Nuclear shape changes are observed during a variety of developmental processes, pathological conditions and ageing. Here, the molecular mechanism is analyzed how the farnesylated nuclear proteins interact with the nuclear envelope and deform the phospholipid bilayer.
Abstract
Nuclear shape changes are observed during a variety of developmental processes, pathological conditions, and ageing. The mechanisms underlying nuclear shape changes in the above-mentioned situations have mostly remained unclear. To address the molecular mechanism behind nuclear shape changes, we analyzed how the farnesylated nuclear envelope proteins Kugelkern and lamin Dm0 affect the structure of the nuclear membrane. We found that Kugelkern and lamin Dm0 affect nuclear shape without requiring filament formation or the presence of a classical nuclear lamina. We also could show that the two proteins do not depend on a group of selected inner nuclear membrane proteins for their localization to the nuclear envelope. Surprisingly, we found that farnesylated Kugelkern and lamin Dm0 protein constructs change the morphology of protein-free liposomes. Based on these findings, we propose that farnesylated proteins of the nuclear membrane induce nuclear shape changes by being asymmetrically inserted into the phospholipid bilayer via their farnesylated C-terminal part.
INTRODUCTION
Nuclear shape and size changes are frequently observed during differentiation, as well as in various pathologies. A prominent example of developmental nuclear morphology changes are the nuclei of granulocytes (Olins and Olins, 2005). The molecular mechanism that defines nuclear shape remains unclear (Webster et al., 2009). An important role in determining nuclear shape is played by the nuclear lamina, as indicated by the abnormal nuclear shapes observed in diseases caused by mutations in lamina proteins (Capell and Collins, 2006; Mattout et al., 2006).
The nuclear lamina is composed of lamins and lamin-related proteins. In vertebrates, there are three lamin genes—LMNA, LMNB1, and LMNB2—that encode the different lamin proteins, whereas in Drosophila there are two lamin genes (coding for lamin Dm0 and lamin C) and in Caenorhabditis elegans only one gene (Goldman et al., 2002). In Saccharomyces cerevisiae, there are no lamins and typical lamin-associated proteins (Erber et al., 1998). It is unknown whether a structural or functional equivalent to the nuclear lamina is found in yeast (Hattier et al., 2007). The absence of lamins from yeast and the higher number of lamins and inner nuclear membrane (INM) proteins in vertebrates suggest an increase in complexity of the nuclear lamina during metazoan evolution (Cohen et al., 2001).
Lamins are nuclear-specific type V intermediate filaments, consisting of an N-terminal head domain followed by an α-helical rod domain and a globular C-terminal tail domain. The C-terminal tail domain contains the nuclear localization signal (NLS), the immunoglobulin fold, and a CaaX motif. Lamin C lacks the CaaX motif. The CaaX motif (where “C” is a cysteine, “a” is an aliphatic amino acid, and “X” is any amino acid), gets farnesylated, thus rendering the protein lipophilic and promoting its association with the nuclear membrane (NM) (Kitten and Nigg, 1991; Krohne, 1998). In A-type lamins, the modified CaaX motif is cleaved during the maturation of the polypeptide, whereas B-type lamins remain permanently farnesylated (Meshorer and Gruenbaum, 2008).
Mutations spanning the entire human LMNA gene lead to a variety of diseases collectively called laminopathies (Capell and Collins, 2006). A striking example is the Hutchinson–Gilford progeria syndrome (HGPS). This premature ageing syndrome is caused by a point mutation in exon 11 of LMNA. The mutation activates a cryptic splice site, which leads to the formation of the permanently farnesylated lamin A variant LaΔ50 (De Sandre-Giovannoli et al., 2003; Eriksson et al., 2003). Due to the accumulation of LaΔ50, the nuclei of the patient's fibroblasts are large and abnormally shaped (Goldman et al., 2004). Farnesylation is required for the activity of LaΔ50 on nuclear shape, because the use of farnesyltransferase inhibitors (FTIs) reverses the nuclear shape defects (Toth et al., 2005; Yang et al., 2005).
Apart from lamins, the only so far known farnesylated nuclear protein is the Drosophila INM protein Kugelkern (Kuk) (Brandt et al., 2006). Kuk protein contains a putative coiled coil motif in its N terminus, an NLS, and a CaaX motif in the C terminus. Apart from these structural similarities, Kuk shares functional similarities with lamins, because overexpression of Kuk and lamin cause comparable phenotypes (Brandt et al., 2006, 2008).
Increase of Kuk or lamin levels lead to the formation of enlarged nuclei with nuclear membrane infoldings. These effects of farnesylated nuclear proteins on the NM seem to be primarily due to their association with the NM via the lipophilic CaaX motif. B type lamins containing a CaaX motif were shown to induce NM growth in cultured cells and zebrafish embryos, whereas Drosophila lamin C that does not contain a CaaX motif and nonfarnesylatable lamin B mutants containing an SaaX are not able to change nuclear shape (Prufert et al., 2004). The use of short green fluorescent protein (GFP)-tagged truncated lamin variants containing only the C-terminal part of lamins in the same study, led to the formation of lobulated, abnormal nuclei, thus showing that the N-terminal part of lamins is not required for their activity at the NM. Similar results were obtained by increased synthesis of lamin B1 and B2 in Xenopus oocytes and by expressing a chimeric GFP-NLS-CaaX construct in HeLa cells (Ralle et al., 2004). According to the above-mentioned studies, the CaaX motif of farnesylated lamins and potentially of Kugelkern seems to be sufficient for inducing NM growth, because even the chimeric construct GFP-NLS-CaaX, where the rod domain of lamin is absent, shows activity on the NM.
In contrast to the filament forming N-terminal part of lamin that has been extensively studied (Wiesel et al., 2008; Ben-Harush et al., 2009; Kapinos et al., 2010), the analysis of the C-terminal part has been rather limited. Given its clear activity on nuclear size and shape, and its connection to HGPS, we analyzed how the farnesylated proteins lamin Dm0 and Kuk induce nuclear shape changes via their C-terminal farnesylated part. According to a previous study, both the coiled coil motif and farnesylation motif of Kuk are required for localization and function of the protein (Brandt et al., 2006). In addition to these two domains, we show that a conserved C-terminal amino acid (aa) sequence of Kuk is required for its function in nuclear elongation in the fly embryo. Strikingly, we observed that Kuk and a truncated lamin Dm0 construct induce abnormal nuclear shapes in yeast, despite the absence of a classical nuclear lamina, which indicates a lamina independent mechanism of activity. This hypothesis is further supported by the fact that both lamin Dm0 and Kuk localize at the NM independently of other known INM proteins, as shown by an RNA interference (RNAi)-mediated depletion of a set of lamina and INM proteins in S2 cells. Taking into account the indications for a lamina-independent mechanism of activity, we performed in vitro liposome binding and deformation assays and we found that farnesylated Kuk and lamin Dm0 constructs can change the morphology of protein-free liposomes.
MATERIALS AND METHODS
Mammalian Cell Culture
NIH-3T3 cells were maintained in DMEM (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS; Invitrogen) and 2 mM l-glutamine (Invitrogen). For transient transfections, the cells were plated on glass coverslips at 50% confluence and transfected using Effectene (QIAGEN, Hilden, Germany) according to the manufacturer's instructions. All constructs used for the transient transfections were cloned in the pCS2-HA vector except GFP-NLS-C-term and GFP-NLS-CaaX that were cloned in the pCS2 vector. The p-LI-GCN4 construct used for making the Kuk-cc+ΔN185 construct was kindly provided by B. Schwappach (University of Manchester). Twenty-four hours after transfection, the cells were fixed and stained as described in Brandt et al. (2008). For time-lapse recordings, NIH-3T3 cells were transfected with pCS2-GFP-Kuk-Δ353-404 in eight-well chamber μ-slides (Ibidi, Martinsried, Germany). Twenty-four hours after transfection, before recording, the medium was changed to Leibovitz's L-15 medium (Invitrogen) supplemented with 10% FBS.
Drosophila Cell Culture
S2 Drosophila cells were maintained at 25°C in Schneider's medium (Invitrogen) supplemented with 10% FBS. S2 cells were transfected with pCS2-HA-Kuk by using Effectene (QIAGEN) according to the manufacturer's instructions for suspension cells. Twenty-four hours after transfection, the cells were seeded on glass coverslips, fixed in 2% formaldehyde (FA) in phosphate-buffered saline (PBS), permeabilized in 0.1% Triton X-100 in PBS, and immunostained. RNAi treatment was preformed as described previously (Worby et al., 2001). Five days after RNAi treatment, the cells were collected for immunostaining or Western blotting.
FTI Treatment
NIH-3T3 cells were transiently transfected in medium supplemented with the FTI ABT-100 (Abbott Laboratories, Abbott Park, IL) at a concentration of 6.25 μg/ml and fixed and immunostained 24 h after transfection. S2 cells were incubated in medium supplemented with ABT-100 at a concentration of 6.25 μg/ml for 5 d.
