Vps4 disassembly of ESCRT-III plays an important role in MVB sorting, viral budding, and cytokinesis. An in vitro system was developed to investigate this process. These studies revealed new insights into the mechanisms of Vps4 function.
Abstract
ESCRT-III undergoes dynamic assembly and disassembly to facilitate membrane exvagination processes including multivesicular body (MVB) formation, enveloped virus budding, and membrane abscission during cytokinesis. The AAA-ATPase Vps4 is required for ESCRT-III disassembly, however the coordination of Vps4 ATP hydrolysis with ESCRT-III binding and disassembly is not understood. Vps4 ATP hydrolysis has been proposed to execute ESCRT-III disassembly as either a stable oligomer or an unstable oligomer whose dissociation drives ESCRT-III disassembly. An in vitro ESCRT-III disassembly assay was developed to analyze Vps4 function during this process. The studies presented here support a model in which Vps4 acts as a stable oligomer during ATP hydrolysis and ESCRT-III disassembly. Moreover, Vps4 oligomer binding to ESCRT-III induces coordination of ATP hydrolysis at the level of individual Vps4 subunits. These results suggest that Vps4 functions as a stable oligomer that acts upon individual ESCRT-III subunits to facilitate ESCRT-III disassembly.
INTRODUCTION
Many endosomal cargoes destined for the lysosome are subject to an additional level of sorting during the formation of multivesicular bodies (MVBs) (Slagsvold et al., 2006; Piper and Luzio, 2007; Raiborg and Stenmark, 2009). Defects in MVB sorting stabilize cargoes normally targeted for degradation via this pathway and can result in prolonged signaling of activated growth factor receptors that fail to enter the MVB (Gruenberg and Stenmark, 2004; Slagsvold et al., 2006; Tanaka et al., 2008; Stuffers et al., 2009). MVB sorting is conserved throughout eukaryotes, as is the cellular machinery responsible for this reaction: the endosomal sorting complexes required for transport (ESCRTs) and associated proteins (Piper and Katzmann, 2007; Hurley, 2008; reviewed in Davies et al., 2009; Raiborg and Stenmark, 2009). ESCRT-0, -I, -II, and -III and associated proteins execute cargo selection and membrane deformation during MVB sorting. The ESCRT machinery also contributes to topologically-similar membrane deformation events including budding of enveloped viruses and membrane abscission during cytokinesis (Morita and Sundquist, 2004; Carlton and Martin-Serrano, 2007; Morita et al., 2007; Carlton et al., 2008; Lee et al., 2008). Thus, ESCRT-mediated membrane deformation impacts cellular physiology at multiple levels.
ESCRT-III is unique among the ESCRTs in that its membrane recruitment and assembly are coincident (Katzmann et al., 2001; Babst et al., 2002a; Babst et al., 2002b; Bilodeau et al., 2003). Yeast ESCRT-III comprises four core subunits essential for MVB sorting (Vps20, Snf7, Vps24, and Vps2) and three accessory subunits (Did2, Ist1, and Vps60) that function in a redundant manner (Babst et al., 2002a; Dimaano et al., 2008; Rue et al., 2008; Bajorek et al., 2009b; Xiao et al., 2009). Although ESCRT-III subunits exhibit a conserved core tertiary structure (Muziol et al., 2006; Bajorek et al., 2009b; Xiao et al., 2009), they display divergence at their carboxyl termini and appear to contribute unique functions to ESCRT-III.
Genetic and biochemical evidence supports a temporal recruitment and assembly of ESCRT-III subunits into a polymer, suggested to take on a fibril form (Babst et al., 2002a; Ghazi-Tabatabai et al., 2008; Hanson et al., 2008; Lata et al., 2008; Teis et al., 2008; Saksena et al., 2009). In vitro addition of core ESCRT-III subunits to giant unilamellar vesicles indicated that ESCRT-III can drive ILV formation; however, continuous ILV formation required the addition of Vps4 to permit repeated ESCRT-III function (Wollert et al., 2009). In vivo studies have implicated a role for Vps4 in mediating ESCRT-III disassembly, but the precise roles of Vps4 and ESCRT-III during ILV formation are not understood (Davies et al., 2009; Hanson et al., 2009). Vps4 is a type 1 AAA-ATPase that contains an amino-terminal Microtubule Interacting and Trafficking (MIT) domain which binds to ESCRT-III (Babst et al., 1997; Scott et al., 2005a; Scott et al., 2005b; Obita et al., 2007; Stuchell-Brereton et al., 2007; Kieffer et al., 2008). The ATPase cycle of Vps4 has been suggested to be impacted by interaction with membrane-associated ESCRT-III polymers: ESCRT-III binding facilitates Vps4 oligomerization; oligomerization stimulates Vps4 ATP hydrolysis; and ATP hydrolysis has been proposed to lead to both Vps4 oligomer dissociation and ESCRT-III disassembly, releasing ESCRT-III subunits into the cytoplasm (Babst et al., 1997; Babst et al., 1998; Scott et al., 2005a; Landsberg et al., 2009). However, an alternative model has proposed that Vps4 functions as a stable oligomer during ESCRT-III disassembly, as suggested by the oligomerization-promoting activity of the Vps4 cofactor Vta1, by mutation of the Vps4 central pore, and by analogy to other AAA-ATPases (Scott et al., 2005a; Azmi et al., 2006; White and Lauring, 2007; Gonciarz et al., 2008; Yu et al., 2008; Landsberg et al., 2009). While Vps4 disassembly of ESCRT-III plays a critical role in MVB sorting, the mechanism by which Vps4 disassembles ESCRT-III and the means by which ESCRT-III assembly and Vps4-mediated disassembly are coordinated are unclear.
To understand the role of Vps4 in ESCRT-III disassembly, an in vitro system was developed for characterizing ESCRT-III and Vps4 contributions. This system recapitulated observations of experiments in yeast, validating its use for examining Vps4 disassembly of ESCRT-III. Our analyses revealed that concerted hydrolysis is not required within the Vps4 oligomer and that not all subunits need to engage ESCRT-III via MIT domains at the same time. However, incorporation of full-length Vps4 subunits incapable of hydrolyzing ATP (Vps4E233Q) disrupted Vps4-mediated ESCRT-III disassembly, uncovering coordination between ESCRT-III binding via the MIT domain and ATP hydrolysis at the subunit level within the Vps4 oligomer. These results further suggest that Vps4 can function as a stable oligomer acting upon individual ESCRT-III subunits to facilitate ESCRT-III polymer disassembly in a processive manner.
MATERIALS AND METHODS
Plasmid Construction and Yeast Strains
Standard molecular biology and yeast genetics were used to generate plasmids and strains as listed in Table 1. Mutation of VPS4 was performed using the GeneTailor Site Directed Mutagenesis System (Invitrogen, Carlsbad, CA). All plasmids were sequenced to identify and exclude unanticipated mutations.
Table 1.
Plasmids and yeast strains
| Strains/plasmids | Description/genotype | Reference |
|---|---|---|
| MBY52 | vps4Δ:TRP pep4Δ:LEU2 prb1Δ:LEU2 | Katzmann, et al., 2003 |
| JPY188 | vps4Δ:TRP pep4Δ:LEU2 vps20Δ:HIS3 vta1Δ:HIS3 | This study |
| JPY315 | vps4Δ:TRP pep4Δ:LEU2 prb1Δ:LEU2 vps2Δ:HIS3 | This study |
| JPY296 | vps4Δ:TRP pep4Δ:LEU2 vps24Δ:HIS3 | This study |
| JPY311 | vps4Δ:TRP pep4Δ:LEU2 prb1Δ:LEU2 did2Δ:HIS3 | This study |
| JPY309 | vps4Δ:TRP pep4Δ:LEU2 prb1Δ:LEU2 vta1Δ:HIS3 | This study |
| JPY137 | vps4Δ:TRP pep4Δ:LEU2 vta1:HIS3 | This study |
| JPY378 | vps4Δ:TRP pep4Δ:LEU2 ist1Δ:HIS3 | This study |
| JPY310 | vps4Δ:TRP pep4Δ:LEU2 vps60Δ:HIS3 | This study |
| JPY459 | vps4Δ:TRP pep4Δ:LEU2 vps24Δ:HIS3 did2Δ:HIS3 vta1Δ:HIS3 | This study |
| JPY373 | vps4Δ:TRP pep4Δ:LEU2 vps20Δ:HIS3 ist1Δ:HIS3 | This study |
| JPY293 | vps4Δ:TRP pep4Δ:LEU2 vps25Δ:HIS3 | This study |
| JPY188 | vps4Δ:TRP pep4Δ:LEU2 vps28Δ:HIS3 | This study |
| JPY140 | vps4Δ:TRP pep4Δ:LEU2 snf7Δ:HIS3 | This study |
| JPY554 | vps4Δ:TRP pep4Δ:LEU2 snf7Δ:HIS3 vps2Δ:HIS3 | This study |
| JPY48 | vta1Δ::HIS3 | Azmi, et al., 2006 |
| JPY45 | vps4Δ:TRP vta1Δ::HIS3 | Azmi, et al., 2006 |
| pMB63 | pGEX-4T Vps4E233Q | Babst, et al., 1998 |
| pMB40 | pGEX-4T Vps4ΔMIT | Babst, et al., 1998 |
| pMB90 | pGEX-4T Vps4ΔMIT,E233Q | Babst, et al., 1998 |
| pGST Vps4I18D | pGEX-4T Vps4I18D | This study |
| pGST Vps4I18D,E233Q | pGEX-4T Vps4I18D | This study |
| pGST Vps4L64D | pGEX-4T Vps4L64D | This study |
| pGST Vps4L64D,E233Q | pGEX-4T Vps4L64D,E233Q | This study |
| pMB168 | pRS416 Vps20-HA | Babst, et al., 2002a |
| pET28Vta1 | pET28b Vta1 | Azmi, et al., 2006 |
| pMB106 | pRS416 Vps24-GFP | This study |
| pCherryDid2 | pRS415 (Vps21 Prom.) Cherry-Did2 | This study |
| pMB97 | pRS426 Vps4E233Q-GFP | This study |
| pMB341 | pRS416 GFP-Vps4ΔMIT,E233Q | This study |
| p416 ChVps4E233Q | pRS415 (Vps21 Prom.) Cherry-Vps4E233Q 1–437 | This study |
| p416 ChVps4ΔMIT,E233Q | pRS415 (Vps21 Prom.) Cherry-Vps4E233Q 118–437 | This study |
| pMB118 | pRS425 GFP-CPS | This study |
| pRS426 Snf7 | pRS426 (Snf7 Prom.) Snf7 1–240 | This study |
| pRS426 Snf7Vps2MIM1 | pRS426 (Snf7 Prom.) Snf7 1–227::Vps2 218–232 | This study |
Protein Expression and Purification
Expression and purification of Vps4, Vps4 mutants, and Vta1 were performed as previously described (Babst et al., 1998; Azmi et al., 2006). Briefly, a short induction (1–2 h) with 500 μM isopropyl-β-D-1-thiogalactopyranoside IPTG was used to express GST-Vps4 fusions, which were subsequently affinity purified with Glutathione Sepharose 4B (GE Healthcare, Piscataway, NJ). Induction of His6-Vta1 was conducted overnight at 22°C, and the protein was purified by Ni+2 affinity chromatography. After elution, the GST and His6 epitopes were removed by treatment with Thrombin, and the proteins were purified using a BioSource Q2 anion exchange column (Bio-Rad, Hercules, CA).
