The localization of active genes to the nuclear periphery is regulated through the cell cycle by Cdk1 phosphorylation of a single nuclear pore protein.
Abstract
Many inducible genes in yeast are targeted to the nuclear pore complex when active. We find that the peripheral localization of the INO1 and GAL1 genes is regulated through the cell cycle. Active INO1 and GAL1 localized at the nuclear periphery during G1, became nucleoplasmic during S-phase, and then returned to the nuclear periphery during G2/M. Loss of peripheral targeting followed the initiation of DNA replication and was lost in cells lacking a cyclin-dependent kinase (Cdk) inhibitor. Furthermore, the Cdk1 kinase and two Cdk phosphorylation sites in the nucleoporin Nup1 were required for peripheral targeting of INO1 and GAL1. Introduction of aspartic acid residues in place of either of these two sites in Nup1 bypassed the requirement for Cdk1 and resulted in targeting of INO1 and GAL1 to the nuclear periphery during S-phase. Thus, phosphorylation of a nuclear pore component by cyclin dependent kinase controls the localization of active genes to the nuclear periphery through the cell cycle.
INTRODUCTION
Chromatin is spatially organized within the nucleus. At a global level, chromosomes fold into stereotypical patterns. In many organisms, chromosomes assume a Rabl conformation in which telomeres cluster together at one pole of the nucleus and centromeres colocalize with the nuclear envelope at the opposite pole (Rabl, 1885; Marshall et al., 1996). In differentiated metazoan cells, chromosomes fold back on themselves and occupy distinct “territories” (Cremer et al., 2006). The spatial organization of the genome is reproducible and correlates with cell type and differentiation status, suggesting either that it is a product of transcriptional state or that it plays a role in regulating transcription.
In addition to the more global organization of chromosomes, the localization of individual genes can be coupled to their transcription. A number of developmentally regulated genes relocalize with respect to nuclear landmarks such as the nuclear periphery during differentiation (Kosak et al., 2002; Chuang et al., 2006; Ragoczy et al., 2006). We have used the recruitment of inducible genes to the nuclear periphery in the yeast S. cerevisiae as a model for these phenomena. Genes such as INO1 and GAL1 relocalize from the nucleoplasm to the nuclear periphery upon activation (Brickner and Walter, 2004; Casolari et al., 2004, 2005; Menon et al., 2005; Dieppois et al., 2006; Taddei et al., 2006). Recruitment to the nuclear periphery is rapid, correlates with a physical interaction with the nuclear pore complex (NPC), and requires protein components of the NPC (Casolari et al., 2004; Cabal et al., 2006; Dieppois et al., 2006; Brickner et al., 2007; Luthra et al., 2007; Ahmed et al., 2010). Potentially related phenomena have been observed in Drosophila and mammalian cells (Mendjan et al., 2006; Kurshakova et al., 2007; Brown et al., 2008). Peripheral localization has been proposed to promote transcription (Brickner and Walter, 2004; Menon et al., 2005; Taddei et al., 2006) or to couple transcription with mRNA processing and export (Blobel, 1985; Casolari et al., 2005; Dieppois et al., 2006). Targeting of the INO1 gene to the nuclear periphery is not dependent on transcription (Brickner et al., 2007), but is controlled by DNA “zip codes” in the promoter and promotes transcription (Ahmed et al., 2010).
An important aspect of gene recruitment to the nuclear periphery that has not been addressed to date is how it changes through the cell cycle. Because yeast undergoes a closed mitosis, it is possible for chromosomal loci to remain associated with the nuclear envelope throughout the cell cycle. However, the spatial organization of the genome might also be regulated to accommodate episodic events such as DNA replication and repair, chromosome condensation, and chromosome segregation. Consistent with this notion, telomeres localize at the nuclear periphery in G1, S-phase, and mitosis, but lose peripheral localization during G2 (Laroche et al., 2000; Hediger et al., 2002; Ebrahimi and Donaldson, 2008). Loss of telomere localization at the nuclear periphery occurs after DNA replication and is due to loss of Ku binding to telomeres (Ebrahimi and Donaldson, 2008). However, it is unclear how the regulators of the cell cycle impinge upon Ku binding.
A number of the nuclear pore proteins that are required for peripheral localization of genes are phosphorylated by the master regulator of the cell cycle, cyclin-dependent kinase (Cdk1; Cdc28 in Saccharomyces cerevisiae). We hypothesized that Cdk1 might regulate gene localization through the cell cycle by phosphorylation of nuclear pore components. We find that, unlike telomeres, active INO1 and GAL1 localize at the nuclear periphery during G1 and G2/M, but localize to the nucleoplasm during S-phase. Loss of peripheral localization of these genes occurs after the initiation of DNA replication and was not observed in mutants lacking the Cdk inhibitor Sic1. Peripheral localization of INO1 and GAL1 during G1 and G2/M requires Cdk1. Phosphorylation of two sites in the nuclear pore protein Nup1 is necessary to promote peripheral targeting of active INO1 and GAL1. Mutations that mimic phosphorylation of either of these sites bypass the requirement for Cdk1 in gene localization. These results provide important insight into the functional significance of Cdk1 phosphorylation of a nuclear pore component and suggest that phosphoregulation of the NPC may function to coordinate DNA localization and transcription of highly expressed genes with DNA replication, repair, and segregation.
MATERIALS AND METHODS
Chemicals and Enzymes
All chemicals unless noted otherwise were from Sigma Aldrich (St. Louis, MO). Alpha factor was from Zymo Research (Orange, CA). Yeast media components were from Sunrise Science Products (San Diego, CA). Unless noted otherwise, restriction enzymes and DNA-modifying enzymes were from New England Biolabs (Ipswich, MA). Rabbit anti-green fluorescent protein (GFP) antibody, goat anti-mouse-Alexafluor 594 and goat anti-rabbit Alexafluor 488 were from Invitrogen (Carlsbad, CA). Anti-myc mouse mAb was from Santa Cruz Biotechnology (Santa Cruz, CA).
Yeast Genetics and Growth Conditions
All yeast strains used in this study are listed in Table 1. Null mutant strains were constructed in W303 strain CRY1 using a PCR-based strategy as described (Longtine et al., 1998). Conditional cdc6-1, cdc20-1, and cdc28-1 mutants were introduced into the W303 background by backcrossing American Type Culture Collection strains 208547 (cdc6-1), 208581 (cdc20-1), and 208591 (cdc28-1) by at least five rounds of backcrossing with W303 strains CRY1 or CRY2. Strains used for microscopy were transformed with pAFS144 (Straight et al., 1996), pRS304-Sec63-myc (Brickner et al., 2007), and either p6LacO128-INO1 (Brickner and Walter, 2004) or p6LacO128-GAL1 (Brickner et al., 2007). For Figure 1E, the strain was transformed with plasmid pGFP-FFAT-LacI (Brickner and Walter, 2004) instead of pAFS144.
Table 1.
Yeast strains used in this study
| Strain | Genotypea | Reference |
|---|---|---|
| CRY1 | MATa ade2-1 can1-100 his3-15,112 trp1-1 ura3-1 | Brickner and Fuller (1997) |
| CRY2 | MATα ade2-1 can1-100 his3-15,112 trp1-1 ura3-1 | Brickner and Fuller (1997) |
| JBY397 | SEC63-13myc:Kanr INO1:LacO128 HIS3:LacI-GFP | Brickner and Walter (2004) |
| DBY32 | SEC63-13myc:Kanr GAL1:LacO128 HIS3:LacI-GFP | Brickner et al. (2007) |
| DBY247 | nup1Δ::Kanr INO1:LacO128 HIS3:LacI-GFP TRP1:Sec63myc | This study |
| DBY178 | clb2Δ::Kanr INO1:LacO128 HIS3:LacI-GFP TRP1:Sec63myc | This study |
| DBY180 | clb5Δ::Kanr INO1:LacO128 HIS3:LacI-GFP TRP1:Sec63myc | This study |
| DBY182 | cln3Δ::Kanr INO1:LacO128 HIS3:LacI-GFP TRP1:Sec63myc | This study |
| DBY184 | sic1Δ::Kanr INO1:LacO128 HIS3:LacI-GFP TRP1:Sec63myc | This study |
| DBY243 | nup1Δ::Kanr INO1:LacO128 HIS3:LacI-GFP TRP1:Sec63myc LEU2:NUP1-PM1 | This study |
| DBY245 | nup1Δ::Kanr INO1:LacO128 HIS3:LacI-GFP TRP1:Sec63myc LEU2:NUP1-PM2 | This study |
| DBY248 | nup1Δ::Kanr INO1:LacO128 HIS3:LacI-GFP TRP1:Sec63myc LEU2:NUP1 | This study |
| DBY249 | nup1Δ::Kanr INO1:LacO128 HIS3:LacI-GFP TRP1:Sec63myc LEU2:nup1-ΔP1 | This study |
| DBY250 | nup1Δ::Kanr INO1:LacO128 HIS3:LacI-GFP TRP1:Sec63myc LEU2:nup1-ΔP2 | This study |
| DBY251 | nup1Δ::Kanr INO1:LacO128 HIS3:LacI-GFP TRP1:Sec63myc LEU2:NUP1 P1ΔP2 | This study |
| DBY252 | SEC63-13myc:Kanr INO1:LacO128 HIS3:LacI-GFP LEU2:NUP1-PM1 | This study |
| DBY253 | SEC63-13myc:Kanr INO1:LacO128 HIS3:LacI-GFP LEU2:NUP1-PM2 | This study |
| DBY263 | cdc6-1 GAL1:LacO128 HIS3:LacI-GFP TRP1:Sec63myc | This study |
| DBY264 | cdc20-1 GAL1:LacO128 HIS3:LacI-GFP TRP1:Sec63myc | This study |
| DBY265 | nup1Δ::Kanr GAL1:LacO128 HIS3:LacI-GFP TRP1:Sec63myc LEU2:NUP1 | This study |
| DBY266 | nup1Δ::Kanr GAL1:LacO128 HIS3:LacI-GFP TRP1:Sec63myc LEU2:nup1-ΔP1 | This study |
| DBY267 | nup1Δ::Kanr GAL1:LacO128 HIS3:LacI-GFP TRP1:Sec63myc LEU2:nup1-ΔP1ΔP2 | This study |
| DBY268 | nup1Δ::Kanr GAL1:LacO128 HIS3:LacI-GFP TRP1:Sec63myc LEU2:NUP1-PM1 | This study |
| DBY269 | nup1Δ::Kanr GAL1:LacO128 HIS3:LacI-GFP TRP1:Sec63myc LEU2:NUP1-PM2 | This study |
| DBY270 | SEC63-13myc:Kanr GAL1:LacO128 HIS3:LacI-GFP LEU2:NUP1-PM1 | This study |
| DBY271 | SEC63-13myc:Kanr GAL1:LacO128 HIS3:LacI-GFP LEU2:NUP1-PM2 | This study |
| DBY273 | cdc28-1 INO1:LacO128 LEU2:LacI-GFP TRP1:Sec63myc | This study |
| DBY274 | cdc28-1 GAL1:LacO128 HIS3:LacI-GFP TRP1:Sec63myc | This study |
| DBY279 | cdc28-1 INO1:LacO128 HIS3:LacI-GFP TRP1:Sec63myc LEU2:NUP1-PM1 | This study |
| DBY280 | cdc28-1 INO1:LacO128 HIS3:LacI-GFP TRP1:Sec63myc LEU2:NUP1-PM2 | This study |
| DBY283 | cdc28-1 INO1:LacO128 HIS3:LacI-GFP TRP1:Sec63myc LEU2:CDC28 | This study |
| DBY284 | cdc28-1 INO1:LacO128 HIS3:LacI-GFP TRP1:Sec63myc LEU2:CDC28 | This study |
| DBY285 | cdc28-1 INO1:LacO128 HIS3:LacI-GFP TRP1:Sec63myc LEU2:NUP1-PM1 | This study |
| DBY286 | cdc28-1 INO1:LacO128 HIS3:LacI-GFP TRP1:Sec63myc LEU2:NUP1-PM2 | This study |
| DBY293 | nup1Δ::Kanr GAL1:LacO128 HIS3:LacI-GFP TRP1:Sec63myc LEU2:nup1-ΔP2 | This study |
| DBY295 | cdc28-1 INO1:LacO128 HIS3:LacI-GFP TRP1:Sec63myc LEU2:CDC28 | This study |
| DBY296 | cdc28-1 GAL1:LacO128 HIS3:LacI-GFP TRP1:Sec63myc LEU2:CDC28 | This study |
| DBY299 | cdc28-1 GAL1:LacO128 HIS3:LacI-GFP TRP1:Sec63myc LEU2:NUP1hyphen]PM2 | This study |
| DBY300 | cdc28-1 GAL1:LacO128 HIS3:LacI-GFP TRP1:Sec63myc LEU2:NUP1-PM1 | This study |
a All strains are isogenic with CRY1, except as indicated.
