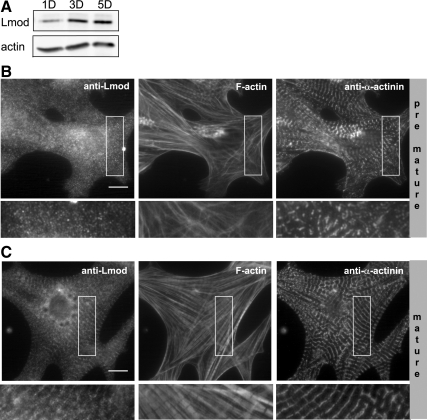
Western blot analysis detected only relatively low levels of Lmod protein in cardiomyocytes 1 d after plating, whereas Lmod levels increased during myofibril maturation (Figure 1A). More importantly, 1 d after plating Lmod did not display detectable localization to the stress fiber-like structures in cardiomyocytes, whereas 3–5 d after plating it displayed striated localization along “mature” myofibrils (Figure 1, B and C). These data suggest that Lmod associates with and plays an important role only in the mature sarcomeres of contractile cardiomyocytes. Therefore, we focused our attention on the study of Lmod in mature myofibrils.
Figure 1.
The expression and sarcomere localization of Lmod are enhanced during myofibril maturation. (A) Western blot analysis showing lower levels of Lmod protein in newly plated neonatal rat cardiomyocytes (1 d of culture; 1D) compared with cells 3 (3D) and 5 (5D) d after plating (top). Actin was used as a loading control (bottom). Comparison of Lmod localizations in (B) nonmature and (C) mature cardiomyocytes. One-day-old cells (nonmature) display punctate cytoplasmic Lmod staining, whereas mature (3D) cells display striated Lmod localization along myofibrils. Cells were costained with phalloidin to visualize F-actin (middle), and with antibodies against the Z-disk protein α-actinin (left). Insets show enlarged views on selected cell regions (rotated 90° counterclockwise). Bars, 10 μm.
Similar Dynamics and Different Subcellular Localizations of Lmod and Tmod in Myofibrils
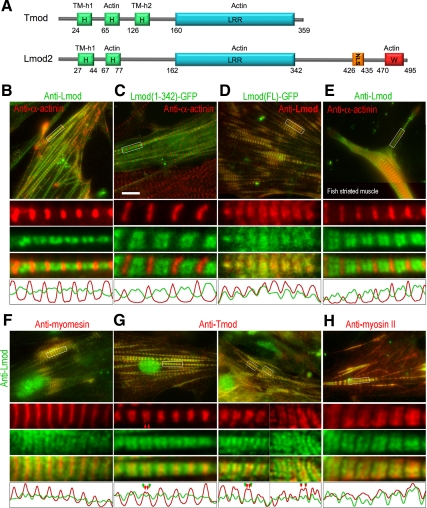
We hypothesized that despite the existing similarities between Lmod and Tmod the two proteins could have different functions and subcellular localizations. As a Tmod-related pointed-end–binding protein, Lmod would be expected to localize as two closely spaced bands on each side from M-lines in sarcomeres, as occasionally observed for Tmod (Castillo et al., 2009). Consistent with this idea, our original studies suggested that LmodFL was enriched near M-lines (Chereau et al., 2008), but filament pointed ends and M-lines could not be resolved in this study. Here, we analyzed the localization of Lmod relative to other sarcomeric proteins in cardiac and skeletal muscle cells in more detail. Costaining with α-actinin and Lmod antibodies showed double bands of endogenous Lmod in the middle of some sarcomeres, well separated from α-actinin in Z-lines (Figure 2, B and E). Ectopically expressed GFP-tagged LmodFL fully colocalized with endogenous Lmod revealed by immunostaining (Figure 2D). GFP-Lmod1-342, a Tmod-like fragment of Lmod lacking the C-terminal extension and having low nucleation activity (Chereau et al., 2008), also showed double bands in low expressing cells (Figure 2C), but in highly overexpressing cells it was additionally enriched in Z-lines. Double staining of Lmod and myomesin in M-lines confirmed that the Lmod bands flanked M-lines (Figure 2F), which is consistent with Lmod localizing to filament pointed ends. Surprisingly, however, Lmod did not show strong colocalization with Tmod (Figure 2G). Although Tmod formed narrow bands in the middle of sarcomeres that were resolved as doublets only in 12.7% of sarcomeres (N = 537), the bands of Lmod were broader and better separated. Moreover, Lmod typically localized farther away from M-lines than Tmod. Even in sarcomeres where the Lmod doublets could not be resolved, Lmod still formed much broader bands than Tmod. Thus, single Tmod bands were positioned in-between the double Lmod bands or were narrower than single Lmod bands in 93.4% of sarcomeres (n = 469). Double Tmod bands were also more frequently found in-between double Lmod bands (57.4%, n = 68), but Lmod and Tmod double bands overlapped in the remaining 42.6% of sarcomeres. The zone of Lmod-positive staining overlapped significantly with the myosin II thick filaments (Figure 2H). It is also important to note that the subcellular localization of Lmod was very similar in both cardiac and skeletal muscle cells (Figure 2), suggesting that this protein plays a similar role in these two cell-types.
