Abstract
Trafficking of AMPA receptors is important for many forms of synaptic plasticity. However, the link between activity and resulting synaptic alterations is not fully understood. Here, we identified a direct interaction between NSF, an ATPase involved in membrane fusion events and stabilization of surface AMPARs, and Plk2, an activity-inducible kinase that homeostatically decreases excitatory synapse number and strength. Plk2 disrupted interaction of NSF with the GluA2 subunit of AMPARs, promoting extensive loss of surface GluA2 in rat hippocampal neurons, greater association of GluA2 with adapter proteins PICK1 and GRIP1, and decreased synaptic AMPAR current. Plk2 engagement of NSF, but not Plk2 kinase activity, was required for this mechanism and occurred through a novel motif within Plk2 independent from canonical polo box interaction sites. These data reveal that heightened synaptic activity, acting through Plk2, leads to homeostatic decreases in surface AMPAR expression via the direct dissociation of NSF from GluA2.
Keywords: Plk2, SNK, NSF, GluA2, GluR2, endocytosis, GluA1, GluR1, homeostatic plasticity, polo kinase
Experience-dependent synaptic plasticity is often expressed by changes in synaptic content of AMPA receptors (AMPARs), heteromeric fast excitatory channels composed of combinations of GluA1–4 subunits. Accordingly, a wide variety of interacting proteins have been identified that affect AMPAR trafficking1. For instance, N-ethylmaleimide-sensitive fusion protein (NSF), a hexameric ATPase universally involved in membrane fusion events2, interacts specifically with GluA23-6, influencing its membrane cycling6-8. Interference with NSF-GluA2 binding results in rapid rundown of AMPAR-mediated currents3,4,6 that occludes long-term depression (LTD)6,9,10, a major mechanism of synaptic weakening. Thus, NSF is important for stabilization of surface AMPARs11. However, the precise functions of NSF and the physiological pathways that regulate its binding to GluA2 are unclear.
Polo-like kinase 2 (Plk2, also called serum-inducible kinase), a member of the polo family of serine/threonine kinases12, is rapidly induced in brain following intense synaptic activity13. Newly synthesized Plk2 preferentially distributes to proximal dendrites and triggers degradation of the dendritic spine-promoting factor SPAR14 through a phosphorylation-dependent ubiquitin-proteasome pathway15,16, causing spine shrinkage and loss of excitatory synapses15. Furthermore, elimination of SPAR by Plk2 is critical for certain forms of homeostatic synaptic downscaling in response to hyperexcitation17,18. Thus, elevated synaptic activity induces Plk2 and leads to homeostatic negative feedback regulation of both synapse morphology and strength.
Here, we show that Plk2 directly interacts with NSF and causes its dissociation from GluA2, resulting in intracellular sequestration of GluA2 in complex with adapter proteins, reduced AMPAR transmission, and increased ratio of surface GluA1 to GluA2 subunits. This mode of regulation uniquely utilizes a kinase-independent mechanism and is mediated by a novel motif in Plk2 distinct from classical polo kinase substrate targeting domains. These results suggest the NSF-Plk2 connection comprises a homeostatic sensor to directly link synaptic overactivity and subsequent downregulation in synaptic strength.
Results
Plk2 target identification
We conducted an affinity chromatography screen using glutathione-S-transferase (GST) fused to the C-terminal half of Plk2 (Plk2c), a region containing the conserved polo box domain (PBD) (Fig. 1a) considered essential for polo-like kinase substrate interactions19. Rat brain lysates were applied to a column of GST-Plk2c or control GST. After extensive washing and elution, proteins recovered in GST-Plk2c eluates were subjected to mass spectrometry. Strikingly, this analysis identified several proteins involved in endocytosis (Supplementary Table 1), suggesting Plk2 may regulate postsynaptic membrane dynamics.
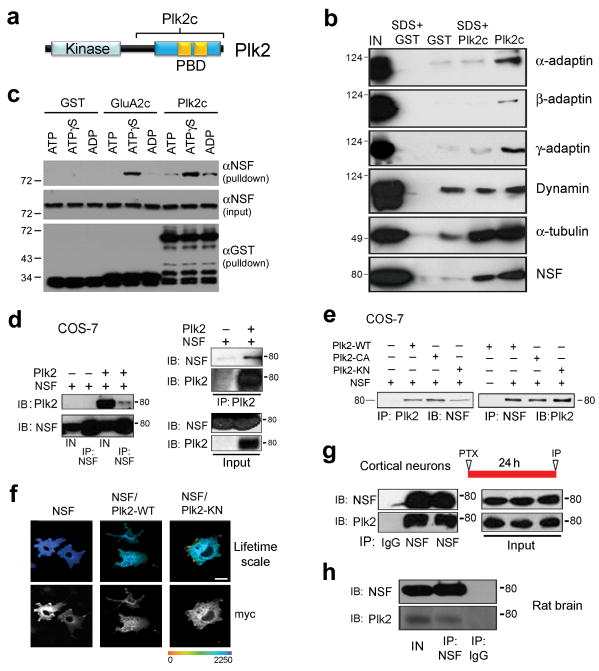
Figure 1. Interaction of Plk2 and NSF.
