The RNA-dependent protein kinase (PKR), initially known as a virus infection response protein, is found to differentially regulate two major players in the insulin signaling pathway, IRS1 and IRS2. PKR up-regulates the inhibitory phosphorylation of IRS1 and the expression of IRS2 at the transcriptional level.
Abstract
Initially identified to be activated upon virus infection, the double-stranded RNA–dependent protein kinase (PKR) is best known for triggering cell defense responses by phosphorylating eIF-2α, thus suppressing RNA translation. We as well as others showed that the phosphorylation of PKR is down-regulated by insulin. In the present study, we further uncovered a novel function of PKR in regulating the IRS proteins. We found that PKR up-regulates the inhibitory phosphorylation of IRS1 at Ser312, which suppresses the tyrosine phosphorylation of IRS1. This effect of PKR on the phosphorylation of IRS1 is mediated by two other protein kinases, JNK and IKK. In contrast, PKR regulates IRS2, another major IRS family protein in the liver, at the transcriptional rather than the posttranslational level, and this effect is mediated by the transcription factor, FoxO1, which has been previously shown to be regulated by insulin and plays a significant role in glucose homeostasis and energy metabolism. In summary, we found for the first time that initially known as a virus infection response gene, PKR regulates the upstream central transmitters of insulin signaling, IRS1 and IRS2, through different mechanisms.
INTRODUCTION
Insulin signaling, a central signaling pathway that regulates many cellular activities, such as glucose and lipid metabolism, protein synthesis and degradation, and cell growth and differentiation (Saltiel and Kahn, 2001), has been extensively studied over the past decades. Insulin signaling is initiated upon binding of insulin to the insulin receptor (IR), a receptor tyrosine kinase (Patti and Kahn, 1998), and transmitted intracellularly by the insulin receptor substrates (IRS; White, 1998). At least four of the IR substrates belong to the IRS group, with IRS1 and IRS2 being predominant and expressed in most tissues, including the liver (White, 1998; Thirone et al., 2006). On phosphorylation of the tyrosine residues catalyzed by IR, the IRS proteins initiate, through different binding mechanisms (White, 1998), various downstream signal transduction cascades, including mitogen-activated protein kinase (MAPK), pathways (c-Jun N-terminal kinase (JNK), extracellular signal–related kinase (ERK), p38 MAPK; Lowenstein et al., 1992; Skolnik et al., 1993), and phosphoinositide 3-kinase (PI3K; Backer et al., 1992), which in turn activates Akt/protein kinase B (Akt/PKB; Alessi et al., 1997), and atypical protein kinase C (aPKC; Standaert et al., 1997).
As one of the central signaling pathways regulating multiple fundamental cellular activities, insulin signaling is sophisticatedly tuned by a large number of regulators. Dysregulation of insulin signaling is closely related to the development of insulin resistance (Sone et al., 2001) and contributes to multiple diseases and disorders, such as type 2 diabetes as well as other metabolic, endocrine, and cardiovascular disorders (Reaven, 1988; Kahn, 1998; Sone et al., 2001). At the molecular level, dysregulation of insulin signaling could occur at several possible stages, e.g., degradation or mutation of IR (McElduff et al., 1984; Imamura et al., 1994), inhibitory phosphorylation or degradation of IRS (Zick, 2005; Herschkovitz et al., 2007), or suppression of down-stream signaling molecules, such as PI-3 kinase or Akt/PKB (reviewed in Taylor and Arioglu, 1998). However, most of the regulation of insulin signaling occurs at the level of the IRS proteins, the hub proteins that transmit signals from IR to down-stream targets of insulin signaling (Zick, 2005; Herschkovitz et al., 2007). The phosphorylations of IRS play critical roles in determining its activity. IRS proteins mediate intracellular insulin signaling, through the tyrosine residues, which facilitate recruitment of IRS substrates and therefore promote insulin signaling (White, 1998), whereas its phosphorylations at the serine residues generally suppress the activities of IRS by blocking the interaction between IRS and IR (Paz et al., 1997), inhibiting the tyrosine phosphorylation of IRS (Hotamisligil et al., 1996) or inducing the degradation of IRS (Pederson et al., 2001). A number of serine residues have been identified to negatively regulate the activity of IRS1, in particular, Ser307 (equivalent to Ser312 in human IRS1). Ser307 has been extensively investigated and characterized as a key indicator of inhibitory serine phosphorylation of IRS1 and insulin resistance and confirmed in insulin-resistant rodent models (Hirosumi et al., 2002). Interestingly, most of the down-stream targets of insulin signaling, such as mammalian target of rapamycin (mTOR; Ozes et al., 2001), PKCζ (Ravichandran et al., 2001), S6 Kinase 1 (S6K1; Tremblay and Marette, 2001), and JNK (Aguirre et al., 2000; Lee et al., 2003), have been shown to function as IRS serine kinases, which are involved in the negative feedback pathways of insulin signaling by promoting the inhibitory serine phosphorylation of IRS1.
Initially identified as an antiviral protein, the double-stranded RNA–dependent protein kinase (PKR) is best known for triggering cell defense responses and initiating innate immune responses by arresting general protein synthesis and inducing apoptosis during virus infection (Proud, 1995). Until recently, PKR had not been reported to be involved in the insulin signaling pathway. However, studies have shown that insulin or insulin-like growth factor-I (IGF-I) suppressed the phosphorylation of PKR in muscle cells (Russell et al., 2007; Eley et al., 2008), by activating protein phosphatase 1 (PP1) through the IRS–PI3K–Akt pathway. We have found a similar inhibitory effect of insulin on PKR in HepG2 cells (Wu et al., 2009). Given the feedbacks involved in insulin signaling, we wanted to explore whether PKR, as a downstream target of insulin signaling, is also able to initiate a feedback pathway that functions on IRS1 phosphorylation. In fact, as a signal integrator of many intracellular signaling events (Williams, 2001), PKR has been shown to activate certain IRS kinases, e.g., IκB kinase (IKK; Bonnet et al., 2000; Gao et al., 2002) and JNK (Zhou et al., 2003; Yang and Chan, 2009). Therefore, we investigated whether PKR is involved in the induction of inhibitory Ser312 phosphorylation of IRS1 and if so, whether this effect is mediated by other IRS kinases.
Because IRS1 and IRS2 are both expressed in liver cells, we also investigated whether PKR affected IRS2. Our results indicated that PKR regulates the gene transcription, rather than the posttranslational modification, of IRS2. The transcription of the IRS2 gene has been shown to be up-regulated by several transcription factors, such as forkhead box O1 (FoxO1; Zhang et al., 2001; Ide et al., 2004) and cAMP response element binding (CREB; Jhala et al., 2003). We found that PKR enhanced the expression of IRS2 through FoxO1, which regulates the transcription of IRS2 but not IRS1. In summary, PKR, initially known as a virus infection response gene, differentially regulates the major IRS proteins, IRS1 and IRS2, which are central hubs in the insulin signaling system.
MATERIALS AND METHODS
Cell Culture and Reagents
Human hepatoblastoma cells (HepG2/C3A) were cultured in Dulbecco's modified Eagle medium (DMEM; Invitrogen, Carlsbad, CA) with 10% fetal bovine serum (FBS; Biomeda, Foster City, CA) and penicillin-streptomycin (penicillin: 10,000 U/ml, streptomycin: 10,000 μg/ml; Invitrogen). Freshly trypsinized HepG2 cells were suspended at 5 × 105 cells/ml in standard HepG2 culture medium and seeded at a density of 106 cells per well in standard six-well tissue culture plates. After seeding, the cells were incubated at 37°C in a 90% air/10% CO2 atmosphere, and 2 ml of fresh medium was supplied every other day to the cultures after removal of the supernatant. The HepG2 cells were cultured in standard medium for 5–6 d to achieve 90% confluence before any treatment. Human insulin and 2-aminopurine (2-AP) were purchased from Sigma-Aldrich (St. Louis, MO), C2 ceramide, tautomycetin, SC-514, okadaic acid (OA), PKR inhibitor, JNK inhibitor (SP600125) and their analogues, used as negative controls, from EMD Biosciences (San Diego, CA).
Insulin Treatment
Human insulin was stocked in HEPES buffer, which was therefore used in controls for all the experiments with the insulin treatment. We treated the cells with insulin at the concentrations lower than 1 nM to mimic the physiological concentrations (Gual et al., 2003). Cells were deprived of serum for 16 h before insulin treatment.
Chemical Inhibitors
In the present study, we used the commercially available 2-AP and PKR inhibitor as one of the tools to elucidate the role of PKR (data not shown). However, even though these chemical have been widely used in the studies of PKR for a variety of systems, the specificity of these inhibitor has not been extensively tested in the literature. Keeping in mind the potential nonspecific targets of these PKR inhibitors, we performed more PKR gene silencing and overexpressing studies to test our hypothesis and draw the conclusions. The JNK inhibitor, SP600125, IKK inhibitor, SC-514, PP1c inhibitor, TMC, and PP2A inhibitor, OA are popularly used and have been proven to be specific to their targets at the concentrations we used (Muranyi et al., 1997; Bennett et al., 2001; Shim et al., 2002; Kishore et al., 2003; Mitsuhashi et al., 2003). All of the inhibitors, except 2-AP, are dissolved in DMSO. 2-AP is dissolved in glacial acetic acid solution in PBS (1:200; GA).