Antibodies Used for Immunostaining and Immunoblotting
Primary antibodies included the following: rabbit-α-Kuk (Brandt et al., 2006), goat-α-LaminA/C (Santa Cruz Biotechnology, Santa Cruz, CA), mAb414 (Sigma-Aldrich, St. Louis, MO), mouse-α-hemagglutinin (HA) (BAbCO. Richmond, CA), rabbit-α-Nup50 (Brandt et al., 2006), mouse-α-Lamin Dm0 (kindly provided by H. Saumweber, Humboldt-Universität, Berlin, Germany), guinea pig-α-LBR, guinea pig-α-Otefin, guinea pig-α-dMAN1 (all three antibodies were kindly provided by G. Krohne, Biozentrum Universität, Wurzburg, Germany), mouse-α-LaminC (Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA), guinea pig-α-Slam (Brandt et al., 2006), rabbit-α-barrier-to-autointegration factor (BAF; kindly provided by P. A. Fisher, Stony Brook University, NY), rabbit-α-p55 (kindly provided by J. T. Kadonaga, University of California San Diego, San Diego, CA), and mouse-α-Tubulin clone B-512 (Sigma-Aldrich). Secondary antibodies included the following: Alexa-coupled goat-α-mouse, goat-α-rabbit, goat-α-guinea pig, and donkey-α-goat (Invitrogen). DNA was stained using 4,6-diamidino-2-phenylindole (DAPI; Sigma-Aldrich). Horseradish peroxidase-coupled goat-α-rabbit, goat-α-guinea pig, and goat-α-mouse secondary antibodies (Sigma-Aldrich) were used for immunoblotting.
mRNA Injections
Capped transcripts were synthesized using the SP6 mMESSAGE mMACHINE high-yield capped RNA transcription kit (Applied Biosystems, Foster City, CA). mRNA injections were performed as described previously (Brandt et al., 2006). Zero- to 30-min-old kukΔ15 embryos (Brandt et al., 2006) were injected and after developing to late cellularization stage, they were fixed and immunostained.
Inducible Protein Expression in Yeast
For all yeast experiments, we used the strain AK725 (MATa ura3-52 leu2Δ1 his3Δ200 trp1Δ63 NUP133-mCherry-KanMX6). NUP133 was tagged endogenously using standard polymerase chain reaction (PCR)-based methods (Janke et al., 2004) in the S288c background. Tagging was confirmed by colony PCR and microscopy. The strain was a gift from A. Khmelinskii and E. Schiebel (ZMBH, Universität Heidelberg, Germany). Yeast cells were grown in YPD or in appropriate synthetic dropout medium supplemented with 2% glucose. For galactose-inducible expression experiments, GFP-LaminDm0ΔN, and GFP-full-length (FL)-Kuk were cloned in the pMM6 vector. Yeast cells transformed with the respective plasmid, grown in synthetic dropout medium supplemented with raffinose to mid-log phase, and induced by the addition of 2% galactose. Sixteen hours later, the cells were fixed in 4% FA, collected in 1.2 M sorbitol, and mounted on poly-l-lysine–coated slides.
Protein Expression and Purification
All Kuk constructs used for protein expression were cloned in the pQE80-H10-zz vector (kindly provided by D. Görlich, Max Planck Institut for Biophysical Chemistry, Göttingen, Germany). Lamin constructs were cloned in the pQE80-H10-GFP vector. Expression of recombinant proteins in Escherichia coli BL21-Rosetta-DE3 was induced with 0.1 mM isopropyl β-d-thiogalactoside for 4 h at 37°C (ZZ-Kuk, ZZ-Kuk-C567S, ZZ-Kuk-ΔN185, and ZZ-Kuk-ΔN437) or for 4 h at 18°C (GFP-LaminDm0ΔN, GFP-NLS-C-term, and GFP-NLS-CaaX). The proteins were purified from the cleared lysate by nickel chelate chromatography (HisTrap, ÄKTAprimeTM Plus; GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom). ZZ-Kuk, ZZ-Kuk-C567S and ZZ-Kuk-ΔN185 were further purified by ion-exchange chromatography using Q-Sepharose (GE Healthcare). For storage, the buffer of all proteins was changed to PBS by using PD-10 desalting columns (GE Healthcare), and 20% glycerol was added before freezing. Expression of rat farnesyltransferase (FT) (plasmids kindly provided by M. P. Mayer, ZMBH, Universität Heidelberg) was induced in MC1061 cells cotransformed with pMPM359 and pMPM369 plasmids, by addition of l-arabinose overnight at room temperature. The protein was purified by nickel chelate chromatography (HisTrap, ÄKTAprimeTM Plus; GE Healthcare) and stored in FT freezing buffer (50 mM Tris, pH 7.5,, 50 μM ZnCl2, 5 mM MgCl2, 10 mM β-mercaptoethanol, and 10% glycerol).
Liposome Assays
Liposomes were prepared from total bovine brain lipids (Folch fraction 1; Sigma-Aldrich) supplemented with 3% (vol/vol) Rhodamine-PE (Avanti Polar Lipids, Alabaster, AL). The lipid mixture was dried under vacuum using a rotary evaporator and resuspended in HK buffer (25 mM HEPES and 150 mM KCl, pH 7.4) supplemented with 10% sucrose. After homogenization by 10 freeze–thaw cycles, liposomes were prepared using a mini extruder (Avanti Polar Lipids) by using 0.1-μm polycarbonate membranes (Avanti Polar Lipids). For the liposome binding assay, 30 μl of liposomes was incubated with 4% bovine serum albumin in HK buffer for 30 min; spun at 15,000 × g for 10 min (at room temperature); and protein, FT, and farnesyl pyrophosphate (FPP) (Sigma-Aldrich) in 100 μl of HK-farnesylation buffer (25 mM HEPES, pH 7.4, 150 mM KCl, 1 mM MgCl2, 20 μM ZnCl2, 5 mM dithiothreitol, and 5 mM NaF) were added to the pellet. FPP was not added to the nonfarnesylated samples. After incubation for 3 h, the samples were spun, the supernatant was collected, and the pellet was washed twice with HK buffer. Supernatant and pellet were used for SDS-polyacrylamide gel electrophoresis (PAGE). For analysis of liposome morphology, 10 μl of liposomes was mixed with 2 μg of protein, FT, FPP, and HK farnesylation buffer in a final volume of 20 μl; incubated for 10 min; and analyzed by fluorescence microscopy. The final protein concentrations in this mixture, referring to the situation where 1× protein amount was used, are shown in Table 2.
Table 2.
Summary of the results from the liposome deformation experiments
| Recombinant protein+farn. | Deformation of liposomes |
||
|---|---|---|---|
| 0.1× protein (∼0.1–0.4 μM) | 1× protein (∼1–4 μM) | 10× protein (∼10–40 μM) | |
| zz-Kuk | − | + | NA |
| zz-Kuk-ΔN185 | − | + | NA |
| zz-Kuk-ΔN437 | − | + | NA |
| GFP-LaminΔN | + | + | +++ |
| GFP-NLS-Cterm | − | − | + |
| GFP-NLS-CaaX | − | − | + |
| GFP | No effect and no colocalization with liposomes | No effect and no colocalization with liposomes | No effect and no colocalization with liposomes |
The table summarizes the results of the liposome deformation experiments (see Figure 8 and Supplemental Figure S4). The effect of the respective constructs on the liposome morphology is shown by +++ (very strong), + (strong), or − (no effect). The proteins were used in 0.1×, 1×, or 10× amounts, except for the three Kuk constructs for which a 10× amount was not possible (NA). All the results shown in this table refer to the sample where the components of the farnesylation reaction were added to the liposome–protein mix. The second column shows the micromolar concentration for each of the proteins in the mixture of protein–liposomes (final volume, 20 μl), where 1× protein amount was used. The 1× protein amount refers to the amount shown on the Coomassie Blue-stained gel in Supplemental Figure S4.
SDS-PAGE and Western Blotting (WB)
For the Coomassie Blue-stained SDS-gel showing the 1× protein amounts used in the liposome deformation assays, total protein was adjusted so that ∼2 μg of protein was loaded from each construct, on a 10% gel. For WB using S2 cell lysates, the equivalent of 105 cells was loaded on a 10% gel. For detection of BAF by WB, a 15% gel was used.
Imaging
An Axiovert 200 M PerkinElmer Ultra-View Spinning Disk confocal microscope (100× numerical aperture [NA] 1.4 oil; Carl Zeiss, Jena, Germany) was used for time-lapse recordings of NIH-3T3 cells and for imaging of yeast cells (flatfield capture mode). For time-lapse recordings, z-stacks of seven images covering a distance of 8 μm were recorded and the layers were subsequently fused. Fluorescent images of fixed and immunostained embryos were obtained using a confocal microscope (model DMIRE2, HCX PL APO 63× NA 1.4 oil; laser at 405, 488, 568, and 633 nm; Leica, Wetzlar, Germany). Images were processed with ImageJ (National Institutes of Health, Bethesda, MD) and Photoshop (Adobe Systems, Mountain View, CA). Fluorescent images of liposomes, NIH-3T3 cells, and S2 cells were obtained using an Axioplan 2 (100×/1.30 oil; Carl Zeiss).
Electron Microscopy (EM)
Liposome samples prepared the same way as for fluorescence microscopy and loaded on Pioloform/carbon-coated grids (100 mesh). The samples were negatively stained using 0.5% uranyl acetate. EM data were obtained using an EM 10 CR transmission electron microscope (Carl Zeiss). Measurements of tubule length were done using ImageJ.