(a) Proteins were recovered from rat brain extracts by GST or GST-Plk2c pulldown, in the presence or absence of brief 1% SDS treatment, and immunoblotted as indicated. (b) Schematic representation of Plk2 domain structure. PBD, polo box domain. Plk2c, C-terminal fragment used for GST pulldown. Tandem polo boxes within PBD are shown in gold. (c) GST pulldown assays following incubation of purified GST-Plk2c or GST-GluA2c with His6-NSF, in the presence of ATP, ADP or ATP-γ-S, and immunoblotted as indicated. (d) COS-7 cells were transfected as indicated at top of blots, and lysates immunoprecipitated (IP) and analyzed by immunoblotting (IB) for proteins as indicated. (e) IP and IB were performed on cell lysates of COS-7 cells transfected with NSF and Plk2 wild-type (WT), constitutively active (CA), or kinase negative (KN) as shown. (f) FLIM analysis of interaction between NSF and Plk2. Myc-tagged NSF and HA-tagged Plk2 were transfected into COS-7 cells and labeled by indirect immunocytochemistry using mouse myc and rabbit HA antibodies, followed by secondary Alexa488 anti-mouse (donor fluorophore) and Cy3-anti-rabbit (acceptor fluorophore). Lifetime range shown in pseudocolor. Scale bar, 10 μm. (g) Interaction of endogenous NSF and Plk2. Lysates from cultured cortical neurons, stimulated with picrotoxin (PTX) for 24 h, were used for IP and IB as indicated. (h) Co-immunoprecipitation of endogenous NSF and Plk2 from rat brain lysates. IP and IB were performed for proteins as indicated. IN, Input, 5% lysate used for IPs. MW shown in kDa. Full-length blots are presented in Supplementary Fig. 9.
For target validation, we performed additional GST-Plk2c pulldowns followed by immunoblotting for candidate interactors. Alpha, beta, and gamma adaptins, components of AP1 and AP2 clathrin adapter complexes, as well as NSF, were recovered by GST-Plk2c but not GST (Fig. 1b). As positive controls, known Plk2-interacting proteins tubulin20 and SPAR15 were specifically retained by GST-Plk2c (Fig. 1b and Supplementary Fig. 1). Dynamin, however, bound equally well to GST and to GST-Plk2c and was not examined further. NSF and tubulin were largely retained even under stringent pulldown conditions incorporating brief treatment with the strong detergent SDS, whereas other candidate interactors were lost (Fig. 1b). Thus, the NSF-Plk2 interaction was specific and robust.
NSF and Plk2 bind directly in an ATP-dependent manner
To test for direct interaction, we conducted in vitro binding assays with purified hexahistidine-tagged NSF (His6-NSF) and GST, GST-Plk2c, or GST fused to the GluA2 C-terminus (GST-GluA2c). Because NSF interactions are often ATP regulated5, binding assays were performed in the presence of ADP, ATP or the non-hydrolyzable analog ATPγS. By GST-pulldown, we observed GST-Plk2c and His6-NSF bind in this completely defined system (Fig. 1c), indicating direct contact. Furthermore, Plk2c and NSF bound most strongly in the presence of ATPγS, similar to the interaction between NSF and GST-GluA2c5 (Fig. 1c). However, neither Plk2c nor GluA2c affected NSF intrinsic ATPase activity (Supplementary Fig. 2).
When coexpressed in COS-7 cells, the full-length NSF and Plk2 proteins could be bidirectionally coimmunoprecipitated (Fig. 1d). Kinase-negative, constitutively active, and wild-type Plk2 bound to NSF equivalently (Fig. 1e), suggesting Plk2 kinase activity does not regulate this interaction. Fluorescence lifetime microscopy, a technique for reporting molecular proximity (<10 nm) between fluorescently tagged proteins (see Methods), also demonstrated significant association of Plk2 and kinase-dead Plk2 with NSF (Fig. 1f and Supplementary Table 2).
To analyze the interaction of endogenous NSF and Plk2, we used cultured cortical neurons (18–21 days in vitro (DIV)). Because basal Plk2 expression is low, we stimulated neuronal activity with the GABAA receptor antagonist picrotoxin (PTX, 100 μM, 24 h), shown previously to robustly induce Plk2 expression15,17. Following stimulation, NSF antibodies, but not control IgG, recovered abundant Plk2 from neuronal lysates (Fig. 1g). These results indicated that Plk2 and NSF strongly interacted in neurons following chronic stimulation of synaptic activity. Lastly, NSF and Plk2 could also be coimmunoprecipitated from rat brain lysate (Fig. 1h), demonstrating that native Plk2 and NSF bound in vivo.