ATPase Assay
WT and mutant Vps4 ATPase activities were analyzed by ATPase assays as previously described (Babst et al., 1998). All reactions were initiated with 1 mM ATP, and quantitation of activity was performed with time-points exhibiting <30% ATP hydrolysis (to avoid complications from product inhibition). When mixings of wild-type and mutant Vps4 were performed in conjunction with the ATPase assay, proteins were preincubated for at least 10 min, and the activity was assessed as ADP generated per wild-type Vps4 per minute. For Figures 4C and 6B, 250 nM Vps4 was used to permit molar excesses of Vps4E233Q or Vps4ΔMIT,E233Q with lesser amounts of these proteins. For the dilution analysis depicted in Supplemental Figure 3, C and D, standard 20-μl reactions were initiated. After removal of the first three time points to evaluate initial activities, reactions were diluted by addition of nine volumes of 1× ATPase buffer with 1 mM ATP at 10 min after initiation of the reaction. ATP to ADP conversion was then evaluated at up to 260 min post dilution. The ADP generated per Vps4 was determined and graphed versus time. Linear regression was used at assess ATPase activities before and after dilution (Prizm, Graphpad Software, La Jolla, CA). A similar procedure was used for the dilution analysis depicted in Figure 7, A and B, except dilution was initiated at 30 min and included or excluded additional ATP during dilution. Readdition of ATP to 1 mM (to the samples diluted without additional ATP) was performed 30 min after dilution. The reactions were performed in triplicate and the experiment performed twice. ADP/Vps4 amounts were determined at 5-min intervals, and linear regression was used to assess ATPase activities. The initial rate of hydrolysis (I) was determined using the 5- to 15-min time points (before 30% ATP hydrolysis). The rate of activity following dilution with ATP (II) was assessed using the 35- to 90-min time points (blue). The rate of activity after dilution without maintaining ATP at 1 mM (III) was assessed using the 35- to 90-min time points (black). The rate of activity after dilution and with ATP readdition to 1 mM (IV) was assessed using the 65- to 90-min time points (red). Activities relative to the initial rate (I) are presented. 500 nM Vps4 was used in these dilution experiments as it is a concentration below the apparent Km, the ATPase activities of 50 nM and 500 nM Vps4 are readily detectable and distinct, these concentrations do not require excessive amounts of Vps4, and adequate time-points can be taken before reaching conditions with apparent product inhibition.
Figure 4.
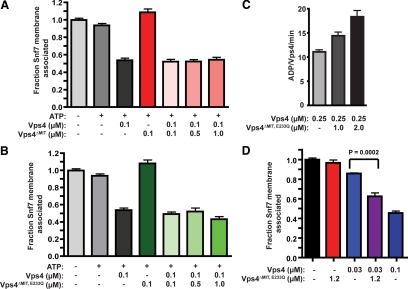
Vps4 oligomer MIT domain composition for ESCRT-III disassembly. (A) ESCRT-III disassembly reactions were performed with vps4Δ membranes (0.5 OD600 equivalents) and 100 nM Vps4, 100 nM Vps4ΔMIT, or 100 nM Vps4 with increasing concentrations of Vps4ΔMIT (100 nM, 500 nM, and 1 μM). (B) ESCRT-III release reactions were performed as in A but with increasing concentrations of Vps4ΔMIT,E233Q (100 nM, 500 nM, and 1 μM). (C) ATPase activity of 250 nM Vps4 alone or with addition of 1 μM or 2 μM Vps4ΔMIT,E233Q. The initial ATP concentration was 1 mM. Activity is presented as ADP generated per wild-type Vps4 per minute. (D) ESCRT-III disassembly reactions were performed as in A but with 30 nM Vps4 alone, 1.2 μM Vps4ΔMIT,E233Q alone, 30 nM Vps4 and 1.2 μM Vps4ΔMIT,E233Q, or 100 nM Vps4. The addition of Vps4ΔMIT,E233Q enhanced 30 nM Vps4-mediated ESCRT-III disassembly (p < 0.0002).
Figure 6.
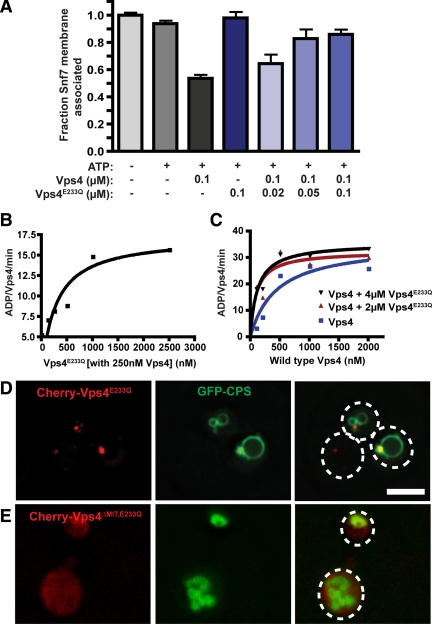
Vps4E233Q inhibition of ESCRT-III disassembly. (A) ESCRT-III disassembly reactions were performed with vps4Δ membranes (0.5 OD600 equivalents) and 100 nM Vps4, 100 nM Vps4E233Q, or 100 nM Vps4 with decreasing concentrations of Vps4E233Q (100 nM, 50 nM, and 20 nM). (B) ATP hydrolysis reaction were performed with 250 nM Vps4 and increasing concentrations of Vps4E233Q. The initial ATP concentration was 1 mM. Rates are presented as ADP generated per wild-type Vps4 per minute. (C) ATP hydrolysis reactions were initiated with 2 μM Vps4E233Q (red), 4 μM Vps4E233Q (black), or Vps4 alone (blue) with Vps4 concentrations from 100 nM to 2 μM. Rates are presented as ADP generated per wild-type Vps4 per minute. (D and E) Distribution of the MVB sorting reporter GFP-CPS in wild-type yeast expressing Cherry-Vps4E233Q (D) or Cherry-Vps4ΔMIT,E233Q (E). Dashed white lines represent the borders of individual yeast cells. Bar, 5 μm.
Figure 7.
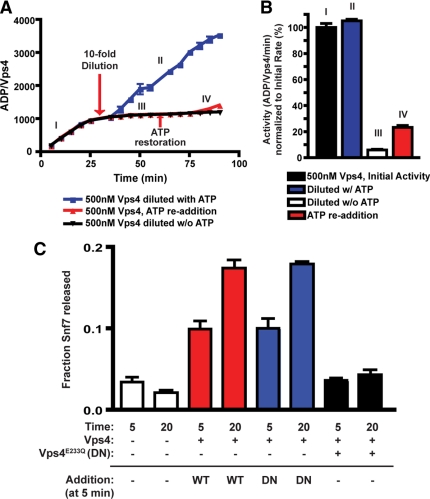
The Vps4 oligomer is functionally stable in vitro. (A) ATP hydrolysis reactions were initiated with 500 nM Vps4 and 1 mM ATP. After 30 min, reactions were diluted 10-fold in the absence of additional ATP (black, red) or with ATP maintained at 1 mM (blue). After an additional 30 min, ATP restoration (to 1 mM, red) was performed. The ADP generated per Vps4 was determined at 5-min intervals and plotted versus time. (B) The activities of ATP hydrolysis (ADP generated/Vps4/min) relative to the initial rate (I) are presented. (C) ESCRT-III release reactions were initiated with 30 nM Vps4 (red, blue) or with 30 nM Vps4 and 100 nM Vps4E233Q (black). After a 5-min incubation, Vps4 (30 nM, red) or Vps4E233Q (100 nM, blue) were added to the reactions initiated with Vps4 only. The fraction of Snf7 released into the soluble material was assessed at both 5 and 20 min.
In Vitro ESCRT-III Release Assay
Membrane Generation.
One liter of yeast cells were grown at 30°C in Yeast Extract Peptone Dextrose medium (YPD) to an optical density at 600 nm (OD600) of 0.5–0.8. Cells were harvested, softened, and spheroplasted as previously described (Vida et al., 1990). Spheroplasted cells were harvested by centrifugation at 3,000g for 5 min and resuspended in chilled lysis buffer [0.2M Sorbitol, 50 mM KOAc, 2 mM EDTA, 20 mM HEPES, pH 6.8 with complete protease inhibitors (Roche)] and lysed by 15 strokes with a glass tissue homogenizer. Intact cells were removed by 500g centrifugation for 5 min. The 500g supernatant was carefully removed and centrifuged at 13,000g for 10 min to generate a pellet (P13) fraction. The P13 was resuspended in 1 ml ATPase reaction buffer (100 mM KOAc, 20 mM HEPES pH 7.4, 5 mM MgOAc) with 1 mM sorbitol and protease inhibitors (Buffer A), and the 13,000g spin was repeated to remove contaminating soluble material. The repelleted P13 fraction was resuspended in Buffer A and passed through an 18-gauge needle three times and a 30-gauge needle five times to homogenize the resuspended membranes. Membranes were diluted to 50 OD600/ml in buffer A and stored in −80°C until further use.
ESCRT-III Disassembly Assay.