RESULTS
Extranuclear Membrane Structures Formed upon Kuk Overexpression Show an Asymmetric Composition and Are Highly Dynamic
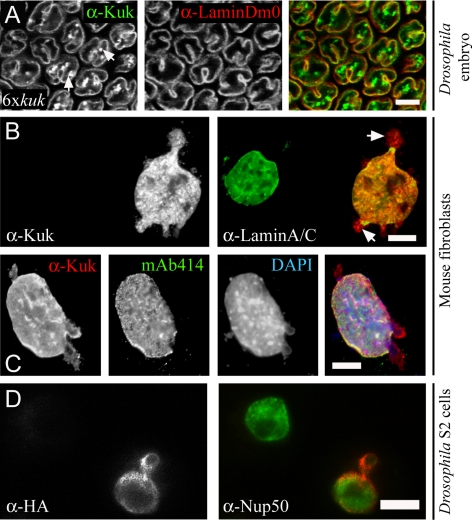
In fibroblasts expressing LaΔ50, the nuclear membrane protrusions contain LaΔ50 but not lamin B, showing a differential composition than the NM of the main body of the nucleus (Goldman et al., 2004). Here, we examined whether Kuk behaves similarly. Increasing Kuk levels in the Drosophila embryo induce changes in nuclear morphology, in a dose-dependent manner (Brandt et al., 2008). In embryos with six genomic copies of kuk, where the cortical nuclei show pronounced apical ruffling, we observed an asymmetry in the composition of the NM invaginations. Although in these nuclei, Kuk generally colocalized with lamin Dm0 at the nuclear envelope, the intranuclear membrane invaginations showed Kuk staining but much less if any lamin Dm0 staining (Figure 1A, arrows).
Figure 1.
Asymmetric composition of the extranuclear membrane structures in Kuk-overexpressing cells. (A) Surface view of a Drosophila embryo with six genomic copies of kuk, in late cellularization. The differential staining of the intranuclear membrane structures is indicated by arrows. Lamin Dm0 (red) is used as a marker of the nuclear membrane. Kuk staining is shown in green. Bar, 10 μm. (B and C) Nuclear morphology upon Kuk overexpression in NIH-3T3 cells. (B) Nucleus of a transiently transfected NIH-3T3 cell expressing Kuk (red), compared with the nucleus of a nontransfected cell. Lamin A/C (green) marks the nuclear membrane. The differential staining of the membrane blebs is indicated by arrows. (C) Kuk (red), nuclear pores (green) and DNA-staining (blue) of a Kuk-transfected NIH-3T3 cell. Bar (B and C), 5 μm. (D) Nucleus of a transiently transfected S2 cell expressing HA-Kuk (red), compared with the nucleus of a nontransfected cell. Nup50 (green) marks the nuclear membrane and the nucleoplasm. Bar, 5 μm.
Similar asymmetry in the composition of the extranuclear membrane structures formed upon Kuk overexpression also was observed in other cell types. Kuk affected nuclear shape in cultured Drosophila cells, as shown in transiently transfected S2 cells (Figure 1D). The bleb formed in the Kuk-transfected cell shows less Nup50 staining than the main body of the nucleus. In Kuk-transfected mouse fibroblasts, where no endogenous Kuk is present, we observed formation of large and abnormal nuclei as described previously (Brandt et al., 2008). The nuclei of the transfected cells showed blebs and extensive intranuclear membrane structures (Figure 1, B and C). Similarly as in the 6xkuk fly embryo, the blebs showed differential staining pattern, with less lamin A/C (Figure 1B, arrows) and less nuclear pores (Figure 1C, mAb414) than the main body of the nucleus. In addition, the blebs generally contained less DNA, as shown by the DAPI staining (Figure 1C, DAPI). This indicates that Kuk behaves similarly to LaΔ50, with respect to the formation of NM abnormalities of a defined composition that differs from the NM composition of the main body of the nucleus.
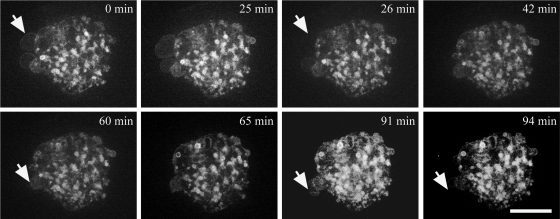
Because we noticed that the NM blebs formed upon Kuk overexpression showed a differential composition, we analyzed the dynamics of these blebs by time-lapse movies of transfected cells. When we compared GFP-Kuk and GFP-Kuk-Δ353-404 (Kuk deletion construct, described in Figure 3) in mouse fibroblasts, they showed the same behavior. Because blebbed nuclei appeared at higher frequency in GFP-Kuk-Δ353-404 transfected cells, this construct was chosen for the time-lapse experiments. In GFP-Kuk-Δ353-404–transfected fibroblasts, we observed that the blebs and the intranuclear structures showed differential behavior (Figure 2 and Supplemental Movie S1). Although the intranuclear structures were quite stable as reported previously (Broers et al., 1999) and remained unchanged throughout the 94 min of time-lapse recording, the nuclear blebs were highly dynamic, forming and disappearing fast, within a few minutes (Figure 2, arrows).
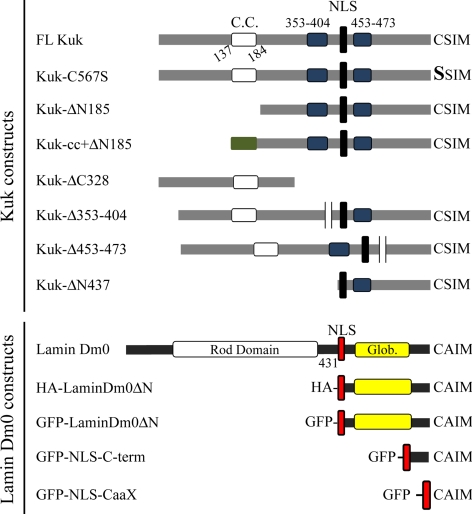
Figure 3.
Constructs used in the study. The structure of Kuk and lamin Dm0 constructs used in the study is schematically shown. The putative coiled-coil motif of Kuk (C.C., aa 137–184) is shown in white. The two highly conserved aa sequences 353–404 and 453–473 are shown in blue and the NLS in black. In Kuk-cc+ΔN185 the coiled coil of the GCN4 leucine zipper protein that has substituted the putative coiled coil of Kuk is shown in green. In the lamin Dm0 constructs, the NLS is shown in red, the rod domain in white, and the globular C-terminal domain in yellow. In LaminDm0ΔN, the N-terminal part consisting of aa 1–430 including the rod domain has been deleted. GFP-NLS-C-term contains the NLS of lamin Dm0 fused to the very C-terminal part of the protein (aa 570–622). GFP-NLS-CaaX consists of GFP fused to the NLS of lamin Dm0 followed by the CaaX motif of lamin Dm0.
Figure 2.
Dynamics of the blebs and intranuclear structures in transiently transfected NIH-3T3 cells. The figure was assembled from selected time points of Supplemental Movie S1, showing a NIH-3T3 cell transiently transfected with GFP-Kuk-Δ353-404.The nuclear membrane blebs formed upon GFP-Kuk-Δ353-404 expression show a very dynamic behavior. Arrows indicate examples of blebs growing and disappearing. (GFP-Kuk and GFP-Kuk-Δ353-404 showed comparable effect on nuclear morphology. Due to the more frequent appearance of blebs in GFP-Kuk-Δ353-404–transfected cells the construct was chosen for the time-lapse experiments shown.) Bar, 7 μm.
A Conserved Domain of Kuk Is Specifically Required for Nuclear Elongation but Not for NM Localization in the Early Drosophila Embryo
Kuk is a 570-aa protein that contains a putative coiled coil motif (aa 137–184), a NLS (aa 440–447), and a C-terminal farnesylation motif (schematic representation of FL-Kuk shown in Figure 3). The putative coiled coil motif and the CaaX box have been shown by experiments in Xenopus A6 cells, to be required for Kuk activity on nuclear size and shape (Brandt et al., 2006). Here, we used an mRNA injection assay using kuk-deficient embryos to test different Kuk constructs for their NM localization and function in nuclear elongation and apical ruffling. In addition to the coiled coil and the CaaX motif, we examined the function of the aa sequences 353–404 and 453–473 that were found by BLAST analysis to show the highest degree of conservation among two different Drosophila species (D. melanogaster and D. grimshawi) and mosquitoes (Anopheles gambiae and Aedes aegypti). All the constructs used in this study are shown in Figure 3.
During cellularization in the wild-type fly embryo, the cortical nuclei elongate and maintain this elongated shape until gastrulation (Supplemental Figure S1B, side view). In addition, the nuclei ruffle apically (Figure S1B, surface view). In kuk-deficient embryos, the nuclei are round and smooth by the end of cellularization (Supplemental Figure S1A). Kuk-deficient embryos, in early blastoderm stage were injected with mRNA in their posterior part, and the nuclear morphology at late cellularization stage was compared with wild-type (wt) nuclei (Supplemental Figure S1B). The different constructs were tested for NM localization and for their ability to induce and maintain nuclear elongation and apical ruffling (phenotypic rescue).