Figure 2.
Localization of Lmod in sarcomeres. (A) Schematic presentation of the domain structures of Tmod and Lmod constructs used in these experiments. (B and E–H) Immunostaining of endogenous Lmod (green) relative to other proteins (red) in chicken cardiomyocytes (B and F–H) or zebra fish skeletal muscle myocytes (E). (C) Localization of expressed Lmod1-342-EGFP (green) relative to α-actinin (red) in chicken cardiomyocytes. (D) Localization of expressed full-length Lmod–EGFP (green) relative to endogenous Lmod revealed by immunostaining (red). Boxed regions in each panel are enlarged below the main panels as individual channels and as a merged image. Line scans through the middle of the selections are shown at the bottom of each panel. Red and green arrowheads in G indicate position of individual bands within doublets of Tmod and Lmod, respectively. Lmod localizes at both sides from myomesin in M-lines, is well separated from α-actinin in Z-disks, frequently flanks single or double bands of Tmod, and partially overlaps with the myosin thick filaments. Bars, 10 μm.
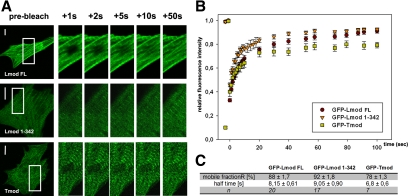
To gain insights into the dynamics of Lmod association with myofibrils, we carried out a FRAP analysis of cells expressing GFP fusions of LmodFL, Lmod1-342, and Tmod. All three GFP-fusion proteins displayed striated localization along myofibrils and displayed dynamic association with myofibrils as detected by FRAP (Figure 3A). Analysis of recovery curves from several cells revealed that both Lmod and Tmod associated with myofibrils with a characteristic half-life of ∼8 s. However, the mobile fraction of Lmod (∼90%) was slightly higher than the mobile fraction of Tmod (∼80%). Removal of the C-terminal 150-aa extension of Lmod did not significantly affect the dynamics of its sarcomere association/dissociation (Figure 3, B and C).
Figure 3.
Dynamics of Lmod association with sarcomeres. (A) Images of cells expressing GFP-tagged LmodFL, Lmod1-342, and Tmod before photobleaching (pre-bleach) and during the fluorescence recovery period. Enlarged views from selected areas of the frames (left panels) are shown. (B) Recovery profiles of bleached zones over the time course of the FRAP experiments. (C) Quantitative analysis of the Lmod and Tmod mobile fraction and fluorescence recovery half-lives in myofibrils. Bars, 10 μm.
The Myofibril Localization of Lmod, But Not Tmod, Is Actin Polymerization-dependent
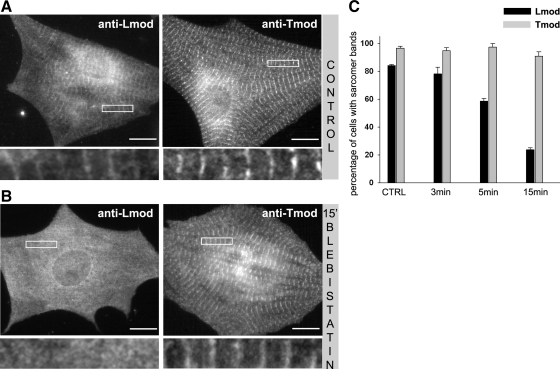
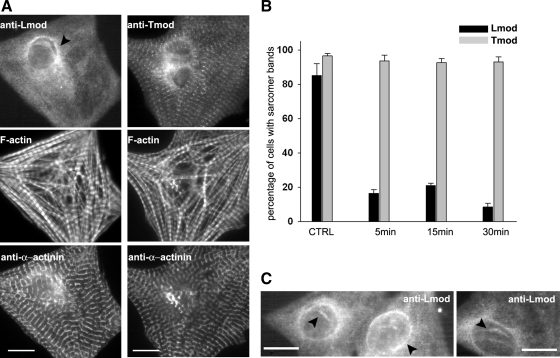
A subset of actin filaments in the sarcomeres of cardiomyocytes undergoes rapid dynamics, which is dependent on myofibril contractility (Skwarek-Maruszewska et al., 2009). We thus tested whether the localization of Lmod to myofibrils is affected by cardiomyocyte contractility. Treatment of cardiomyocytes with the myosin II inhibitor blebbistatin efficiently inhibited cell contractility within 60 s. Halting myosin contractility did not immediately affect the striated localization of Lmod in myofibrils. However, after longer blebbistatin treatments (3–15 min), the localization of Lmod to myofibrils was gradually reduced in the majority of the cells, whereas that of Tmod was mostly unchanged (Figure 4). These results suggest that whereas the localization of Lmod to myofibrils is not directly dependent on the activity of myosin II, the inhibition of contractility induces progressive and relatively fast Lmod delocalization.