Plk2 exchanges GluA2-bound NSF for PICK1/GRIP1
An important question was whether Plk2 regulated NSF interaction with GluA2. We first verified that NSF and GluA2 could be coimmunoprecipitated in both directions when expressed by themselves in COS-7 cells (Fig. 2a). However, coexpression of Plk2 completely prevented NSF and GluA2 binding, with concomitant association of Plk2 with NSF (Fig. 2a). No binding of Plk2 was observed with GluA2 (Fig. 2a; also see 2d,e). Therefore, Plk2 and GluA2 bound NSF in a mutually exclusive fashion, with Plk2 being dominant. Interestingly, Plk2 coprecipitation with NSF was significantly enhanced with GluA2 coexpression (Fig. 2b), suggesting Plk2 has higher affinity for NSF prebound to GluA2.
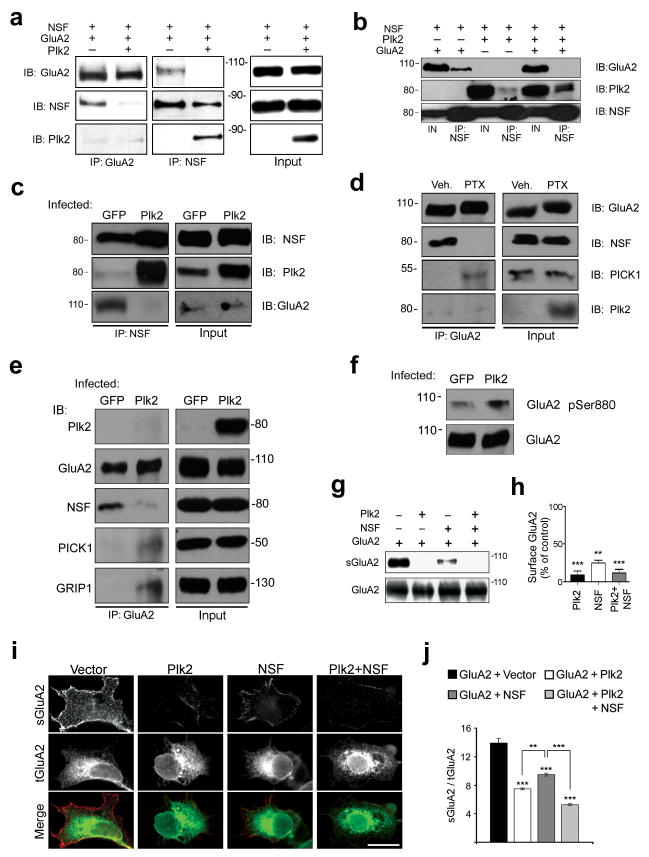
Figure 2. Plk2 binding disrupts NSF association with GluA2.
(a, b) Lysates from COS-7 cells transfected as shown at top were analyzed by IP and IB for proteins as indicated. For (b), densitometry of Plk2 associated with NSF plus GluA2: 3.45±0.34 (arbitrary units); minus GluA2: 1.57±0.15; p=0.03. (c) Lysates from cultured hippocampal neurons, infected for 18–20 h with Sindbis virus expressing Plk2 or GFP, were subjected to IP and IB as indicated. (d) Cultured hippocampal neurons were treated with PTX or vehicle (Veh.) for 24 h, and cell lysates used for IP and IB as indicated. (e) Lysates from cultured hippocampal neurons, infected for 18-20 h with Sind-Plk2 or -GFP, were subjected to IP and IB as indicated. Full-length blots are presented in Supplementary Fig. 9. All MW in kDa. IN, Input, 5% lysate used for IPs. (f) Lysates of cultured hippocampal neurons infected with Sind-GFP or -Plk2 for 18–20 h were immunoblotted as indicated. Densitometry of phospho-ser880 with GFP: 0.25±0.09 (arbitrary units); with Plk2: 0.89±0.16; p=0.008. (g) COS-7 cells were transfected as shown at top, and cell surface proteins were biotin-labeled, isolated with avidin-beads, and immunoblotted for total and surface GluA2 (sGluA2). (h) Quantification of data in (g); surface GluA2 was normalized to total GluA2, then plotted as percent of control. (i) HEK cells were transfected as indicated across top row. Cells were immunostained for sGluA2 (top row, red) and total GluA2 (tGluA2, middle row, green); overlay is shown in yellow (bottom row). Scale bar, 10 μm. (j) Quantification of surface to total GluA2 ratio from (i). **p<0.01; ***p<0.001.
To ask if Plk2 could disrupt native GluA2-NSF complexes, we infected hippocampal cultures (18–21 DIV) at saturating titers with Sindbis virus expressing Plk2 (Sind-Plk2) or control GFP (Sind-GFP). In GFP-expressing neurons, NSF coimmunoprecipitated readily with GluA2, but modestly with the limited pool of basal Plk2 (Fig. 2c). In contrast, NSF interaction with GluA2 was abolished in Sind-Plk2-infected neurons, in exchange for strong association with overexpressed Plk2 (Fig. 2c). Thus, Plk2 attachment to NSF efficiently displaced the interaction of NSF with GluA2 in neurons. Remarkably, PTX induction of Plk2 also completely disrupted NSF-GluA2 interaction, while association of GluA2 and the adapter PICK1 markedly increased in the same stimulated cultures (Fig. 2d).