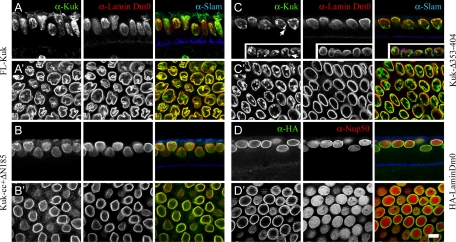
FL-Kuk localized to the NM, rescued the Kuk phenotype, and induced apical ruffling of the nuclei (Figure 4, A and A′, and Table 1), whereas a differential staining of the extra membrane structures was observed (Figure 4A′) similarly as in the 6xkuk embryo (Figure 1A). The nonfarnesylatable Kuk-C567S mutant showed nucleoplasmic localization and was unable to restore the Kuk phenotype (Supplemental Figure S1, D and D′, and Table 1). Kuk-ΔN185 that lacks the putative coiled coil motif accumulated in the nucleoplasm where it formed punctate structures (Supplemental Figure S1E and Table 1) and showed no activity on nuclear shape (Supplemental Figure S1E′). To gain further insight in the role of the putative coiled coil motif of Kuk, we added the coiled coil motif of the GCN4 leucine zipper (Harbury et al., 1993; Yuan et al., 2003) to Kuk-ΔN185. The resulting Kuk-cc+ΔN185 construct was able to localize to the NM but could not rescue the mutant phenotype (Figure 4, B and B′, and Table 1). This suggests that the coiled coil motif mediates Kuk localization at the INM. However, the GCN4 coiled coil did not restore Kuk function, suggesting that specific features of the endogenous coiled coil cannot be substituted by the coiled coil motif we used. The results of this experiment also indicate that NM localization of Kuk is not sufficient for its function.
Figure 4.
A conserved domain is required for the function of Kuk in nuclear elongation in the early Drosophila embryo. Nuclear morphology of kukΔ15 embryos injected with mRNA of different constructs, at the end of cellularization. The membrane front, stained by Slam (blue) indicates the embryonic stage. Lamin Dm0 (red) or Nup50 (red; D) are used as markers of the NM. (A) FL-Kuk–injected embryo, side view. (A′), surface view. (B) Kuk-cc+ΔN185–injected embryo, side view. (B′) Surface view. (C) Kuk-Δ353-404–injected embryo, side view. The NM blebs are indicated by arrows. (C′) Surface view. (D) HA-LaminDm0–injected embryo, side view. (D′) Surface view. Bar, 10 μm.
Table 1.
Summary of the structure–function analysis of Kuk in the fly embryo
| NM localization | Activity on nuclear shape | Rescue of the kuk phenotype | |
|---|---|---|---|
| FL-Kuk | + | + | + |
| Kuk-ΔC328 | − | − | − |
| Kuk-C567S | − | − | − |
| Kuk-ΔN185 | − | − | − |
| Kuk-cc+ΔN185 | + | − | − |
| Kuk-Δ353-404 | + | + | − |
| Kuk-Δ453-473 | + | + | + |
| HA-LaminDm0 | + | − | − |
Kuk-ΔC328 was mostly cytoplasmic (Supplemental Figure S1C and Table 1), primarily due to the lack of the main NLS, and not functional (Supplemental Figure S1C′). Kuk-Δ453-473 showed the same behavior as FL-Kuk (Supplemental Figure S1, F and F′, and Table 1), indicating that this conserved sequence is not required for Kuk function. In contrast, when Kuk-Δ353-404 was injected in Kuk-deficient embryos, we observed that even though the construct localized at the NM and induced nuclear membrane ruffling (Figure 4C′), it was unable to rescue the nuclear elongation defect (Figure 4C and Table 1). This suggests that the domain is specifically required for nuclear elongation. The construct induced formation of intranuclear structures (Figure 4C′), similar to the ones observed upon Kuk overexpression and also nuclear membrane blebs (Figure 4C, arrows). Such blebs were not observed with the full-length Kuk, even under very high overexpression conditions (6× kuk and 8× kuk embryos; data not shown). Kuk-Δ353-404 and Kuk-Δ453-473 also were transfected in mouse fibroblasts, where they showed NM localization and affected nuclear shape (Supplemental Figure S2, B and C).
In addition, we tested whether the kuk phenotype in the blastoderm embryo could be rescued by overexpressing the other farnesylated protein of the INM, lamin Dm0. When HA-LaminDm0 was injected in kuk-deficient embryos, even though it properly localized at the NM, the nuclei were neither elongated nor apically ruffled in late cellularization (Figure 4, D and D′, and Table 1). Therefore, it seems that the activity of Kuk in elongating the cortical nuclei is specific and cannot be complemented by additional lamin Dm0.
In conclusion, our analysis led to the identification of a conserved aa sequence that is required for the function of Kuk in nuclear elongation. The inability of Kuk-Δ353-404 to rescue the mutant phenotype suggests that this conserved motif is specifically required for maintaining nuclear elongation during cellularization, whereas it does not seem to play a role in NM localization and induction of apical ruffling of the nuclei. Kuk-Δ353-404 was the only construct tested where the function of Kuk on nuclear elongation could be separated from its effect on apical ruffling. Furthermore, our results from Kuk-Δ353-404 and Kuk-cc+ΔN185 suggest that NM localization of Kuk is not sufficient for function of the protein.
Inhibition of Farnesylation Affects the Localization of Ectopically Expressed and Endogenous Kuk
Farnesylation renders proteins lipophilic, resulting in their association to membranes. To further analyze the role of farnesylation, we tested the behavior of selected Kuk constructs and LaminDm0ΔN in transiently transfected fibroblasts, in the presence of the FTI ABT-100. When FL-Kuk was transfected in mouse fibroblasts, it localized at the NM and induced formation of abnormally shaped nuclei (Figure 5A), as shown previously (Brandt et al., 2008). Deletion of the N-terminal part of Kuk results in proteins that behave differently compared with FL-Kuk. Kuk-ΔN185, lacking the coiled coil motif, formed punctate intranuclear structures (Figure 5B). Kuk-ΔN437, lacking the entire N-terminal part, formed dot-like cytoplasmic structures (Figure 5C) that did not colocalize with ER markers (data not shown). This shows that deletion of the N-terminal part of Kuk has a different effect than the deletion of the N-terminal part of lamin. The globular domain of lamins was shown to be sufficient to properly localize and induce NM overproliferation in cultured cells (Prufert et al., 2004; Ralle et al., 2004) and can here be observed in fibroblasts transfected with LaminDm0ΔN (Figure 5D).
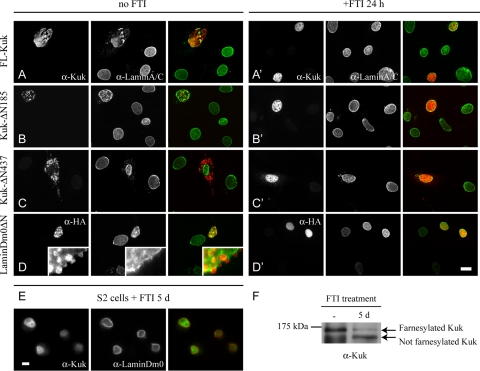
Figure 5.
Role of farnesylation in localization and activity of Kuk constructs and LaminDm0ΔN in cultured cells. (A–D) NIH-3T3 cells transiently transfected with different constructs. (A′–D′) Transiently transfected NIH-3T3 cells, treated with the FTI ABT-100 for 24 h. Lamin A/C staining (green) marks the NM (A–D′). Localization pattern of the transfected constructs is shown in red (Kuk in A–C′ and HA in D and D′). Bar, 10 μm. (E) S2 cells treated with the FTI ABT-100 for 5 d. Lamin Dm0 staining (red) marks the NM and Kuk is shown in green. Bar, 5 μm. (F) FTI treatment increases the electrophoretic mobility of Kuk. In total extract of nontreated S2 cells a band of ∼120 kDa, corresponding to farnesylated Kuk is detected. In the total extract of FTI-treated cells, the intensity of the ∼120-kDa band is reduced and an ∼110-kDa band representing not farnesylated Kuk is also detected. Kuk runs at a significantly higher MW than predicted (Brandt et al., 2006).
Treatment with the FTI resulted in nucleoplasmic localization of FL-Kuk and HA-LaminDm0ΔN and eliminated their ability to alter nuclear shape (Figure 5, A′ and D′). In the presence of the FTI, the two constructs behaved similarly as the nonfarnesylatable Kuk-C567S (Supplemental Figure S2A). Farnesylation seems to be an absolute requirement for Kuk and LaminDm0ΔN localization and function because treatment with the FTI dissociates the proteins from the NM and abolishes their activity on nuclear shape. In addition, the FTI promoted the nucleoplasmic localization of the constructs Kuk-ΔN185 and Kuk-ΔN437 (Figure 5, B′ and C′).
After showing that localization of ectopically expressed Kuk is affected by FTIs, we tested whether the localization of endogenous Kuk also could be affected by such a treatment. Indeed we observed that the FTI ABT-100 promoted nucleoplasmic localization of endogenous Kuk in Drosophila S2 cells (Figure 5E). The inhibitor failed to affect the NM localization of lamin Dm0 (Figure 5E, lamin Dm0 staining). This is not surprising, because lamin Dm0 is expected to have a slow turnover as indicated by photobleaching experiments using GFP-Lamin fusion proteins (Broers et al., 1999). The inhibition of Kuk farnesylation was confirmed by Western blot (Figure 5F). After treatment with the FTI, the amount of farnesylated Kuk, represented by an ∼120-kDa band, is significantly reduced. A second band of ∼110 kDa corresponding to nonfarnesylated Kuk is present in the FTI-treated sample. The shift in Kuk molecular weight (MW) induced by the FTI corresponds to the size difference of in vitro farnesylated and nonfarnesylated recombinant Kuk (Figure 8A) or nonfarnesylatable Kuk-C567S (data not shown).