Figure 1.
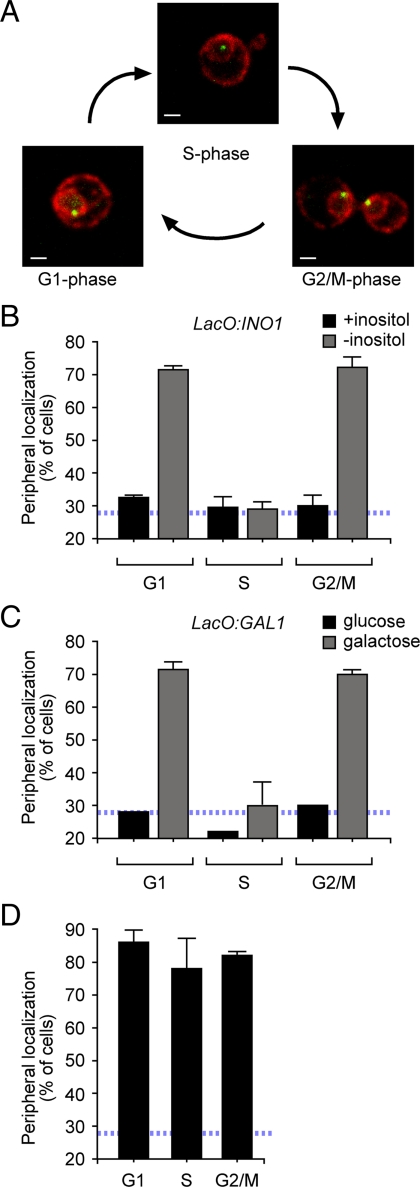
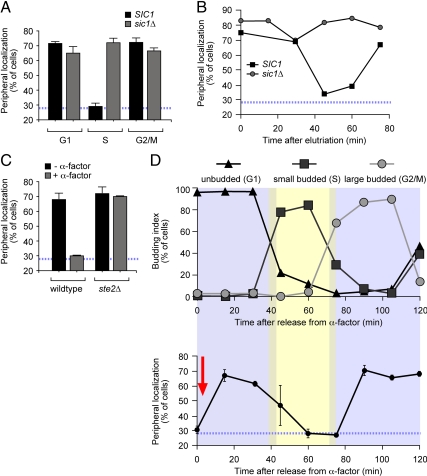
Gene localization through the cell cycle. (A) Representative images of unbudded, small-, and large-budded cells stained for GFP-Lac I (green) and Sec63-myc (red). Scale bar, 1 μm. The localization of the INO1 gene (B) and the GAL1 gene (C) was quantified under either repressing (■) or activating (▩) conditions in unbudded (G1), small- (S), and large-budded (G2/M) cells from an asynchronous culture. (D) Localization of artificially tethered INO1 through the cell cycle. Localization of tethered INO1 was performed as in B and C. In B–D, the blue, hatched line represents the level of colocalization of the lac repressor spot with the nuclear envelope predicted by chance (Brickner and Walter, 2004).
For all experiments, cells were grown in synthetic, defined medium (SDC; Burke et al., 2000). Cells grown under activating conditions for INO1 were grown in SDC-inositol. Cells grown under activating conditions for GAL1 were grown in SGC. Cells grown under repressing conditions for either INO1 or GAL1 were grown in SDC. Except for experiments involving temperature-sensitive mutants, cells were grown at 30°C. For experiments with temperature-sensitive strains, the permissive temperature was 22°C and the restrictive temperature was 37°C.
Molecular Biology
All oligonucleotides used in this study are listed in Table 2. The NUP1 gene and ∼500 base pairs 5′ and 3′ of the coding sequence was amplified by PCR using primers NUP1F and NUP1R from yeast genomic DNA. The PCR product was TA TOPO-cloned (Invitrogen) and then moved as a BamHI-NotI fragment into pRS305 (Sikorski and Hieter, 1989). The mutant versions of NUP1 were made using PCR-based mutagenesis in pRS305-NUP1. Mutations were confirmed by DNA sequencing. These plasmids were integrated at the LEU2 locus in nup1Δ strain (see Figures 5 and 6) or a NUP1 strain (Figure S4) by digestion with AflII and transformation into yeast. Transformants were selected on plated lacking leucine.
Table 2.
Primers used in this study
| Name | Sequence (5′ to 3′) |
|---|---|
| CDC28F | CCCTTACTGGGCGCACGCAGTGTATC |
| CDC28R | GAAATAAAAACAATACCCGTCTCGGTTATTC |
| CLB2confirm | GCCAATTTAGCTAGACTTTTGAAAAG |
| CLB2KOF1 | AGCCTTTTATTGATTACCCCCTCTCTCTCTTCATTGATCTTATAGCGGATCCCCGGGTTAATTAA |
| CLB2KOR1 | TTTATCGATTATCGTTTTAGATATTTTAAGCATCTGCCCCTCTTCGAATTCGAGCTCGTTTAAAC |
| CLB5confirm | CTTGTCATACTGTGGGTATCATATTCTG |
| CLB5KOF1 | GCGCTTTTCCCTGTATTTAAAGCCGCTGAACACCTTTACTGAACACGGATCCCCGGGTTAATTAA |
| CLB5KOR1 | AAAATGTAAAGAGTATGCGAATTCATGAGCATTACTAGTACTAATGAATTCGAGCTCGTTTAAAC |
| CLN3confirm | GGCCTTGGGTTTTTGCCCTCATC |
| CLN3KNOF1 | CTTTTACTCTCGTTCAAGACACTGATTTGATACGCTTTCTGTACGCGGATCCCCGGGTTAATTAA |
| CLN3KOR1 | AAATTTTAATTTATTTGTTGTTAAATGCATTTTTTTTTTGTCGTTGAATTCGAGCTCGTTTAAAC |
| KanB | CTGCAGCGAGGAGCCGTAAT |
| NUP1F | GACACGAAGTCTCTCTGGAGTGCTTTG |
| NUP1R | CATCATTGTTGTGAATACGCACCCTAC |
| NUP1ΔP1F | CATCTACTCCTTGCCCTATTAAAAAC |
| NUP1ΔP1R | GTTTTTAATAGGGCAAGGAGTAGATG |
| NUP1ΔP2F | CAAGGACAATGAGTGTCCATCTAAGAAAACTTC |
| NUP1ΔP2R | GAAGTTTTCTTAGATGGACACTCATTGTCCTTG |
| NUP1PM1F | CATCTACTCCTGACCCTATTAAAAAC |
| NUP1PM1R | GTTTTTAATAGGGTCAGGAGTAGATG |
| NUP1PM2F | CAAGGACAATGAGGATCCATCTAAGAAAACTTC |
| NUP1PM2R | GAAGTTTTCTTAGATGGATCCTCATTGTCCTTG |
| SIC1confirm | GTGAGACCAGAGTTGCAGAGATGGAC |
| SIC1KOF1 | CCTCTACGGAATTTTGACCCTTGAAGCAGGGACTATTACACGAAACGGATCCCCGGGTTAATTAA |
| SIC1KOR1 | GTAAATAAAATATAATCGTTCCAGAAACTTTTTTTTTTCATTTCTGAATTCGAGCTCGTTTAAAC |
| STE2confirm | GCATAGAACGAACTGTAGAATAGTCCGG |
| STE2KOF1 | GTTACTTAAAAATGCACCGTTAAGAACCATATCCAAGAATCAAAACGGATCCCCGGGTTAATTAA |
| STE2KOR1 | ATACCGAAGGTCACGAAATTACTTTTTCAAAGCCGTAAATTTTGAGAATTCGAGCTCGTTTAAAC |
Figure 5.