Twenty-microliter reactions were set up with 0.5 OD600 equivalent of resuspended P13 membranes. This amount of membranes is anticipated to yield an Snf7/ESCRT-III concentration in the low nM range (∼1–3 nM), as assessed both by Western blotting with dilutions of purified Snf7 (data not shown) and by calculations based on the number of molecules of Snf7 per cell (Ghaemmaghami et al., 2003). The ATP regeneration system (10 mM phosphocreatine, 10 U/ml creatine phosphokinase, and 1 mM ATP) and purified Vps4 were added in the form of drops on the side of the tube to prevent mixing before reaction initiation. The reaction was initiated by a brief spin (∼5 s at 4°) and was incubated at 30°C in a rotator for 10 min. The reaction was terminated by centrifugation at 13,000g for 10 min at 4°C. The soluble fraction from the 13,000g spin (S13) was combined with 5 μL of 5× sample buffer, and the P13 was resuspended in 25 μL of 5× sample buffer. Five microliters of the reaction was run on a 15% gel, and Western blotting was performed with anti-Snf7 polyclonal antibody (1:10,000) (Babst et al., 1998). Reactions were performed in triplicate within an experiment, and experiments were performed at least thrice. Error bars indicate the SE of the mean. The use of 100 nM as the standard Vps4 concentration in the ESCRT-III disassembly assay was empirically determined as the concentration providing 50% Snf7 release from the membrane in reactions with 0.5 OD600 equivalents of membrane incubated for 10 min at 30°.
Biochemical Analyses.
Immunoprecipitation (IP) experiments were performed as described (Babst et al., 2002a) using ESCRT-III release material. Briefly, ESCRT-III release assays were performed as above except additional substrate (10 OD600 equivalents), Vps4 (500 nM) and time (30 min) were used. Total reactions (for Snf7-Vps20HA coimmunoprecipitations, as Vps20HA release from the membrane was complicated by its myristolation) or the pelleted material (for Snf7-Vps24 coimmunoprecipitations) were then extracted with 1% TritonX-100 for 10 min on ice. Samples were diluted 10-fold, and the insoluble material was removed by centrifugation. Immunoprecipitation with either Snf7 or Vps24 antisera was performed, and antibody complexes were isolated with Protein A Sepharose. Isolated material and input fractions were then analyzed by Western blotting with HA.11 mAb (Covance, Princeton, NJ) or polyclonal antisera against Snf7 (Babst et al., 1998). Quantitation of Vps20HA (Figure 1D) represents the mean from three independent experiments. The immunoprecipitation of Snf7 under native conditions was more efficient after release from the membrane (Figure 1C), suggesting that epitopes recognized by the Snf7 antisera are occluded in the ESCRT-III assembled state.
Figure 1.
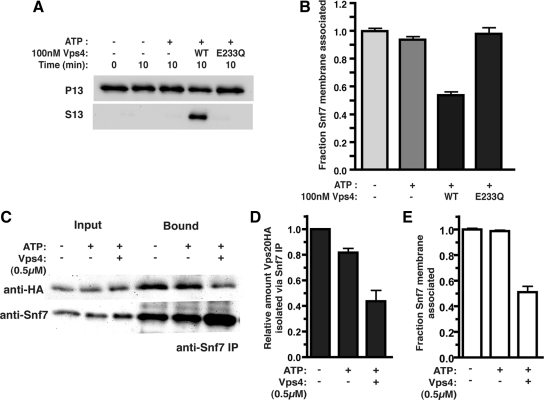
In vitro ESCRT-III release and disassembly. (A) ESCRT-III release reactions were initiated with resuspended P13 membranes (0.5 OD600 equivalents) generated from vps4Δ cells, in the presence (+) or absence (−) of 1 mM ATP and an ATP regeneration system (ATP), 100 nM Vps4 (WT), or 100 nM Vps4E233Q (E233Q). After a 10 min incubation at 30°C, the membrane-associated (P13) and soluble (S13) materials were separated. Western blotting with Snf7 antiserum was performed. (B) Quantitation of ESCRT-III release reaction. Data are presented as the fraction of Snf7 remaining membrane associated and represent the mean of at least three independent experiments with reactions performed in triplicate within each experiment. (C) Vps20HA-containing membranes (10 OD600 equivalents) were incubated for 30 min with 500 nM Vps4 and ATP or ATP alone. Snf7 immunoprecipitations were then performed after TritonX-100 extraction of the total reaction. Vps20HA was detected with the HA.11 mAb and Snf7 with the polyclonal antiserum. (D) Quantitation of Vps20HA isolation from the Snf7 immunoprecipitation. (E) Quantitation of the fraction of Snf7 remaining membrane associated under the reaction conditions used in C.
Subcellular Localization
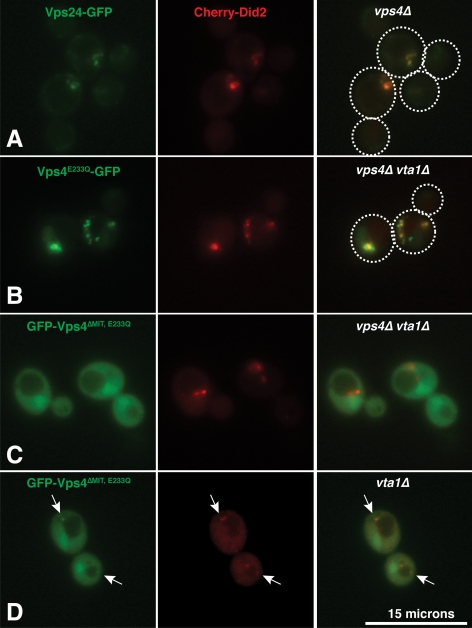
Microscopy was performed on living cells using the DeltaVisionRT System (Applied Precision, Issaquah, WA) including a fluorescence microscope (Nikon) equipped with EGFP and Cherry filter sets and a digital camera (Coolsnap HQ, Phototmetrix). Images were processed using the DeltaVision software package (Applied Precision). The Cherry-Did2 fusion used for colocalization is a partially functional form of Did2 (data not shown). Strains lacking the Vps4 cofactor Vta1 (vta1Δ and vta1Δ vps4Δ) were used in the colocalization study to avoid complications from Vta1-dependent recruitment to the endosome (Shestakova et al., 2010) as well as to enhance Cherry-Did2 endosomal localization (Nickerson et al., 2010). Cherry-Did2 colocalized with Vps24-GFP in vps4Δ cells (Figure 5A), confirming the Cherry-Did2 compartment as an ESCRT-III-positive endosome.
Figure 5.
Vps4ΔMIT,E233Q colocalization with Did2 is Vps4-dependent. (A) In yeast lacking Vps4 (vps4Δ), Cherry-Did2 exhibits colocalization with the core ESCRT-III subunit Vps24-GFP. Dashed white lines represent the borders of individual yeast cells. Bar, 15 μm. (B) Vps4E233Q-GFP exhibits punctate distribution and colocalizes extensively with Cherry-Did2 in vps4Δ vta1Δ cells. (C) GFP-Vps4ΔMIT,E233Q exhibits a diffuse distribution and little colocalization with Cherry-Did2 in vps4Δ vta1Δ cells. (D) In the presence of Vps4 (vta1Δ cells), GFP-Vps4ΔMIT,E233Q displays some punctate association that colocalizes with Cherry-Did2 (indicated by arrows). Additional micrographs demonstrating colocalization of GFP-Vps4ΔMIT,E233Q with Cherry-Did2 in the presence of wild-type Vps4 are presented in Supplemental Figure 4.
RESULTS
Reconstitution of Vps4-Dependent ESCRT-III Disassembly and Membrane Release
Two general models for Vps4 function have been proposed: 1) Vps4 undergoes a dynamic cycle of oligomerization and dissociation to facilitate ESCRT-III disassembly in mass; and 2) Vps4 functions as a stable oligomer disassembling the ESCRT-III polymer. To explore the mechanism of Vps4 function, an in vitro assay was developed that reconstituted Vps4-mediated ESCRT-III membrane release. This assay monitored the redistribution of ESCRT-III subunits from membrane into soluble fractions.
To validate the assay, the Vps4-, energy-, and time-dependences of ESCRT-III subunit (Snf7 or Vps24) release from the membrane fraction were examined. Ten-minute incubation of ESCRT-III-containing membranes at 30°C with 100 nM Vps4, 1 mM ATP, and an ATP regeneration system resulted in a 50% decrease in membrane-associated Snf7 (P13) with a commensurate increase of Snf7 in the soluble fraction (S13) (Figure 1, A and B). Snf7 release into the S13 was dependent on both ATP and Vps4 (Figure 1, A and B) as was Vps24 (Supplemental Figure 1A). Consistent with in vivo studies (Babst et al., 1998), the catalytically inactive Vps4E233Q failed to support Snf7 release into the S13 fraction (Figure 1, A and B). Time dependence and Vps4-concentration dependence of Snf7 release were also observed (Supplemental Figure 1B). Ten-minute incubation at 30°C with 100 nM Vps4 and 1 mM ATP was selected as standard reaction conditions as these parameters generated ∼50% Snf7 release from the membrane fraction.
To address whether ESCRT-III release from the membrane corresponded to disassembly of the ESCRT-III polymer, the association of the ESCRT-III subunits Vps20 and Snf7 was examined after the release reaction. Vps20HA-containing membranes were incubated with ATP alone or with Vps4 and ATP, and Vps20HA association with Snf7 was assessed by coimmunoprecipitation from detergent-solubilized reactions. Vps4 reduced the association of Vps20HA with Snf7 (Figure 1, C and D) commensurate with Snf7 release from the membrane fraction (Figure 1E). Similarly, Snf7 coimmunoprecipitation with Vps24 was abrogated upon ESCRT-III release from the membrane (Supplemental Figure 1C). These results demonstrate that the ESCRT-III membrane release is coincident with disassembly of ESCRT-III. As such, this in vitro assay will be referred to as the ESCRT-III disassembly assay or the ESCRT-III membrane release assay interchangeably. These results demonstrate that Vps4 disassembles the ESCRT-III polymer, as suggested by observations correlating ESCRT-III assembly state with Vps4 function (Babst et al., 1998; Babst et al., 2002a).