Figure 4.
The inhibition of myosin II–based contractility leads to the delocalization of Lmod, but not Tmod. (A) In control cells, Lmod and Tmod display striated localization along myofibrils. (B) After blebbistatin treatment (50 μM for 15 min), Lmod displayed mainly diffuse cytoplasmic localization, whereas Tmod still localized to myofibrils in a striated pattern. Selected enlarged areas of the cells are shown below each picture. Bars, 10 μm. (C) Fraction of cells with detectable myofibril localization of Tmod and Lmod. Inhibition of contractility gradually displaced Lmod from sarcomeres, whereas the localization of Tmod was not affected. Values represent the mean percent of the cells displaying clear myofibril localization of the two proteins; error bars, ±SEM. Data represent averages from three independent experiments. In each experiment >150 cells were analyzed.
Because contractility promotes actin dynamics in myofibrils (Skwarek-Maruszewska et al., 2009), it is possible that the delocalization of Lmod from sarcomeres after blebbistatin treatment resulted from decreased actin dynamics. Thus, we examined the effect of LatB, a drug that binds actin monomers and inhibits their ability to polymerize, on the subcellular localization of Lmod in cardiomyocytes. Treatment with LatB rapidly and efficiently removed Lmod from myofibrils, suggesting that proper Lmod localization requires the availability of polymerization competent actin monomers (Figure 5). This effect was fully reversible, because after “wash-out” of LatB, Lmod relocalized rapidly to myofibrils (data not shown). In contrast to Lmod, the myofibril localization of Tmod was mostly unaffected by LatB (Figure 5). To reveal the role of the unique C-terminal extension of Lmod in actin monomer–dependent myofibril localization, we compared the localization of GFP-LmodFL and GFP-Lmod1-342 after LatB treatment in cardiomyocytes. Interestingly, the localization of Lmod1-342 was significantly less sensitive to the treatment with LatB than that of LmodFL. This result suggests that the C-terminal extension of Lmod, which is absolutely required for its strong nucleation activity (Chereau et al., 2008), is at least partly responsible for the dependence of Lmod localization on the availability of polymerization competent actin monomers (Supplemental Figure S2).
Figure 5.
The absence of polymerization-competent actin monomers leads to the delocalization of Lmod, but not Tmod. (A). Representative immunolocalization of Lmod (left panel) and Tmod (right panel) in cardiomyocytes after 15-min treatment with 20 μM LatB. Cells were costained with phalloidin to visualize F-actin and with an antibody against the Z-disk protein α-actinin. Bars, 10 μm. (B) Analysis of Tmod and Lmod localization 5/15/30 min after treatment with 20 μM LatB. Incubation of cardiomyocytes with LatB induces rapid dissociation of Lmod from sarcomeres, whereas the localization of Tmod was not affected. For each experiment, >200 cells were analyzed. Data represent averages from three independent experiments; error bars, ±SEM. (C) LatB treatment induces Lmod delocalization to the perinuclear region of cardiomyocytes, as well as to rods inside the nucleus (arrowheads). Bars, 10 μm.