RNA Interference and Reverse Transfection
Silencer-validated small interfering RNA (siRNA) targeting human PKR, JNK1, and JNK2 mRNAs and Silencer predesigned siRNAs targeting human FoxO1, IRS1, and IRS2 mRNAs were purchased from Ambion (Austin, TX). Prevalidated siRNA targeting human IKK mRNAs were purchased from Qiagen (Valencia, CA). The synthesized oligonucleotides are 5′-GGUGAAGGUAGAUCAAAGATT-3′ and 5′-UCUUUGAUCUACCUUCACCTT-3′ for siRNA of PKR, 5′-GGGAUUUGUUAUCCAAAAUTT-3′ and 5′-AUUUUGGAUAACAAAUCCCTT-3′ for siRNA of JNK1, 5′-GGGAUUGUUUGUGCUGCAUTT-3′ and 5′-AUGCAGCACAAACAAUCCCTT-3′ for siRNA of JNK2, 5′-GCUCAAAUGCUAGUACUAUTT-3′ and 5′-AUAGUACUAGCAUUUGAGCTA-3′ for siRNA of FoxO1, 5′-CCCAAGAGCAUGCACAAACTT-3′ and 5′-GUUUGUGCAUGCUCUUGGGTT-3′ for siRNA of IRS1, 5′-GCGAGUACAUCAACAUCGATT-3′ and 5′-UCGAUGUUGAUGUACUCGCCG-3′ for siRNA of IRS2. The prevalidated siRNA of IKKβ was designed to target the mRNA sequence AAACCGAGTTTGGGATCACAT. As described previously (Yang and Chan, 2009), reverse transfection of siRNA was performed with the transfection reagent, Lipofectamine RNAiMAX (Invitrogen). The scrambled nontargeting siRNA was used as a negative control. Titration of the siRNA and the transfection reagent was performed (not shown), and the lowest working amounts of the siRNA and the transfection reagent were applied in the loss-of-function (LOF) experiments in the present study.
Overexpression of PKR and Forward Transfection
The PKR plasmid, pCMV6-XL5-hPKR, and the empty vector, pCMV6-XL5, were purchased from Origene (Rockville, MD). In general, as described previously (Yang and Chan, 2009), regular HepG2 cells (see Figure 5C) or cells with the other gene silenced by the siRNAs (see Figures 3B and 5E) were subjected to forward transfections of the plasmids according to the Lipofectamine 2000 (Invitrogen) method. After 6 h of transfection, the cells were then cultured in regular media for 42 h, and harvested (Figure 5E) or treated with other chemicals (Figures 3B and 5C) subsequently.
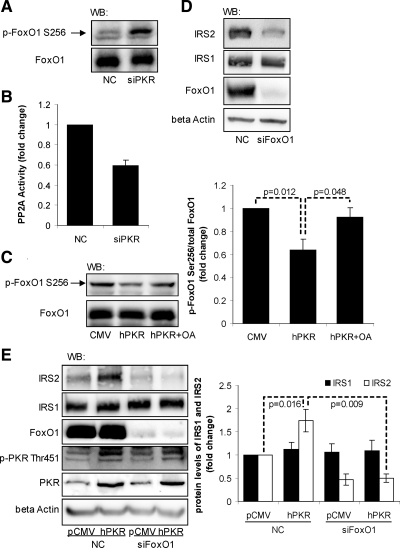
Figure 5.
Involvement of FoxO1 in mediating the effect of PKR on IRS2. Reverse transfection of suspended HepG2 cells was performed with scrambled siRNA (control) or siRNA of PKR (A and B) or siRNA of FoxO1 (D) for 24 h, and the transfected cells were cultured in regular media for another 24 h. The forward transfection of empty vector pCMV6-XL5 (pCMV6) or plasmid containing PKR cDNA sequence (pCMV6-hPKR) was performed, and the cells were then treated with OA (2 nM) or its vehicle, ethanol, as a control, for 1 h (C). Reverse transfection of scramble siRNA (negative control, lanes 1 and 2) or siRNA of FoxO1 (siPKR, lanes 3 and 4) was performed, followed by the forward transfection of empty vector pCMV6-XL5 (pCMV, lanes 1 and 3) or the plasmid containing PKR cDNA sequence (hPKR, lanes 2 and 4; E). After treatment, the cells were harvested, and Western blot analysis was performed to detect the total and phosphorylated levels of FoxO1 (A and C), the total levels of IRS1, IRS2, FoxO1 and β-actin (D and E), and the total and phosphorylated levels of PKR (E). PP2A activity assay was performed to detect the phosphatase activity of PP2A (B). The phosphorylations of FoxO1 at Ser256 normalized to the total FoxO1 protein levels (C) and the protein levels of IRS1 and IRS2 normalized to β-actins (E) are expressed as the average of three samples ± SD from three independent experiments. Student's t test was performed, and p values were calculated for analyzing the differences between the indicated samples.
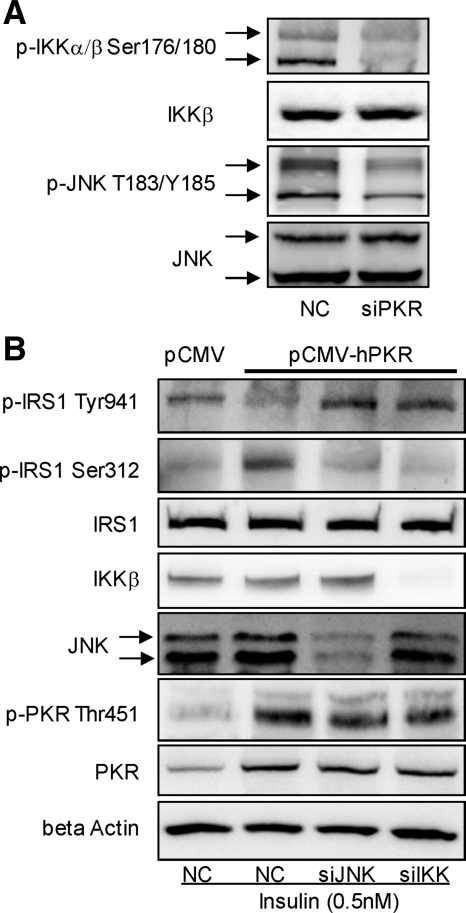
Figure 3.
Involvement of JNK and IKK in regulating the phosphorylation of IRS1. Reverse transfection of suspended HepG2 cells was performed with scramble siRNA (negative control) or siRNA of PKR (siPKR) for 24 h, and the transfected cells were cultured in regular medium for another 24 h (A). Cells were harvested, and Western blot analysis was performed to detect the total and phosphorylated levels of JNK and IKK (A). Reverse transfection of suspended HepG2 cells was performed with scrambled siRNA (control) or siRNAs of JNK1 and JNK2 together or siRNA of IKK for 24 h, and the transfected cells were cultured in regular media for another 24 h. Next, the forward transfection of empty vector pCMV6-XL5 (pCMV6) or plasmid containing PKR cDNA sequence (pCMV6-hPKR) was performed, followed by insulin treatment (0.5 nM) for 15 min (B). After treatment, cells were then harvested, and Western blot analysis was performed to detect the protein level of IKK and JNK and total and phosphorylated levels of PKR and IRS1 (B).
Western Blot Analysis and Immunoprecipitation
HepG2 cells were lysed as described previously (Yang and Chan, 2009). Total protein levels were quantified by bicinchoninic acid (BCA) assay kit from Pierce Biotechnology (Rockford, IL). Twenty to 40 μg of total protein was resolved by SDS-PAGE gels from Bio-Rad (Hercules, CA), transferred to nitrocellulose membranes, and probed with primary and secondary antibodies. Biotinylated protein ladders (Cell Signaling, Beverly, MA) were loaded to one well of each SDS-PAGE gel, and anti-biotin antibody was used to detect the protein ladders on Western blots. Antibody detection was performed using the enhanced chemiluminescence kit from Pierce Biotechnology and imaged on the Molecular Imager ChemiDoc XRS System from Bio-Rad. Immunoprecipitation was performed as described previously (Yang and Chan, 2009). The Western blots were quantified using the Quantity One software (Bio-Rad). Phospho site-specific anti-IRS1 (Tyr941), PPP1A (Thr320), and anti-PPP1A antibodies were purchased from Abcam (Cambridge, MA); phospho site-specific anti-IKKα/β (Ser176/180), FoxO1 (Ser256), anti-biotin, anti-IKKβ, and anti-FoxO1 antibodies from Cell Signaling; and phospho site-specific anti-IRS1 (Ser312), IRS2 (Ser731), PKR (Thr451), JNK (T183/Y185), anti-IRS1, anti-IRS2, anti-PKR, anti-JNK, and anti-β-actin antibodies from Sigma-Aldrich. Secondary anti-rabbit and anti-mouse antibodies were purchased from Pierce Biotechnology.