In vitro ESCRT-III Disassembly Depends on Did2, Vps24, and Vps2
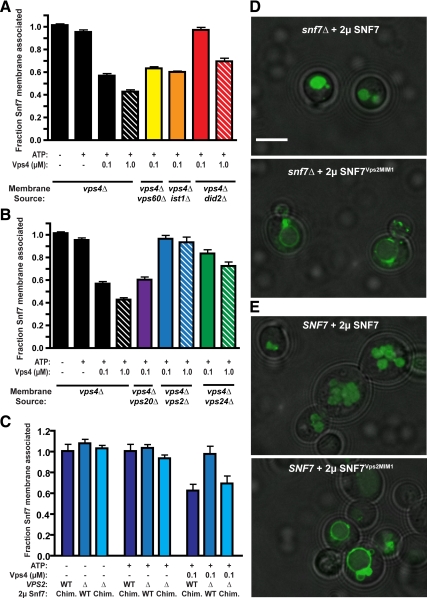
Vps4 disassembly of ESCRT-III appears to be impacted by the ESCRT-III subunit composition as yeast lacking Vps24, Vps2, or Did2 exhibit stabilized ESCRT-III membrane association (Babst et al., 2002a; Nickerson et al., 2006; Teis et al., 2008). To examine these ESCRT-III subunit contributions, Vps4 ESCRT-III disassembly reactions were performed with membranes isolated from yeast lacking Vps4 and individual ESCRT-III subunits (i.e., vps4Δ vps60Δ). Membrane fractions from each strain contained Snf7; however, loss of Vps20 partially compromised Snf7 abundance (Supplemental Figure 2A). Losses of the ESCRT-III subunits Vps60, Ist1, or Vps20 or of ESCRT-I (Vps28) or ESCRT–II (Vps25) subunits did not perturb Vps4-mediated Snf7 membrane release (Figure 2, A and B, and data not shown). By contrast, losses of Vps24, Vps2, or Did2 abrogated Snf7 membrane release catalyzed by 100 nM Vps4 (Figure 2, A and B). However, increased Vps4 concentration (1 μM Vps4) partially restored ESCRT-III disassembly in the absences of Vps24 or Did2 but not in the absence of Vps2 (Figure 2, A and B) or in the combined absence of Vps24 and Did2 (Supplemental Figure 2B). These results implicated distinct roles for Did2, Vps24, and Vps2 in promoting Vps4 disassembly of ESCRT-III. However, we cannot distinguish whether these contributions are related to assembly of ESCRT-III polymers or as participants in the disassembly reaction. Regardless, Vps2 contributes to Vps4 disassembly of ESCRT-III to a more significant extent while Vps24 and Did2 make partially redundant contributions.
Figure 2.
ESCRT-III subunit contributions to Vps4 disassembly of ESCRT-III. (A) ESCRT-III–containing membranes were generated from yeast lacking the accessory ESCRT-III subunits Did2 (red), Ist1 (orange), and Vps60 (red). Membranes (0.5 OD600 equivalent) were incubated with ATP and 100 nM or 1 μM Vps4, and the fraction of Snf7 remaining membrane associated was determined. (B) ESCRT-III membrane release assays performed with membranes generated from yeast lacking the core ESCRT-III subunits Vps20 (purple), Vps24 (green), and Vps2 (blue). (C) ESCRT-III membrane release assays performed with membranes generated from yeast lacking Snf7 and Vps2 (Δ) or Snf7 alone (WT) and expressing either Snf7 or the Snf7Vps2MIM1 chimera. (D and E) Distribution of the MVB sorting reporter GFP-CPS in snf7Δ (D) or wild-type yeast (E) expressing Snf7 or the Snf7Vps2MIM1 chimera. Bar, 5 μm.
Vps2, Vps24, and Did2 contain a carboxyl-terminal sequence motif (MIT-interacting motif 1, MIM1) that mediates association with Vps4 (Obita et al., 2007; Stuchell-Brereton et al., 2007). The distinct contributions of Vps2, Vps24, and Did2 suggested their MIM1 elements are not functionally equivalent and that the Vps2 MIM1 contributes a critical function in ESCRT-III disassembly. To evaluate this contribution, a chimeric ESCRT-III subunit (Snf7Vps2MIM1) was generated by replacing the Snf7 α6 with the Vps2 α6 and its MIM1 (Snf7 1–227 fused to Vps2 218–232). Snf7 or Snf7Vps2MIM1 were expressed in vps4Δ snf7Δ or vps4Δ snf7Δ vps2Δ yeast, and membranes generated from these strains were analyzed for ESCRT-III disassembly by Vps4. Whereas loss of Vps2 abrogated Vps4 release of Snf7 from these membranes, release of Snf7Vps2MIM1 occurred independent of Vps2 (Figure 2C). This result implicated the Vps2 MIM1 as playing an important role in ESCRT-III disassembly by Vps4 and suggests that the remainder of Vps2 is dispensable. To extend this analysis, the localization of the MVB cargo GFP-Carboxypeptidase S (GFP-CPS) was examined in snf7Δ yeast expressing Snf7 or Snf7Vps2MIM1. Whereas Snf7 reexpression restored GFP-CPS localization to the lumen of the vacuole, the Snf7Vps2MIM1 chimera failed to complement the snf7Δ MVB sorting defects, as GFP-CPS localized to the vacuolar limiting membrane (Figure 2D). Moreover, Snf7Vps2MIM1 disrupted GFP-CPS localization to the vacuolar lumen of wild type cells (SNF7), demonstrating a dominant negative effect on MVB sorting (Figure 2E). These results indicated that while the Vps2 MIM1 element can bypass the requirement for Vps2 in Vps4-mediated Snf7 membrane release, the Snf7Vps2MIM1 chimera does not support MVB sorting.
ESCRT-III Disassembly Is Dependent on Vps4 MIM1 and MIM2 Interactions
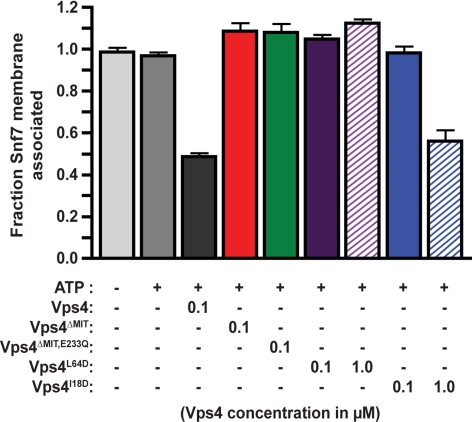
Vps4 interacts with ESCRT-III subunits via its amino-terminal MIT domain (Obita et al., 2007; Stuchell-Brereton et al., 2007; Kieffer et al., 2008). Although ATPase activity is not affected (Supplemental Figure 3A and Babst et al., 1998; Azmi et al., 2006), deletion of this domain (Vps4ΔMIT) abrogates MVB sorting (Vajjhala et al., 2007). Similarly, Vps4ΔMIT failed to support ESCRT-III disassembly (Figure 3), indicating that the MIT domain is required for ESCRT-III disassembly and that Vps4 ATP hydrolysis is not sufficient for this activity. Vps4 mutants predicted to be defective for either MIM2 interaction (Vps4I18D) or MIM1 interaction preferentially (Vps4L64D) (Supplemental Figure 3, A and B and Kieffer et al., 2008) were also analyzed to examine contributions of these two modes of MIT-ESCRT-III association. Vps4L64D was incapable of supporting ESCRT-III disassembly (Figure 3), consistent with Vps4 ESCRT-III disassembly assay dependence on Vps2, Vps24, and Did2 (Figure 2, A and B). While 100 nM Vps4I18D did not support Snf7 release from the membrane, 1 μM Vps4I18D exhibited ESCRT-III disassembly activity (Figure 3). The inability of 100 nM Vps4I18D to support disassembly contrasts with the findings that this reaction was not perturbed by individual or combined losses of MIM2-containing ESCRT-III subunits Vps20 and Ist1 (Figure 2, A and B, and data not shown), suggesting that ESCRT-III subunit redundancy may exist for the MIM2-Vps4 interactions. These results also indicated that Vps4 MIT interactions via both MIM1 and MIM2 binding surfaces contribute to ESCRT-III disassembly.
Figure 3.
ESCRT-III disassembly depends on Vps4 MIT associations with MIM1 and MIM2. ESCRT-III disassembly reactions were performed with inclusion of 100 nM Vps4 (black), 100 nM Vps4ΔMIT (red), 100 nM Vps4ΔMIT,E233Q (green), 100 nM and 1 μM Vps4L64D (purple, MIM1 binding surface), or 100 nM and 1 μM Vps4I18D (blue, MIM2 binding surface).
Coordination within the Vps4 Oligomer During ESCRT-III Disassembly
Vps4 functions as an oligomer, likely comprising 12 subunits (Hanson and Whiteheart, 2005; White and Lauring, 2007; Yu et al., 2008; Landsberg et al., 2009). To examine the relationships among Vps4 subunits, their ATPase activity and engagement with ESCRT-III within the oligomer, ESCRT-III disassembly assays were performed with mixtures of wild-type and mutant Vps4 subunits.
To test whether a full complement of MIT domains is required within the Vps4 oligomer, Vps4ΔMIT subunits were mixed with wild-type subunits (Vps4) in the ESCRT-III disassembly assay. Mixing of Vps4 (100 nM) with Vps4ΔMIT (100 nM, 500 nM, and 1 μM) generated disassembly activities indistinguishable from Vps4 alone (Figure 4A). Similar results were observed upon mixing Vps4ΔMIT,E233Q subunits with Vps4 (Figure 4B). These results suggested that the Vps4 oligomer does not require a full complement of MIT domains to be functional.