The fate of GluA2 dissociated from NSF was further probed using hippocampal cultures infected with Sind-Plk2 or Sind-GFP and immunoprecipitated for GluA2. Accompanying Plk2-induced disruption of GluA2 and NSF binding, GluA2 association was dramatically increased with both PICK1 and GRIP1 (Fig. 2e), adapter proteins implicated in binding GluA2 intracellularly10,21,22. Additionally, Plk2 significantly enhanced GluA2 serine 880 phosphorylation (Fig. 2f), a PKC-dependent biochemical signature of PICK1 binding, GluA2 endocytosis, and LTD10,23 (but see24). As total GluA2 levels always remained constant (Fig. 2c–f), these data suggest disruption of GluA2-NSF binding by Plk2 promotes redistribution of GluA2 among adapter proteins and intracellular sequestration.
Plk2 reduces surface GluA2 expression via NSF
We next used surface biotinylation assays to assess the effect of Plk2 on GluA2 surface expression in COS-7 cells. As expected, Plk2 (or Plk2 and NSF together) caused a large decrease in surface levels of GluA2 (sGluA2) compared to GluA2 transfected alone (Fig. 2g,h). The effect of Plk2 by itself on sGluA2 was likely mediated through endogenous NSF present in COS-7 cells. Similar results were obtained by surface immunostaining of GluA2 expressed in HEK cells (with cotransfected Plk2 and/or NSF) using an extracellular epitope-directed GluA2 antibody under non-permeabilizing conditions (Fig. 2i,j). Unexpectedly, NSF itself decreased sGluA2 in heterologous cells (Fig. 2g–j) and in NSF-overexpressing hippocampal neurons (Supplementary Fig. 3a), albeit less potently than Plk2, possibly due to excess intracellular NSF impeding GluA2 membrane trafficking to the cell surface.
To examine the effect of Plk2 on endogenous GluA2 surface expression, we infected primary hippocampal cultured neurons (18–21 DIV) with Sind-Plk2 and live-labeled for sGluA2 with N-terminal antibodies. Steady-state sGluA2 was profoundly decreased in secondary dendrites by Sind-Plk2 compared to Sind-GFP (Fig. 3a,c) whereas surface GluA1 (sGluA1) levels were only modestly decreased (Fig. 3a,d). No change was observed in total GluA1 or GluA2 under any conditions (Fig. 3b,c,d), suggesting an alteration in receptor trafficking rather than metabolism. NSF levels and distribution were also unchanged (Supplementary Fig. 3b,c), consistent with the lack of degradation of NSF by Plk2 in COS-7 cells and neuronal lysates. We observed an even greater effect of Plk2 on neuronal cell bodies that displayed virtually undetectable levels of sGluA2, whereas somatic sGluA1 trended to a slight decrease by Plk2 that did not reach significance (Fig. 3e). Thus, Plk2 can specifically and potently remove all surface GluA2-containing receptors.
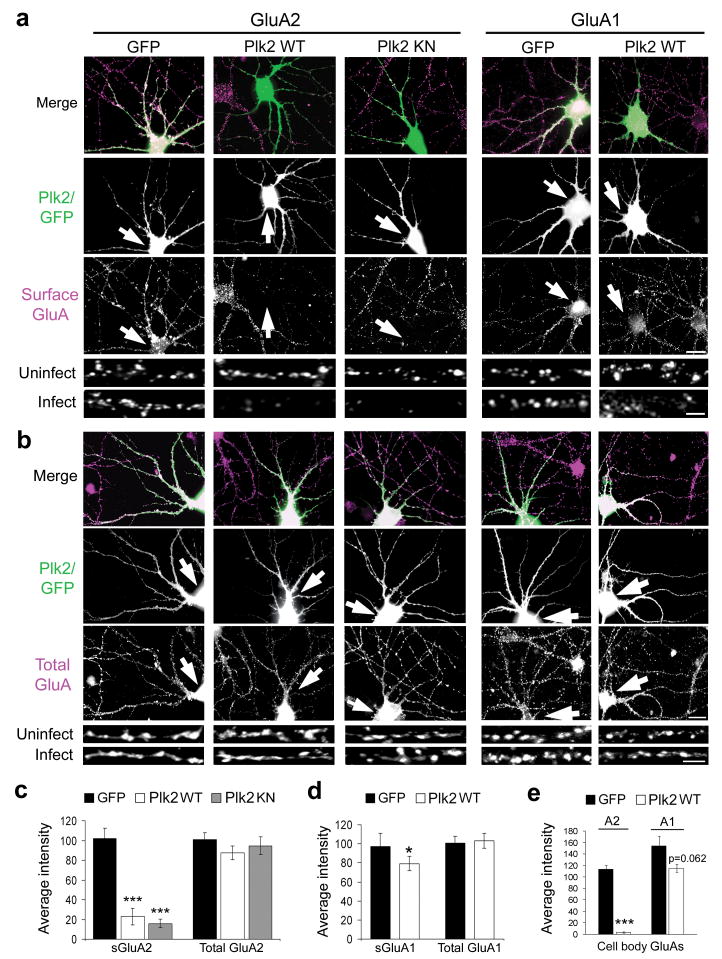
Figure 3. Plk2 decreases neuronal surface GluA2.