Real-Time Quantitative RT-PCR Analysis
Total RNA was extracted from cells with the RNeasy mini kit (Qiagen) and depleted of contaminating DNA with RNase-free DNase (Qiagen). Equal amounts of total RNA (1 μg) were reverse-transcribed using an iScript cDNA synthesis kit (Bio-Rad). The first-strand cDNA was used as a template. The primers used for quantitative RT-PCR analyses of human IRS1 (5′-TCCACCTCGGATTGTCTCTT-3′ and 5′-AGGGACTGGAGCCATACTCA-3′), human IRS2 (5′-CCACTCGGACAGCTTCTTCT-3′ and 5′-AGGATGGTCTCGTGGATGTT-3′), and human GAPDH (5′-AACTTTGGTATCGTGGAAGGA-3′ and 5′-CAGTAGAGGCAGGGATGATGT-3′) were synthesized by Operon Biotechnologies (Huntsville, AL). RT-PCR was performed As described previously, and normalized to GAPDH expression levels (Yang and Chan, 2009).
PP2A Phosphatase Activity Assay
The PP2A immunoprecipitation phosphatase assay kit purchased from Millipore (Temecula, CA) was used to measure dephosphorylation of a phosphopeptide as an index of phosphatase activity. Briefly, the cells were lysed using the phosphatase extraction buffer specified by the assay kit, and the catalytic subunit of PP2A (PP2A/C) was immunoprecipitated with anti-PP2A-C supplied in the assay kit. Agarose-bound immune complexes were collected and resuspended in 80 μl of Ser/Thr buffer with 750 μM of phosphopeptide (KRpTIRR; obtained from the kit). The reaction was conducted for 10 min at 30°C in a shaking incubator. Supernatants (25 μl) were transferred in a 96-well plate, and released phosphate was measured by adding 100 μl of malachite green phosphate detection solution. Color was developed for 10 min before reading the plate at 650 nm. The absorbance of the reactions was corrected by subtracting the absorbance in samples treated without Ab. Results were expressed as fold change of PP2A activity compared with control cells.
Statistical Analysis
All experiments were performed at least three times, and representative results are shown. All data, unless specified, are shown as the mean ± SD for indicated number of experiments. One-way ANOVA with Student's t test was used to evaluate statistical significances between different treatment groups.
RESULTS
PKR Induces the Phosphorylation of IRS1 at Ser312 and Mediates the Effects of Ceramide on IRS1 Phosphorylation
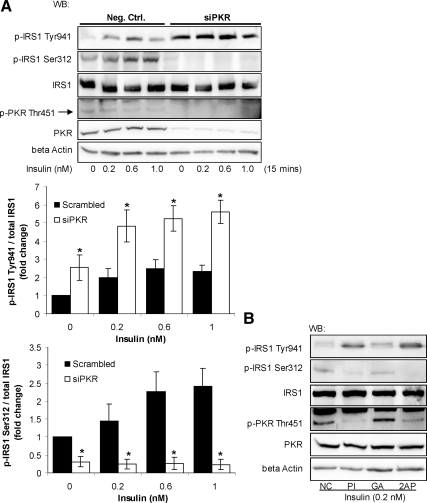
We as well as others showed that the phosphorylation of PKR is down-regulated by insulin through the IRS proteins (Russell et al., 2007; Eley et al., 2008; Wu et al., 2009). We then further studied whether feedback exists between PKR and the IRS proteins. To evaluate the potential function of PKR on the phosphorylation of IRS1, we silenced PKR with a previously validated siRNA, which significantly reduced the PKR mRNA, protein, and phosphorylation levels (Yang and Chan, 2009; Figure 1A). PKR, known as an eIF-2α kinase, directly binds to eIF-2α and induces the phosphorylation of eIF-2α at Ser51 (Taylor et al., 2005). Therefore the phosphorylation of eIF-2α at Ser51 is considered to directly indicate the kinase activity of PKR (Woldehawariat et al., 1999; Pataer et al., 2002; Zhang et al., 2008). In fact, the phosphorylation of eIF-2α at Ser51 was shown to be highly correlated with the phosphorylation of PKR at Thr451 or its activity (Woldehawariat et al., 1999; Pataer et al., 2002; Morimoto et al., 2004; Zhang et al., 2008). We confirmed that gene silencing of PKR significantly reduced the phosphorylation of eIF-2α at Ser51 (Supplementary Figure 1), indicating that gene silencing indeed reduced the overall kinase activity of PKR. Silencing PKR in HepG2 cells significantly blocked the Ser312 and amplified the Tyr941 phosphorylation of IRS1 induced by insulin (Figure 1A). To further confirm a catalytic role of PKR in regulating apoptosis, we inhibited the activity of PKR with pharmaceutical inhibitors of PKR, PKR inhibitor (PI; Jammi et al., 2003) and 2-AP (Ben-Asouli et al., 2002). They suppressed the Ser312 and amplified the Tyr941 phosphorylation of IRS1 (Figure 1B), similar to the siRNA of PKR. These results suggest that PKR induces the inhibitory phosphorylation of IRS1 at Ser312 in HepG2 cells, thereby suppressing the phosphorylation at Tyr941.
Figure 1.
Involvement of PKR in regulating the phosphorylation of IRS1. Reverse transfection of suspended HepG2 cells was performed with scrambled siRNA (negative control) or siRNA of PKR for 24 h, and the transfected cells were cultured in regular media for another 24 h (A). Cells were then treated with different concentrations of insulin for 15 min and harvested after the treatment (A). HepG2 cells were exposed to 5 μM PKR inhibitor (PI) or its analogue as a negative control (NC) or 10 mM 2-AP dissolved in PBS:glacial acetic acid (200:1; GA, control) for 12 h followed by the treatment of 0.2 nM insulin for 15 min (B). Western blot analysis was performed to detect the levels of β-actin and PKR and the total and phosphorylated levels of PKR and IRS1. The phosphorylation levels of IRS1 at Ser312 and Tyr941 were quantified by normalizing to total IRS1 levels and are expressed as the average of three samples ± SD from three independent experiments (A, middle and bottom). Student's t test was performed for analyzing the differences between samples transfected with siPKR and scrambled siRNA (negative control; A). Significantly higher (Tyr941) or lower (Ser312) than negative control; i.e., scrambled siRNA; *p < 0.01.
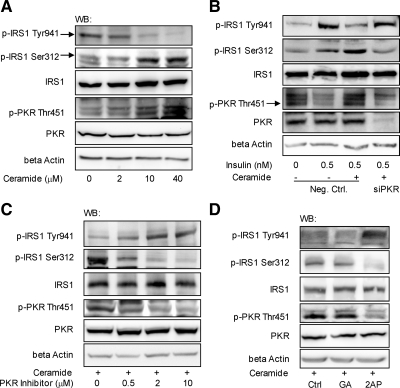
Indeed, a PKR activator (Ruvolo et al., 2001), ceramide, has been shown to inhibit IRS1 activity by up-regulating the Ser phosphorylation and blocking the Tyr phosphorylation of IRS1 (Kanety et al., 1996; Miura et al., 2003). Given the effects of PKR on the phosphorylation of IRS1, we further asked whether PKR mediates the up-regulation of the serine and down-regulation of the tyrosine phosphorylation of IRS1 by ceramide through gene silencing and inhibition studies.
We first confirmed that ceramide induces the phosphorylation of PKR at Thr451, promotes phosphorylation of IRS1 at Ser312, and suppresses phosphorylation at Tyr941 in HepG2 cells (Figure 2, A and B). Furthermore, in ceramide-treated cells, silencing the gene expression of PKR reduced the phosphorylation of IRS1 at Ser312 to control level (Figure 2B), which is consistent with the recovery IRS1 phosphorylation at Tyr941 in response to insulin (Figure 2B). Inhibiting PKR with PI or 2-AP had a similar effect as PKR silencing in blocking the effects of ceramide on the phosphorylation of IRS1 (Figure 2, C and D).
Figure 2.
PKR mediates the effects of ceramide on the phosphorylation of IRS1. HepG2 cells were exposed to different levels of ceramide for 12 h (A). Reverse transfection of suspended HepG2 cells was performed with scrambled siRNA (negative control) or siRNA of PKR for 24 h, and the transfected cells were cultured in regular media for another 12 h (B). Cells were then treated with ceramide (10 μM) for 12 h followed by insulin (0.5 nM) treatment for 15 min (B). Pretreated with 10 μM ceramide for 12 h, HepG2 cells were exposed to different levels of PKR inhibitor dissolved in DMSO (control; C) or 10 mM 2-AP dissolved in PBS:glacial acetic acid (200:1; GA, control; D) for another 12 h. After treatment, the cells were harvested, and Western blot analysis was performed to detect the level of β-actin and the total and phosphorylated levels of PKR and IRS1.
Taken together, both the gene silencing and inhibition studies suggest that PKR may be involved in regulating insulin signaling by inducing phosphorylation of IRS1 at Ser312 and suppressing phosphorylation at Tyr941. This function of PKR provides a potential mechanism by which ceramide regulates IRS1 phosphorylation. However, it is unclear how PKR regulates the phosphorylation of IRS1. Coimmunoprecipitation showed that PKR did not directly interact with IRS1 (Supplementary Figure 2), even upon activation of PKR by ceramide (data not shown), suggesting other intermediate signaling molecules must mediate the effect of PKR on the phosphorylation of IRS1. IRS1 Ser kinases, which directly phosphorylate the serine residues of IRS1, include IKK (Gao et al., 2002), mTOR (Ozes et al., 2001), PKCζ (Ravichandran et al., 2001), S6K1 (Tremblay and Marette, 2001), and JNK (Aguirre et al., 2000; Lee et al., 2003). PKR has been reported to positively activate IKK (Bonnet et al., 2000) and the MAPKs, in particular JNK (Zhou et al., 2003; Yang and Chan, 2009). Therefore, we hypothesize that PKR induces phosphorylation of IRS1 at Ser312 through IRS serine kinases: JNK, IKK, or both.