However, an alternative explanation was that Vps4ΔMIT and Vps4ΔMIT,E233Q subunits were not incorporated into Vps4 oligomers in this assay. To address this possibility, the ability of Vps4ΔMIT,E233Q to promote ATPase activity of Vps4 subunits was examined. Vps4 ATP hydrolysis exhibits concentration-dependent specific activity, attributed to the role of oligomerization in Vps4 ATP hydrolysis (Babst et al., 1998; Azmi et al., 2006). While Vps4ΔMIT,E233Q did not exhibit ATPase activity by itself (Supplemental Figure 3A), addition of 1 or 2 μM Vps4ΔMIT,E233Q to 250 nM Vps4 enhanced the ATPase activity of the Vps4 subunits (Figure 4C), consistent with formation of Vps4-Vps4ΔMIT,E233Q hetero-oligomers in vitro. To further address this issue, the ESCRT-III disassembly assay was performed with 30 nM Vps4 in the presence of excess Vps4ΔMIT,E233Q (1.2 μM). The addition of 1.2 μM Vps4ΔMIT,E233Q significantly enhanced ESCRT-III disassembly mediated by 30 nM Vps4 (Figure 4D). This result supports the model that Vps4ΔMIT,E233Q forms hetero-oligomers with Vps4 in vitro that support ESCRT-III disassembly. To address whether hetero-oligomer formation occurs in vivo, the association of GFP-Vps4ΔMIT,E233Q with ESCRT-III-positive endosomes was examined in yeast lacking Vta1 (see Materials and Methods). Vps4E233Q-GFP colocalized with Cherry-Did2 (Figure 5B), indicating Vps4E233Q-GFP recruitment to ESCRT-III–positive endosomes. GFP-Vps4ΔMIT,E233Q exhibited partial colocalization with Did2-positive endosomes in vta1Δ yeast (Figure 5D, Supplemental Figure 4A), but this association was no longer evident in vta1Δ vps4Δ cells (Figure 5C, Supplemental Figure 4B). The Vps4-dependence of GFP-Vps4ΔMIT,E233Q localization to ESCRT-III–positive endosomes supported the assertion that Vps4ΔMIT,E233Q forms hetero-oligomers with Vps4, consistent with recent observations (Shestakova et al., 2010). These analyses suggested that the Vps4-Vps4ΔMIT,E233Q hetero-oligomer forms both in vivo and in vitro and supported the conclusion that the Vps4 oligomer does not require a full complement of MIT domains to support ESCRT-III disassembly.
To assess the contribution of Vps4 subunit ATPase activity in ESCRT-III disassembly, mixing experiments were performed with wild-type and Vps4E233Q subunits. Addition of equal molar Vps4E233Q inhibited 100 nM Vps4 ESCRT-III disassembly (Figure 6A). Moreover, reduction of the molar ratio to 1:2 (Vps4E233Q:Vps4) also inhibited Snf7 membrane release. These results suggested that incorporation of subunits that cannot hydrolyze ATP disrupted Vps4 oligomer activity. However, addition of 1 μM Vps4ΔMIT,E233Q to 100 nM Vps4 (10:1) failed to inhibit ESCRT-III disassembly (Figure 4B). This contrast indicated that Vps4E233Q inhibition of ESCRT-III disassembly was dependent on the MIT domain (Figure 8). The inability of Vps4ΔMIT,E233Q subunits to inhibit the Vps4-Vps4ΔMIT,E233Q hetero-oligomer also indicated that every subunit of the Vps4 oligomer does not require catalytic activity to permit ESCRT-III disassembly. However, the inhibitory activity of Vps4E233Q suggested that when a subunit engages ESCRT-III via its MIT domain, ATP hydrolysis by that subunit is required. These observations refine the understanding of subunit composition and action of the Vps4 oligomer by uncovering an intimate link between the engagement of ESCRT-III via the MIT domain and ATP hydrolysis by the engaged Vps4 subunit during ESCRT-III disassembly.
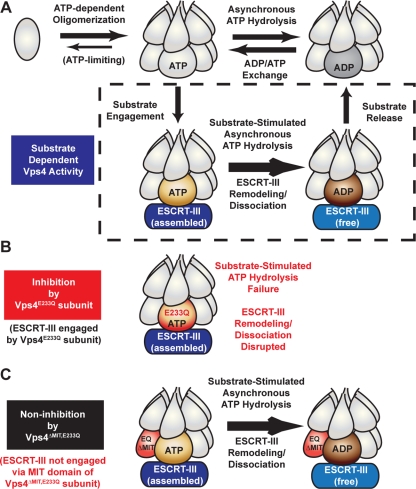
Figure 8.
Coordination of Vps4 ESCRT-III engagement and ATP hydrolysis at the subunit level. (A) Vps4 ATP hydrolysis cycle in the presence or absence of the substrate ESCRT-III as suggested by in vitro analyses. Vps4 oligomerizes in an ATP-dependent manner, suggested to form a two-ring dodecamer (Yu et al., 2008; Landsberg et al., 2009). Neither concerted hydrolysis nor dissociation appear requisite for ATPase activity (upper row) or ESCRT-III disassembly (lower row), suggesting that individual subunits within the oligomer may hydrolyze and reload ATP. The presence of the substrate ESCRT-III can enhance Vps4 ATPase activity in an MIT-dependent manner, suggesting that engagement via the MIT domain potentiates ATP hydrolysis at the Vps4 subunit level to remodel and release ESCRT-III subunits (lower row). [Please note that the orientation of the asymmetric Vps4 oligomer relative to substrate is depicted with artistic license rather than adherence to supportive biochemical observations.] (B) If the engaged Vps4 subunit cannot hydrolyze ATP (Vps4E233Q), then ESCRT-III disassembly is arrested. However, Vps4E233Q does not disrupt substrate-independent ATPase activity (upper row in A). (C) The inhibition of ESCRT-III disassembly by Vps4E233Q is dependent upon the MIT domain, suggesting that coordination between ESCRT-III engagement via the MIT domain and ATP hydrolysis occurs at the subunit level within the Vps4 oligomer.
To further explore Vps4E233Q inhibitory activity, Vps4E233Q effects on ATPase activity of Vps4 subunits was examined. Vps4E233Q addition enhanced the ATPase activity of 250 nM Vps4 in a concentration-dependent manner (Figure 6B). Furthermore, titration of Vps4 subunits in the presence of 2 or 4 μM Vps4E233Q indicated that Vps4E233Q addition reduced the Vps4 apparent Km while the Vps4 Vmax was not enhanced (Figure 6C). These effects are consistent with Vps4E233Q supporting hetero-oligomer formation without disrupting wild-type subunit ATPase activity, as observed with Vps4ΔMIT,E233Q-Vps4 mixtures (Figure 4C). The dissimilar effects of Vps4E233Q and Vps4ΔMIT,E233Q subunits on Vps4 disassembly of ESCRT-III (Figures 4B and 6A) suggested that the presence of ESCRT-III itself plays a critical role in MIT-dependent Vps4E233Q inhibitory activity.
To examine the role of the MIT domain in Vps4E233Q inhibition in vivo, GFP-CPS localization was examined in yeast expressing Vps4E233Q or Vps4ΔMIT,E233Q. Cherry-Vps4E233Q expression resulted in GFP-CPS delivery to the limiting vacuolar membrane, indicating defective MVB sorting (Figure 6D). By contrast, GFP-CPS localization to the vacuolar lumen was not perturbed by Cherry-Vps4ΔMIT,E233Q expression (Figure 6E), indicating that the Vps4E233Q dominant negative effect on MVB sorting is dependent on the MIT domain. This conclusion is consistent with recently reported observations (Shestakova et al., 2010) and is also supported by the largely diffuse distribution, with only limited punctate localization, of Cherry-Did2 upon GFP-Vps4ΔMIT,E233Q expression in vta1Δ cells (Figure 5D). This Did2 pattern is similar to the distribution in vta1Δ cells and is distinct from the extensive Cherry-Did2 endosomal accumulation in vps4Δ cells or in cells expressing Vps4E233Q-GFP (Figure 5A and data not shown). These observations support the conclusion that the Vps4E233Q dominant negative effect on MVB sorting is dependent on the MIT domain, consistent with observations using the ESCRT-III disassembly assay.
Vps4 Functions as a Stable Oligomer In Vitro
Vps4 dissociation upon ATP hydrolysis has been suggested to contribute to the disassembly of ESCRT-III (Babst et al., 1998; Landsberg et al., 2009); however, Vps4 has also been suggested to function as a stable AAA-ATPase oligomer (Scott et al., 2005a; Gonciarz et al., 2008). Because the stability of the Vps4 oligomer is relevant for the mechanism of ESCRT-III disassembly, it was important to determine whether dissociation of the Vps4 oligomer is an implicit step within the ATP hydrolysis cycle.
Reaction conditions of the ATP hydrolysis assay were modified to test Vps4 functional stability. Vps4 exhibits concentration dependent activity (Figure 6C and Babst et al., 1998; Azmi et al., 2006). If dissociation of the Vps4 oligomer is implicit within the ATP hydrolysis cycle, then Vps4 ATPase activity should be sensitive to dilution. However, if the Vps4 oligomer is stable through multiple rounds of ATP hydrolysis, then ATPase activity should be resistant to dilution once the Vps4 oligomer forms. ATPase reactions were initiated with 500 nM Vps4 in the presence of 1 mM ATP. After 30 min, reactions were diluted 10-fold (to 50 nM Vps4) and the continued hydrolysis of ATP was assessed (Figure 7A). Dilution of Vps4 10-fold in the presence of 1 mM ATP resulted in ATPase activity equivalent to the initial rate (Figure 7B), and this activity was undiminished over an additional 60 min. However, if Vps4 was diluted without maintaining the ATP level and then ATP was subsequently restored to 1 mM, the resulting specific activity was reduced by 78% (Figure 7B). This analysis suggested that the Vps4 oligomer dissociates when ATP becomes limiting but is functionally stable when ATP is present. Furthermore, similar dilution experiments revealed that the functional stability of Vps4 ATPase activity is maintained for more than 4 h post dilution in the presence of ATP (Supplemental Figure 3, C and D). These observations support the conclusion that Vps4 can function as a stable oligomer rather than a model in which Vps4 oligomer dissociation is implicit within the ATP hydrolysis cycle.