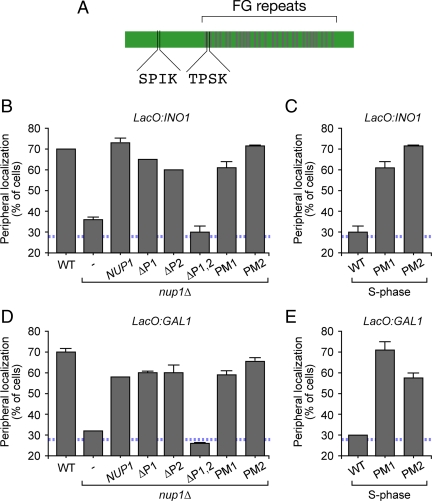
Phosphorylation of Nup1 at two Cdk consensus sites is required for gene recruitment to the nuclear periphery. (A) Schematic of the Nup1 protein. Two Cdk consensus sites with the phospho-acceptor sites corresponding to amino acids 161 and 344 are highlighted, and the FG repeats are indicated as gray bars. (B and D) Effect of loss of phospho-acceptor sites on INO1 (B) or GAL1 (D) localization at the nuclear periphery in an asynchronous population grown under activating conditions. nup1Δ cells were transformed with integrating plasmids expressing the indicated alleles of NUP1. ΔP1 = S161C, ΔP2 = T344C, PM1 = S161D, and PM2 = T344D. (C and E) Effect of phosphomimetic NUP1 mutations on localization of INO1 (C) or GAL1 (E) in small-budded cells grown under activating conditions. Cells expressing either wild-type NUP1, NUP1-PM1, or NUP1-PM2 were scored for localization of INO1 (C) or GAL1 (E) in small-budded cells. The blue, hatched line represents the level of colocalization of the lac repressor spot with the nuclear envelope predicted by chance (Brickner and Walter, 2004).
Figure 6.
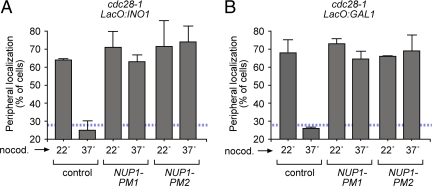
Phosphomimetic mutations in Nup1 bypass the requirement for Cdk1 in gene targeting to the nuclear periphery. cdc28-1 cells having the lac repressor array integrated at INO1 (A) or GAL1 (B) were transformed with integrating plasmids expressing NUP1-PM1 or NUP1-PM2. Cells were grown under activating conditions at 22°C and treated with nocodazole for 2 h as in Figure 4. Cells were then either maintained at 22°C or shifted to 37°C for 30 min before localization of INO1 (A) or GAL1 (B) was quantified. For comparison, untransformed cells were also scored (control). The blue, hatched line represents the level of colocalization of the lac repressor spot with the nuclear envelope predicted by chance (Brickner and Walter, 2004).
The CDC28 gene and ∼500 base pairs 5′ and 3′ of the coding sequence was amplified by PCR using primers CDC28F and CDC28R from yeast genomic DNA and TA TOPO-cloned. The gene was then moved as a BamHI-NotI fragment into pRS305 to create pRS305-CDC28. This plasmid was integrated at the LEU2 locus by digestion with BsrGI and transformation into yeast. Transformants were selected on plated lacking leucine.
Chromatin Localization Assay
Chromatin localization assay was performed as described (Brickner et al., 2010; Brickner and Walter, 2004). Briefly, cells having the lac repressor array integrated at either INO1 or GAL1 (Brickner et al., 2007) and expressing both the lac repressor fused to GFP (Straight et al., 1996) and Sec63-myc were grown under activating conditions (SDC-inositol for INO1 or SGC for GAL1) or repressing conditions (SDC for both INO1 and GAL1). Cells were harvested by centrifugation, resuspended in cold 100% methanol and incubated at −20°C for ≥20 min. Cells were spheroplasted with lyticase, detergent-extracted, and placed in polylysine-treated wells. Fixed spheroplasts were stained with anti-myc and anti-GFP antibodies. After staining with secondary antibodies, the cells were visualized on a Zeiss Pascal line-scanning confocal microscope (Thornwood, NY).
For experiments in which we examined all of the cells, the budding morphology was ignored and the localization of the GFP spot was scored in each cell with respect to the nuclear envelope (Brickner and Walter, 2004). For cells in which we scored each budding class, the localization of the GFP spot with respect to the nuclear envelope was scored and then the size of the bud was determined by scanning through the entire cell in the z dimension and examining the Sec63-myc staining. Cells lacking a visible bud were classified as unbudded G1 cells. Cells having a small, spherical bud with a diameter one third or less of the long axis of the mother cell were classified as small-budded S-phase cells. Cells having an ovoid bud with a diameter more than one-third of the long axis of the mother cell but having only one nucleus were classified as G2/M cells. For each experiment and each class, 30–50 cells were scored. For all experiments, three or more biological replicates were performed.
Centrifugal Elutriation
Wild-type and sic1Δ cells were grown in SDC-inositol to OD600 ∼0.7 at 30°C and shifted for 1 h to 20°C. Using a Masterflex L/S peristaltic pump (Cole Parmer, Chicago, IL), cells were flowed into a Beckman TE-5.0 elutrator chamber, spinning at 3000 rpm in Beckman J-6M/E centrifuge (Fullerton, CA). Fresh SDC-inositol was then pumped through the chamber, and the flow rate was slowly increased until unbudded cells were visualized under the microscope. Fractions were collected over ∼15 min. The cells were concentrated by filtration and resuspended in 10 ml of fresh SDC-inositol. Elutriated cells (1 ml) were collected at various times, fixed in methanol, and processed for immunofluorescence.
RESULTS
Localization of Active INO1 and GAL1 Is Regulated through the Cell Cycle
We used a quantitative chromatin localization assay to monitor the localization of genes with respect to the nuclear periphery in yeast (Brickner and Walter, 2004; Brickner et al., 2010). This assay uses a gene-marking system developed by Belmont and colleagues (Robinett et al., 1996; Straight et al., 1996). In this approach, an array of lac repressor binding sites is integrated adjacent to the gene of interest in a yeast strain expressing the lac repressor fused to GFP. We then perform immunofluorescence and confocal microscopy to quantify the fraction of the population in which the GFP spot colocalized with the nuclear envelope (Brickner and Walter, 2004). Nucleoplasmically localized genes like URA3 colocalize with the nuclear envelope in ∼27% of the cells in the population (this baseline is denoted by the blue hatched line in Figures 1–6; Brickner and Walter, 2004). Before activation, the INO1 and GAL1 genes localize in a pattern similar to URA3; they colocalize with the nuclear envelope in ∼30% of cells in the population (Brickner and Walter, 2004; Casolari et al., 2004; Brickner et al., 2007). However, upon activation, these genes are rapidly recruited to the nuclear periphery and colocalize with the nuclear envelope in ∼65% of the cells in the population (Brickner and Walter, 2004; Brickner et al., 2007). More stably tethered loci such as telomeres or artificially tethered genes localize at the nuclear periphery in ∼80% of cells using this assay (our unpublished results; Brickner and Walter, 2004). Therefore, the dynamic range of this assay for localization of any given locus to the nuclear periphery in yeast is 25–80% of the cells in the population (Brickner et al., 2007).
Figure 2.
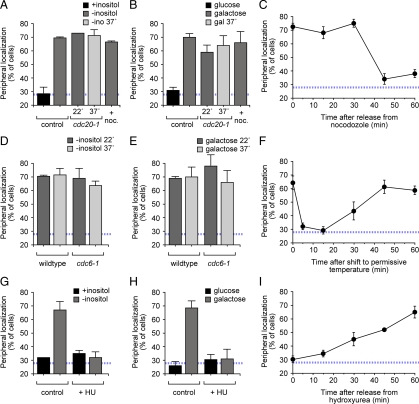
Loss of peripheral localization during S-phase. (A and B) Localization of INO1 (A) or GAL1 (B) arrested during mitosis. Wild-type (control) or cdc20-1 cells were grown under repressing conditions (■), activating conditions (dark gray bars) or activating conditions at 37°C (2 h; light gray bars) before scoring for localization. The right-most bar is the wild-type strain grown under activating conditions treated with nocodazole (+noc) for 2 h (dark gray bars). (C) Nocodazole-arrested cells having the lac operator array at INO1 were washed into fresh medium, and the localization of INO1 was scored at the indicated times. (D and E) Localization of INO1 (D) or GAL1 (E) arrested at the G1/S transition. Wild-type (control) or cdc6-1 cells were grown under activating conditions (dark gray bars) or shifted to the restrictive temperature of 37°C for 2 h (light gray bars) before scoring for localization. (F) cdc6-1 mutant cells arrested at the restrictive temperature were washed into fresh medium at 25°C and the localization of INO1 was scored at the indicated times. (G and H) Localization of INO1 (G) and GAL1 (H) in cells arrested during S-phase. Cells were grown under repressing conditions (■) or activating conditions (dark gray bars) with or without 0.1 M hydroxyurea (+HU) for 2 h and scored for localization. (I) Cells arrested with hydroxyurea were washed into activating medium without drug and the localization of INO1 was scored at the indicated times. The blue, hatched line represents the level of colocalization of the lac repressor spot with the nuclear envelope predicted by chance (Brickner and Walter, 2004).
Figure 3.