(a, b) Hippocampal cultured neurons were infected with Sind-Plk2 (WT or KN) or Sind-GFP, as indicated at top. Immunostaining is shown for exogenous protein (green) and either surface GluA2, surface GluA1, total GluA2, or total GluA1 (violet) as indicated. Colocalization appears white in merged images (upper rows). Higher magnification views of representative dendrites are shown below for infected and nearby uninfected control neurons. Arrows indicate infected neurons. (c, d) Quantification of results from (a, b). Average immunofluorescence intensity for surface and total GluA2 (c) and for GluA1 (d); N = 40–50 neurons per condition. (e) Quantification of surface GluA2 and GluA1 immunofluorescence intensity measured on the cell body for same neurons as above. ***p<0.001, *p<0.05. Scale bars, 20 μm (wide view), 5 μm (magnified images).
We previously reported that Plk2 causes synapse elimination, primarily within proximal dendrites15. Although the preferential effects on sGluA2 vs. sGluA1 were unlikely to be secondary to synapse loss, we analyzed PSD-95 puncta in Plk2-infected neurons and found no significant changes in secondary dendrites where we observed strong depletion of sGluA2 (Supplementary Fig. 4a,b). These data suggested loss of surface AMPARs with otherwise intact postsynaptic sites. Furthermore, we confirmed removal of PSD-95 immunofluorescence in proximal primary dendrites due to Sind-Plk2 (Supplementary Fig. 4a), as previously shown15. Loss of proximal dendritic PSD-95 required Plk2 kinase activity, as we found significantly increased proximal PSD-95 levels with expression of Sind-Plk2-KN (Supplementary Fig. 4a,b), a construct known to exert dominant negative effects15. Surprisingly, Plk2 reduction of sGluA2 did not require kinase activity and occurred equally with both Plk2 and Plk2-KN (Fig. 3a,c). Thus, the actions of Plk2 on synapse elimination and GluA2 are mechanistically dissociable by dendritic locus and kinase dependence.
To ask whether endogenous Plk2 regulates sGluA2 levels, we used PTX stimulation to induce Plk2 expression. We observed that basal Plk2 levels as well as Plk2 induction displayed considerable cell-to-cell variation, likely as a function of heterogeneity of synaptic input in dissociated cell culture. Nevertheless, the level of sGluA2 in secondary dendrites was, on average, significantly decreased after PTX treatment (Fig. 4a,b). Quantifying Plk2 expression together with sGluA2 intensity in individual neurons revealed a strong negative correlation (Fig. 4c), suggesting that endogenous Plk2 downregulates sGluA2 in a dose-dependent manner. However, a ceiling effect could be seen in the flattening of the best-fit line for induced cultures, approaching a maximal value of ∼75% loss of sGluA2 (Fig. 4c), similar to values obtained with Plk2 overexpression. There was also significant but more modest loss of sGluA1 with PTX stimulation in secondary dendrites (Fig. 4a,b), but no change in total GluA2 (Fig. 4a,b) or PSD-95 (Supplementary Fig. 4c,d).
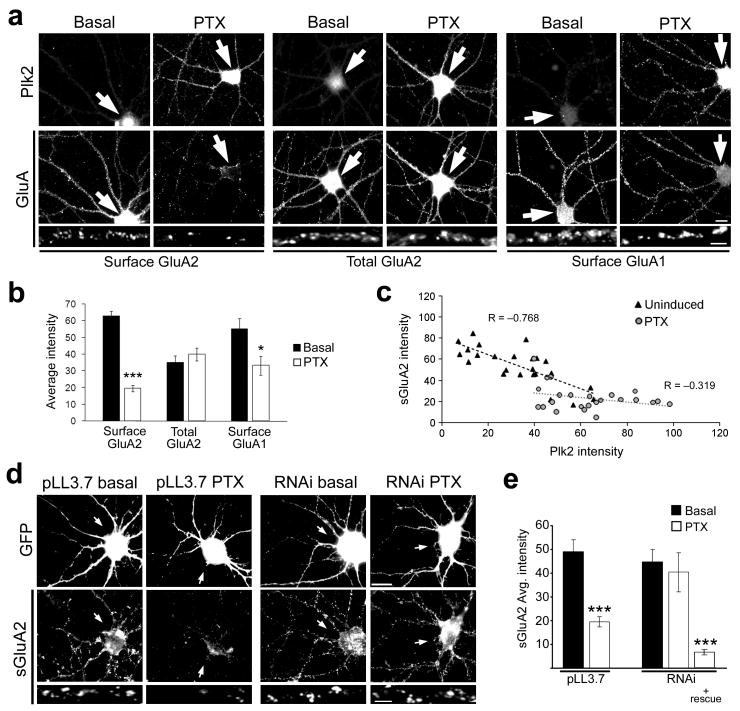
Figure 4. Plk2 induction is required for activity-dependent decreases in surface GluA2.