To address the functional stability of the Vps4 oligomer in the presence of ESCRT-III, the ability of Vps4E233Q to inhibit Vps4 disassembly activity (Figure 6A) was exploited. If Vps4 undergoes dynamic oligomerization and dissociation during ESCRT-III disassembly, then addition of Vps4E233Q subunits subsequent to initiation of the reaction should arrest the ESCRT-III disassembly process; however, if Vps4 functions as a stable oligomer, then Vps4 disassembly activity may be resistant to subsequent addition of Vps4E233Q. Reactions were initiated with Vps4 and ATP, then Vps4E233Q was added 5 min after initiation, and the effect on ESCRT-III disassembly was assessed. Whereas premixing of Vps4 and Vps4E233Q inhibited ESCRT-III disassembly, addition of Vps4E233Q subsequent to initiation failed to arrest Vps4 activity (Figure 7C). This result indicated that Vps4E233Q inhibition of ESCRT-III disassembly was sensitive to the time of addition. This result also suggested that the active Vps4 oligomer was resistant to Vps4E233Q inhibition and that exchange of Vps4 subunits with the soluble pool did not occur once the active oligomer formed. While we cannot exclude that additional factors modulate the dynamics of Vps4 oligomerization in vivo, these results are consistent with the conclusion that the Vps4 oligomer is stable during ATP hydrolysis both in the absence and presence of ESCRT-III and suggest that the Vps4 oligomer remains intact during ESCRT-III disassembly.
DISCUSSION
ESCRT-III disassembly by Vps4 ATP hydrolysis plays a key role in MVB sorting, although how this process occurs and how this reaction contributes to ILV formation remain poorly defined. In vitro reconstitutions of ESCRT-III subunits binding to membranes have demonstrated the principles that the ESCRT-III subunits themselves can deform the membrane and accomplish membrane scission even in the absence of Vps4 or the other ESCRTs (Saksena et al., 2009; Wollert et al., 2009). Whether Vps4 functions subsequent to ESCRT-III–mediated ILV budding to recycle ESCRT-III subunits (Wollert et al., 2009) or whether Vps4 also plays an active role rearranging ESCRT-III during ILV budding are issues that remain to be resolved. Two general models for Vps4 remodeling of ESCRT-III have emerged: 1) Vps4 undergoes a dynamic cycle of oligomerization upon ATP binding and hydrolysis, with ESCRT-III disassembly linked to Vps4 dissociation; or 2) Vps4 functions as a stable oligomer with ATP hydrolysis facilitating ESCRT-III function independent of Vps4 dissociation. The functional studies presented here support models in which Vps4 functions as a stable oligomer during ESCRT-III disassembly. In addition, these studies demonstrated that neither a full complement of MIT domains nor concerted ATP hydrolysis by all subunits are required for the Vps4 oligomer to support ESCRT-III disassembly. These studies suggest a model wherein ESCRT-III engagement (via the MIT domain) and ATP hydrolysis are coordinated at the subunit level within the Vps4 oligomer, leading to disassembly of the ESCRT-III polymer by the stable Vps4 oligomer in a processive manner.
Vps4 is a type 1 AAA-ATPase that forms an active oligomer in the presence of ATP (Babst et al., 1998). Reconstructions of electron micrographs have suggested that the Vps4E233Q oligomer consists of two conformationally distinct hexameric rings (Yu et al., 2008; Landsberg et al., 2009), analogous to type 2 AAA-ATPases including p97 and NSF (White and Lauring, 2007). Size exclusion chromatography of Vps4 and Vps4E233Q has implicated ATP hydrolysis in rapidly dissociating the Vps4 oligomer (Babst et al., 1998). This feature along with the potential of the Vps4 oligomer to concurrently engage multiple ESCRT-III subunits via MIT domains have suggested a possible mechanism for Vps4-mediated ESCRT-III disassembly (Babst et al., 1998; Landsberg et al., 2009). However, another model has emerged through investigations of the central pore of the Vps4 oligomeric ring (Scott et al., 2005a; Gonciarz et al., 2008). Mutational analyses of residues lining this central cavity have suggested that the Vps4 pore facilitates substrate remodeling in a manner analogous to other AAA-ATPases such as ClpX, Hsp104 and NSF (Hanson and Whiteheart, 2005; White and Lauring, 2007). This model implies that Vps4 functions as a stable oligomer facilitating ESCRT-III disassembly via the pore, although neither the ability of Vps4 to alter ESCRT-III subunit conformations nor the stability of the Vps4 oligomer during ESCRT-III disassembly had been demonstrated. To explore these two distinct models of Vps4 function and to gain insights into coordination among subunits within the Vps4 oligomer, the Vps4-mediated ESCRT-III disassembly reaction was reconstituted in vitro using ESCRT-III polymers generated in yeast. This system also permitted examination of the ESCRT-III subunit-dependence of this disassembly process.
ESCRT-III subunits have been observed to assemble into fibril-like polymers in vivo that may spiral to facilitate membrane deformation and ILV budding (Hanson et al., 2008). Snf7 appears to be the predominant component of these ESCRT-III fibrils (Hanson et al., 2008; Teis et al., 2008) and was selected as the primary indicator of ESCRT-III membrane association and assembly in this reconstituted system. While the individual losses of Vps20, Ist1, or Vps60 did not perturb Snf7 membrane release, the absences of Vps24, Vps2, or Did2 compromised the ESCRT-III disassembly reaction with the loss of Vps2 associated with the most severe effect. These observations recapitulate prior studies indicating that Snf7 recycling in vivo is dependent upon Vps24, Vps2, and Did2 (Babst et al., 2002a; Nickerson et al., 2006; Teis et al., 2008), supporting the suitability of this system for investigating Vps4 disassembly of ESCRT-III. While Vps24, Vps2, and Did2 may directly contribute to the recruitment, activation, and/or progression of Vps4 during ESCRT-III disassembly, it is also possible that the observed effects are indirect. For instance, the absences of these subunits (Vps24, Vps2 or Did2) may permit Snf7 subunits to assemble in an aberrant manner recalcitrant to Vps4 disassembly. Another possibility is that losses of these subunits may permit other factors to bind to ESCRT-III that impinge on Vps4 activity. While further investigation of this system is required to address these possibilities and to define the individual contributions, the experiments presented herein demonstrate that Vps24, Vps2 and Did2 facilitate Vps4 disassembly of ESCRT-III.
These studies also imply that the Vps20-Snf7 oligomer is resistant to Vps4 dissociation in spite of the presence of a Vps4 interaction motif (MIM2). This suggests that the MIM1-mediated activation of Vps4 ATPase activity is critical for ESCRT-III dissociation. Thus, maturation of the ESCRT-III polymer with the incorporation of MIM1-containing subunits may serve both to permit completion of ILV formation (Wollert et al., 2009) and to facilitate Vps4-mediated ESCRT-III disassembly, thereby providing a timing component for ESCRT-III function (Supplemental Figure 4C). The role of the Vps2 MIM1 element in ESCRT-III function was investigated, as loss of Vps2 could not be suppressed by increasing the Vps4 concentration. A Snf7-Vps2 chimera (Snf7Vps2MIM1) was examined for function in the ESCRT-III disassembly system as well as for effects on MVB sorting in vivo. Snf7Vps2MIM1 was competent as a substrate for disassembly by Vps4 and could bypass the requirement for Vps2 in this system; this observation is consistent with the ability of the Vps2 MIM1 to enable Vps4 disassembly of Vps24Vps2MIM1 homo-oligomers (Ghazi-Tabatabai et al., 2008). However, Snf7Vps2MIM1 was not able to complement the MVB sorting defects of snf7Δ yeast and perturbed MVB sorting of wild-type cells. These results indicate that it is possible to separate the requirements for ESCRT-III function in MVB sorting from the requirements for Vps4-mediated ESCRT-III disassembly, although this distinction is accomplished via a nonnative protein. Snf7Vps2MIM1 may enable the premature disassembly of ESCRT-III, disrupting the coordination of ESCRT-III assembly and disassembly necessary for MVB sorting. However, alternative possibilities, including indirect effects on ESCRT-III assembly or the altered recruitment of ESCRT-III effectors, may also contribute to the disruptive effects of Snf7Vps2MIM1. Further investigation will be required to understand the coordination of ESCRT-III assembly and disassembly that facilitates MVB sorting.
The ESCRT-III disassembly assay was also used to examine the functional requirements for Vps4. Both ATP hydrolysis and ESCRT-III engagement via the MIT domain were required for Vps4 to mediate ESCRT-III disassembly. The in vitro system permitted these requirements to be examined further through mixing wild-type and mutant subunits, addressing functional aspects of the Vps4 oligomer. Addition of equimolar or excess Vps4ΔMIT or Vps4ΔMIT,E233Q subunits did not compromise the ESCRT-III disassembly function of wild-type Vps4 subunits. Moreover, an excess of Vps4ΔMIT,E233Q was able to enhance the activity of a limiting amount of Vps4. These observations indicate that subunits lacking the MIT domain can be incorporated into a Vps4 oligomer without abolishing oligomer activity and that the Vps4 oligomer does not require a full complement of MIT domains. The observations with Vps4ΔMIT,E233Q subunits also indicate that concerted ATP hydrolysis by all subunits of the Vps4 oligomer is not required for ESCRT-III disassembly. The demonstration that even a single catalytically active subunit can enable function of the hexameric AAA-ATPase ClpX (Hersch et al., 2005; Martin et al., 2005) is consistent with this conclusion that concerted ATP hydrolysis by all subunits is not required for Vps4 function; however, additional refinement of the hydrolysis requirements for the Vps4 oligomer will be required to permit further comparisons with the enzymology and mechanisms of ClpX (Glynn et al., 2009). Consistent with these in vitro observations of Vps4, Vps4ΔMIT,E233Q was observed to localize to endosomes in the presence of wild-type Vps4 subunits without disrupting MVB sorting, consistent with recently reported studies (Shestakova et al., 2010). While the incorporation of Vps4ΔMIT or Vps4ΔMIT,E233Q subunits are anticipated to alter the Vps4 oligomer affinity for ESCRT-III and/or the efficiency of ESCRT-III disassembly, these effects were not apparent under the conditions examined. Further examination of these properties is warranted, but the results presented here establish the principles that neither a full complement of MIT domains nor concerted ATP hydrolysis by all subunits are required for ESCRT-III disassembly by the Vps4 oligomer.