PKR Positively Regulates JNK and IKK, Both of Which Mediate the Effect of PKR on the Ser Phosphorylation of IRS1
We previously showed that PKR coimmunoprecipitates with JNK and activates JNK in HepG2 cells (Yang and Chan, 2009). Here, we show that IKK also is activated by PKR. Silencing PKR (Yang and Chan, 2009) significantly reduced the phosphorylation of IKKα/β at Ser176/180 (Figure 3A), which indicates IKK activity. To further confirm the involvement of JNK and IKK in mediating the effect of PKR on the phosphorylation of IRS1 at Ser312 and Tyr941, we overexpressed PKR in HepG2 cells and silenced or inhibited JNK or IKK. We confirmed that overexpression of PKR increased the phosphorylation of eIF-2α (Supplementary Figure 1), indicating that overexpression indeed up-regulates the overall kinase activity of PKR. Overexpressing PKR by transfecting the plasmid pCMV6-hPKR into HepG2 cells enhanced phosphorylation of IRS1 at Ser312 and suppressed phosphorylation at Tyr941 (Figure 3B), supporting our previous results that PKR induces serine phosphorylation of IRS1 at Ser312 (Figures 1 and 2). Furthermore, in PKR-overexpressing cells, silencing the gene expression of JNK1/2 or IKK significantly reduced Ser312 phosphorylation and restored Tyr941 phosphorylation of IRS1 in response to insulin (Figure 3B), suggesting that both kinases, JNK and IKK, mediate the effect of PKR on the phosphorylation of IRS1 at Ser312 and in turn the suppression of the tyrosine phosphorylation of IRS1. This was also supported by inhibiting JNK or IKK using their respective specific chemical inhibitor, SP600125 or SC-514 (quantified Western blots results are shown in Supplementary Figure 3; Wu et al., 2009).
In addition to IRS1, we also investigated the potential effect of PKR on another major IRS family protein, IRS2, which also mediates insulin signaling in the liver (Thirone et al., 2006).
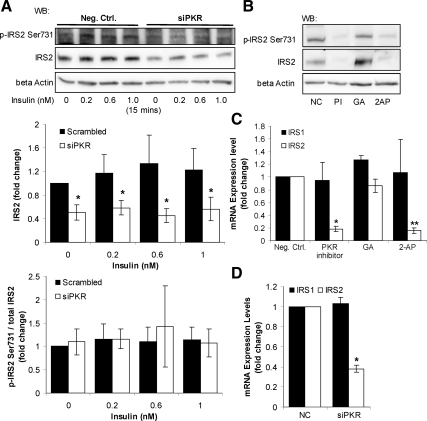
PKR Up-Regulates the Protein Expression Level of IRS2
Silencing the gene expression of PKR (Figure 1) down-regulated the protein level of IRS2 (Figure 4A), but not IRS1 (Figure 1), suggesting that PKR is required for cells to maintain proper protein expression level of IRS2. Similarly, both PKR inhibitors (Figure 1), PKR inhibitor (PI) and 2-AP, also down-regulated the protein level of IRS2 (Figure 4B), further supporting the effect of PKR on the protein expression level of IRS2. Therefore, PKR exploits a regulatory role on the protein level of IRS2, but not on IRS1. As discussed above, PKR affects IRS1 serine phosphorylation (Figures 1–3). Notably, upon inhibiting or silencing PKR, the phosphorylation of IRS2 at Ser731 varied proportionally to its total protein level (Figure 4, A and B), suggesting that PKR is not affecting the phosphorylation of IRS2 at Ser731. As expected, the general tyrosine phosphorylation of IRS2, normalized to the total protein level of IRS2, is not affected by PKR silencing or inhibition, as measured by Western blotting of the IRS2 immunoprecipitates with anti-phospho tyrosine antibody (data not shown). Next, to determine whether PKR transcriptionally regulates the protein level of IRS2, we measured the mRNA expression levels of IRS1 and IRS2 upon PKR inhibition and gene silencing. Both the PKR inhibitors and the siRNA of PKR down-regulated the mRNA expression of IRS2, but not of IRS1 (Figure 4, C and D), suggesting that PKR regulates IRS2 at the transcriptional level.
Figure 4.
Involvement of PKR in regulating IRS2. Reverse transfection of suspended HepG2 cells was performed with scrambled siRNA (negative control) or siRNA of PKR for 24 h, and the transfected cells were cultured in regular media for another 24 h. Cells were then harvested (D) or treated with different concentrations of insulin for 15 min and harvested after the treatment (A). Confluent HepG2 cells were treated with 5 μM PKR inhibitor (PI) or its analogue as a negative control (NC) or 10 mM 2-AP dissolved in PBS:glacial acetic acid (200:1) (GA) for 12 h (B and C). Western blot analysis was performed to detect the total and phosphorylated levels of IRS2 at Ser731 (A and B). RT-PCR was performed to detect the gene expression levels of IRS1 and IRS2 in response to the PKR inhibitors (C) or siRNA of PKR (D). Gene expression data were expressed as the average of nine samples ± SD from three independent experiments. The protein levels of IRS2 were quantified by normalizing to β-actin, and the phosphorylation levels of IRS2 at Ser731 were quantified by normalizing to total IRS2 levels. Both the protein and phosphorylation levels of IRS2 are expressed as the average of three samples ± SD from three independent experiments. Student's t test was performed for analyzing the differences between siPKR and scrambled siRNA (negative control). Significantly lower than negative control; i.e., scrambled siRNA (A and D) or chemical analogue of the PKR inhibitor (B and C); *p < 0.05. Significantly lower than control; GA, solvent of 2-AP; **p < 0.05.
PKR Up-Regulates the Protein Level of IRS2 through the Transcription Factor FoxO1
PKR as a protein kinase, activates several transcription factors, such as IRF-1, p53, and nuclear factor kappa B (NF-κB; Kumar et al., 1997; Cuddihy et al., 1999), but these transcription factors have not been shown to regulate the transcription of IRS2. However, the transcription of IRS2 has been shown to be dependent on the transcription factor FoxO1 in the liver (Zhang et al., 2001; Ide et al., 2004) or CREB in pancreatic β-cells (Jhala et al., 2003). It is not known whether PKR interacts with either of these two transcription factors. We found that PKR had no effect on the activity or translocation of CREB in HepG2 cells (not shown). However, silencing the PKR gene significantly increased the phosphorylation of FoxO1 at Ser256 (Figure 5A). Phosphorylation at Ser256 inhibits the DNA-binding activity of FoxO1 and its nuclear import by suppressing the nuclear targeting signal on its DNA-binding domain (Rena et al., 2001, 2002). We also performed nuclear extraction and measured the nuclear level of p-FoxO1 Ser256 in response to PKR silencing (Supplementary Figure 4A). The nuclear level of the p-FoxO1 Ser256 was significantly increased upon silencing of PKR, further confirming that PKR regulates FoxO1 activity by regulating its phosphorylation and translocation. Our results suggest, for the first time, that PKR reduces the phosphorylation of FoxO1 and thereby activates it. As a protein kinase, PKR does not have the phosphatase activity that is required to dephosphorylate FoxO1. However, PKR is known to phosphorylate B56α, the regulatory subunit of PP2A, which then activates the catalytic subunit of PP2A (Xu and Williams, 2000). PP2A, in turn, is known to dephosphorylate FoxO1 at Ser256 (Yan et al., 2008). Indeed, silencing PKR significantly suppressed the activity of PP2A (Figure 5B), thereby confirming that PKR activates PP2A in HepG2. To further investigate the potential involvement of PP2A in mediating the dephosphorylation of Ser256 on FoxO1 by PKR, we overexpressed PKR in HepG2 cells and inhibited the activity of PP2A with OA. Overexpressing PKR by transfecting the plasmid pCMV6-hPKR into HepG2 cells reduced the phosphorylation of FoxO1 at Ser256 (Figure 5C), supporting our results that PKR induces dephosphorylation of FoxO1 at Ser256 (Figure 5A). More importantly, OA, a specific PP2A inhibitor, restored the serine phosphorylation level of FoxO1 in PKR overexpressed cells (Figure 5C). Similarly overexpressing PKR also decreased the Ser256 phosphorylation of FoxO1 in the nucleus, and OA restored the nuclear level of p-FoxO1 Ser256 in PKR overexpressed cells (Supplementary Figure 4B). Taken together, PKR dephosphorylates and activates FoxO1, mediated by PP2A.