(a) Immunolabeling as indicated for endogenous Plk2 and surface or total GluAs in cultured hippocampal neurons under basal conditions and after stimulation with picrotoxin (PTX) for 24 h. Higher magnification views of representative dendrites are shown below. Arrows indicate cell body of neuron shown. (b) Quantification of data in (a). ***p<0.001, *p<0.05; N=25 neurons per condition. (c) Inverse correlation between Plk2 expression and surface GluA2 levels in individual cells under basal and PTX induced conditions. (d) Hippocampal neurons were transfected with pLL3.7 empty vector or Plk2 RNAi as indicated, along with pEGFP to mark transfected cells. Neurons were treated with PTX or vehicle (basal) and immunostained for GFP and sGluA2 as indicated. Arrows indicate transfected neuron cell body. (e) Quantification of data from (d). N=10–15 neurons per condition; ***p<0.001. Scale bars, 10 μm (wide view), 5 μm (magnified images).
To perturb Plk2 function in this process, we developed an RNA interference (RNAi) construct that decreased basal Plk2 immunoreactivity in neurons and prevented Plk2 induction by PTX, and also efficiently knocked down Plk2 expression in heterologous cells (Supplementary Fig. 5). In vector control-transfected neurons, sGluA2 immunofluorescence was again strongly reduced with PTX treatment (Fig. 4d,e). However, PTX-induced loss of sGluA2 was fully blocked with Plk2 knockdown (Fig. 4d,e) and restored by cotransfection of a Plk2 rescue construct (Fig. 4e) insensitive to the RNAi (Supplementary Fig. 5b,c). Combined with earlier data, we conclude that Plk2 is both necessary and sufficient for overexcitation-induced downregulation of sGluA2.
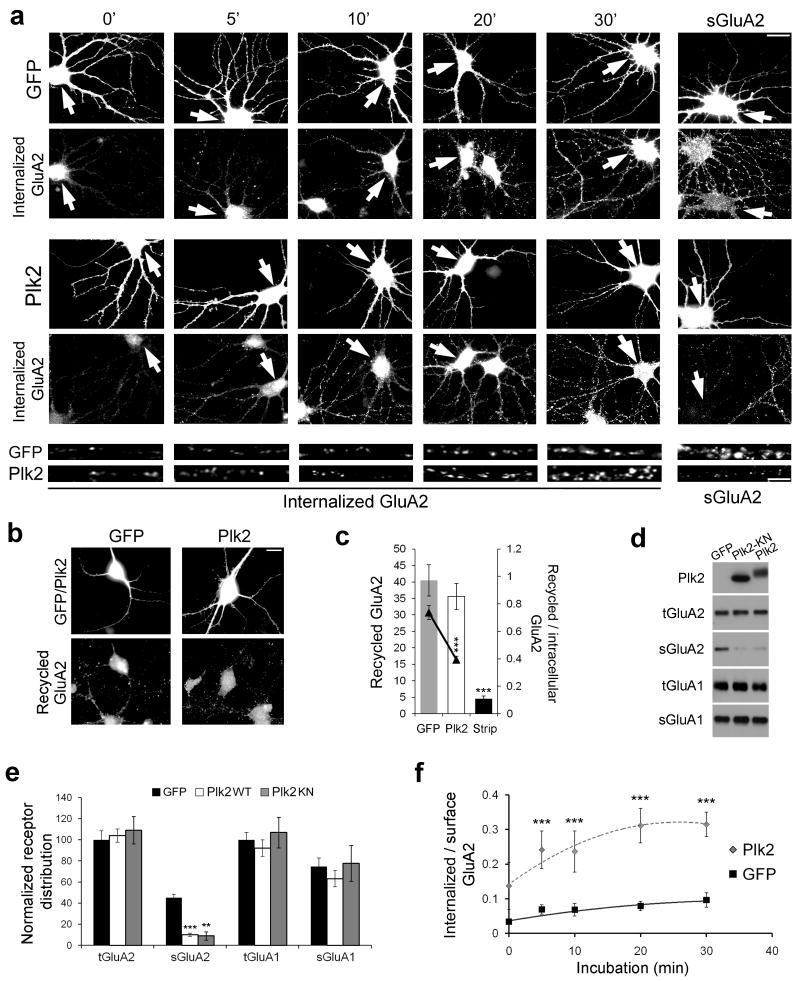
Decreased surface expression of GluA2 could result from impaired cell surface delivery, increased internalization, or both. To distinguish these possibilities, we live-labeled sGluA2 and conducted internalization assays to analyze endocytosis of GluA2-containing AMPARs, as well as recycling assays to measure their rate of return to the cell surface (see Methods). Plk2 slightly increased the absolute rate of GluA2 internalization (Fig. 5a; at 30 min, GFP=10.3±2.1, Plk2=12.2±2.2; p=0.35) and decreased its recycling (Fig. 5b,c; GFP=40.5±4.7, Plk2=35.6±4.0, p=0.19), although neither effect was significant. However, these data did not account for the greatly reduced pool of available surface GluA2 and consequently increased pool of intracellular GluA2 in Plk2-expressing cells, as shown by both surface staining and biotinylation (Fig. 5a,d,e). When expressed as the ratio of internalized to available surface receptors, the rate of sGluA2 internalization per receptor was significantly increased in Sind-Plk2 infected neurons vs. Sind-GFP at all time points examined (Fig. 5f). Similarly, Plk2 significantly decreased the rate of sGluA2 recycling per available intracellular receptor (Fig. 5c). Thus, Plk2 increases the fraction of surface GluA2 receptors undergoing endocytosis, while decreasing the fraction subsequently reinserted.