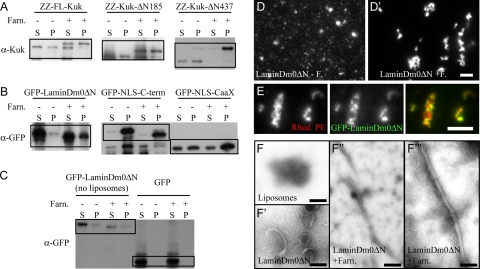
Figure 8.
Farnesylated Kuk and lamin Dm0 constructs bind to protein-free liposomes and affect their morphology. (A–C) Western blots showing the results of the lipid binding assay. Lanes labeled by − correspond to the samples with nonfarnesylated proteins, where no FPP was added. Samples labeled by + show the samples where all the components of the farnesylation reaction were added. (FT enzyme was added to all samples, to keep the protein concentration equal between − and + samples.) S, supernatant or unbound protein fraction. P, pellet or bound protein fraction. (A) α-Kuk WB showing the distribution of FL-Kuk, Kuk-ΔN185, and Kuk-ΔN437 between supernatant and pellet depending on farnesylation. (B) α-GFP WB showing the distribution of GFP-LaminDm0ΔN, GFP-NLS-C-term, and GFP-NLS-CaaX between supernatant and pellet depending on farnesylation. (C) α-GFP WB showing the distribution of GFP-LaminDm0ΔN between supernatant and pellet when no liposomes were added to the sample. The distribution of the control GFP protein, which cannot be farnesylated, is shown in the right half of C. (D–D″) Liposome morphology as observed by fluorescence microscopy. (D) Liposomes after addition of nonfarnesylated GFP-LaminDm0ΔN. (D′) Liposomes upon addition of farnesylated GFP-LaminDm0ΔN. (E) Colocalization of farnesylated GFP-LaminDm0ΔN (green) and Rhodamine-PE–labeled liposomes (red). Bar (D and E), 15 μm. (F–F′′′ EM analysis of liposome morphology. (F) Protein-free liposomes. (F′) Liposomes after addition of nonfarnesylated GFP-LaminDm0ΔN. (F″) Liposomes after addition of farnesylated GFP-LaminDm0ΔN. (F′′′) Liposomes after addition of farnesylated GFP-LaminDm0ΔN shown in a higher magnification. Bar, 100 nm.
In conclusion, farnesylation was found to be essential for protein targeting to the NM or in case of the Kuk-ΔN185 and Kuk-ΔN437 constructs for accumulation in nucleoplasmic or cytoplasmic structures. This may be caused by simply increasing the lipophilicity of the proteins. Another possibility is that association of the farnesylated C terminus with the NM has specific effects, such as orienting the protein toward the lipid bilayer or positioning the protein in a way that it can participate in protein–protein interactions.
Kuk and Lamin Dm0 Localize at the INM Independently of a Group of Selected INM Proteins
Farnesylated proteins normally only have a weak affinity for membranes. In some cases, this affinity becomes higher by a second modification i.e., in the case of Ras by palmitoylation (Hancock et al., 1990; Wright and Philips, 2006). Because a second modification after farnesylation has not been described for Kuk and lamin Dm0, we wondered whether the two proteins would be stabilized at the NM with the contribution of other lamina or INM proteins. For this purpose, we depleted selected lamina and INM proteins by RNAi in Drosophila S2 cells. Kuk and lamin Dm0 localization did not seem to depend on one another (Figure 6A and Supplemental S3C), as shown previously (Brandt et al., 2006). When LBR, MAN1, or lamin C were down-regulated, localization of neither lamin Dm0 nor Kuk was affected (Figure 6B and Supplemental S3, A and B). In contrast, Otefin localization was dependent on lamin Dm0 (Figure 6E), as described previously (Wagner et al., 2004).
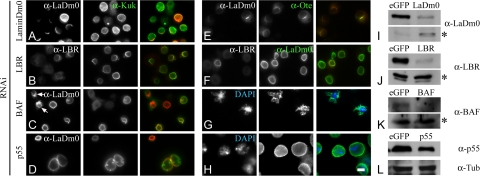
Figure 6.
Kuk and lamin Dm0 localize at the NM independently of a group of selected INM proteins. Localization of Kuk and lamin Dm0 after depletion of selected INM proteins by RNAi in Drosophila S2 cells. (A–D) Kuk staining (green) shows Kuk localization in RNAi-treated cells. In red, the staining for the respective depleted protein is shown. Arrows in C indicate deformed nuclei, due to BAF depletion. (E) Otefin staining (green) in lamin Dm0 RNAi-treated cells. (F) Lamin Dm0 staining (green) in LBR RNAi-treated cells. LBR staining is shown in red. (G) Deformed nuclei after depletion of BAF (blue, DNA; green, lamin Dm0). (H) Enlarged nuclei after p55 depletion (blue, DNA; green, lamin Dm0). Bar, 5 μm. (I–L) Western blots from total cell extracts, showing the reduced amounts of the respective RNAi-depleted proteins. Cells treated with RNAi against eGFP were used as control samples. Asterisks in I–K indicate cross-reacting bands used as a loading control. In L, the immunoblot against α-Tubulin is shown as a control for protein amounts.
Surprisingly, under our experimental conditions, the NM localization of Kuk but not of lamin Dm0 was dependent on the chromatin-binding protein BAF and on the chromatin remodeling complex component p55. Kuk failed to localize at the NM when BAF was depleted (Figure 6C, arrows). On down-regulation of p55, Kuk localized at the NM, but it also formed intranuclear structures where no lamin Dm0 was detected by immunostaining (Figure 6D). In the nuclei that were severely deformed due to the depletion of BAF lamin Dm0 localization was affected but not as much as Kuk localization (Figure 6C, arrows; and G). The observation comes in agreement with the data shown in BAF RNAi-treated C. elegans embryos, where Ce-Lamin and three nuclear membrane proteins fail to assemble properly (Margalit et al., 2005). Altered lamin Dm0 distribution has also been found to correlate with the loss of detectable BAF amounts in fly imaginal discs (Furukawa et al., 2003).
Another finding of our RNAi experiment was that down-regulation of both BAF and p55 severely affected nuclear morphology, potentially due to indirect effects. In BAF RNAi treated cells, we observed severely deformed nuclei (Figure 6C, arrows; and G). On down-regulation of p55, the nuclei were found to be particularly enlarged (Figure 6, D and H). Although depletion of BAF has already been described to result in NM distortion (Furukawa et al., 2003), nothing has been so far reported about an effect of p55 on nuclear morphology.
Our experiments reveal an unexpected function of chromatin-interacting proteins for the NM localization of Kuk and might suggest that the interaction of chromatin with components of the nuclear envelope is involved in proper organization of the INM and lamina. In addition, the results indicate a lamina independent mechanism of activity for lamin Dm0 and Kuk.
Kuk and LaminDm0ΔN Affect Nuclear Morphology Even in the Absence of a Classical Nuclear Lamina
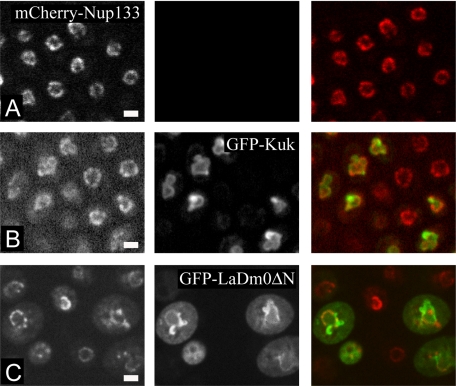
Because we observed that Kuk and lamin Dm0 do not seem to depend on other lamina and candidate INM proteins for their localization, we examined whether they would also be able to localize properly in the complete absence of the lamina. For this purpose we expressed Kuk and LaminDm0ΔN in yeast, which does not have a classical nuclear lamina. Ectopically expressed human lamin B has been shown to localize at the nuclear envelope of S. cerevisiae (Smith and Blobel, 1994), but an activity on nuclear shape has not been reported. The nuclei of S. cerevisiae cells in log phase are generally round, as seen in a yeast strain expressing mCherry-Nup133, which marks the NM (Figure 7A). Inducible GFP-Kuk expression resulted in the formation of abnormal, lobulated nuclei (Figure 7B). Abnormally shaped nuclei were also formed upon inducible expression of GFP-LaminDm0ΔN, a truncated lamin Dm0 construct (described in Figure 3) where the N-terminal filament-forming rod domain has been deleted (Figure 7C). In contrast to GFP-Kuk that seemed to localize exclusively at the NM, GFP-LaminDm0ΔN could partially be found in the cytoplasm as we have also observed when the construct is expressed in mammalian or Drosophila cells (data not shown).
Figure 7.
Kuk and LaminDm0ΔN affect nuclear morphology even in the absence of a classical nuclear lamina. (A–C) Nuclear morphology of S. cerevisiae cells in mid-log phase. mCherry-Nup133 marks the nuclear membrane. (A) Nontransformed cells. (B) GFP-Kuk–expressing S. cerevisiae. (C) GFP-LaminDm0ΔN–expressing S. cerevisiae. Bar, 2.5 μm.