To confirm the positive effect of FoxO1 on the expression of IRS2 in HepG2 cells, we performed gene silencing of FoxO1. Silencing the gene expression of FoxO1 significantly reduced the protein level of IRS2, but not IRS1 (Figure 5D), suggesting that FoxO1 controls the expression of IRS2 in HepG2 cells. To further confirm that FoxO1 mediates the effect of PKR on IRS2 protein level, we overexpressed PKR in HepG2 cells as well as silenced FoxO1. Overexpressing PKR in HepG2 cells increased the protein level of IRS2 (lanes 1 vs. 2 in Figure 5E), whereas silencing FoxO1 in control and PKR overexpressed cells significantly reduced the protein level of IRS2 (Figure 5E), confirming that PKR up-regulates the protein level of IRS2 through the transcription factor FoxO1.
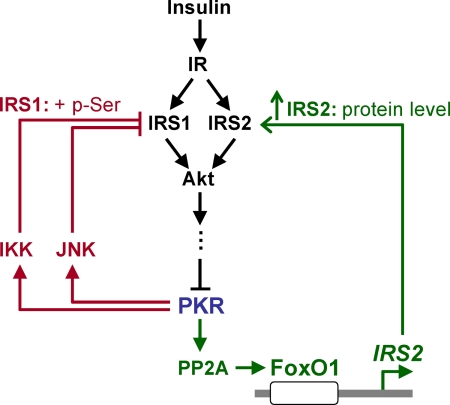
In summary, our results suggest PKR differentially regulates IRS proteins (Figure 6). First, PKR induces phosphorylation of IRS1 at Ser312 and suppresses tyrosine phosphorylation of IRS1, mediated by the IRS kinases, JNK and IKK. Second, PKR activates a transcription factor, FoxO1, which up-regulates the gene expression of IRS2. In addition, we as well as others have identified PKR as a downstream substrate of insulin signaling. Therefore taken together, our results suggest that PKR is involved in insulin signaling through a feedback mechanism and regulates the central transmitters of intracellular insulin signaling in the liver, IRS1, and IRS2, through different pathways (Figure 6).
Figure 6.
Proposed signaling pathways through which PKR is involved in insulin signaling network in HepG2 cells. Insulin activates insulin signaling by IR and IRS, leading to the suppression of PKR phosphorylation at Thr451. PKR induces the phosphorylation of IRS1 at Ser312 through two other kinases, JNK and IKK. In addition, by activating PP2A, PKR dephosphorylates a transcription factor, FoxO1, which up-regulates the gene expression of IRS2.
DISCUSSION
In the present study, we identified the effects of PKR on two major IRS proteins, IRS1 and IRS2, in HepG2 cells. First, PKR up-regulates the phosphorylation of IRS1 at Ser312, which in turn suppresses the tyrosine phosphorylation of IRS1. This effect of PKR is mediated by JNK and IKK (Figure 3). It is well known that PKR stimulates the transcription factor NF-κB by activating IKK (Bonnet et al., 2000), and this process does not require the catalytic activity of PKR. Instead, The N-terminus of PKR is responsible for the activation of IKK (Bonnet et al., 2006). As discussed previously (Yang and Chan, 2009), PKR has been reported also to play a role in the phosphorylation of the three MAPKs: JNK, ERK, and p38 MAPK, in the rank order of JNK > p38 MAPK > ERK (Zhou et al., 2003). Among the three MAPKs, JNK has been suggested to play a central role in inducing the inhibitory serine phosphorylation of IRS1 (Aguirre et al., 2000; Hirosumi et al., 2002). We did not test the effects of PKR on the other two less responsive MAPK proteins, ERK and p38 MAPK, which were also suggested to induce phosphorylation of IRS1 at Ser residues (Rui et al., 2001; Fujishiro et al., 2003). Thus, although JNK may be an important intermediate, it is not likely the only one involved in mediating the signaling pathway from PKR to IRS1 phosphorylation.
The phosphorylation of IRS1 at Ser312 by PKR provides a potential mechanism through which ceramide, an activator of PKR, promotes the inhibitory serine phosphorylation of IRS1. Indeed, other activators of PKR have also been shown to function on IRS1 in a similar manner. For example, HCV core protein, which directly binds and activates PKR (Yan et al., 2007), has been shown to induce the phosphorylation of IRS1 at Ser312 (Banerjee et al., 2008). The ability of PKR to promote serine phosphorylation of IRS1 provides a possible mechanism by which PKR mediates HCV infection and the inhibition of the IRS1 activity (Aytug et al., 2003; Banerjee et al., 2008).
A novel function of PKR that we uncovered is its regulation of the protein level of IRS2 through the transcription factor FoxO1. The effect of PKR on FoxO1 has not been studied previously. We show that PKR dephosphorylates and activates FoxO1 mediated by the protein phosphatase PP2A. FoxO1 directs the expression of genes involved in a wide variety of cellular responses, one of which is the regulation of glucose homeostasis and energy metabolism (Gross et al., 2008). Insulin induces the phosphorylation of FoxO1, thereby inhibiting its activity (Barthel et al., 2005). In healthy states, the low insulin level during fasting sustains the activity of FoxO1, which facilitates the transcription of key enzymes involved in gluconeogenesis (Puigserver et al., 2003). FoxO1 also can up-regulate IRS2 gene expression through a feedback loop (Zhang et al., 2001; Ide et al., 2004), which occurs in HepG2 cells (Figure 5). However in insulin resistance models, the persistent activation of FoxO1, due to disruption of the IRS–PI3K–Akt pathway, contributes to the development of hyperglycemia and glucose intolerance (Samuel et al., 2006; Dong et al., 2008). Therefore, FoxO1, which becomes activated during insulin resistance, is believed to serve as a dominant regulator of hepatic gene expression. The roles of FoxO1 in regulating fasting glucose homeostasis and enhancing hyperglycemia and glucose intolerance in insulin resistance models begs the question of whether PKR activation is involved in the regulation of glucose metabolism mediated by FoxO1.
Given the inhibitory effect of insulin on PKR and the regulation of FoxO1 phosphorylation by PKR, we hypothesize that insulin-stimulated FoxO1 phosphorylation is mediated by PKR (Figure 6). However, it has been previously shown that insulin stimulates the FoxO1 phosphorylation through Akt, which directly induces the phosphorylation of FoxO family of transcription factors, including FoxO1 (Brunet et al., 1999; Guo et al., 1999; Rena et al., 1999; Barthel et al., 2005). As shown in Figure 6, the effect of PKR on FoxO1 phosphorylation stimulated by insulin is a complementary pathway to the direct phosphorylation of FoxO1 by Akt. Indeed we found that insulin is also able to stimulate the phosphorylation of FoxO1, even in PKR-overexpressing cells (data not shown). Therefore, the PKR pathway identified in this study does not preclude the strong direct phosphorylation of FoxO1 by Akt induced by insulin.
In the present study, we identified that PKR regulates IRS1 and IRS2 through different mechanisms. Although the protein structures of the IRS proteins are highly conserved, both animal and cell studies indicate that IRS1 and IRS2 serve complementary, rather than redundant, roles in insulin signaling (Araki et al., 1994; Tamemoto et al., 1994; Withers et al., 1998; Kido et al., 2000). It has been suggested that hepatic IRS1 and IRS2 control different aspects of hepatic metabolism, with IRS1 more closely related to glucose homeostasis and IRS2 more closely related to lipid metabolism (Taniguchi et al., 2005). However more recently, using specific knockouts of liver IRS1 or IRS2, researchers demonstrated that the two proteins may overlap in their insulin action (Dong et al., 2008). Nevertheless, the different functions of IRS1 and IRS2 on hepatic insulin signaling have not been completely elucidated. Investigators have claimed that IRS1 and IRS2 are expressed differently and exert distinct functions under fasting and refeeding conditions (Kubota et al., 2008). IRS1 is stably expressed in the postprandial state, while IRS2 is highly expressed in the fasted state and down-regulated in the fed state (Dong et al., 2008; Kubota et al., 2008). This has been proposed to be mainly due to the different circulating insulin levels during fasted and fed states (Dong et al., 2008; Kubota et al., 2008). High levels of insulin, in the fed state, suppress the IRS2 mRNA and protein expressions at the transcriptional level, through the PI3K-Akt pathway (Hirashima et al., 2003). Because Akt serves as a kinase that directly induces the phosphorylation of FoxO1 (Guo et al., 1999; Barthel et al., 2005), the down-regulation of Akt due to the low insulin levels, during fasting, results in dephosphorylation and therefore the translocation and activation of FoxO1 (Rena et al., 2001; Rena et al., 2002). This then contributes to the up-regulation of IRS2 gene transcription during fasting.
Along with the differential expression patterns of IRS1 and IRS2, a functional relay exists between IRS1 and IRS2 in mediating hepatic insulin signaling during fasting and after refeeding. In the fasted state, especially the late stage of fasting, the major downstream target of insulin signaling, PI3K, is more associated with IRS2 than IRS1 (Kubota et al., 2008). Therefore, the elevated expression of IRS2 in the fasted state may serve as a complementary mechanism to compensate for the decreased level of circulating insulin (Dong et al., 2008; Kubota et al., 2008). On the other hand, after refeeding, the IRS1-associated PI3K starts to rise, whereas the IRS2-associated PI3K decreases (Kubota et al., 2008). This suggests a switch of hepatic insulin signaling hub protein, from IRS2 in the fasting state to IRS1 in the fed state. Thus, insulin signaling is mediated primarily through IRS1 after refeeding, whereas IRS2 plays a more predominant role during fasting, compensating for the loss in insulin signaling activity due to the low insulin level (Kubota et al., 2008).