Cdk inhibitors negatively regulate gene recruitment to the nuclear periphery. (A) Wild-type (■) and sic1Δ mutant (▩) cells grown under activating conditions were scored for INO1 localization in cells with no buds (G1), small buds (S) or large buds (G2/M) as in Figure 1. (B) Newly born wild-type and sic1Δ mutant cells grown under activating conditions were recovered by centrifugal elutriation, and the localization of INO1 was scored at the indicated times. (C) Wild-type and ste2Δ mutant cells were grown under activating conditions in the presence (▩) or absence (■) of alpha factor. The population was scored for INO1 localization with respect to the nuclear periphery. (D) Alpha factor–arrested cells were washed into fresh activating medium lacking alpha factor at time 0 (red arrow) and INO1 localization (bottom panel) and budding index (top panel) were scored at the indicated times. The blue, hatched line represents the level of colocalization of the lac repressor spot with the nuclear envelope predicted by chance (Brickner and Walter, 2004).
Figure 4.
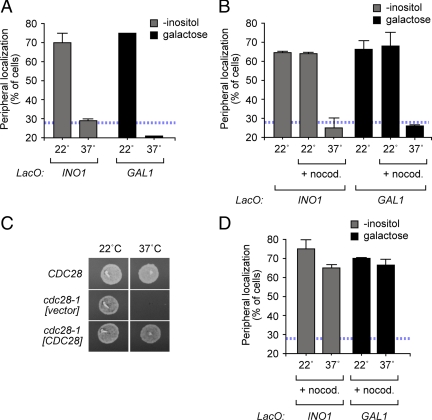
Cdk1 promotes gene recruitment to the nuclear periphery. (A) cdc28-1 mutant cells having the lac operator array at INO1 (■) or at GAL1 (▩) were grown under activating conditions at 22°C or shifted to 37°C for 2 h. (B) cdc28-1 mutant cells having the lac operator array at INO1 (■) or at GAL1 (▩) were grown under activating conditions at 22°C. Cells were then treated with nocodazole for 2 h and then either maintained at 22°C or shifted to 37°C for 30 min before quantifying gene localization. (C) Complementation of the cdc28-1 mutation by cloned CDC28. The cdc28-1 mutant was transformed with the indicated plasmids and tested for growth at either 22 or 37°C. (D) The cdc28-1 mutant transformed with CDC28 was tested for localization as in B. In A, B, and D, the blue, hatched line represents the level of colocalization of the lac repressor spot with the nuclear envelope predicted by chance (Brickner and Walter, 2004).
Using a modified version of this assay, we asked if the localization of active genes to the nuclear periphery was constant through the cell cycle. In budding yeast, the morphology of the cells correlates with the cell cycle stage (Hartwell et al., 1970). Recently separated daughter and mother cells in G1 do not have visible buds. S-phase cells possess a small, spherical bud with a diameter one-third or less of the long axis of the mother cell. Before cell division, G2 cells possess a large, ovoid bud with a long axis one-third or more of the long axis of the mother. Thus, within a mixed population of cells, bud size serves as a reasonable proxy for cell cycle stage. Because Sec63 marks both the nuclear envelope and the cell cortex (i.e., the cortical endoplasmic reticulum), we were able to determine both the position of the gene with respect to the nuclear envelope and the budding morphology for each cell (Figure 1A). We used this approach to quantify the fraction of unbudded (G1), small- (S), or large-budded (G2/M) cells in which INO1 or GAL1 colocalized with the nuclear envelope (Figure 1, B and C). Both INO1 and GAL1 localized at the nuclear periphery in ∼70% of the unbudded and large-budded cells under activating conditions (Figure 1, B and 1C). However, INO1 and GAL1 localized at the nuclear periphery in only ∼30% of small-budded cells (Figure 1, B and C), suggesting that active INO1 and GAL1 were not targeted to the nuclear periphery during S-phase. The change in localization between unbudded and small-budded cells was highly significant (using a two-tailed t test, p < 0.0001 for INO1; p = 0.0047 for GAL1), as was the change between small- and large-budded cells (p = 0.003 for INO1; p = 0.005 for GAL1).
The loss of peripheral localization during S-phase could be due to either a specific regulation of gene recruitment or to a dominant, nonspecific effect of DNA replication. We therefore tested if DNA replication nonspecifically disrupts peripheral localization by examining the localization of a form of INO1 that is artificially tethered to the nuclear envelope during S-phase. In a strain having a lac repressor array beside INO1 and expressing a nuclear envelope-bound lac repressor, the INO1 gene is very efficiently tethered to the nuclear envelope (GFP-FFAT-LacI; Supplementary Figure S1; Brickner and Walter, 2004; Brickner et al., 2007). The tethered INO1 gene localized at the nuclear periphery in ∼80% of cells during all cell cycle stages (Figure 1E), indicating that DNA replication is not sufficient to disrupt (artificial) peripheral localization. This is consistent with the observation that telomeres can be replicated at the nuclear periphery (Ebrahimi and Donaldson, 2008). Therefore, loss of peripheral localization of active genes is more likely due to regulation of gene targeting by the cell cycle machinery.
Loss of Peripheral Localization Occurs after the Initiation of DNA Replication
To more precisely determine when gene recruitment to the nuclear periphery is lost, we examined the localization of active INO1 and GAL1 in cells arrested at three points in the cell cycle: during mitosis, at the G1/S transition and during S phase. To arrest cells during mitosis, we treated cells with the tubulin depolymerizing drug nocodazole. This treatment results in activation of the spindle checkpoint cell cycle arrest (Kunkel, 1980; Hardwick and Murray, 1995). In cells arrested with nocodazole, both INO1 and GAL1 localized at the nuclear periphery in ∼65% of the cells (Figure 2, A and 2B).
We also arrested cells in metaphase using a cdc20-1 temperature-sensitive mutant. Cdc20 is an essential activator of the anaphase-promoting complex and is required for cells to proceed through metaphase (Visintin et al., 1997; Hwang et al., 1998). At the permissive temperature, INO1 and GAL1 localized at the nuclear periphery in the cdc20-1 mutant. Consistent with the nocodazole arrest, when cdc20-1 cells were arrested at the restrictive temperature, both INO1 and GAL1 localized at the nuclear periphery in >65% of the cells in the population (Figure 2, A and B). Thus, INO1 and GAL1 localize at the nuclear periphery during mitosis.
We next examined the localization of INO1 and GAL1 in cells arrested at the G1/S transition. To arrest cells, we used a cdc6-1 temperature-sensitive mutant. Cdc6 is a component of the prereplication complex that is essential for the initiation of DNA replication (Cocker et al., 1996). Inactivation of Cdc6 results in cell cycle arrest immediately before the initiation of DNA replication (Cocker et al., 1996). At the both the permissive temperature and the restrictive temperature, INO1 and GAL1 localized at the nuclear periphery in cdc6-1 mutant cells (Figure 2, D and E). This suggested that the loss of peripheral localization occurs after the G1/S transition.
To arrest cells during S-phase, we treated with hydroxyurea, which inhibits ribonucleotide reductase, depleting deoxynucleotide pools and activating the DNA damage checkpoint after DNA replication has initiated (Young and Hodas, 1964; Hartwell and Weinert, 1989; Hartwell et al., 1994). In cells treated with hydroxyurea, both INO1 and GAL1 localized to the nucleoplasm (Figure 2, G and H). Thus, consistent with the experiments in which we classified cells from an asynchronous population, the loss of peripheral localization correlates with DNA replication.
To confirm that the loss of peripheral targeting of active genes occurred at a unique moment of the cell cycle, we also monitored the localization of INO1 over time in synchronized populations after release from nocodazole (mitotic arrest), cdc6-1 arrest (G1/S arrest), or hydroxyurea (S-phase arrest). In cells released from mitotic arrest, the INO1 gene remained peripheral for ∼40 min before localizing to the nucleoplasm (Figure 2C). In cells released from G1/S arrest, the localization of INO1 to the nuclear periphery was immediately lost; the gene localized to the nucleoplasm within 5 min of release, remained nucleoplasmic for ∼20 min before returning to the nuclear periphery (Figure 2F). Finally, in cells released from S-phase arrest, we observed a gradual relocalization to the nuclear periphery (Figure 2I). The relocalization in cells released from hydroxyurea was slower than in cells released from a cdc6-1 arrest, perhaps reflecting time required to synthesize dNTPs to allow the completion of DNA replication. Taken together, these observations show that loss of peripheral targeting of INO1 and GAL1 occurred during a specific, ∼20–30-min period during S-phase after the initiation of DNA replication.
Cdk Inhibitors Impact Gene Localization through the Cell Cycle
To probe the mechanism that regulates the peripheral localization of active genes through the cell cycle, we characterized the effects of loss of a number of nonessential cell cycle regulators on the localization of INO1. Mutants lacking these proteins had no effect on the targeting of INO1 to the nuclear periphery in an asynchronous population (Figure S2). However, in mutants lacking the Cdk inhibitor Sic1, the INO1 gene localized at the nuclear periphery in small-budded cells (Figure 3A). This suggested that the loss of peripheral localization during S-phase did not occur in the absence of Sic1. However, because sic1Δ mutants have a very short G1 phase, it was also possible that, in these mutants, the correlation between cell cycle stage and bud morphology was disrupted (Schneider et al., 1996). Therefore, as a complementary approach, we followed the localization of INO1 in cells synchronized in G1 by centrifugal elutriation (Gordon and Elliott, 1977). This approach allows isolation of newborn daughter cells by their size and shape. In wild-type daughters purified by elutriation, INO1 localized to the nuclear periphery in >65% of the cells for ∼40 min, after which it relocalized to the nucleoplasm for ∼20 min (Figure 3B). This is consistent with the expected duration of G1 for newborn daughters (∼37 min; Brewer et al., 1984) and with the time course observed after release from nocodazole (Figure 2C). However, in the sic1Δ strain, the INO1 gene localized at the nuclear periphery in >65% of the cells throughout the time course (Figure 3B). Thus, loss of Sic1 led to loss of cell cycle regulation of INO1 gene targeting to the nuclear periphery.