The activity of the truncated GFP-LaminDm0ΔN construct indicates that the C-terminal farnesylated part of NM proteins is sufficient for inducing nuclear shape changes in yeast, whereas filament formation does not seem to be required. Overall, our observations in yeast, suggest that the farnesylated proteins Kuk and lamin Dm0 exert their activity on NMs independently of intermediate filament formation and regardless of the absence of lamina proteins.
Farnesylation Is Required for Deformation of Protein-free Liposomes by Kuk and Lamin Dm0 Constructs
Because our results from the yeast and the RNAi experiment indicate a lamina-independent mechanism of activity for lamin Dm0 and Kuk, we wondered whether these farnesylated proteins could directly affect the structure of the lipid bilayer of the NM. If the proteins were asymmetrically incorporated in the bilayer, this would lead in expansion of the surface area of the inner layer of the membrane, thus leading in abnormal shapes. To address this possibility, we tested whether recombinant FL-Kuk, Kuk-ΔN185, Kuk-ΔN437, LaminDm0ΔN, GFP-NLS-C-term, and GFP-NLS-CaaX can bind to protein-free liposomes. A protein gel showing the purity and the amounts of the proteins used is shown in Supplemental Figure S4. GFP-NLS-C-term and GFP-NLS-CaaX also affect nuclear morphology in mouse fibroblasts (Supplemental Figure S2, D and E), but only after up to 72 h after transfection, indicating a lower activity than HA-LaminDm0ΔN which shows activity already at 24 h after transfection.
The results of the binding assay are in agreement with what has been described for the binding of small farnesylated peptides to liposomes. According to Silvius and l'Heureux (1994) and Rowat et al. (2004), even though farnesylated peptides are lipophilic, they are not quantitatively bound to the lipid vesicles. A part of the total peptide amount remains in the aqueous phase. As shown in Figure 8A, farnesylation slightly improved liposome binding of FL-Kuk and Kuk-ΔN185 (Figure 8A). The same was observed for LaminDm0ΔN and GFP-NLS-CaaX (Figure 8B). The control protein GFP (Figure 8C) remained in the supernatant regardless of the addition of farnesylation reaction components. Kuk-ΔN437 shifted almost exclusively to the bound fraction when it was farnesylated (Figure 8A). GFP-NLS-C-term bound to liposomes regardless of farnesylation (Figure 8B). This unusual behavior of GFP-NLS-C-term might be due to the positively charged NLS motif binding to the negatively charged liposomes. The fact that even the nonfarnesylated proteins were able to partially bind to the liposomes, implies that there are other parts of the protein that mediate lipid binding in addition to the farnesylation motif. The increased amounts of protein found in the bound fraction (liposome pellet) were not due to protein precipitation because it can be seen in the samples were no liposomes were added (Figure 8C). Farnesylation of Kuk induced a shift to higher MW, as also shown in Figure 5F. Farnesylation of LaminDm0ΔN, GFP-NLS-C-term, and GFP-NLS-CaaX increases the electrophoretic mobility of the proteins, in agreement with what has been shown for LaΔ50 (Capell et al., 2005).
Interestingly, the farnesylated recombinant protein constructs changed the morphology of the liposomes, because we could observe by incubation of the liposomes with the proteins, followed by fluorescence microscopy. The morphology of the liposomes (Supplemental Figure S5A) was not affected by the addition of nonfarnesylated LaminDm0ΔN (Figure 8D) or other nonfarnesylated proteins (Supplemental Figure S5A), despite that they bound to the liposomes as shown in the binding assays. Addition of farnesylated LaminDm0ΔN resulted in the formation of large lipid structures (Figure 8D), morphologically different from the protein-free liposomes. In these large liposome structures, accumulation of GFP-LaminDm0ΔN could be observed (Figure 8E).
Because the resolution of fluorescence microscopy was not sufficient for analyzing in detail the structural changes in the liposomes, we performed the same lipid binding assay by using 5× LaminDm0ΔN protein amounts (Supplemental Figure S4) and analyzed the negatively stained samples by EM. The EM analysis showed that a certain proportion of the spherical liposomes formed tubules when incubated with farnesylated LaminDm0ΔN (Supplemental Figure 8, F″ and F′′′). The elongated structures corresponded to <5% of the total area of the sample, in nonrandomly taken pictures. Even though the morphological changes in the liposomes were found to be very pronounced by fluorescence microscopy, not many structures showing an altered morphology could be observed by EM. This might be due to not stable attachment of the morphologically altered liposome structures on the grid during preparation of the EM samples. If a sphere of a given diameter is deformed in a way that it gives rise to a cylindrical shape, the length of the cylinder depends on the diameter of the sphere, because the volume of the shape is not expected to change. When we measured the average length of the tubules formed by the deformation of the liposomes, we found that it came in agreement with the expected length of a tubule (with a diameter of ∼30 nm) formed by the deformation of spherical liposome with an ∼100 nm diameter (Supplemental Figure S6).
Apart from LaminDm0ΔN, we also tested the effect of all other farnesylated recombinant proteins on liposome morphology by fluorescence microscopy. The results are summarized in Table 2. All Kuk constructs could induce liposome deformation when farnesylated (Supplemental Figure S5A, left), even though the effect was weaker than the effect of LaminDm0ΔN, probably due to the lower protein concentration of the constructs (Supplemental Figure S4). GFP-NLS-C-term and GFP-NLS-CaaX showed activity on liposome morphology, even though this activity is noticeable only at 10× higher protein amounts than LaminDm0ΔN (Supplemental Figure S5A and Table 2). As mentioned, the two constructs also showed lower activity than LaminDm0ΔN when expressed in fibroblasts (Supplemental Figure S2, D and E). These observations suggest that the globular part of the C terminus of lamin Dm0 might have a role in deforming membranes. In addition, the experiments indicate that binding to the liposomes, which was observed for all the constructs upon farnesylation, is not sufficient for deforming the liposomes, at least when the 1× protein amount is used.
The control protein GFP did not induce any morphological changes to the liposomes and did not show any accumulation on them regardless of its high concentration and of the addition of farnesylation reaction components (Supplemental Figure S5B). Overall, the morphological changes observed in the liposome assays indicate that farnesylated NM proteins could have a direct activity on the lipid bilayer of the NM, which would result in the observed nuclear shape changes.
DISCUSSION
The analysis of lamin function and activity has been mostly focused on the N-terminal filament forming part of the protein (Wiesel et al., 2008; Ben-Harush et al., 2009; Kapinos et al., 2010). Triggered by the observations of Ralle et al. (2004) and Prufert et al. (2004), we have investigated the relation of farnesylated NM proteins with the NM and the phospholipid bilayer of the membrane, focusing on the C-terminal part containing the CaaX motif.
From our structure–function analysis in the fly embryo, we confirmed that both the coiled coil motif and the CaaX motif are required for Kuk NM localization and activity, as reported previously (Brandt et al., 2006). Substitution of the putative coiled coil motif of Kuk by the coiled coil of GCN4, results in a construct that localizes at the NM but does not have the wt activity on nuclear shape. This indicates that NM localization is not sufficient for Kuk function. In addition, we found that the conserved motif consisting of aa 353–404 is required for the nuclear elongation induced by Kuk during cellularization. Kuk-Δ353-404 localizes at the NM and induces nuclear shape changes both in fibroblasts and fly embryos but fails to elongate the cortical nuclei in the embryo. Kuk-Δ353-404 and Kuk-cc+ΔN185 were the only constructs for which localization at the NM was not combined with rescue of the nuclear elongation phenotype in the embryo. Considering the above mentioned observations, it is conceivable that the conserved part 353–404 is essential for maintaining nuclear elongation but not for inducing nuclear ruffling. It has been shown that microtubules are required for maintaining the elongated shape of nuclei in the fly embryo, because upon their destruction the nuclei fail to elongate even though they still ruffle apically (Brandt et al., 2006). One possibility could be that the domain 353–404 takes part in an interaction of microtubules with the NM that could contribute in the maintenance of the elongated nuclear shape.
We showed that ectopic expression of Kuk and LaminDm0ΔN in yeast results in the formation of abnormally shaped nuclei. The activity of farnesylated proteins in yeast, where there is no classical nuclear lamina, suggests that the nuclear shape changes could be induced due to the incorporation of the proteins at the NM via their farnesylated moiety and would therefore be independent of filament formation. The latter is further supported by there being so far no evidence that Kuk can form filaments as well as by the activity of the truncated, nonfilament forming construct LaminDm0ΔN in yeast. The yeast nuclear morphology upon Kuk or LaminDm0ΔN expression is similar to the morphology observed when Esc1p, a yeast protein of the nuclear periphery is overexpressed (Hattier et al., 2007). Esc1p is a nonmembrane and nonfilament forming protein of the nuclear periphery and despite its coiled coil motifs and its activity on nuclear shape it is not comparable with lamins. Solubilization properties of the protein suggest that it bears a lipid modification via which it associates to membranes (Taddei et al., 2004). Therefore, Esc1p could be another example of a lipid-modified protein that changes nuclear shape when overexpressed, without depending on filament formation or the presence of a classical nuclear lamina. Because Esc1p can function as an anchor for chromatin (Taddei et al., 2004), it is possible that chromatin-nuclear periphery interactions can also participate in nuclear shape changes. A potential role of chromatin is also suggested by the nuclear shape changes that were observed upon depletion of the chromatin interacting protein BAF in S2 cells. Together, we suggest that nuclear shape changes due to the overexpression of Kuk or LaminDm0ΔN are independent of filament formation and might also be related to altered chromatin–NM interactions.