While concerted hydrolysis by all subunits is not required, the coordination of ESCRT-III binding via the MIT domain with ATP hydrolysis by the engaged subunit appears to be a critical aspect of Vps4 function. Hydrolysis-defective mutants of Vps4, such as Vps4E233Q, have previously been exploited as dominant negative mutants disrupting MVB sorting, viral budding and abscission (Babst et al., 1997; Garrus et al., 2001; Carlton and Martin-Serrano, 2007). However, the basis of compromised Vps4 oligomer function was not previously addressed. In contrast to the dominant negative effect in vivo, Vps4E233Q subunits enhanced the ATPase activity of wild-type subunits in vitro. A similar stimulation by noncatalytic subunits has been observed with Hsp104/ClpB AAA-ATPases by treating with a nonhydrolyzable ATP analog, ATPγS (Doyle et al., 2007); however, ATPγS treatment inhibited Vps4 activity in a manner comparable to the behavior of the AAA-ATPase ClpA rather than the stimulation observed with Hsp104 and ClpB (data not shown and Doyle et al., 2007). Whereas Vps4E233Q subunits enhanced ATPase activity of wild-type subunits, Vps4E233Q addition disrupted Vps4 ESCRT-III disassembly even with Vps4 in excess. The distinct effects of Vps4E233Q and Vps4ΔMIT,E233Q on ESCRT-III disassembly indicated that the inhibitory effect of Vps4E233Q is dependent on the MIT domain, and this dependence is similarly observed for Vps4E233Q inhibition of MVB sorting (Figure 6D and Shestakova et al., 2010). These results suggest that coordination between MIT engagement and ATP hydrolysis is required at the subunit level within the Vps4 oligomer. One possibility is that ESCRT-III binding via the MIT domain induces a conformational change within the engaged Vps4 subunit that must be resolved by ATP hydrolysis for the Vps4 oligomer to continue to disassemble ESCRT-III. Another nonexclusive possibility is that ESCRT-III binding via the MIT domain facilitates transfer of ESCRT-III to the central pore and that ATP hydrolysis by the MIT-engaged Vps4 subunit must occur to prevent occlusion of the pore. Moreover, these two possibilities may be related and serve to orchestrate substrate engagements via the MIT domain and the central pore with ATP hydrolysis. While the analysis presented here indicates that coordination of MIT domain engagement and ATP hydrolysis occurs at the subunit level within the Vps4 oligomer, the mechanisms by which this occurs remains to be determined.
The ability of Vps4E233Q to inhibit ESCRT-III disassembly enabled the dynamics of Vps4 oligomerization to be examined. Whereas premixing of Vps4E233Q with Vps4 inhibited ESCRT-III disassembly, addition of Vps4E233Q subsequent to initiation of the reaction prevented the inhibitory effect. This result suggests that dynamic exchange of Vps4 subunits does not occur during the in vitro ESCRT-III disassembly reaction and that Vps4 functions as a stable oligomer. Moreover, these results suggest that a Vps4 oligomer may remain associated with an ESCRT-III polymer as it disassembles ESCRT-III subunits in a processive manner (rather than the Vps4 oligomer releasing and rebinding to the ESCRT-III polymer). The stability of the Vps4 oligomer during ESCRT-III disassembly is consistent with the dilution-resistance of ATPase activity in vitro. While we cannot exclude the possibility that the dynamics of Vps4 oligomerization are altered by additional factors in vivo and we acknowledge the absence of direct biophysical demonstration of Vps4 oligomer stability in vitro, these results support a model wherein Vps4 assembles into an oligomer and facilitates ESCRT-III disassembly without itself dissociating. This model is consistent with asynchronous/nonconcerted ATP hydrolysis among Vps4 subunits, whereby the remaining ATP-bound subunits stabilize the oligomer to permit ATP reloading after hydrolysis without dissociation (Figure 8A). This model is supported by the observation that catalytically inactive subunits (Vps4E233Q) stimulate the ATPase activity of wild-type subunits in vitro in the absence of substrate. ESCRT-III binding has been demonstrated to enhance Vps4 ATPase activity in a MIT-dependent manner (Azmi et al., 2008). This observation in conjunction with the MIT-dependence of Vps4E233Q inhibition suggests that ESCRT-III binding via the MIT domain induces an activated state of the Vps4 oligomer with stimulation occurring at the level of the engaged subunit (Figure 8A). ATP hydrolysis by the engaged subunit is then required to facilitate ESCRT-III disassembly and permit continued Vps4 function (Figure 8B). The observation that the Vps4 oligomer does not require a full complement of MIT domains suggests that this cycle of ESCRT-III polymer binding, Vps4 subunit activation, ATP hydrolysis, and ESCRT-III subunit release occurs in a progressive manner to disassemble the ESCRT-III polymer rather than in mass. This cycle appears consistent with Vps4 remodeling ESCRT-III subunits via the central pore in a subunit-by-subunit manner to facilitate ESCRT-III disassembly, although experimental evidence will be required to confirm this model.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Zhaohui Xu (U. Michigan) for discussions regarding the Vps4 oligomer, Drs. Wesley Sundquist and Jack J. Skalicky (U. Utah) for discussions concerning Vps4 MIT-MIM2 interactions, members of the Katzmann and Horazdovsky labs for helpful scientific discussions, and Sandra Severson for technical support. This work has been supported by a Fraternal Order of Eagles postdoctoral fellowship (to B.A.D.), an American Heart Association predoctoral fellowship (AHA07-155882 to I.F.A.), and by grants R01 GM073024 (to D.J.K.) and R01 GM074171 (to M. B.) from the National Institutes of Health.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-06-0512) on August 4, 2010.
REFERENCES
- Agromayor M., Carlton J. G., Phelan J. P., Matthews D. R., Carlin L. M., Ameer-Beg S., Bowers K., Martin-Serrano J. Essential role of hIST1 in cytokinesis. Mol. Biol. Cell. 2009;20:1374–1387. doi: 10.1091/mbc.E08-05-0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azmi I., Davies B., Dimaano C., Payne J., Eckert D., Babst M., Katzmann D. J. Recycling of ESCRTs by the AAA-ATPase Vps4 is regulated by a conserved VSL region in Vta1. J. Cell Biol. 2006;172:705–717. doi: 10.1083/jcb.200508166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azmi I. F., Davies B. A., Xiao J., Babst M., Xu Z., Katzmann D. J. ESCRT-III family members stimulate Vps4 ATPase activity directly or via Vta1. Dev. Cell. 2008;14:50–61. doi: 10.1016/j.devcel.2007.10.021. [DOI] [PubMed] [Google Scholar]
- Babst M., Katzmann D. J., Estepa-Sabal E. J., Meerloo T., Emr S. D. Escrt-III: an endosome-associated heterooligomeric protein complex required for mvb sorting. Dev. Cell. 2002a;3:271–282. doi: 10.1016/s1534-5807(02)00220-4. [DOI] [PubMed] [Google Scholar]
- Babst M., Katzmann D. J., Snyder W. B., Wendland B., Emr S. D. Endosome-associated complex, ESCRT-II, recruits transport machinery for protein sorting at the multivesicular body. Dev. Cell. 2002b;3:283–289. doi: 10.1016/s1534-5807(02)00219-8. [DOI] [PubMed] [Google Scholar]
- Babst M., Sato T. K., Banta L. M., Emr S. D. Endosomal transport function in yeast requires a novel AAA-type ATPase, Vps4p. EMBO J. 1997;16:1820–1831. doi: 10.1093/emboj/16.8.1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst M., Wendland B., Estepa E. J., Emr S. D. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 1998;17:2982–2993. doi: 10.1093/emboj/17.11.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajorek M., Morita E., Skalicky J. J., Morham S. G., Babst M., Sundquist W. I. Biochemical analyses of human IST1 and its function in cytokinesis. Mol. Biol. Cell. 2009a;20:1360–1373. doi: 10.1091/mbc.E08-05-0475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajorek M., Schubert H. L., McCullough J., Langelier C., Eckert D. M., Stubblefield W. M., Uter N. T., Myszka D. G., Hill C. P., Sundquist W. I. Structural basis for ESCRT-III protein autoinhibition. Nat. Struct. Mol. Biol. 2009b;16:754–762. doi: 10.1038/nsmb.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilodeau P. S., Winistorfer S. C., Kearney W. R., Robertson A. D., Piper R. C. Vps27-Hse1 and ESCRT-I complexes cooperate to increase efficiency of sorting ubiquitinated proteins at the endosome. J. Cell Biol. 2003;163:237–243. doi: 10.1083/jcb.200305007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton J. G., Agromayor M., Martin-Serrano J. Differential requirements for Alix and ESCRT-III in cytokinesis and HIV-1 release. Proc. Natl. Acad. Sci. USA. 2008;105:10541–10546. doi: 10.1073/pnas.0802008105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton J. G., Martin-Serrano J. Parallels between cytokinesis and retroviral budding: a role for the ESCRT machinery. Science. 2007;316:1908–1912. doi: 10.1126/science.1143422. [DOI] [PubMed] [Google Scholar]
- Davies B. A., Lee J. R., Oestreich A. J., Katzmann D. J. Membrane protein targeting to the MVB/lysosome. Chem. Rev. 2009;109:1575–1586. doi: 10.1021/cr800473s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimaano C., Jones C. B., Hanono A., Curtiss M., Babst M. Ist1 regulates Vps4 localization and assembly. Mol. Biol. Cell. 2008;19:465–474. doi: 10.1091/mbc.E07-08-0747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle S. M., Shorter J., Zolkiewski M., Hoskins J. R., Lindquist S., Wickner S. Asymmetric deceleration of ClpB or Hsp104 ATPase activity unleashes protein-remodeling activity. Nat. Struct. Mol. Biol. 2007;14:114–122. doi: 10.1038/nsmb1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrus J. E., von Schwedler U. K., Pornillos O. W., Morham S. G., Zavitz K. H., Wang H. E., Wettstein D. A., Stray K. M., Cote M., Rich R. L., Myszka D. G., Sundquist W. I. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell. 2001;107:55–65. doi: 10.1016/s0092-8674(01)00506-2. [DOI] [PubMed] [Google Scholar]