Figure 5. Plk2 controls GluA2 endocytosis and recycling.
(a) Time-course of GluA2 internalization following 18–20 h expression of Sindbis virus-expressed GFP or Plk2. Immunolabeling of infected neurons (GFP or Plk2) and internalized GluA2 or steady-state sGluA2 are indicated. Bottom, higher magnification views of representative dendrites for Sind-Plk2- or Sind-GFP-infected neurons. Arrows indicate infected neurons. Scale bars, 10 μm (wide view), 5 μm (magnified). (b) GluA2 recycling shown 30 min after acid strip of remaining surface labeling. Immunolabeling of infected cells (GFP or Plk2, top row) and recycled GluA2 (bottom row) are shown. (c) Quantification of (b); control with acid strip and no incubation time to allow for recycling (‘Strip’) demonstrates successful stripping of remaining surface primary antibody. Ratio of recycled receptor to available intracellular receptor is shown by line graph on secondary axis. (d) Surface biotinylation of cultured hippocampal neurons. Sister cultures were infected with Sind-GFP, Sind-Plk2-KN, or Sind-Plk2 for 20 h and cell surface proteins were biotin-labeled, isolated with avidin-beads, and immunoblotted for total and surface GluA2 (tGluA2 and sGluA2), total and surface GluA1 (tGluA1 and sGluA1), and Plk2 as indicated. No Plk2 was detected in surface fractions (not shown). Both Plk2-KN and Plk2 significantly reduced sGluA2. (e) Quantification of data from (d), normalized to total receptor expression in GFP control cells. (f) Quantification of (a) plotted as ratio of internalized to available surface GluA2 immunofluorescence; N=18–25 neurons per condition. **p<0.01; ***p<0.001.
To test the involvement of NSF in Plk2-mediated loss of sGluA2, we performed occlusion experiments using N-ethylmalemide (NEM), a thiol-reactive compound that inhibits NSF function. NEM greatly reduced sGluA2, quantitatively resembling the effect of Plk2; however, in Plk2-infected neurons, NEM treatment was largely occluded (Supplementary Fig. 6). Because NEM does not act specifically against NSF, we also used previously characterized peptide inhibitors of NSF (Fig. 6a–c). Pep2m contains the NSF binding site on the GluA2 C-terminus and inhibits NSF-GluA2 interaction, causing loss of surface AMPARs and rundown of AMPAR-mediated mEPSCs3,4,6,11. However, pep2m also blocks GluA2 binding to AP225. In contrast, pepR845A (pepRA) specifically inhibits NSF binding to GluA2 without affecting AP2, while control peptide pepK844A (pepKA) does not interfere with either NSF or AP225. As with NEM, pepRA dramatically lowered sGluA2, and coexpression of pepRA and Plk2 caused no further effect than Plk2 or pepRA alone (Fig. 6a,b); similar results were observed for pep2m (not shown; quantified in Fig. 6c). The pepKA control did not affect GluA2 surface expression or removal of sGluA2 by Plk2 (Fig. 6a,b). Occlusion of the effects of Plk2 by pep2m and pepRA further supports the idea that Plk2 decreases sGluA2 through NSF.
Figure 6. Plk2 effects on surface GluA2 are occluded by NSF disruption.
(a) Cultured hippocampal neurons were transfected with GFP or Plk2 as indicated and cotransfected with pepRA to block NSF-GluA2 interaction or control peptide pepKA. Cells were immunolabeled as indicated for sGluA2 (violet) and for GFP or Plk2 (green). Colocalization appears white in merged images (upper row). Arrows indicate transfected cells. Higher magnification views of representative dendrites are shown below wide view images. Scale bars, 10 μm (wide view), 5 μm (magnified images). (b) Quantification of data from (a) including data from pep2m expression. N=10–15 neurons per condition; ***p<0.001. (c) Amino acid sequences of peptides used.