Considering the distinct expressions and functions of IRS1 and IRS2 during fasted versus fed states, we propose that PKR may regulate insulin signaling activity differentially during the fasted versus fed states. Taking into account the inhibitory effect of insulin on the phosphorylation of PKR, in the present manuscript we propose that insulin suppresses the activity of PKR in the fed state, resulting in the suppression of the negative feedbacks that functions on IRS1 Ser phosphorylation and thereby promoting insulin signaling activity through IRS1. On the other hand, when the circulating insulin is at a low level, PKR remains at a relatively higher activity level during the fasting (vs. fed) state, enhancing IRS2 gene expression through FoxO1, and promoting hepatic insulin signaling to compensate for the low insulin levels during the fasted state. Taken together, the disparate effects of PKR on the IRS1 and IRS2 proteins may serve as a potential mechanism by which the two IRS proteins are differentially regulated.
In summary, we identified that PKR functions as a key regulator of the central transmitters of insulin signaling in liver cells, IRS1 and IRS2 (Figure 6). PKR induces the inhibitory phosphorylation of IRS at Ser312 (Figures 1–3) and activates the transcription factor, FoxO1, which up-regulates the protein expression level of IRS2 (Figures 4 and 5). Taken together, PKR appears to be an important player in the regulation of insulin signaling through the major transmitters of insulin signaling, IRS1 and IRS2.
Supplementary Material
ACKNOWLEDGMENTS
We thank Linxia Zhang and Linsey Christine Seitz for their help with the western blotting evaluation of the effect of PKR on CREB. The work was supported in part by the National Science Foundation (CBET 0941055), the National Institutes of Health (R01GM079688, R21CA126136 and R21RR024439), and the MSU Foundation and the Center for Systems Biology.
Abbreviations used:
- eIF-2α
eukaryotic initiation factor 2-alpha
- ERK
extracellular signal–regulated kinase
- FoxO1
forkhead box O1
- HCV
hepatitis C virus
- HepG2
human hepatocellular carcinoma cell
- IGF-1
insulin-like growth factor 1
- IR
insulin receptor
- IKK
I-κB kinase
- IRS
insulin receptor substrate
- JNK
c-Jun N-terminal kinase
- MAPK
mitogen-activated protein kinase
- NF-κB
nuclear factor kappa B
- PKR
double-stranded RNA–dependent protein kinase
- PP2A
protein phosphatase 2A.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-06-0481) on August 4, 2010.
REFERENCES
- Aguirre V., Uchida T., Yenush L., Davis R., White M. F. The c-Jun NH(2)-terminal kinase promotes insulin resistance during association with insulin receptor substrate-1 and phosphorylation of Ser(307) J. Biol. Chem. 2000;275:9047–9054. doi: 10.1074/jbc.275.12.9047. [DOI] [PubMed] [Google Scholar]
- Alessi D. R., James S. R., Downes C. P., Holmes A. B., Gaffney P. R., Reese C. B., Cohen P. Characterization of a 3-phosphoinositide-dependent protein kinase which phosphorylates and activates protein kinase Balpha. Curr. Biol. 1997;7:261–269. doi: 10.1016/s0960-9822(06)00122-9. [DOI] [PubMed] [Google Scholar]
- Araki E., Lipes M. A., Patti M. E., Bruning J. C., Haag B., 3rd, Johnson R. S., Kahn C. R. Alternative pathway of insulin signalling in mice with targeted disruption of the IRS-1 gene. Nature. 1994;372:186–190. doi: 10.1038/372186a0. [DOI] [PubMed] [Google Scholar]
- Aytug S., Reich D., Sapiro L. E., Bernstein D., Begum N. Impaired IRS-1/PI3-kinase signaling in patients with HCV: a mechanism for increased prevalence of type 2 diabetes. Hepatology. 2003;38:1384–1392. doi: 10.1016/j.hep.2003.09.012. [DOI] [PubMed] [Google Scholar]
- Backer J. M., Schroeder G. G., Kahn C. R., Myers M. G., Jr, Wilden P. A., Cahill D. A., White M. F. Insulin stimulation of phosphatidylinositol 3-kinase activity maps to insulin receptor regions required for endogenous substrate phosphorylation. J. Biol. Chem. 1992;267:1367–1374. [PubMed] [Google Scholar]
- Banerjee S., Saito K., Ait-Goughoulte M., Meyer K., Ray R. B., Ray R. Hepatitis C virus core protein upregulates serine phosphorylation of insulin receptor substrate-1 and impairs the downstream akt/protein kinase B signaling pathway for insulin resistance. J. Virol. 2008;82:2606–2612. doi: 10.1128/JVI.01672-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthel A., Schmoll D., Unterman T. G. FoxO proteins in insulin action and metabolism. Trends Endocrinol. Metab. 2005;16:183–189. doi: 10.1016/j.tem.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Ben-Asouli Y., Banai Y., Pel-Or Y., Shir A., Kaempfer R. Human interferon-gamma mRNA autoregulates its translation through a pseudoknot that activates the interferon-inducible protein kinase PKR. Cell. 2002;108:221–232. doi: 10.1016/s0092-8674(02)00616-5. [DOI] [PubMed] [Google Scholar]
- Bennett B. L., et al. SP600125, an anthrapyrazolone inhibitor of Jun N-terminal kinase. Proc. Natl. Acad. Sci. USA. 2001;98:13681–13686. doi: 10.1073/pnas.251194298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet M. C., Daurat C., Ottone C., Meurs E. F. The N-terminus of PKR is responsible for the activation of the NF-kappaB signaling pathway by interacting with the IKK complex. Cell Signal. 2006;18:1865–1875. doi: 10.1016/j.cellsig.2006.02.010. [DOI] [PubMed] [Google Scholar]
- Bonnet M. C., Weil R., Dam E., Hovanessian A. G., Meurs E. F. PKR stimulates NF-kappaB irrespective of its kinase function by interacting with the IkappaB kinase complex. Mol. Cell. Biol. 2000;20:4532–4542. doi: 10.1128/mcb.20.13.4532-4542.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet A., Bonni A., Zigmond M. J., Lin M. Z., Juo P., Hu L. S., Anderson M. J., Arden K. C., Blenis J., Greenberg M. E. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96:857–868. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- Cuddihy A. R., Li S., Tam N. W., Wong A. H., Taya Y., Abraham N., Bell J. C., Koromilas A. E. Double-stranded-RNA-activated protein kinase PKR enhances transcriptional activation by tumor suppressor p53. Mol. Cell. Biol. 1999;19:2475–2484. doi: 10.1128/mcb.19.4.2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X. C., Copps K. D., Guo S., Li Y., Kollipara R., DePinho R. A., White M. F. Inactivation of hepatic Foxo1 by insulin signaling is required for adaptive nutrient homeostasis and endocrine growth regulation. Cell Metab. 2008;8:65–76. doi: 10.1016/j.cmet.2008.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eley H. L., Russell S. T., Tisdale M. J. Role of the dsRNA-dependent protein kinase (PKR) in the attenuation of protein loss from muscle by insulin and insulin-like growth factor-I (IGF-I) Mol. Cell Biochem. 2008;313:63–69. doi: 10.1007/s11010-008-9742-4. [DOI] [PubMed] [Google Scholar]
- Fujishiro M., et al. Three mitogen-activated protein kinases inhibit insulin signaling by different mechanisms in 3T3–L1 adipocytes. Mol. Endocrinol. 2003;17:487–497. doi: 10.1210/me.2002-0131. [DOI] [PubMed] [Google Scholar]
- Gao Z., Hwang D., Bataille F., Lefevre M., York D., Quon M. J., Ye J. Serine phosphorylation of insulin receptor substrate 1 by inhibitor kappa B kinase complex. J. Biol. Chem. 2002;277:48115–48121. doi: 10.1074/jbc.M209459200. [DOI] [PubMed] [Google Scholar]
- Gross D. N., van den Heuvel A. P., Birnbaum M. J. The role of FoxO in the regulation of metabolism. Oncogene. 2008;27:2320–2336. doi: 10.1038/onc.2008.25. [DOI] [PubMed] [Google Scholar]
- Gual P., Gonzalez T., Gremeaux T., Barres R., Le Marchand-Brustel Y., Tanti J. F. Hyperosmotic stress inhibits insulin receptor substrate-1 function by distinct mechanisms in 3T3–L1 adipocytes. J. Biol. Chem. 2003;278:26550–26557. doi: 10.1074/jbc.M212273200. [DOI] [PubMed] [Google Scholar]
- Guo S., Rena G., Cichy S., He X., Cohen P., Unterman T. Phosphorylation of serine 256 by protein kinase B disrupts transactivation by FKHR and mediates effects of insulin on insulin-like growth factor-binding protein-1 promoter activity through a conserved insulin response sequence. J. Biol. Chem. 1999;274:17184–17192. doi: 10.1074/jbc.274.24.17184. [DOI] [PubMed] [Google Scholar]