Because the relocalization of INO1 to the nucleoplasm was lost in the absence of a Cdk inhibitor, we hypothesized that Cdk-mediated phosphorylation of a particular substrate might promote gene targeting to the nuclear periphery. To further explore this possibility, we asked if induction of a different Cdk inhibitor would have the same effect. Yeast cells exposed to mating pheromone (alpha factor) induce a signal transduction cascade that results in cell cycle arrest (Herskowitz, 1995; Schwartz and Madhani, 2004). Pheromone-treated cells arrest at “Start” in G1 due to the activation of Far1, a Cdk inhibitor that inhibits Cdk1/Cln activity (Peter and Herskowitz, 1994). We asked if activation of Far1 at a point in the cell cycle when INO1 normally localizes at the nuclear periphery would disrupt peripheral targeting. In cells arrested at Start with alpha factor, the INO1 gene did not localize to the nuclear periphery (Figure 3C). This effect required the alpha factor receptor, Ste2 (Figure 3C) and Far1 (not shown). Localization of the GAL1 gene to the nuclear periphery was also lost in cells arrested with alpha factor (Supplementary Figure S3). Thus, activation of a Cdk inhibitor at a stage of the cell cycle when INO1 and GAL1 would normally localize to the nuclear periphery resulted in relocalization of both genes to the nucleoplasm.
We next monitored the localization of INO1 after release from mating pheromone arrest (Figure 3D, bottom and top panels, respectively). Fifteen minutes after the cells were released from pheromone, INO1 again localized to the nuclear periphery. However, as the population shifted from unbudded G1 cells to small-budded S-phase cells, the INO1 gene lost peripheral localization for ∼20 min (Figure 3D). Finally, as the population transitioned from small- to large-budded cells, the INO1 gene was targeted again to the nuclear periphery. These results suggest that inhibition of Cdk activity by either Far1 or Sic1 leads to loss of localization to the nuclear periphery, suggesting that Cdk activity promotes peripheral targeting. However, because Sic1 protein levels drop during S-phase (Donovan et al., 1994), we think it is unlikely that Sic1 directly antagonizes Cdk to inactivate peripheral targeting (see Discussion). Rather, we think that the loss of Sic1 leads to elevated Cdk activity and elevated phosphorylation of Cdk substrates and that this leads to increased targeting of INO1 to the nuclear periphery during S phase.
Cdk1 Activity Is Required for Targeting of Genes to the Nuclear Periphery
To test if Cdk activity promotes targeting of INO1 and GAL1 to the nuclear periphery, we examined the effect of inactivation of a temperature-sensitive allele of Cdk1, encoded by CDC28 in budding yeast. At the permissive temperature of 22°C, INO1 and GAL1 localized at the nuclear periphery in the cdc28-1 mutant (Figure 4A). However, at the restrictive temperature of 37°C, INO1 and GAL1 localized to the nucleoplasm in the cdc28-1 mutant (Figure 4A). Therefore, Cdc28 activity is essential for maintaining INO1 and GAL1 at the nuclear periphery.
Cdk1 activity is required for passage through both G1 and mitosis (Piggott et al., 1982; Rudner et al., 2000). Thus, the loss of peripheral localization upon inactivation of cdc28-1 at stages of the cell cycle during which INO1 and GAL1 normally localize at the nuclear periphery suggests that Cdk activity promotes peripheral localization. However, it was also possible that the arrested state in the cdc28-1 mutant, or the inactivation of the kinase for >2 h might indirectly affect targeting of genes to the nuclear periphery. To examine the more immediate effects of inactivating Cdc28 at a stage of the cell cycle when INO1 and GAL1 were localized at the nuclear periphery, we first trapped the cdc28-1 cells in mitosis using nocodazole before shifting them for 30 min to the restrictive temperature. In nocodazole-arrested cdc28-1 cells at the permissive temperature INO1 and GAL1 localized to the nuclear periphery (Figure 4B). However, when nocodazole-arrested cdc28-1 cells were shifted to 37°C to inactivate Cdc28, localization of INO1 and GAL1 to the nuclear periphery was lost (Figure 4B). This loss of peripheral localization was not observed in a cdc28-1 mutant complemented with a plasmid expressing wild-type CDC28 (Figure 4, C and D). Therefore, Cdk1 activity is essential for localizing INO1 and GAL1 at the nuclear periphery.
Phosphorylation of Nup1 by Cdk1 Controls Cell Cycle–regulated Targeting of Genes to the Nuclear Periphery
A number of proteins within, or associated with, the NPC are required for targeting of active INO1 and GAL1 to the nuclear periphery (Casolari et al., 2004; Schmid et al., 2006; Brickner et al., 2007; Luthra et al., 2007; Ahmed et al., 2010). We hypothesized that phosphorylation of one or more of these proteins might regulate this function during the cell cycle. Nup1 is required for peripheral targeting of both INO1 and GAL1 (Cabal et al., 2006; Ahmed et al., 2010). Nup1 is an FXFG repeat protein that associates with the nucleoplasmic face of the NPC. Cdk1 phosphorylates Nup1 in vitro and in vivo (Figure 5A; Ubersax et al., 2003; Holt et al., 2009). However, Nup1 is phosphorylated on at least 31 sites in vivo by several kinases (Li et al., 2007; Smolka et al., 2007; Albuquerque et al., 2008), making it difficult to unambiguously assess the phosphorylation state of the full-length protein. Furthermore, in no case is the functional significance of these modifications known. We identified two sites in the amino terminus of Nup1 that match the Cdk consensus: S/TPXK. To determine if phosphorylation of these sites is important for targeting of INO1 and GAL1, we introduced cysteine residues in place of the phosphoacceptor sites (ΔP1 = S161C; ΔP2 = T344C). Neither mutation alone had an effect on localization of INO1 or GAL1 (Figure 5, B and D). However, the ΔP1ΔP2 double mutant form of NUP1 failed to complement the defect in peripheral targeting of both INO1 and GAL1 observed in the nup1Δ mutant (Figure 5, B and D). This mutant form of Nup1 was able to complement the temperature-sensitive phenotype of the nup1Δ mutant (Supplementary Figure S4), suggesting that it was not simply a null mutation. This suggests that phospho-Nup1, modified at either S161 or T344 is essential for localization of active genes to the nuclear periphery.
Loss of phosphorylation sites 1 and 2, loss of Cdk1 activity and induction of the Cdk inhibitor Far1 disrupts targeting of active INO1 and GAL1 to the nuclear periphery. These results suggest that dephosphorylation of sites 1 and 2 during S-phase leads to loss of peripheral targeting. If so, then constitutive phosphorylation of either site 1 or 2 should support targeting of INO1 and GAL1 to the nuclear periphery, even during S-phase. To test this idea, we introduced aspartic acid residues in place of each of the phosphorylated residues. Introduction of phosphomimetic (PM) aspartic acid residues in place of either S161 (PM1) or T344 (PM2) in Nup1 complemented the nup1Δ mutant defect in localization of INO1 and GAL1 (Figure 5, B and D). Furthermore, expression of either Nup1-PM1 or Nup1-PM2 resulted in INO1 and GAL1 localization at the nuclear periphery in small-budded cells (Figure 5, C and E). This effect was dominant when the PM mutant forms of Nup1 were expressed in a strain also expressing wild-type NUP1 (Supplementary Figure S4). Thus, mimicking phosphorylation of either S161 or T344 is sufficient to override the cell cycle regulation of gene recruitment, allowing peripheral localization of INO1 and GAL1 during S-phase.
If S161 and T344 in Nup1 is the only target of Cdc28 phosphorylation that affects gene targeting, then the defect in gene localization in the cdc28-1 mutant should be suppressed by either of the dominant PM mutations in Nup1. To test this idea, we transformed a cdc28-1 strain with plasmids expressing either NUP1-PM1 or NUP1-PM2. Cells were arrested in mitosis with nocodazole and then shifted to the restrictive temperature for 30 min to inactivate Cdc28. In the cdc28-1 control, localization of INO1 and GAL1 to the nuclear periphery was lost after cells were shifted to the restrictive temperature (Figure 6, A and B). However, in cdc28-1 cells expressing either Nup1-PM1 or Nup1-PM2, localization of INO1 and GAL1 to the nuclear periphery was maintained (Figure 6, A and B). Therefore, phosphomimetic mutations at either of the Cdk sites in Nup1 are sufficient to bypass the requirement of Cdc28 in peripheral targeting. This suggests that phosphorylation of Nup1 by Cdc28 is required for peripheral targeting of active INO1 and GAL1 and this phosphorylation is lost during S-phase. Furthermore, Nup1 is apparently the only substrate of the kinase that is essential for localization of these genes at the nuclear periphery.