Modulation of Lmod Activity and Localization by Tropomyosin
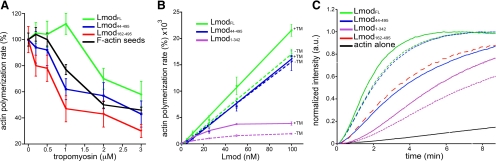
Generally, TM is known to stabilize actin filaments by inhibiting fragmentation, which reduces the number of fast-growing barbed ends and thereby polymerization (Hitchcock-DeGregori et al., 1988; Figure 6A, black line). However, our previous results suggested that TM had a stimulatory effect on the nucleation activity of LmodFL, but not Lmod162-495, which lacks both the TM-binding site and the N-terminal actin-binding site (Chereau et al., 2008). Because only one of the two TM-binding helices of Tmod is conserved in Lmod, located within the N-terminal 43-aa of the protein, we decided to test more precisely the role of TM binding on Lmod function by studying construct Lmod44-495. Note that this construct contains all three actin-binding sites. All TM concentrations inhibited polymerization of 2 μM actin induced by Lmod44-495 (Figure 6A). In contrast, polymerization by LmodFL was stimulated at low TM concentrations, but was inhibited by TM concentrations above 1 μM. At the optimal concentration of 1 μM, TM promoted polymerization with a range of concentrations of LmodFL and Lmod1-342, but not Lmod44-495 (Figure 6B). Importantly, in the absence of TM the nucleation activities of Lmod44-495 and LmodFL are very similar, whereas it is significantly lower for Lmod162-495, which also lacks the N-terminal actin-binding site (Figure 6C). Therefore, the N-terminal 43 amino acids of Lmod appear to play a role in Lmod's nucleation activity almost exclusively through their ability to recruit TM. It must be noted that LmodFL cosediments with actin filaments in vitro, possibly mediated by interaction of Lmod's C-terminal extension with the sides of actin filaments, but TM does not affect cosedimentation (Supplemental Figure S3).
Figure 6.
Modulation of Lmod nucleation activity by interaction with TM. (A) Effect of TM on the nucleation activities of Lmod constructs. TM inhibits polymerization of 2 μM actin induced by 0.5 μM F-actin seeds. However, low TM concentrations stimulate polymerization by 25 nM LmodFL, but not Lmod162-495 nor Lmod44-495, which lack the TM-binding site. Polymerization rates were determined from the slope of the curves at 50% polymerization and expressed as percentage of the rates without TM. Data reported are mean of three or more experiments; error bars, ±SD. (B) Polymerization rates of 2 μM actin with increasing concentrations of LmodFL, Lmod44-495, or Lmod1-342 in the absence (dashed lines) or the presence (solid lines) of 1 μM TM. Rates are expressed as percentages of the polymerization rate of actin alone in the absence or the presence of 1 μM TM. Data reported are mean of three experiments; error bars, ±SD. (C) Time course of fluorescence increase upon polymerization of 2 μM actin (6% pyrene-labeled) alone (black) or with addition of 25 nM Lmod constructs, and in the presence (solid lines) or the absence (dashed lines) of 1 μM TM. Each measurement was performed at least three times (one representative curve is shown).
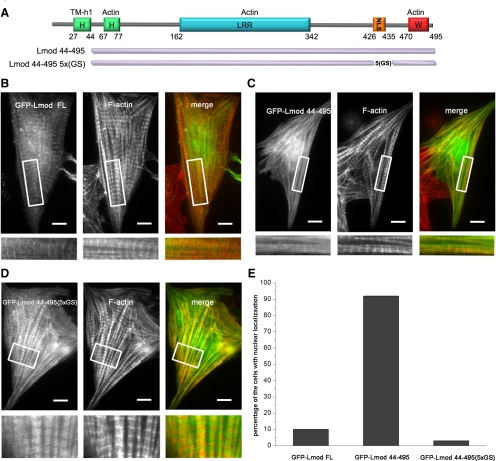
To study the role of the Lmod–TM interaction on Lmod localization, we compared the subcellular localizations of GFP fusions of LmodFL and Lmod44-495 in rat neonatal cardiomyocytes. LmodFL displayed a characteristic striated localization pattern. In contrast, Lmod44-495 was nearly uniformly distributed along myofibrils and did not display striated localization (Figure 7, B and C). Therefore, the interaction with TM appears to play a crucial role in the localization of Lmod to filament pointed ends in sarcomeres.
Figure 7.
Subcellular localization of Lmod mutants lacking the tropomyosin-binding site and the basic patch. (A) Schematic representation of the GFP-Lmod constructs used in this study. Localization in cardiomyocytes of GFP-tagged LmodFL (B), Lmod44-495 (C), and Lmod44-495(5xGS) (D). Insets display selected regions of the cells at higher magnification, highlighting the localization pattern of each protein in myofibrils. LmodFL shows striated localization along myofibrils, with distinguishable accumulation near M-lines. Lmod44-495 shows diffuse localization along myofibrils, with a large fraction localizing to the nucleus (E). Lmod44(5xGS) displays a striated pattern along myofibrils but, in contrast to LmodFL, it does not accumulate near M-lines. The cells were also stained with rhodamine-phalloidin to visualize F-actin. Bars, 10 μm.