The hypothesis for a lamina-independent activity of Kuk is consistent with the results of our RNAi experiment that indicate that localization of Kuk and lamin Dm0 does not depend on other known INM proteins. For lamin Dm0, this result was not unexpected because the protein is itself required for structural integrity of the nucleus as well as for the localization of many INM proteins. In contrast to the LEM domain proteins otefin, bocksbeutel, and dMAN1, and several newly identified nuclear envelope transmembrane proteins that depend on lamins for their INM localization (Wagner et al., 2004, 2006; Malik et al., 2010), Kuk localizes at the INM even when lamin Dm0 is down-regulated, which indicates that Kuk is tethered at the NM without depending on lamin Dm0. The dependence of Kuk NM localization on BAF and p55 was an unexpected finding that suggests that the organization of the NM might depend at a certain extent on its direct or indirect interaction with chromatin. Nevertheless, so far we do not have any evidence of direct Kuk-chromatin or Kuk-chromatin binding proteins interactions. The abnormal nuclear morphologies observed upon BAF down-regulation, have been attributed to the disruption of interactions between BAF, LEM domain proteins and lamin Dm0 (Furukawa et al., 2003). However, this hypothesis cannot explain the nuclear phenotype of the p55 RNAi-treated S2 cells, because p55 is a nucleoplasmic WD-repeat component of histone modifying and chromatin assembly complexes (Tyler et al., 1996, 2001), which is not expected to interact with NM proteins. Therefore, it seems that changes in chromatin organization induced by down-regulation of chromatin associated proteins can affect nuclear shape in a direct or indirect way.
In the in vitro liposome binding assay, we observed that farnesylation results in the enrichment of the respective protein in the lipid bound fraction but not in exclusion of the protein from the aqueous phase. This result is consistent with experiments using farnesylated peptides (Silvius and l'Heureux, 1994; Rowat et al., 2004), where it was shown that the peptides partition between the aqueous and lipid phases. Because we did not use peptides but large protein fragments and even the FL protein in the case of Kuk, we cannot exclude the contribution of other domains in addition to the CaaX motif, to the determination of the final lipid binding properties. Nevertheless, the binding affinity of all the protein constructs we tested was rather moderate, as expected for farnesylated proteins. Farnesylation is a rather weak lipid modification, and although for other proteins, i.e., Ras, a second modification, palmitoylation, follows in order to render the binding more stable (Hancock et al., 1990) nothing similar is known to take place in the case of lamins or Kuk.
The effect of farnesylated proteins on nuclear shape has been extensively described but the mechanism through which these morphological changes are induced has remained unclear. Changes in membrane structure can be primarily achieved by two mechanisms (Kozlov, 2010). One mechanism involves contraction of boundaries between domains of different lipid phases. In this model, protein molecules can act as scaffolds and stabilize membrane curvature. The second mechanism involves asymmetric insertion of lipid or protein molecules in one of the two layers of the membrane. Considering our liposome assay results, which showed that farnesylated recombinant proteins bind to liposomes and change their morphology, we suggest that farnesylated NM proteins change nuclear shape using the second mechanism, involving asymmetric insertion. Because we do not have evidence of altered lipid composition of the bilayer, we rather propose that the membrane shape changes are due to asymmetric insertion of farnesylated protein molecules to the lipid monolayer, which would lead to membrane deformation. Nevertheless, the contribution of lipid asymmetry cannot be excluded. It is possible that the farnesylated moiety of the proteins preferentially associates with specific categories of lipids that would cluster asymmetrically when the protein is inserted to the lipid layer. Contraction of boundaries (first suggested mechanism) might also in part take place, because it has been shown that prenylated peptides are excluded from lipid raft domains (Zacharias et al., 2002). The exclusion of farnesylated proteins from specific membrane domains could subsequently lead in boundary contraction and altered membrane morphology.
In conclusion, the results of our study demonstrate for the first time that farnesylated proteins of the NM affect nuclear shape in absence of a classical lamina and induce morphological changes to protein-free liposomes depending on their farnesylation. The reduction of activity on liposome deformation when the Ig-fold globular part of LaminDm0ΔN is removed suggests that this part of the protein plays a role in the activity of the protein. The requirement of farnesylation for activity on NMs and liposomes suggests that nuclear morphology changes might be directly arising from the insertion of the proteins to the lipid bilayer of the NM via their lipophilic farnesylated moiety. Nevertheless, the contribution of other mechanisms, including the alteration of NM–chromatin interactions that could consequently affect nuclear structure cannot be excluded.
Supplementary Material
ACKNOWLEDGMENTS
We thank P. A. Fisher, J. T. Kadonaga, G. Krohne, H. Saumweber, A. Khmelinskii, E. Schiebel, D. Görlich, M. P. Mayer, and B. Schwappach for sharing antibodies, yeast strains, and plasmids; H. Bading (Department of Neurobiology, University of Heidelberg, Heidelberg, Germany) for providing the facilities for transmission electron microscopy; and C. Wenzl for critically reading the manuscript. We also thank Abbott Laboratories for supplying ABT-100. The work was funded by the Deutsche Forschungsgemeinschaft and Landesstiftung Baden-Württemberg. M. P. is supported by the Netzwerk AlternsfoRschung, University of Heidelberg.
Abbreviations used:
- aa
amino acids
- FL
full length
- FT
farnesyltransferase
- FTI
farnesyltransferase inhibitor
- GFP
green fluorescent protein
- INM
inner nuclear membrane
- Kuk
Kugelkern
- NLS
nuclear localization signal
- NM
nuclear membrane.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-03-0230) on August 4, 2010.
REFERENCES
- Ben-Harush K., Wiesel N., Frenkiel-Krispin D., Moeller D., Soreq E., Aebi U., Herrmann H., Gruenbaum Y., Medalia O. The supramolecular organization of the C. elegans nuclear lamin filament. J. Mol. Biol. 2009;386:1392–1402. doi: 10.1016/j.jmb.2008.12.024. [DOI] [PubMed] [Google Scholar]
- Brandt A., Krohne G., Grosshans J. The farnesylated nuclear proteins KUGELKERN and LAMIN B promote aging-like phenotypes in Drosophila flies. Aging Cell. 2008;7:541–551. doi: 10.1111/j.1474-9726.2008.00406.x. [DOI] [PubMed] [Google Scholar]
- Brandt A., et al. Developmental control of nuclear size and shape by Kugelkern and Kurzkern. Curr. Biol. 2006;16:543–552. doi: 10.1016/j.cub.2006.01.051. [DOI] [PubMed] [Google Scholar]
- Broers J. L., Machiels B. M., van Eys G. J., Kuijpers H. J., Manders E. M., van Driel R., Ramaekers F. C. Dynamics of the nuclear lamina as monitored by GFP-tagged A-type lamins. J. Cell Sci. 1999;112:3463–3475. doi: 10.1242/jcs.112.20.3463. [DOI] [PubMed] [Google Scholar]
- Capell B. C., Collins F. S. Human laminopathies: nuclei gone genetically awry. Nat. Rev. Genet. 2006;7:940–952. doi: 10.1038/nrg1906. [DOI] [PubMed] [Google Scholar]
- Capell B. C., Erdos M. R., Madigan J. P., Fiordalisi J. J., Varga R., Conneely K. N., Gordon L. B., Der C. J., Cox A. D., Collins F. S. Inhibiting farnesylation of progerin prevents the characteristic nuclear blebbing of Hutchinson-Gilford progeria syndrome. Proc. Natl. Acad. Sci. USA. 2005;102:12879–12884. doi: 10.1073/pnas.0506001102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen M., Lee K. K., Wilson K. L., Gruenbaum Y. Transcriptional repression, apoptosis, human disease and the functional evolution of the nuclear lamina. Trends Biochem. Sci. 2001;26:41–47. doi: 10.1016/s0968-0004(00)01727-8. [DOI] [PubMed] [Google Scholar]
- De Sandre-Giovannoli A., Bernard R., Cau P., Navarro C., Amiel J., Boccaccio I., Lyonnet S., Stewart C. L., Munnich A., Le Merrer M., Levy N. Lamin a truncation in Hutchinson-Gilford progeria. Science. 2003;300:2055. doi: 10.1126/science.1084125. [DOI] [PubMed] [Google Scholar]
- Erber A., Riemer D., Bovenschulte M., Weber K. Molecular phylogeny of metazoan intermediate filament proteins. J. Mol. Evol. 1998;47:751–762. doi: 10.1007/pl00006434. [DOI] [PubMed] [Google Scholar]