Recently Repressed Genes Localize to the Nuclear Periphery throughout the Cell Cycle
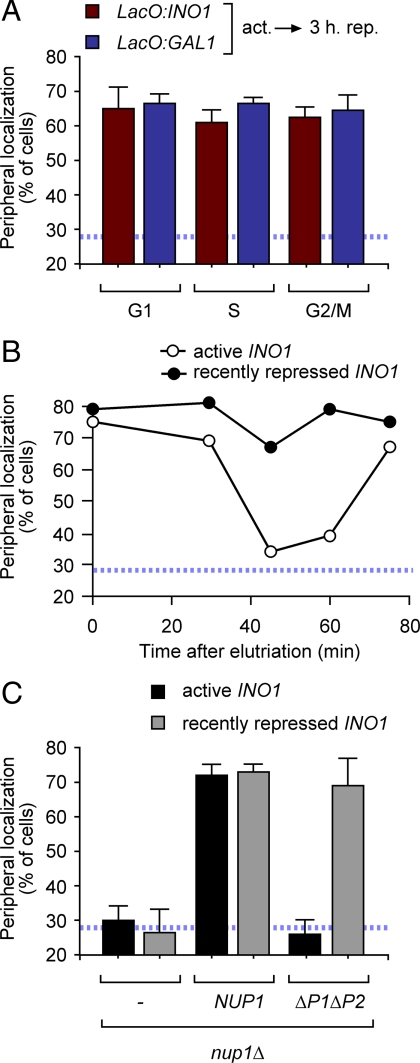
After repression, INO1 and GAL1 remain at the nuclear periphery for several generations, primed for reactivation (Brickner et al., 2007). We have recently discovered that the mechanism controlling localization of active INO1 to the nuclear periphery is different from the mechanism controlling localization of recently repressed INO1 to the nuclear periphery. The DNA zip codes responsible for targeting of active INO1 to the nuclear periphery are not required to maintain the gene at the nuclear periphery after repression. Retention of recently repressed INO1 requires a distinct DNA element that confers a distinct interaction with the NPC (Light et al., in press). We next asked if the localization of recently repressed INO1 and GAL1 at the nuclear periphery was also regulated through the cell cycle. We shifted cells from activating conditions to repressing conditions for 3 h, classified them according to their bud morphology and quantified the fraction of cells in which INO1 or GAL1 localized at the nuclear periphery. Unlike active INO1 and GAL1, recently repressed INO1 and GAL1 localized to the nuclear periphery in all stages of the cell cycle (Figure 7A). Likewise, in a synchronized population of newly born daughter cells (purified by centrifugal elutriation), whereas active INO1 lost peripheral localization after ∼45 min, recently repressed INO1 remained at the nuclear periphery (Figure 7B). Finally, we asked if Cdk1 phosphorylation of Nup1 affected the localization of recently repressed INO1 at the nuclear periphery. Localization of recently repressed INO1 at the nuclear periphery required Nup1 (Figure 7C). However, loss of the two Cdk phosphorylation sites in Nup1 that are essential for targeting active INO1 to the nuclear periphery had no effect on the localization of recently repressed INO1 to the nuclear periphery (Figure 7C). Thus, the phosphorylation of these sites plays a specific role in the cell cycle–regulated targeting of active genes to the nuclear periphery.
Figure 7.
Recently repressed genes localize to the nuclear periphery throughout the cell cycle. (A) Strains having the lac operator array integrated either at INO1 or GAL1 were shifted from activating conditions to repressing conditions. After 3 h, cells were harvested, fixed, and processed for immunofluorescence. Cells were classified according to their bud morphology as in Figure 1 and, within each class, the percentage of cells in which the gene localized at the nuclear periphery was determined. (B) Newly born daughter cells having the lac operator array integrated at INO1 were purified by centrifugal elutriation under activating conditions or after 3 h of repression. Cells were harvested at the indicated times after elutriation, fixed, and processed for immunofluorescence. (C) nup1Δ cells having the lac operator array integrated at INO1 were transformed with either a vector, wild-type NUP1 or nup1ΔP1ΔP2. Cells were harvested either under activating conditions or after 3 h of repression, fixed, and processed for immunofluorescence. The blue, hatched line represents the level of colocalization of the lac repressor spot with the nuclear envelope predicted by chance (Brickner and Walter, 2004).
DISCUSSION
Genomes of eukaryotic organisms are spatially organized. How spatial organization is achieved, how it impacts genome function, and how it is regulated are not well understood. We have used the recruitment of active genes to the nuclear periphery in yeast as a general model to understand how the localization of individual genes can be determined and controlled. The localization of genes like INO1 to the nuclear periphery is controlled by cis-acting DNA elements that confer a physical interaction with the NPC (Ahmed et al., 2010). Here we show that phosphorylation of a component of the NPC by cyclin-dependent kinase (Cdk1) is necessary for targeting of active INO1 and GAL1 to the nuclear periphery. Furthermore, our results suggest that Nup1 phosphorylation is lost during S-phase, resulting in relocalization of active INO1 and GAL1 to the nucleoplasm. Therefore cyclic phosphorylation of the NPC by Cdk1 regulates the localization of genes at the nuclear periphery through the cell cycle, perhaps to allow coordination of transcription with DNA replication, DNA repair, and chromosome segregation.
Phosphorylation of the NPC Controls Gene Localization
The complete list of in vivo targets of yeast Cdk1 has begun to be elucidated by both in vitro kinase assays (Ubersax et al., 2003) and in vivo mass spectrometric approaches (Holt et al., 2009). Cdk1 phosphorylates proteins involved in every aspect of cell biology (Holt et al., 2009). However, the role of phosphorylation has been described for only a small number of these proteins. Proteins in the NPC and associated transport factors are targets of Cdk phosphorylation in yeast and vertebrates (Favreau et al., 1996; Glavy et al., 2007; Lusk et al., 2007). Phosphorylation of the NPC regulates nuclear transport of mRNA and proteins (Kehlenbach and Gerace, 2000; Makhnevych et al., 2003) and the disassembly of the NPC during mitosis (Onischenko et al., 2005). Here we show that Cdk phosphorylation of an NPC protein also regulates the localization of genes within the nucleus.
It is not clear how the NPC promotes gene targeting to the nuclear periphery. Previous work suggests that many proteins associated with the nucleoplasmic “basket” are essential for gene recruitment to the nuclear periphery (Cabal et al., 2006; Brickner et al., 2007; Luthra et al., 2007; Ahmed et al., 2010). Proteins in this structure such as Nup1, Nup2, Nup60, Mlp1, Mlp2, and associated factors may constitute the structure to which genes are targeted by specific adaptor proteins that recognize DNA zip codes. Nup1 is a member of a family of proteins composed of natively unstructured FxFG repeat domains that determine the selective permeability barrier of the NPC (Denning et al., 2003; Strawn et al., 2004; Frey and Gorlich, 2007). We find that phosphorylation of this protein on either of two sites is required for targeting of active genes to the nuclear periphery. This suggests either that the adaptor proteins that are responsible for targeting active genes to the nuclear periphery interact with Nup1 in a phosphorylation-dependent manner or that the structure to which these genes is recruited is phosphorylation-dependent. Importantly, targeting of recently repressed INO1 to the nuclear periphery, which also requires Nup1 but is not regulated through the cell cycle, did not require the phosphoacceptor sites in Nup1. Therefore, the function of the NPC in targeting genes under some conditions, but not others, requires Nup1 phosphorylation by Cdk.
Cdk1 Phosphorylation of Nup1
It is somewhat surprising that phosphorylation of S161 and T344 is apparently lost during S-phase. Cdk1 activity is relatively high during S-phase and peaks during mitosis (Morgan, 1997). This suggests either that S-phase cyclins (Clb5 and Clb6 in yeast) do not efficiently promote Cdk1 phosphorylation of Nup1 on these sites or that a phosphatase is active during S-phase. Two results support the former possibility. First, activation of the Far1 Cdk inhibitor was sufficient to cause INO1 to localize in the nucleoplasm during G1 phase. Second, inactivation of Cdk1 in cells arrested during mitosis caused INO1 and GAL1 to lose peripheral localization. Therefore, phosphatases capable of dephosphorylating Nup1 are active during G1 and M. This suggests that the G1 cyclins (Cln1, Cln2, and Cln3) and the G2/M cyclins (Clb1, Clb2 and Clb3) promote Cdk1 phosphorylation of Nup1, whereas the S-phase cyclins do not. In combination with a constitutive level of phosphatase activity, this scenario would result in a drop in the net phosphorylation of S161 and T344 during S-phase.
If Cdk1-mediated control of Nup1 function in gene recruitment were important for fitness, then the S161 and T344 sites should be conserved. We compared the sequences of Nup1 from several closely related or more divergent Saccharomyces species. Nup1, like many natively unstructured proteins, is not highly conserved, even among Saccharomyces species (Cliften et al., 2003; Kellis et al., 2003). However, the Cdk consensus sequences at S161 and T344 were conserved both among closely related species (S. bayanus, S. paradoxus, and S. mikatae) and a more distantly related species (S. castellii; Figure S5). The conservation of these sites suggests that Cdk1 phosphorylation and the regulation of gene recruitment is adaptive.
In mutant cells lacking Sic1, we did not observe cycling in the localization of INO1 at the nuclear periphery (Figure 3). As mentioned above, because Sic1 levels drop to allow cells to enter S-phase, we do not think that this phenotype reflects a direct role for Sic1 in regulating phosphorylation of Nup1. We propose that, in cells lacking Sic1, the levels of Cdk activity are elevated throughout the cell cycle, leading to increased phosphorylation of Cdk targets, perhaps reducing the fraction of Nup1 that is dephosphorylated during S-phase. Alternatively, the duration of the period during which Nup1 is ordinarily dephosphorylated could be much shorter in the sic1Δ mutant, making it difficult to observe in a population of cells.
Genome-Wide Changes in Spatial Organization during the Cell Cycle
Tethering of yeast telomeres to the nuclear envelope is lost during G2 (Laroche et al., 2000; Hediger et al., 2002; Ebrahimi and Donaldson, 2008). Here we have identified a new type of cell cycle regulation of DNA localization. We find that active genes lose their peripheral localization during a 20 min period during S-phase, corresponding to about one-fifth of the cell cycle. This observation provides a partial explanation for the fact that active genes localize at the nuclear periphery in fewer cells than an artificially tethered locus (Brickner and Walter, 2004). This may also explain why previous work using high-throughput localization of the active GAL1 gene found that it had two distinct patterns: one in the nucleoplasm and one at the nuclear periphery in populations of asynchronous cells (Berger et al., 2008).
The mechanisms leading to loss of peripheral localization of telomeres and active genes are related but distinct. The loss of peripheral localization of telomeres follows DNA replication and is due to the loss of Ku binding (Laroche et al., 2000; Hediger et al., 2002; Ebrahimi and Donaldson, 2008). The timing of telomere release from the nuclear envelope is determined by the timing of DNA replication, but is not caused by DNA replication (Ebrahimi and Donaldson, 2008). The localization of active genes to the nucleoplasm during S-phase follows the initiation of DNA replication, but is not simply a consequence of DNA replication. Loss of peripheral localization of active genes represents specific regulation of gene targeting through the cell cycle. Cyclic phosphorylation of a single nucleoporin by Cdk1 provides a mechanistic connection between the master regulator of the cell cycle and gene targeting.