- Herschkovitz A., Liu Y. F., Ilan E., Ronen D., Boura-Halfon S., Zick Y. Common inhibitory serine sites phosphorylated by IRS-1 kinases, triggered by insulin and inducers of insulin resistance. J. Biol. Chem. 2007;282:18018–18027. doi: 10.1074/jbc.M610949200. [DOI] [PubMed] [Google Scholar]
- Hirashima Y., Tsuruzoe K., Kodama S., Igata M., Toyonaga T., Ueki K., Kahn C. R., Araki E. Insulin down-regulates insulin receptor substrate-2 expression through the phosphatidylinositol 3-kinase/Akt pathway. J. Endocrinol. 2003;179:253–266. doi: 10.1677/joe.0.1790253. [DOI] [PubMed] [Google Scholar]
- Hirosumi J., Tuncman G., Chang L., Gorgun C. Z., Uysal K. T., Maeda K., Karin M., Hotamisligil G. S. A central role for JNK in obesity and insulin resistance. Nature. 2002;420:333–336. doi: 10.1038/nature01137. [DOI] [PubMed] [Google Scholar]
- Hotamisligil G. S., Peraldi P., Budavari A., Ellis R., White M. F., Spiegelman B. M. IRS-1-mediated inhibition of insulin receptor tyrosine kinase activity in TNF-alpha- and obesity-induced insulin resistance. Science. 1996;271:665–668. doi: 10.1126/science.271.5249.665. [DOI] [PubMed] [Google Scholar]
- Ide T., et al. SREBPs suppress IRS-2-mediated insulin signalling in the liver. Nat. Cell Biol. 2004;6:351–357. doi: 10.1038/ncb1111. [DOI] [PubMed] [Google Scholar]
- Imamura T., et al. Two naturally occurring mutations in the kinase domain of insulin receptor accelerate degradation of the insulin receptor and impair the kinase activity. J. Biol. Chem. 1994;269:31019–31027. [PubMed] [Google Scholar]
- Jammi N. V., Whitby L. R., Beal P. A. Small molecule inhibitors of the RNA-dependent protein kinase. Biochem. Biophys. Res. Commun. 2003;308:50–57. doi: 10.1016/s0006-291x(03)01318-4. [DOI] [PubMed] [Google Scholar]
- Jhala U. S., Canettieri G., Screaton R. A., Kulkarni R. N., Krajewski S., Reed J., Walker J., Lin X., White M., Montminy M. cAMP promotes pancreatic beta-cell survival via CREB-mediated induction of IRS2. Genes Dev. 2003;17:1575–1580. doi: 10.1101/gad.1097103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn B. B. Type 2 diabetes: when insulin secretion fails to compensate for insulin resistance. Cell. 1998;92:593–596. doi: 10.1016/s0092-8674(00)81125-3. [DOI] [PubMed] [Google Scholar]
- Kanety H., Hemi R., Papa M. Z., Karasik A. Sphingomyelinase and ceramide suppress insulin-induced tyrosine phosphorylation of the insulin receptor substrate-1. J. Biol. Chem. 1996;271:9895–9897. doi: 10.1074/jbc.271.17.9895. [DOI] [PubMed] [Google Scholar]
- Kido Y., Burks D. J., Withers D., Bruning J. C., Kahn C. R., White M. F., Accili D. Tissue-specific insulin resistance in mice with mutations in the insulin receptor, IRS-1, and IRS-2. J. Clin. Invest. 2000;105:199–205. doi: 10.1172/JCI7917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishore N., et al. A selective IKK-2 inhibitor blocks NF-kappa B-dependent gene expression in interleukin-1 beta-stimulated synovial fibroblasts. J. Biol. Chem. 2003;278:32861–32871. doi: 10.1074/jbc.M211439200. [DOI] [PubMed] [Google Scholar]
- Kubota N., et al. Dynamic functional relay between insulin receptor substrate 1 and 2 in hepatic insulin signaling during fasting and feeding. Cell Metab. 2008;8:49–64. doi: 10.1016/j.cmet.2008.05.007. [DOI] [PubMed] [Google Scholar]
- Kumar A., Yang Y. L., Flati V., Der S., Kadereit S., Deb A., Haque J., Reis L., Weissmann C., Williams B. R. Deficient cytokine signaling in mouse embryo fibroblasts with a targeted deletion in the PKR gene: role of IRF-1 and NF-kappaB. EMBO J. 1997;16:406–416. doi: 10.1093/emboj/16.2.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y. H., Giraud J., Davis R. J., White M. F. c-Jun N-terminal kinase (JNK) mediates feedback inhibition of the insulin signaling cascade. J. Biol. Chem. 2003;278:2896–2902. doi: 10.1074/jbc.M208359200. [DOI] [PubMed] [Google Scholar]
- Lowenstein E. J., Daly R. J., Batzer A. G., Li W., Margolis B., Lammers R., Ullrich A., Skolnik E. Y., Bar-Sagi D., Schlessinger J. The SH2 and SH3 domain-containing protein GRB2 links receptor tyrosine kinases to ras signaling. Cell. 1992;70:431–442. doi: 10.1016/0092-8674(92)90167-b. [DOI] [PubMed] [Google Scholar]
- McElduff A., Hedo J. A., Taylor S. I., Roth J., Gorden P. Insulin receptor degradation is accelerated in cultured lymphocytes from patients with genetic syndromes of extreme insulin resistance. J. Clin. Invest. 1984;74:1366–1374. doi: 10.1172/JCI111547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuhashi S., Shima H., Tanuma N., Matsuura N., Takekawa M., Urano T., Kataoka T., Ubukata M., Kikuchi K. Usage of tautomycetin, a novel inhibitor of protein phosphatase 1 (PP1), reveals that PP1 is a positive regulator of Raf-1 in vivo. J. Biol. Chem. 2003;278:82–88. doi: 10.1074/jbc.M208888200. [DOI] [PubMed] [Google Scholar]
- Miura A., Kajita K., Ishizawa M., Kanoh Y., Kawai Y., Natsume Y., Sakuma H., Yamamoto Y., Yasuda K., Ishizuka T. Inhibitory effect of ceramide on insulin-induced protein kinase Czeta translocation in rat adipocytes. Metabolism. 2003;52:19–24. doi: 10.1053/meta.2003.50011. [DOI] [PubMed] [Google Scholar]
- Morimoto H., Okamura H., Yoshida K., Kitamura S., Haneji T. Okadaic acid induces apoptosis through double-stranded RNA-dependent protein kinase/eukaryotic initiation factor-2alpha pathway in human osteoblastic MG63 cells. J. Biochem. 2004;136:433–438. doi: 10.1093/jb/mvh144. [DOI] [PubMed] [Google Scholar]
- Muranyi A., Gergely P., Nagy G. M., Fekete M. I. The possible role of protein phosphatase 2A in the sodium sensitivity of the receptor binding of opiate antagonists naloxone and naltrindole. Brain Res. Bull. 1997;44:273–279. doi: 10.1016/s0361-9230(97)00136-6. [DOI] [PubMed] [Google Scholar]
- Ozes O. N., Akca H., Mayo L. D., Gustin J. A., Maehama T., Dixon J. E., Donner D. B. A phosphatidylinositol 3-kinase/Akt/mTOR pathway mediates and PTEN antagonizes tumor necrosis factor inhibition of insulin signaling through insulin receptor substrate-1. Proc. Natl. Acad. Sci. USA. 2001;98:4640–4645. doi: 10.1073/pnas.051042298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pataer A., et al. Adenoviral transfer of the melanoma differentiation-associated gene 7 (mda7) induces apoptosis of lung cancer cells via up-regulation of the double-stranded RNA-dependent protein kinase (PKR) Cancer Res. 2002;62:2239–2243. [PubMed] [Google Scholar]
- Patti M. E., Kahn C. R. The insulin receptor—a critical link in glucose homeostasis and insulin action. J. Basic Clin. Physiol. Pharmacol. 1998;9:89–109. doi: 10.1515/jbcpp.1998.9.2-4.89. [DOI] [PubMed] [Google Scholar]
- Paz K., Hemi R., LeRoith D., Karasik A., Elhanany E., Kanety H., Zick Y. A molecular basis for insulin resistance. Elevated serine/threonine phosphorylation of IRS-1 and IRS-2 inhibits their binding to the juxtamembrane region of the insulin receptor and impairs their ability to undergo insulin-induced tyrosine phosphorylation. J. Biol. Chem. 1997;272:29911–29918. doi: 10.1074/jbc.272.47.29911. [DOI] [PubMed] [Google Scholar]
- Pederson T. M., Kramer D. L., Rondinone C. M. Serine/threonine phosphorylation of IRS-1 triggers its degradation: possible regulation by tyrosine phosphorylation. Diabetes. 2001;50:24–31. doi: 10.2337/diabetes.50.1.24. [DOI] [PubMed] [Google Scholar]
- Proud C. G. PKR: a new name and new roles. Trends Biochem. Sci. 1995;20:241–246. doi: 10.1016/s0968-0004(00)89025-8. [DOI] [PubMed] [Google Scholar]
- Puigserver P., et al. Insulin-regulated hepatic gluconeogenesis through FOXO1-PGC-1alpha interaction. Nature. 2003;423:550–555. doi: 10.1038/nature01667. [DOI] [PubMed] [Google Scholar]
- Ravichandran L. V., Esposito D. L., Chen J., Quon M. J. Protein kinase C-zeta phosphorylates insulin receptor substrate-1 and impairs its ability to activate phosphatidylinositol 3-kinase in response to insulin. J. Biol. Chem. 2001;276:3543–3549. doi: 10.1074/jbc.M007231200. [DOI] [PubMed] [Google Scholar]