Lmod Shuttles between the Nucleus and the Cytoplasm
In addition to defects in sarcomeric localization, a relatively large fraction of Lmod44-495 localized to the nucleus (in ∼90% of the cells), where it induced the formation of actin filament rods (Figure 7, C and E). Similar nuclear rods were observed for endogenous Lmod after treatment of cardiomyocytes with LatB or DMSO, or after heat shock, suggesting that endogenous Lmod may also shuttle between the nucleus and the cytoplasm (Figure 5, A and C, and Supplemental Figure S1).
To separate the two phenotypes observed with construct Lmod44-495 (i.e., nuclear localization vs. reduced accumulation near M-lines), we made a new construct, Lmod44-495(5xGS), in which the 10 residues corresponding to the basic patch in the C-terminal extension, a putative NLS, were replaced by a glycine-serine repeat (Figure 7A). This mutation abolished the nuclear localization of Lmod44-495 (Figure 7E). Indeed, Lmod44-495(5xGS) localized to sarcomers, where it displayed a periodic staining pattern (Figure 7D). However, compared with LmodFL, construct Lmod44-495(5xGS) appeared to localize uniformly throughout the entire sarcomere, except for Z-disks (Figure 7, B and D). This is consistent with the importance of the Lmod–TM interaction for proper localization of Lmod to a subset of filament pointed ends. Furthermore, these data suggest that the N-terminal region of Lmod may contain a nuclear export signal (NES), whereas the basic patch in the C-terminal extension may constitute an NLS. However, the physiological relevance, if any, of the nuclear localization is still unclear.
DISCUSSION
Lmod is a powerful actin filament nucleator that was shown to localize to myofibrils and contribute to their organization in cultured rat cardiomyocytes (Chereau et al., 2008). However, the exact localization pattern of Lmod in myofibrils and its role in dynamic myofibril assembly and/or maintenance were unknown. By carrying out a cell biological and biochemical analysis of Lmod, we have now shown that Lmod displays highly dynamic localization to specific sarcomeric regions near M-lines in mature myofibrils. Furthermore, we have provided conclusive evidence that the actin nucleation activity and subcellular localization of Lmod are both positively regulated by interaction with TM. Quite interestingly, differences in the ways Lmod and Tmod interact with TM may also explain their different subcellular localizations. Finally, we show that Lmod, but not Tmod, localization to sarcomeres is sensitive to actin monomer sequestration by LatB, suggesting that the correct localization of Lmod to myofibrils is dependent on the availability of polymerization-competent actin monomers.
Myofibrillogenesis begins with the formation of stress fiber-like premyofibrils, which compared with mature myofibrils display a less regular α-actinin/myosin II organization, and lack periodic F-actin staining and clearly recognizable M-lines (Sanger et al., 2005, 2009). Because Lmod is mostly absent from the stress fiber-like precursors of myofibrils in rat neonatal cardiomyocytes, we suggest that it is unlikely to contribute to actin filament nucleation at the initial stages of premyofibril assembly. Premyofibrils are believed to assemble through a similar mechanism to that of stress fibers in nonmuscle cells (Sparrow and Schöck, 2009), which are generated by formin- and Arp2/3 complex–mediated actin filament nucleation (Hotulainen and Lappalainen, 2006). It is thus possible that formins and/or the Arp2/3 complex mediate filament nucleation during the initial stages of premyofibril formation. In support of this idea, a recent study reported that a member of the formin family, Fhod3, plays an important role in myofibril organization (Taniguchi et al., 2009). Intriguingly, however, Fhod3 does not promote, but inhibits, actin polymerization in vitro and may thus have a different function in myofibrils than the classical nucleation/elongation functions associated with mDia formins (Dominguez, 2010). Importantly, our work revealed that Lmod localizes to sarcomeres of mature, contractile myofibrils in cardiomyocytes, suggesting that Lmod contributes to the maintenance (or repair) of sarcomeric actin arrays. In line with this hypothesis, our recent studies revealed that a subpopulation of actin filaments in cardiomyocyte myofibrils undergo relatively rapid turnover, which may contribute to the maintenance of correct thin filament organization in mature myofibrils (Skwarek-Maruszewska et al., 2009). It is also possible that Lmod additionally contributes to the conversion of premyofibrils into mature myofibrils. Attempts to specifically examine the role of Lmod in contractility-dependent actin dynamics were unsuccessful because of inefficient cotransfection of neonatal cardiomyocytes with Lmod RNAi oligonucleotides/constructs and GFP-actin, and the development of new methods will be needed to address this specific question. Nevertheless, the mechanism that is emerging is that the formation and maintenance of muscle cell myofibrils may require several, biochemically distinct, actin filament nucleators.