These two examples suggest that the localization of different parts of the genome within the nucleus is carefully orchestrated during different phases of the cell cycle. The function of these cell cycle–regulated changes in genome architecture is unclear. Collapsed or paused DNA replication forks are enriched over highly transcribed genes like GAL1 (Azvolinsky et al., 2009), presumably because of physical interference between transcription and DNA replication. Furthermore, sites of DNA damage are targeted to the NPC (Nagai et al., 2008). Therefore, it is possible that loss of peripheral localization of highly transcribed genes like INO1 and GAL1 allows better access of damaged parts of the genome to the repair machinery at the NPC during S-phase. In this way, loss of peripheral localization of transcriptionally active genes during S-phase might allow better coordination of transcription with DNA replication and repair.
Supplementary Material
ACKNOWLEDGMENTS
We thank members of the Brickner laboratory, Dr. Richard Gaber, Dr. Eric Weiss, and Dr. David O. Morgan for helpful comments on the manuscript and Dr. Robert Lamb for generously sharing his confocal microscope. This work was supported by National Institutes of Health Grant GM080484.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-01-0065) on August 11, 2010.
REFERENCES
- Ahmed S., Brickner D. G., Light W. H., McDonough M., Froyshteter A. B., Volpe T., Brickner J. H. DNA zip codes control an ancient mechanism for targeting genes to the nuclear periphery. Nat. Cell Biol. 2010;12:111–118. doi: 10.1038/ncb2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albuquerque C. P., Smolka M. B., Payne S. H., Bafna V., Eng J., Zhou H. A multidimensional chromatography technology for in-depth phosphoproteome analysis. Mol. Cell Proteom. 2008;7:1389–1396. doi: 10.1074/mcp.M700468-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azvolinsky A., Giresi P. G., Lieb J. D., Zakian V. A. Highly transcribed RNA polymerase II genes are impediments to replication fork progression in Saccharomyces cerevisiae. Mol. Cell. 2009;34:722–734. doi: 10.1016/j.molcel.2009.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger A. B., Cabal G. G., Fabre E., Duong T., Buc H., Nehrbass U., Olivo-Marin J. C., Gadal O., Zimmer C. High-resolution statistical mapping reveals gene territories in live yeast. Nat. Methods. 2008;5:1031–1037. doi: 10.1038/nmeth.1266. [DOI] [PubMed] [Google Scholar]
- Blobel G. Gene gating: a hypothesis. Proc. Natl. Acad. Sci. USA. 1985;82:8527–8529. doi: 10.1073/pnas.82.24.8527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer B. J., Chlebowicz-Sledziewska E., Fangman W. L. Cell cycle phases in the unequal mother/daughter cell cycles of Saccharomyces cerevisiae. Mol. Cell. Biol. 1984;4:2529–2531. doi: 10.1128/mcb.4.11.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickner D. G., Cajigas I. C., Fondufe-Mittendorf Y., Ahmed S., Lee P.-C., Widom J., Brickner J. H. H2A.Z-mediated localization of genes at the nuclear periphery confers epigenetic memory of previous transcriptional state. PLoS Biol. 2007;5:e81. doi: 10.1371/journal.pbio.0050081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickner D. G., Light W. H., Brickner J. H. Quantitative localization of chromosomal loci by immunofluorescence. In: Weissman J., Guthrie C., Fink G. R., editors. Methods in Enzymology. vol. 470. Burlington: Academic Press; 2010. pp. 569–580. [DOI] [PubMed] [Google Scholar]
- Brickner J. H., Fuller R. S. SOI1 encodes a novel, conserved protein that promotes TGN-endosomal cycling of Kex2p and other membrane proteins by modulating the function of two TGN localization signals. J. Cell Biol. 1997;139:23–36. doi: 10.1083/jcb.139.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickner J. H., Walter P. Gene recruitment of the activated INO1 locus to the nuclear membrane. PLoS Biol. 2004;2:e342. doi: 10.1371/journal.pbio.0020342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C. R., Kennedy C. J., Delmar V. A., Forbes D. J., Silver P. A. Global histone acetylation induces functional genomic reorganization at mammalian nuclear pore complexes. Genes Dev. 2008;22:627–639. doi: 10.1101/gad.1632708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke D., Dawson D., Stearns T. Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Press; 2000. [Google Scholar]
- Cabal G. G., et al. SAGA interacting factors confine sub-diffusion of transcribed genes to the nuclear envelope. Nature. 2006;441:770–773. doi: 10.1038/nature04752. [DOI] [PubMed] [Google Scholar]
- Casolari J. M., Brown C. R., Drubin D. A., Rando O. J., Silver P. A. Developmentally induced changes in transcriptional program alter spatial organization across chromosomes. Genes Dev. 2005;19:1188–1198. doi: 10.1101/gad.1307205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casolari J. M., Brown C. R., Komili S., West J., Hieronymus H., Silver P. A. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell. 2004;117:427–439. doi: 10.1016/s0092-8674(04)00448-9. [DOI] [PubMed] [Google Scholar]
- Chuang C. H., Carpenter A. E., Fuchsova B., Johnson T., de Lanerolle P., Belmont A. S. Long-range directional movement of an interphase chromosome site. Curr. Biol. 2006;16:825–831. doi: 10.1016/j.cub.2006.03.059. [DOI] [PubMed] [Google Scholar]
- Cliften P., Sudarsanam P., Desikan A., Fulton L., Fulton B., Majors J., Waterston R., Cohen B. A., Johnston M. Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science. 2003;301:71–76. doi: 10.1126/science.1084337. [DOI] [PubMed] [Google Scholar]
- Cocker J. H., Piatti S., Santocanale C., Nasmyth K., Diffley J. F. An essential role for the Cdc6 protein in forming the pre-replicative complexes of budding yeast. Nature. 1996;379:180–182. doi: 10.1038/379180a0. [DOI] [PubMed] [Google Scholar]
- Cremer T., Cremer M., Dietzel S., Muller S., Solovei I., Fakan S. Chromosome territories—a functional nuclear landscape. Curr. Opin. Cell Biol. 2006;18:307–316. doi: 10.1016/j.ceb.2006.04.007. [DOI] [PubMed] [Google Scholar]
- Denning D. P., Patel S. S., Uversky V., Fink A. L., Rexach M. Disorder in the nuclear pore complex: the FG repeat regions of nucleoporins are natively unfolded. Proc. Natl. Acad. Sci. USA. 2003;100:2450–2455. doi: 10.1073/pnas.0437902100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieppois G., Iglesias N., Stutz F. Cotranscriptional recruitment to the mRNA export receptor Mex67p contributes to nuclear pore anchoring of activated genes. Mol. Cell. Biol. 2006;26:7858–7870. doi: 10.1128/MCB.00870-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan J. D., Toyn J. H., Johnson A. L., Johnston L. H. P40SDB25, a putative CDK inhibitor, has a role in the M/G1 transition in Saccharomyces cerevisiae. Genes Dev. 1994;8:1640–1653. doi: 10.1101/gad.8.14.1640. [DOI] [PubMed] [Google Scholar]
- Ebrahimi H., Donaldson A. D. Release of yeast telomeres from the nuclear periphery is triggered by replication and maintained by suppression of Ku-mediated anchoring. Genes Dev. 2008;22:3363–3374. doi: 10.1101/gad.486208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favreau C., Worman H. J., Wozniak R. W., Frappier T., Courvalin J. C. Cell cycle-dependent phosphorylation of nucleoporins and nuclear pore membrane protein Gp210. Biochemistry. 1996;35:8035–8044. doi: 10.1021/bi9600660. [DOI] [PubMed] [Google Scholar]
- Frey S., Gorlich D. A saturated FG-repeat hydrogel can reproduce the permeability properties of nuclear pore complexes. Cell. 2007;130:512–523. doi: 10.1016/j.cell.2007.06.024. [DOI] [PubMed] [Google Scholar]
- Glavy J. S., Krutchinsky A. N., Cristea I. M., Berke I. C., Boehmer T., Blobel G., Chait B. T. Cell-cycle-dependent phosphorylation of the nuclear pore Nup107–160 subcomplex. Proc. Natl. Acad. Sci. USA. 2007;104:3811–3816. doi: 10.1073/pnas.0700058104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon C. N., Elliott S. C. Fractionation of Saccharomyces cerevisiae cell populations by centrifugal elutriation. J. Bacteriol. 1977;129:97–100. doi: 10.1128/jb.129.1.97-100.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwick K. G., Murray A. W. Mad1p, a phosphoprotein component of the spindle assembly checkpoint in budding yeast. J. Cell Biol. 1995;131:709–720. doi: 10.1083/jcb.131.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell L., Weinert T., Kadyk L., Garvik B. Cell cycle checkpoints, genomic integrity, and cancer. Cold Spring Harb. Symp. Quant. Biol. 1994;59:259–263. doi: 10.1101/sqb.1994.059.01.030. [DOI] [PubMed] [Google Scholar]
- Hartwell L. H., Culotti J., Reid B. Genetic control of the cell-division cycle in yeast. I. Detection of mutants. Proc. Natl. Acad. Sci. USA. 1970;66:352–359. doi: 10.1073/pnas.66.2.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell L. H., Weinert T. A. Checkpoints: controls that ensure the order of cell cycle events. Science. 1989;246:629–634. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- Hediger F., Neumann F. R., Van Houwe G., Dubrana K., Gasser S. M. Live imaging of telomeres: yKu and Sir proteins define redundant telomere-anchoring pathways in yeast. Curr. Biol. 2002;12:2076–2089. doi: 10.1016/s0960-9822(02)01338-6. [DOI] [PubMed] [Google Scholar]