- Eriksson M., et al. Recurrent de novo point mutations in lamin A cause Hutchinson-Gilford progeria syndrome. Nature. 2003;423:293–298. doi: 10.1038/nature01629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa K., Sugiyama S., Osouda S., Goto H., Inagaki M., Horigome T., Omata S., McConnell M., Fisher P. A., Nishida Y. Barrier-to-autointegration factor plays crucial roles in cell cycle progression and nuclear organization in Drosophila. J. Cell Sci. 2003;116:3811–3823. doi: 10.1242/jcs.00682. [DOI] [PubMed] [Google Scholar]
- Goldman R. D., Gruenbaum Y., Moir R. D., Shumaker D. K., Spann T. P. Nuclear lamins: building blocks of nuclear architecture. Genes Dev. 2002;16:533–547. doi: 10.1101/gad.960502. [DOI] [PubMed] [Google Scholar]
- Goldman R. D., et al. Accumulation of mutant lamin A causes progressive changes in nuclear architecture in Hutchinson-Gilford progeria syndrome. Proc. Natl. Acad. Sci. USA. 2004;101:8963–8968. doi: 10.1073/pnas.0402943101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock J. F., Paterson H., Marshall C. J. A polybasic domain or palmitoylation is required in addition to the CAAX motif to localize p21ras to the plasma membrane. Cell. 1990;63:133–139. doi: 10.1016/0092-8674(90)90294-o. [DOI] [PubMed] [Google Scholar]
- Harbury P. B., Zhang T., Kim P. S., Alber T. A switch between two-, three-, and four-stranded coiled coils in GCN4 leucine zipper mutants. Science. 1993;262:1401–1407. doi: 10.1126/science.8248779. [DOI] [PubMed] [Google Scholar]
- Hattier T., Andrulis E. D., Tartakoff A. M. Immobility, inheritance and plasticity of shape of the yeast nucleus. BMC Cell Biol. 2007;8:47. doi: 10.1186/1471-2121-8-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C., et al. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 2004;21:947–962. doi: 10.1002/yea.1142. [DOI] [PubMed] [Google Scholar]
- Kapinos L. E., Schumacher J., Mucke N., Machaidze G., Burkhard P., Aebi U., Strelkov S. V., Herrmann H. Characterization of the head-to-tail overlap complexes formed by human Lamin A, B1 and B2 “half-minilamin” dimers. J. Mol. Biol. 2010;396:719–731. doi: 10.1016/j.jmb.2009.12.001. [DOI] [PubMed] [Google Scholar]
- Kitten G. T., Nigg E. A. The CaaX motif is required for isoprenylation, carboxyl methylation, and nuclear membrane association of lamin B2. J. Cell Biol. 1991;113:13–23. doi: 10.1083/jcb.113.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlov M. M. Biophysics: joint effort bends membrane. Nature. 2010;463:439–440. doi: 10.1038/463439a. [DOI] [PubMed] [Google Scholar]
- Krohne G. Lamin assembly in vivo. Subcell. Biochem. 1998;31:563–586. [PubMed] [Google Scholar]
- Malik P., Korfali N., Srsen V., Lazou V., Batrakou D. G., Zuleger N., Kavanagh D. M., Wilkie G. S., Goldberg M. W., Schirmer E. C. Cell-specific and lamin-dependent targeting of novel transmembrane proteins in the nuclear envelope. Cell Mol. Life Sci. 2010;67:1353–1369. doi: 10.1007/s00018-010-0257-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margalit A., Segura-Totten M., Gruenbaum Y., Wilson K. L. Barrier-to-autointegration factor is required to segregate and enclose chromosomes within the nuclear envelope and assemble the nuclear lamina. Proc. Natl. Acad. Sci. USA. 2005;102:3290–3295. doi: 10.1073/pnas.0408364102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattout A., Dechat T., Adam S. A., Goldman R. D., Gruenbaum Y. Nuclear lamins, diseases and aging. Curr. Opin. Cell Biol. 2006;18:335–341. doi: 10.1016/j.ceb.2006.03.007. [DOI] [PubMed] [Google Scholar]
- Meshorer E., Gruenbaum Y. Gone with the Wnt/Notch: stem cells in laminopathies, progeria, and aging. J. Cell Biol. 2008;181:9–13. doi: 10.1083/jcb.200802155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olins A. L., Olins D. E. The mechanism of granulocyte nuclear shape determination: possible involvement of the centrosome. Eur. J. Cell Biol. 2005;84:181–188. doi: 10.1016/j.ejcb.2004.12.021. [DOI] [PubMed] [Google Scholar]
- Prufert K., Vogel A., Krohne G. The lamin CxxM motif promotes nuclear membrane growth. J. Cell Sci. 2004;117:6105–6116. doi: 10.1242/jcs.01532. [DOI] [PubMed] [Google Scholar]
- Ralle T., Grund C., Franke W. W., Stick R. Intranuclear membrane structure formations by CaaX-containing nuclear proteins. J. Cell Sci. 2004;117:6095–6104. doi: 10.1242/jcs.01528. [DOI] [PubMed] [Google Scholar]
- Rowat A. C., Brask J., Sparrman T., Jensen K. J., Lindblom G., Ipsen J. H. Farnesylated peptides in model membranes: a biophysical investigation. Eur. Biophys. J. 2004;33:300–309. doi: 10.1007/s00249-003-0368-x. [DOI] [PubMed] [Google Scholar]
- Silvius J. R., l'Heureux F. Fluorimetric evaluation of the affinities of isoprenylated peptides for lipid bilayers. Biochemistry. 1994;33:3014–3022. doi: 10.1021/bi00176a034. [DOI] [PubMed] [Google Scholar]
- Smith S., Blobel G. Colocalization of vertebrate lamin B and lamin B receptor (LBR) in nuclear envelopes and in LBR-induced membrane stacks of the yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 1994;91:10124–10128. doi: 10.1073/pnas.91.21.10124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei A., Hediger F., Neumann F. R., Bauer C., Gasser S. M. Separation of silencing from perinuclear anchoring functions in yeast Ku80, Sir4 and Esc1 proteins. EMBO J. 2004;23:1301–1312. doi: 10.1038/sj.emboj.7600144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth J. I., Yang S. H., Qiao X., Beigneux A. P., Gelb M. H., Moulson C. L., Miner J. H., Young S. G., Fong L. G. Blocking protein farnesyltransferase improves nuclear shape in fibroblasts from humans with progeroid syndromes. Proc. Natl. Acad. Sci. USA. 2005;102:12873–12878. doi: 10.1073/pnas.0505767102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler J. K., Bulger M., Kamakaka R. T., Kobayashi R., Kadonaga J. T. The p55 subunit of Drosophila chromatin assembly factor 1 is homologous to a histone deacetylase-associated protein. Mol. Cell. Biol. 1996;16:6149–6159. doi: 10.1128/mcb.16.11.6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler J. K., Collins K. A., Prasad-Sinha J., Amiott E., Bulger M., Harte P. J., Kobayashi R., Kadonaga J. T. Interaction between the Drosophila CAF-1 and ASF1 chromatin assembly factors. Mol. Cell. Biol. 2001;21:6574–6584. doi: 10.1128/MCB.21.19.6574-6584.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner N., Kagermeier B., Loserth S., Krohne G. The Drosophila melanogaster LEM-domain protein MAN1. Eur J. Cell Biol. 2006;85:91–105. doi: 10.1016/j.ejcb.2005.10.002. [DOI] [PubMed] [Google Scholar]
- Wagner N., Schmitt J., Krohne G. Two novel LEM-domain proteins are splice products of the annotated Drosophila melanogaster gene CG9424 (Bocksbeutel) Eur. J. Cell Biol. 2004;82:605–616. doi: 10.1078/0171-9335-00350. [DOI] [PubMed] [Google Scholar]
- Webster M., Witkin K. L., Cohen-Fix O. Sizing up the nucleus: nuclear shape, size and nuclear-envelope assembly. J. Cell Sci. 2009;122:1477–1486. doi: 10.1242/jcs.037333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesel N., Mattout A., Melcer S., Melamed-Book N., Herrmann H., Medalia O., Aebi U., Gruenbaum Y. Laminopathic mutations interfere with the assembly, localization, and dynamics of nuclear lamins. Proc. Natl. Acad. Sci. USA. 2008;105:180–185. doi: 10.1073/pnas.0708974105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worby C. A., Simonson-Leff N., Dixon J. E. RNA interference of gene expression (RNAi) in cultured Drosophila cells. Sci. STKE. 2001;2001:pl1. doi: 10.1126/stke.2001.95.pl1. [DOI] [PubMed] [Google Scholar]
- Wright L. P., Philips M. R. Thematic review series: lipid posttranslational modifications. CAAX modification and membrane targeting of Ras. J. Lipid Res. 2006;47:883–891. doi: 10.1194/jlr.R600004-JLR200. [DOI] [PubMed] [Google Scholar]
- Yang S. H., Bergo M. O., Toth J. I., Qiao X., Hu Y., Sandoval S., Meta M., Bendale P., Gelb M. H., Young S. G., Fong L. G. Blocking protein farnesyltransferase improves nuclear blebbing in mouse fibroblasts with a targeted Hutchinson-Gilford progeria syndrome mutation. Proc. Natl. Acad. Sci. USA. 2005;102:10291–10296. doi: 10.1073/pnas.0504641102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan H., Michelsen K., Schwappach B. 14–3-3 dimers probe the assembly status of multimeric membrane proteins. Curr. Biol. 2003;13:638–646. doi: 10.1016/s0960-9822(03)00208-2. [DOI] [PubMed] [Google Scholar]
- Zacharias D. A., Violin J. D., Newton A. C., Tsien R. Y. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science. 2002;296:913–916. doi: 10.1126/science.1068539. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.