In addition to the presence of a WH2-containing C-terminal extension and strong in vitro nucleation activity, Lmod presents significant cellular differences compared with Tmod. Although both proteins display dynamic localization to myofibrils (half-life of ∼8 s), they exhibit distinct localization patterns and behave differently upon treatment with latrunculin or blebbistatin. Lmod localizes as two relatively broad bands on both sides from M-lines, whereas Tmod localizes closer to M-lines. Plating cardiomyocytes on laminin-coated coverslips and depleting soluble pools of proteins by detergent extraction before fixation helped us to resolve double bands of Lmod and Tmod, which could not be resolved in our previous study (Chereau et al., 2008). Furthermore, Lmod localization to myofibrils depends on sustained muscle contractility and the availability of polymerization-competent actin monomers, whereas that of Tmod does not. The inhibition of muscle contraction leads to reduced filament breakdown, which eliminates the need for de novo polymerization, and could thus explain the delocalization of Lmod produced by treatment with blebbistatin. Similarly, the formation of polymerization nuclei by Lmod–TM may require the presence of polymerization competent actin monomers, thus explaining why their removal by treatment with LatB results in fast Lmod delocalization, whereas the localization of the weak nucleator fragment Lmod1-342 is less affected.
F-actin cosedimentation assays suggested that Lmod may interact with the sides of actin filaments (Supplemental Figure S3). TM has no obvious effect on this behavior. The interaction site is located within the C-terminal extension of Lmod as indicated by the fact that Lmod1-342 does not significantly cosediment with F-actin. However, filament side binding does not appear to play a role in the myofibril localization of Lmod, because Lmod1-342, which does not cosediment with actin filaments in vitro, displays similar myofibril localization and dynamics as LmodFL.
Our data also provide evidence that Lmod can shuttle between the nucleus and the cytoplasm. We mapped the putative nuclear export and import signal sequences of Lmod, respectively, to its N-terminus and to a basic patch within the C-terminal extension. Interestingly, other sarcomeric proteins, most notably Tmod (Kong and Kedes, 2004), have been shown to shuttle between the nucleus and the cytoplasm, but the physiological significance of the nuclear localizations of these proteins remains unclear.
Because the localization of Lmod on each side from M-lines is relatively broad compared with that of Tmod, it appears that Lmod is at the pointed ends of actin filaments of different lengths. As an actin filament nucleator, Lmod could therefore be implicated in the formation of new actin filaments that originate in Z-discs. This would be consistent with the enriched localization of construct Lmod1-342 to Z-disks. This Lmod fragment is a weak actin nucleator (Chereau et al., 2008) and thus cannot effectively initiate new filaments from Z-discs. According to this model, Lmod in association with a single TM molecule would remain bound at the pointed ends of the newly formed filaments as they elongate to attain their regular sarcomeric length, at which point Lmod would be replaced by Tmod near M-lines. Because Tmod recruits a second TM molecule, it is possible that its affinity for filament pointed ends is higher than that of Lmod, consistent with Tmod's main role as a filament pointed-end–capping protein. The trigger for the replacement of Lmod by Tmod is probably located near or at M-lines. The emerging role of Lmod would then be the de novo polymerization of actin filaments during dynamic sarcomere remodeling or repair by filament replenishing.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the Academy of Finland and the Finnish Heart Research Foundation to P.L. and by National Institutes of Health Grants GM073791 to R.D. and GM70898 to T.S. A.S-M. was supported by a fellowship from the Viikki Graduate School in Biosciences.
Abbreviations used:
- FRAP
fluorescent-recovery-after-photobleaching
- Lmod
leiomodin
- NES
nuclear export signal
- NLS
nuclear localization signal
- TM
tropomyosin
- Tmod
tropomodulin.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-02-0109) on August 4, 2010.