- Ghaemmaghami S., Huh W. K., Bower K., Howson R. W., Belle A., Dephoure N., O'Shea E. K., Weissman J. S. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- Ghazi-Tabatabai S., Saksena S., Short J. M., Pobbati A. V., Veprintsev D. B., Crowther R. A., Emr S. D., Egelman E. H., Williams R. L. Structure and disassembly of filaments formed by the ESCRT-III subunit Vps24. Structure. 2008;16:1345–1356. doi: 10.1016/j.str.2008.06.010. [DOI] [PubMed] [Google Scholar]
- Glynn S. E., Martin A., Nager A. R., Baker T. A., Sauer R. T. Structures of asymmetric ClpX hexamers reveal nucleotide-dependent motions in a AAA+ protein-unfolding machine. Cell. 2009;139:744–756. doi: 10.1016/j.cell.2009.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonciarz M. D., Whitby F. G., Eckert D. M., Kieffer C., Heroux A., Sundquist W. I., Hill C. P. Biochemical and structural studies of yeast Vps4 oligomerization. J. Mol. Biol. 2008;384:878–895. doi: 10.1016/j.jmb.2008.09.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenberg J., Stenmark H. The biogenesis of multivesicular endosomes. Nat. Rev. Mol. Cell Biol. 2004;5:317–323. doi: 10.1038/nrm1360. [DOI] [PubMed] [Google Scholar]
- Hanson P. I., Roth R., Lin Y., Heuser J. E. Plasma membrane deformation by circular arrays of ESCRT-III protein filaments. J. Cell Biol. 2008;180:389–402. doi: 10.1083/jcb.200707031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson P. I., Shim S., Merrill S. A. Cell biology of the ESCRT machinery. Curr. Opin. Cell Biol. 2009;21:568–574. doi: 10.1016/j.ceb.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson P. I., Whiteheart S. W. AAA+ proteins: have engine, will work. Nat. Rev. Mol. Cell Biol. 2005;6:519–529. doi: 10.1038/nrm1684. [DOI] [PubMed] [Google Scholar]
- Hersch G. L., Burton R. E., Bolon D. N., Baker T. A., Sauer R. T. Asymmetric interactions of ATP with the AAA+ ClpX6 unfoldase: allosteric control of a protein machine. Cell. 2005;121:1017–1027. doi: 10.1016/j.cell.2005.05.024. [DOI] [PubMed] [Google Scholar]
- Hurley J. H. ESCRT complexes and the biogenesis of multivesicular bodies. Curr. Opin. Cell Biol. 2008;20:4–11. doi: 10.1016/j.ceb.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzmann D. J., Babst M., Emr S. D. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell. 2001;106:145–155. doi: 10.1016/s0092-8674(01)00434-2. [DOI] [PubMed] [Google Scholar]
- Kieffer C., Skalicky J. J., Morita E., De Domenico I., Ward D. M., Kaplan J., Sundquist W. I. Two distinct modes of ESCRT-III recognition are required for VPS4 functions in lysosomal protein targeting and HIV-1 budding. Dev. Cell. 2008;15:62–73. doi: 10.1016/j.devcel.2008.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- Landsberg M. J., Vajjhala P. R., Rothnagel R., Munn A. L., Hankamer B. Three-dimensional structure of AAA ATPase Vps 4, advancing structural insights into the mechanisms of endosomal sorting and enveloped virus budding. Structure. 2009;17:427–437. doi: 10.1016/j.str.2008.12.020. [DOI] [PubMed] [Google Scholar]
- Lata S., Schoehn G., Jain A., Pires R., Piehler J., Gottlinger H. G., Weissenhorn W. Helical structures of ESCRT-III are disassembled by VPS4. Science. 2008;321:1354–1357. doi: 10.1126/science.1161070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. H., Elia N., Ghirlando R., Lippincott-Schwartz J., Hurley J. H. Midbody targeting of the ESCRT machinery by a noncanonical coiled coil in CEP55. Science. 2008;322:576–580. doi: 10.1126/science.1162042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A., Baker T. A., Sauer R. T. Rebuilt AAA + motors reveal operating principles for ATP-fuelled machines. Nature. 2005;437:1115–1120. doi: 10.1038/nature04031. [DOI] [PubMed] [Google Scholar]
- Morita E., Sandrin V., Chung H. Y., Morham S. G., Gygi S. P., Rodesch C. K., Sundquist W. I. Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis. EMBO J. 2007;26:4215–4227. doi: 10.1038/sj.emboj.7601850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita E., Sundquist W. I. Retrovirus budding. Annu. Rev. Cell Dev. Biol. 2004;20:395–425. doi: 10.1146/annurev.cellbio.20.010403.102350. [DOI] [PubMed] [Google Scholar]
- Muziol T., Pineda-Molina E., Ravelli R. B., Zamborlini A., Usami Y., Gottlinger H., Weissenhorn W. Structural basis for budding by the ESCRT-III factor CHMP3. Dev. Cell. 2006;10:821–830. doi: 10.1016/j.devcel.2006.03.013. [DOI] [PubMed] [Google Scholar]
- Nickerson D. P., West M., Henry R., Odorizzi G. Regulators of vps4 ATPase activity at endosomes differentially influence the size and rate of formation of intralumenal vesicles. Mol. Biol. Cell. 2010;21:1023–1032. doi: 10.1091/mbc.E09-09-0776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickerson D. P., West M., Odorizzi G. Did2 coordinates Vps4-mediated dissociation of ESCRT-III from endosomes. J. Cell Biol. 2006;175:715–720. doi: 10.1083/jcb.200606113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obita T., Saksena S., Ghazi-Tabatabai S., Gill D. J., Perisic O., Emr S. D., Williams R. L. Structural basis for selective recognition of ESCRT-III by the AAA ATPase Vps4. Nature. 2007;449:735–739. doi: 10.1038/nature06171. [DOI] [PubMed] [Google Scholar]
- Piper R. C., Katzmann D. J. Biogenesis and function of multivesicular bodies. Annu. Rev. Cell Dev. Biol. 2007;23:519–547. doi: 10.1146/annurev.cellbio.23.090506.123319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper R. C., Luzio J. P. Ubiquitin-dependent sorting of integral membrane proteins for degradation in lysosomes. Curr. Opin. Cell Biol. 2007;19:459–465. doi: 10.1016/j.ceb.2007.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raiborg C., Stenmark H. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature. 2009;458:445–452. doi: 10.1038/nature07961. [DOI] [PubMed] [Google Scholar]
- Rue S. M., Mattei S., Saksena S., Emr S. D. Novel Ist1-Did2 complex functions at a late step in multivesicular body sorting. Mol. Biol. Cell. 2008;19:475–484. doi: 10.1091/mbc.E07-07-0694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saksena S., Wahlman J., Teis D., Johnson A. E., Emr S. D. Functional reconstitution of ESCRT-III assembly and disassembly. Cell. 2009;136:97–109. doi: 10.1016/j.cell.2008.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott A., Chung H. Y., Gonciarz-Swiatek M., Hill G. C., Whitby F. G., Gaspar J., Holton J. M., Viswanathan R., Ghaffarian S., Hill C. P., Sundquist W. I. Structural and mechanistic studies of VPS4 proteins. EMBO J. 2005a;24:3658–3669. doi: 10.1038/sj.emboj.7600818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott A., Gaspar J., Stuchell-Brereton M. D., Alam S. L., Skalicky J. J., Sundquist W. I. Structure and ESCRT-III protein interactions of the MIT domain of human VPS4A. Proc. Natl. Acad. Sci. USA. 2005b;102:13813–13818. doi: 10.1073/pnas.0502165102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shestakova A., Hanono A., Drosner S., Curtiss M., Davies B. A., Katzmann D. J., Babst M. Assembly of the AAA ATPase Vps4 on ESCRT-III. Mol. Biol. Cell. 2010;21:1059–1071. doi: 10.1091/mbc.E09-07-0572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slagsvold T., Pattni K., Malerod L., Stenmark H. Endosomal and non-endosomal functions of ESCRT proteins. Trends Cell Biol. 2006;16:317–326. doi: 10.1016/j.tcb.2006.04.004. [DOI] [PubMed] [Google Scholar]
- Stuchell-Brereton M. D., Skalicky J. J., Kieffer C., Karren M. A., Ghaffarian S., Sundquist W. I. ESCRT-III recognition by VPS4 ATPases. Nature. 2007;449:740–744. doi: 10.1038/nature06172. [DOI] [PubMed] [Google Scholar]
- Stuffers S., Brech A., Stenmark H. ESCRT proteins in physiology and disease. Exp. Cell Res. 2009;315:1619–1626. doi: 10.1016/j.yexcr.2008.10.013. [DOI] [PubMed] [Google Scholar]
- Tanaka N., Kyuuma M., Sugamura K. Endosomal sorting complex required for transport proteins in cancer pathogenesis, vesicular transport, and non-endosomal functions. Cancer Sci. 2008;99:1293–1303. doi: 10.1111/j.1349-7006.2008.00825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teis D., Saksena S., Emr S. D. Ordered assembly of the ESCRT-III complex on endosomes is required to sequester cargo during MVB formation. Dev. Cell. 2008;15:578–589. doi: 10.1016/j.devcel.2008.08.013. [DOI] [PubMed] [Google Scholar]
- Vajjhala P. R., Catchpoole E., Nguyen C. H., Kistler C., Munn A. L. Vps4 regulates a subset of protein interactions at the multivesicular endosome. FEBS J. 2007;274:1894–1907. doi: 10.1111/j.1742-4658.2007.05736.x. [DOI] [PubMed] [Google Scholar]
- Vida T. A., Graham T. R., Emr S. D. In vitro reconstitution of intercompartmental protein transport to the yeast vacuole. J. Cell Biol. 1990;111:2871–2884. doi: 10.1083/jcb.111.6.2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White S. R., Lauring B. AAA+ ATPases: achieving diversity of function with conserved machinery. Traffic. 2007;8:1657–1667. doi: 10.1111/j.1600-0854.2007.00642.x. [DOI] [PubMed] [Google Scholar]
- Wollert T., Wunder C., Lippincott-Schwartz J., Hurley J. H. Membrane scission by the ESCRT-III complex. Nature. 2009;458:172–177. doi: 10.1038/nature07836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J., Chen X.W., Davies B.A., Saltiel A.R., Katzmann D.J., Xu Z. Structural Basis of Ist1 Function and Ist1-Did2 Interaction in the MVB Pathway and Cytokinesis. Mol. Biol. Cell. 2009;20:3514–3524. doi: 10.1091/mbc.E09-05-0403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z., Gonciarz M. D., Sundquist W. I., Hill C. P., Jensen G. J. Cryo-EM structure of dodecameric Vps4p and its 2, 1 complex with Vta1p. J. Mol. Biol. 2008;377:364–377. doi: 10.1016/j.jmb.2008.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.