- Herskowitz I. MAP kinase pathways in yeast: for mating and more. Cell. 1995;80:187–197. doi: 10.1016/0092-8674(95)90402-6. [DOI] [PubMed] [Google Scholar]
- Holt L. J., Tuch B. B., Villen J., Johnson A. D., Gygi S. P., Morgan D. O. Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science. 2009;325:1682–1686. doi: 10.1126/science.1172867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang L. H., Lau L. F., Smith D. L., Mistrot C. A., Hardwick K. G., Hwang E. S., Amon A., Murray A. W. Budding yeast Cdc20, a target of the spindle checkpoint. Science. 1998;279:1041–1044. doi: 10.1126/science.279.5353.1041. [DOI] [PubMed] [Google Scholar]
- Kehlenbach R. H., Gerace L. Phosphorylation of the nuclear transport machinery down-regulates nuclear protein import in vitro. J. Biol. Chem. 2000;275:17848–17856. doi: 10.1074/jbc.M001455200. [DOI] [PubMed] [Google Scholar]
- Kellis M., Patterson N., Endrizzi M., Birren B., Lander E. S. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature. 2003;423:241–254. doi: 10.1038/nature01644. [DOI] [PubMed] [Google Scholar]
- Kosak S. T., Skok J. A., Medina K. L., Riblet R., Le Beau M. M., Fisher A. G., Singh H. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science. 2002;296:158–162. doi: 10.1126/science.1068768. [DOI] [PubMed] [Google Scholar]
- Kunkel W. Effects of the antimicrotubular cancerostatic drug nocodazole on the yeast Saccharomyces cerevisiae. Z. Allg. Mikrobiol. 1980;20:315–324. [PubMed] [Google Scholar]
- Kurshakova M. M., Krasnov A. N., Kopytova D. V., Shidlovskii Y. V., Nikolenko J. V., Nabirochkina E. N., Spehner D., Schultz P., Tora L., Georgieva S. G. SAGA and a novel Drosophila export complex anchor efficient transcription and mRNA export to NPC. EMBO J. 2007;26:4956–4965. doi: 10.1038/sj.emboj.7601901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laroche T., Martin S. G., Tsai-Pflugfelder M., Gasser S. M. The dynamics of yeast telomeres and silencing proteins through the cell cycle. J. Struct. Biol. 2000;129:159–174. doi: 10.1006/jsbi.2000.4240. [DOI] [PubMed] [Google Scholar]
- Li X., Gerber S. A., Rudner A. D., Beausoleil S. A., Haas W., Villen J., Elias J. E., Gygi S. P. Large-scale phosphorylation analysis of alpha-factor-arrested Saccharomyces cerevisiae. J. Proteome Res. 2007;6:1190–1197. doi: 10.1021/pr060559j. [DOI] [PubMed] [Google Scholar]
- Light W., Brickner D. G., Brand V., Brickner J. H. Interaction of a DNA zip code with the nuclear pore complex promotes H2A.Z incorporation and INO1 transcriptional memory. Mol. Cell. 2010 doi: 10.1016/j.molcel.2010.09.007. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Lusk C. P., Waller D. D., Makhnevych T., Dienemann A., Whiteway M., Thomas D. Y., Wozniak R. W. Nup53p is a target of two mitotic kinases, Cdk1p and Hrr25p. Traffic. 2007;8:647–660. doi: 10.1111/j.1600-0854.2007.00559.x. [DOI] [PubMed] [Google Scholar]
- Luthra R., Kerr S. C., Harreman M. T., Apponi L. H., Fasken M. B., Ramineni S., Chaurasia S., Valentini S. R., Corbett A. H. Actively transcribed GAL genes can be physically linked to the nuclear pore by the SAGA chromatin modifying complex. J. Biol. Chem. 2007;282:3042–3049. doi: 10.1074/jbc.M608741200. [DOI] [PubMed] [Google Scholar]
- Makhnevych T., Lusk C. P., Anderson A. M., Aitchison J. D., Wozniak R. W. Cell cycle regulated transport controlled by alterations in the nuclear pore complex. Cell. 2003;115:813–823. doi: 10.1016/s0092-8674(03)00986-3. [DOI] [PubMed] [Google Scholar]
- Marshall W. F., Dernburg A. F., Harmon B., Agard D. A., Sedat J. W. Specific interactions of chromatin with the nuclear envelope: positional determination within the nucleus in Drosophila melanogaster. Mol. Biol. Cell. 1996;7:825–842. doi: 10.1091/mbc.7.5.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendjan S., et al. Nuclear pore components are involved in the transcriptional regulation of dosage compensation in Drosophila. Mol. Cell. 2006;21:811–823. doi: 10.1016/j.molcel.2006.02.007. [DOI] [PubMed] [Google Scholar]
- Menon B. B., Sarma N. J., Pasula S., Deminoff S. J., Willis K. A., Barbara K. E., Andrews B., Santangelo G. M. Reverse recruitment: the Nup84 nuclear pore subcomplex mediates Rap1/Gcr1/Gcr2 transcriptional activation. Proc. Natl. Acad. Sci. USA. 2005;102:5749–5754. doi: 10.1073/pnas.0501768102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan D. O. Cyclin-dependent kinases: engines, clocks, and microprocessors. Annu. Rev. Cell Dev. Biol. 1997;13:261–291. doi: 10.1146/annurev.cellbio.13.1.261. [DOI] [PubMed] [Google Scholar]
- Nagai S., Dubrana K., Tsai-Pflugfelder M., Davidson M. B., Roberts T. M., Brown G. W., Varela E., Hediger F., Gasser S. M., Krogan N. J. Functional targeting of DNA damage to a nuclear pore-associated SUMO-dependent ubiquitin ligase. Science. 2008;322:597–602. doi: 10.1126/science.1162790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onischenko E. A., Gubanova N. V., Kiseleva E. V., Hallberg E. Cdk1 and okadaic acid-sensitive phosphatases control assembly of nuclear pore complexes in Drosophila embryos. Mol. Biol. Cell. 2005;16:5152–5162. doi: 10.1091/mbc.E05-07-0642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter M., Herskowitz I. Direct inhibition of the yeast cyclin-dependent kinase Cdc28-Cln by Far1. Science. 1994;265:1228–1231. doi: 10.1126/science.8066461. [DOI] [PubMed] [Google Scholar]
- Piggott J. R., Rai R., Carter B. L. A bifunctional gene product involved in two phases of the yeast cell cycle. Nature. 1982;298:391–393. doi: 10.1038/298391a0. [DOI] [PubMed] [Google Scholar]
- Rabl C. Über Zeillteilung. Morpholog. Jahrbuch. 1885;10:214–330. [Google Scholar]
- Ragoczy T., Bender M. A., Telling A., Byron R., Groudine M. The locus control region is required for association of the murine beta-globin locus with engaged transcription factories during erythroid maturation. Genes Dev. 2006;20:1447–1457. doi: 10.1101/gad.1419506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinett C. C., Straight A., Li G., Willhelm C., Sudlow G., Murray A., Belmont A. S. In vivo localization of DNA sequences and visualization of large-scale chromatin organization using lac operator/repressor recognition. J. Cell Biol. 1996;135:1685–1700. doi: 10.1083/jcb.135.6.1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudner A. D., Hardwick K. G., Murray A. W. Cdc28 activates exit from mitosis in budding yeast. J. Cell Biol. 2000;149:1361–1376. doi: 10.1083/jcb.149.7.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M., Arib G., Laemmli C., Nishikawa J., Durussel T., Laemmli U. K. Nup-PI: the nucleopore-promoter interaction of genes in yeast. Mol. Cell. 2006;21:379–391. doi: 10.1016/j.molcel.2005.12.012. [DOI] [PubMed] [Google Scholar]
- Schneider B. L., Yang Q. H., Futcher A. B. Linkage of replication to start by the Cdk inhibitor Sic1. Science. 1996;272:560–562. doi: 10.1126/science.272.5261.560. [DOI] [PubMed] [Google Scholar]
- Schwartz M. A., Madhani H. D. Principles of MAP kinase signaling specificity in Saccharomyces cerevisiae. Annu. Rev. Genet. 2004;38:725–748. doi: 10.1146/annurev.genet.39.073003.112634. [DOI] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolka M. B., Albuquerque C. P., Chen S. H., Zhou H. Proteome-wide identification of in vivo targets of DNA damage checkpoint kinases. Proc. Natl. Acad. Sci. USA. 2007;104:10364–10369. doi: 10.1073/pnas.0701622104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight A. F., Belmont A. S., Robinett C. C., Murray A. W. GFP tagging of budding yeast chromosomes reveals that protein-protein interactions can mediate sister chromatid cohesion. Curr. Biol. 1996;6:1599–1608. doi: 10.1016/s0960-9822(02)70783-5. [DOI] [PubMed] [Google Scholar]
- Strawn L. A., Shen T., Shulga N., Goldfarb D. S., Wente S. R. Minimal nuclear pore complexes define FG repeat domains essential for transport. Nat. Cell Biol. 2004;6:197–206. doi: 10.1038/ncb1097. [DOI] [PubMed] [Google Scholar]
- Taddei A., Van Houwe G., Hediger F., Kalck V., Cubizolles F., Schober H., Gasser S. M. Nuclear pore association confers optimal expression levels for an inducible yeast gene. Nature. 2006;441:774–778. doi: 10.1038/nature04845. [DOI] [PubMed] [Google Scholar]
- Ubersax J. A., Woodbury E. L., Quang P. N., Paraz M., Blethrow J. D., Shah K., Shokat K. M., Morgan D. O. Targets of the cyclin-dependent kinase Cdk1. Nature. 2003;425:859–864. doi: 10.1038/nature02062. [DOI] [PubMed] [Google Scholar]
- Visintin R., Prinz S., Amon A. CDC20 and CDH1, a family of substrate-specific activators of APC-dependent proteolysis. Science. 1997;278:460–463. doi: 10.1126/science.278.5337.460. [DOI] [PubMed] [Google Scholar]
- Young C. W., Hodas S. Hydroxyurea: inhibitory Effect on DNA Metabolism. Science. 1964;146:1172–1174. doi: 10.1126/science.146.3648.1172. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.