REFERENCES
- Agarkova I., Perriard J. C. The M-band: an elastic web that crosslinks thick filaments in the center of the sarcomere. Trends Cell Biol. 2005;15:477–485. doi: 10.1016/j.tcb.2005.07.001. [DOI] [PubMed] [Google Scholar]
- Castillo A., Nowak R., Littlefield K. P., Fowler V. M., Littlefield R. S. A nebulin ruler does not dictate thin filament lengths. Biophys. J. 2009;96:1856–1865. doi: 10.1016/j.bpj.2008.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chereau D., Boczkowska M., Skwarek-Maruszewska A., Fujiwara I., Hayes D. B., Renowski G., Lappalainen P., Pollard T. D., Dominguez R. Leiomodin is an actin filament nucleator in muscle cells. Science. 2008;320:239–243. doi: 10.1126/science.1155313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhabra E. S., Higgs H. N. The many faces of actin: matching assembly factors with cellular structures. Nat. Cell Biol. 2007;9:1110–1121. doi: 10.1038/ncb1007-1110. [DOI] [PubMed] [Google Scholar]
- Clark K. A., McElhinny A. S., Beckerle M. C., Gregorio C. C. Striated muscle cytoarchitecture: an intricate web of form and function. Annu. Rev. Cell Dev. Biol. 2002;18:637–706. doi: 10.1146/annurev.cellbio.18.012502.105840. [DOI] [PubMed] [Google Scholar]
- Conley C. A., Fritz-Six K. L., Almenar-Queralt A., Fowler V. M. Leiomodins: larger members of the tropomodulin (Tmod) gene family. Genomics. 2001;73:127–139. doi: 10.1006/geno.2000.6501. [DOI] [PubMed] [Google Scholar]
- Cooper J. A., Walker S. B., Pollard T. D. Pyrene actin: documentation of the validity of a sensitive assay for actin polymerization. J. Muscle Res. Cell Motil. 1983;4:253–262. doi: 10.1007/BF00712034. [DOI] [PubMed] [Google Scholar]
- Cooper J. A., Sept D. New insights into mechanism and regulation of actin capping protein. Int. Rev. Cell. Mol. Biol. 2008;267:183–206. doi: 10.1016/S1937-6448(08)00604-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabiri G. A., Ayoob J. C., Turnacioglu K. K., Sanger J. M., Sanger J. W. Use of green fluorescent proteins linked to cytoskeletal proteins to analyze myofibrillogenesis in living cells. Methods Enzymol. 1999;302:171–186. doi: 10.1016/s0076-6879(99)02017-0. [DOI] [PubMed] [Google Scholar]
- Dominguez R. Structural insights into de novo actin polymerization. Curr. Opin. Struct. Biol. 2010;20:217–225. doi: 10.1016/j.sbi.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer R. S., Fowler V. M. Tropomodulins: life at the slow end. Trends Cell Biol. 2003;13:593–601. doi: 10.1016/j.tcb.2003.09.007. [DOI] [PubMed] [Google Scholar]
- Fowler V. M., Greenfield N. J., Moyer J. Tropomodulin contains two actin filament pointed end-capping domains. J. Biol. Chem. 2003;278:40000–40009. doi: 10.1074/jbc.M306895200. [DOI] [PubMed] [Google Scholar]
- Fritz-Six K. L., Cox P. R., Fischer R. S., Xu B., Gregorio C. C., Zoghbi H, Y, Fowler V. M. Aberrant myofibril assembly in tropomodulin1 null mice leads to aborted heart development and embryonic lethality. J. Cell Biol. 2003;163:1033–1044. doi: 10.1083/jcb.200308164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunst S. J., Zhang W. Actin cytoskeletal dynamics in smooth muscle: a new paradigm for the regulation of smooth muscle contraction. Am. J. Physiol. Cell Physiol. 2008;295:576–587. doi: 10.1152/ajpcell.00253.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris E. S., Higgs H. N. Biochemical analysis of mammalian formin effects on actin dynamics. Methods Enzymol. 2006;406:190–214. doi: 10.1016/S0076-6879(06)06015-0. [DOI] [PubMed] [Google Scholar]
- Hitchcock-DeGregori S. E., Sampath P., Pollard T. D. Tropomyosin inhibits the rate of actin polymerization by stabilizing actin filaments. Biochemistry. 1988;27:9182–9185. doi: 10.1021/bi00426a016. [DOI] [PubMed] [Google Scholar]