- Reaven G. M. Banting lecture 1988. Role of insulin resistance in human disease. Diabetes. 1988;37:1595–1607. doi: 10.2337/diab.37.12.1595. [DOI] [PubMed] [Google Scholar]
- Rena G., Guo S., Cichy S. C., Unterman T. G., Cohen P. Phosphorylation of the transcription factor forkhead family member FKHR by protein kinase B. J. Biol. Chem. 1999;274:17179–17183. doi: 10.1074/jbc.274.24.17179. [DOI] [PubMed] [Google Scholar]
- Rena G., Prescott A. R., Guo S., Cohen P., Unterman T. G. Roles of the forkhead in rhabdomyosarcoma (FKHR) phosphorylation sites in regulating 14-3-3 binding, transactivation and nuclear targetting. Biochem. J. 2001;354:605–612. doi: 10.1042/0264-6021:3540605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rena G., Woods Y. L., Prescott A. R., Peggie M., Unterman T. G., Williams M. R., Cohen P. Two novel phosphorylation sites on FKHR that are critical for its nuclear exclusion. EMBO J. 2002;21:2263–2271. doi: 10.1093/emboj/21.9.2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rui L., Aguirre V., Kim J. K., Shulman G. I., Lee A., Corbould A., Dunaif A., White M. F. Insulin/IGF-1 and TNF-alpha stimulate phosphorylation of IRS-1 at inhibitory Ser307 via distinct pathways. J. Clin. Invest. 2001;107:181–189. doi: 10.1172/JCI10934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell S. T., Eley H., Tisdale M. J. Mechanism of attenuation of angiotensin-II-induced protein degradation by insulin-like growth factor-I (IGF-I) Cell Signal. 2007;19:1583–1595. doi: 10.1016/j.cellsig.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Ruvolo P. P., Gao F., Blalock W. L., Deng X., May W. S. Ceramide regulates protein synthesis by a novel mechanism involving the cellular PKR activator RAX. J. Biol. Chem. 2001;276:11754–11758. doi: 10.1074/jbc.M011400200. [DOI] [PubMed] [Google Scholar]
- Saltiel A. R., Kahn C. R. Insulin signalling and the regulation of glucose and lipid metabolism. Nature. 2001;414:799–806. doi: 10.1038/414799a. [DOI] [PubMed] [Google Scholar]
- Samuel V. T., et al. Targeting foxo1 in mice using antisense oligonucleotide improves hepatic and peripheral insulin action. Diabetes. 2006;55:2042–2050. doi: 10.2337/db05-0705. [DOI] [PubMed] [Google Scholar]
- Shim J. H., Lee H. K., Chang E. J., Chae W. J., Han J. H., Han D. J., Morio T., Yang J. J., Bothwell A., Lee S. K. Immunosuppressive effects of tautomycetin in vivo and in vitro via T cell-specific apoptosis induction. Proc. Natl. Acad. Sci. USA. 2002;99:10617–10622. doi: 10.1073/pnas.162522099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skolnik E. Y., et al. The SH2/SH3 domain-containing protein GRB2 interacts with tyrosine-phosphorylated IRS1 and Shc: implications for insulin control of ras signalling. EMBO J. 1993;12:1929–1936. doi: 10.1002/j.1460-2075.1993.tb05842.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sone H., Suzuki H., Takahashi A., Yamada N. Disease model: hyperinsulinemia and insulin resistance. Part A-targeted disruption of insulin signaling or glucose transport. Trends Mol. Med. 2001;7:320–322. doi: 10.1016/s1471-4914(01)02041-x. [DOI] [PubMed] [Google Scholar]
- Standaert M. L., Galloway L., Karnam P., Bandyopadhyay G., Moscat J., Farese R. V. Protein kinase C-zeta as a downstream effector of phosphatidylinositol 3-kinase during insulin stimulation in rat adipocytes. Potential role in glucose transport. J. Biol. Chem. 1997;272:30075–30082. doi: 10.1074/jbc.272.48.30075. [DOI] [PubMed] [Google Scholar]
- Tamemoto H., et al. Insulin resistance and growth retardation in mice lacking insulin receptor substrate-1. Nature. 1994;372:182–186. doi: 10.1038/372182a0. [DOI] [PubMed] [Google Scholar]
- Taniguchi C. M., Ueki K., Kahn R. Complementary roles of IRS-1 and IRS-2 in the hepatic regulation of metabolism. J. Clin. Invest. 2005;115:718–727. doi: 10.1172/JCI23187. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Taylor S. I., Arioglu E. Syndromes associated with insulin resistance and acanthosis nigricans. J. Basic Clin. Physiol. Pharmacol. 1998;9:419–439. doi: 10.1515/jbcpp.1998.9.2-4.419. [DOI] [PubMed] [Google Scholar]
- Taylor S. S., Haste N. M., Ghosh G. PKR and eIF2alpha: integration of kinase dimerization, activation, and substrate docking. Cell. 2005;122:823–825. doi: 10.1016/j.cell.2005.09.007. [DOI] [PubMed] [Google Scholar]
- Thirone A. C., Huang C., Klip A. Tissue-specific roles of IRS proteins in insulin signaling and glucose transport. Trends Endocrinol. Metab. 2006;17:72–78. doi: 10.1016/j.tem.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Tremblay F., Marette A. Amino acid and insulin signaling via the mTOR/p70 S6 kinase pathway. A negative feedback mechanism leading to insulin resistance in skeletal muscle cells. J. Biol. Chem. 2001;276:38052–38060. doi: 10.1074/jbc.M106703200. [DOI] [PubMed] [Google Scholar]
- White M. F. The IRS-signalling system: a network of docking proteins that mediate insulin action. Mol. Cell Biochem. 1998;182:3–11. [PubMed] [Google Scholar]
- Williams B. R. Signal integration via PKR. Sci. STKE. 2001;2001:RE2. doi: 10.1126/stke.2001.89.re2. [DOI] [PubMed] [Google Scholar]
- Withers D. J., et al. Disruption of IRS-2 causes type 2 diabetes in mice. Nature. 1998;391:900–904. doi: 10.1038/36116. [DOI] [PubMed] [Google Scholar]
- Woldehawariat G., Nekhai S., Petryshyn R. Differential phosphorylation of PKR associates with deregulation of eIF-2alpha phosphorylation and altered growth characteristics in 3T3–F442A fibroblasts. Mol. Cell Biochem. 1999;198:7–17. doi: 10.1023/a:1006978622240. [DOI] [PubMed] [Google Scholar]
- Wu M., Yang X., Chan C. A dynamic analysis of IRS-PKR signaling in liver cells: a discrete modeling approach. PLoS One. 2009;4:e8040. doi: 10.1371/journal.pone.0008040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Williams B. R. The B56alpha regulatory subunit of protein phosphatase 2A is a target for regulation by double-stranded RNA-dependent protein kinase PKR. Mol. Cell. Biol. 2000;20:5285–5299. doi: 10.1128/mcb.20.14.5285-5299.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L., Lavin V. A., Moser L. R., Cui Q., Kanies C., Yang E. PP2A regulates the pro-apoptotic activity of FOXO1. J. Biol. Chem. 2008;283:7411–7420. doi: 10.1074/jbc.M708083200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X. B., Battaglia S., Boucreux D., Chen Z., Brechot C., Pavio N. Mapping of the interacting domains of hepatitis C virus core protein and the double-stranded RNA-activated protein kinase PKR. Virus Res. 2007;125:79–87. doi: 10.1016/j.virusres.2006.12.010. [DOI] [PubMed] [Google Scholar]
- Yang X., Chan C. Repression of PKR mediates palmitate-induced apoptosis in HepG2 cells through regulation of Bcl-2. Cell Res. 2009;19:469–486. doi: 10.1038/cr.2009.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Ou J., Bashmakov Y., Horton J. D., Brown M. S., Goldstein J. L. Insulin inhibits transcription of IRS-2 gene in rat liver through an insulin response element (IRE) that resembles IREs of other insulin-repressed genes. Proc. Natl. Acad. Sci. USA. 2001;98:3756–3761. doi: 10.1073/pnas.071054598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P., Jacobs B. L., Samuel C. E. Loss of protein kinase PKR expression in human HeLa cells complements the vaccinia virus E3L deletion mutant phenotype by restoration of viral protein synthesis. J. Virol. 2008;82:840–848. doi: 10.1128/JVI.01891-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H. R., Lau A. S., Pestka J. J. Role of double-stranded RNA-activated protein kinase R (PKR) in deoxynivalenol-induced ribotoxic stress response. Toxicol. Sci. 2003;74:335–344. doi: 10.1093/toxsci/kfg148. [DOI] [PubMed] [Google Scholar]
- Zick Y. Ser/Thr phosphorylation of IRS proteins: a molecular basis for insulin resistance. Sci. STKE. 2005;2005:pe4. doi: 10.1126/stke.2682005pe4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.