- Hotulainen P., Paunola E., Vartiainen M. K., Lappalainen P. Actin-depolymerizing factor and cofilin-1 play overlapping roles in promoting rapid F-actin depolymerization in mammalian nonmuscle cells. Mol. Biol. Cell. 2005;16:649–664. doi: 10.1091/mbc.E04-07-0555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotulainen P., Lappalainen P. Stress fibers are generated by two distinct actin assembly mechanisms in motile cells. J. Cell Biol. 2006;173:383–394. doi: 10.1083/jcb.200511093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong K. Y., Kedes L. Cytoplasmic nuclear transfer of the actin-capping protein tropomodulin. J. Biol. Chem. 2004;279:30856–30864. doi: 10.1074/jbc.M302845200. [DOI] [PubMed] [Google Scholar]
- Krieger I., Kostyukova A., Yamashita A., Nitanai Y., Maeda Y. Crystal structure of the C-terminal half of tropomodulin and structural basis of actin filament pointed-end capping. Biophys J. 2002;83:2716–2725. doi: 10.1016/S0006-3495(02)75281-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korobova F., Svitkina T. Molecular architecture of synaptic actin cytoskeleton in hippocampal neurons reveals a mechanism of dendritic spine morphogenesis. Mol. Biol. Cell. 2010;21:165–176. doi: 10.1091/mbc.E09-07-0596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostyukova A. S., Hitchcock-Degregori S. E., Greenfield N. J. Molecular basis of tropomyosin binding to tropomodulin, an actin-capping protein. J. Mol. Biol. 2007;372:608–618. doi: 10.1016/j.jmb.2007.05.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littlefield R. S., Fowler V. M. Thin filament length regulation in striated muscle sarcomeres: pointed-end dynamics go beyond a nebulin ruler. Semin. Cell Dev. Biol. 2008;19:511–519. doi: 10.1016/j.semcdb.2008.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudry R. E., Perry C. N., Richards M., Fowler V. M., Gregorio C. C. The interaction of tropomodulin with tropomyosin stabilizes thin filaments in cardiac myocytes. J. Cell Biol. 2003;162:1057–1068. doi: 10.1083/jcb.200305031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger J. W., Kang S., Siebrands C. C., Freeman N., Du A., Wang J., Stout A. L., Sanger J. M. How to build a myofibril. J. Muscle Res. Cell Motil. 2005;26:343–354. doi: 10.1007/s10974-005-9016-7. [DOI] [PubMed] [Google Scholar]
- Sanger J. M., Sanger J. W. The dynamic Z bands of striated muscle cells. Sci Signal. 2008;1:pe37. doi: 10.1126/scisignal.132pe37. [DOI] [PubMed] [Google Scholar]
- Sanger J. W., Wang J., Holloway B., Du A., Sanger J. M. Myofibrillogenesis in skeletal muscle cells in zebrafish. Cell Motil. Cytoskelet. 2009;66:556–566. doi: 10.1002/cm.20365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skwarek-Maruszewska A., Hotulainen P., Mattila P. K., Lappalainen P. Contractility-dependent actin dynamics in cardiomyocyte sarcomeres. J. Cell Sci. 2009;122:2119–2126. doi: 10.1242/jcs.046805. [DOI] [PubMed] [Google Scholar]
- Sparrow J. C., Schöck F. The initial steps of myofibril assembly: integrins pave the way. Nat. Rev. Mol. Cell Biol. 2009;10:293–298. doi: 10.1038/nrm2634. [DOI] [PubMed] [Google Scholar]
- Taniguchi K., Takeya R., Suetsugu S., Kan-O M., Narusawa M., Shiose A., Tominaga R., Sumimoto H. Mammalian formin fhod3 regulates actin assembly and sarcomere organization in striated muscles. J. Biol. Chem. 2009;284:29873–29881. doi: 10.1074/jbc.M109.059303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vartiainen M., Ojala P. J., Auvinen P., Peränen J., Lappalainen P. Mouse A6/twinfilin is an actin monomer-binding protein that localizes to the regions of rapid actin dynamics. Mol. Cell. Biol. 2000;20:1772–1783. doi: 10.1128/mcb.20.5.1772-1783.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Shaner N., Mittal B., Zhou Q., Chen J., Sanger J. M., Sanger J. W. Dynamics of Z-band based proteins in developing skeletal muscle cells. Cell Motil. Cytoskelet. 2005;61:34–48. doi: 10.1002/cm.20063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashiro S., Cox E. A., Baillie D. L., Hardin J. D., Ono S. Sarcomeric actin organization is synergistically promoted by tropomodulin, ADF/cofilin, AIP1 and profilin in C. elegans. J. Cell Sci. 2008;121:3867–3877. doi: 10.1242/jcs.040477. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.