NSF binds a novel Plk2 interaction motif
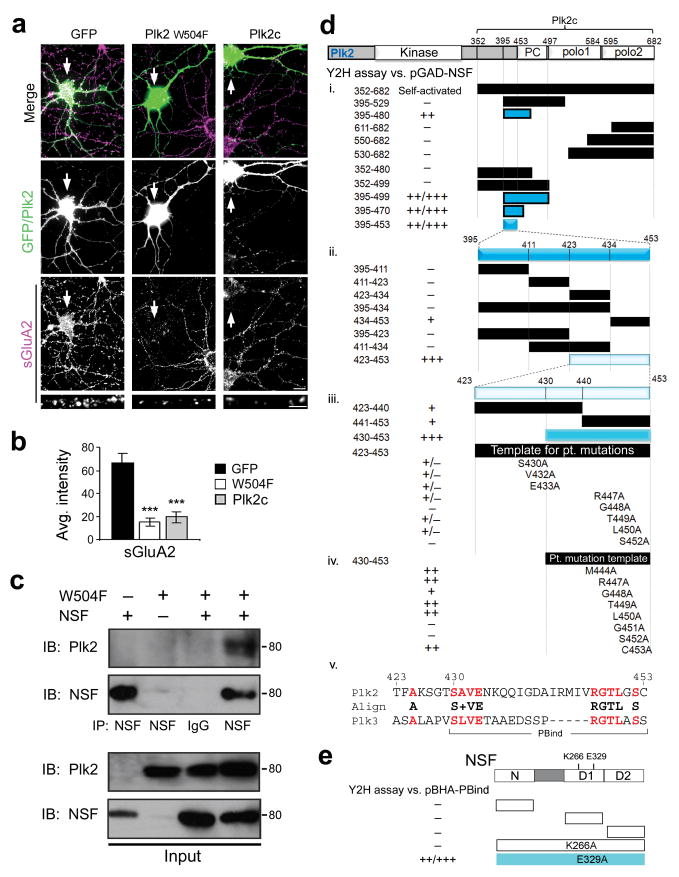
Based on Plk2's atypical kinase-independent regulation of sGluA2, we sought to characterize the NSF-Plk2 interaction in greater detail. First, we confirmed that the Plk2c C-terminal fragment, which lacks the kinase domain but contains the PBD and is sufficient to bind NSF (Fig. 1a–c), caused a reduction in sGluA2 immunofluorescence intensity similar to full-length Plk2 (Fig. 7a,b). In contrast, Plk2c had no effect on postsynapse number, as suggested by unchanged PSD-95 signal (Supplementary Fig. 7). Surprisingly, mutation of a critical residue (W504F) required for PBD-mediated interactions19,26, including Plk2 binding to SPAR17, failed to prevent co-immunoprecipitation with NSF from COS-7 cells (Fig. 7c). Furthermore, Plk2-W504F transfected into neurons decreased sGluA2 with similar potency to wild-type Plk2 (Fig. 7a,b). Thus, the ability of Plk2 to bind NSF and to regulate sGluA2 were both independent of the PBD, implying the existence of an unknown interaction site.
Figure 7. Plk2 binds NSF through a novel interaction motif.
(a) Cultured hippocampal neurons transfected with GFP, Plk2-W504F or Plk2c as indicated were immunostained for sGluA2 (bottom panels, violet) and for exogenous proteins (middle panels, green). Colocalization appears white in merged images (upper panels). Magnified views of representative dendrites shown at bottom. Arrows indicate transfected neurons. Scale bars, 10 μm (wide), 5 μm (magnified). (b) Quantification of data from (a); N=15–20 neurons per condition; ***p<0.001. (c) COS-7 cells transfected with NSF and/or Plk2 W504F were used for immunoprecipitation (IP) with NSF antibodies or control IgG and immunoblotted (IB) as indicated. Full-length blots are presented in Supplementary Fig. 9. (d) Deletion analysis of Plk2c-pBHA using yeast two hybrid (Y2H) assays against pGAD-NSF (i, ii). Strongly positive interactions, colored bars. Interaction strength is reported by X-gal reaction time (+++, <10 min; ++, 10–45 min; +, 45–75 min; +/−, 75–120 min; −, >120 min). Numbering represents amino acid residues. (iii, iv) Site-directed mutagenesis targeting conserved residues between Plk2 and Plk3 (bold red in panel v). (v) Amino acid sequence of the PBind site. (e) Y2H analysis of Plk2 binding site on NSF. PBind in pBHA was tested against pGAD constructs of individual NSF domains (N, D1 and D2 as shown) or point mutations in D1 ATPase domain (K266A or E329A) in full-length NSF.
To map this putative site, we conducted deletion analysis of the NSF-binding Plk2c region using yeast two-hybrid (Y2H) assays. PBD primary structure consists of two homologous polo boxes, along with a linker region (termed PC) between the kinase domain and the PBD (Fig. 7d). We tested NSF interaction with various fragments of the two polo boxes, the PC linker, or their flanking regions. Interestingly, NSF bound a region (aa395–480) near the N-terminal boundary of Plk2c but including neither polo box (Fig. 7di).
A series of additional deletions revealed a minimal 24-residue polypeptide of Plk2 (aa430–453) sufficient for binding NSF (Fig. 7dii,iii). This region included no portion of the PBD and thus represents an entirely new polo kinase interaction site; we have termed this element the polo box independent (PBind) site. Alignment of Plk2 and the highly related Plk3, which also binds NSF (data not shown), identified two clusters of conserved residues (Fig. 7dv). Site-directed mutagenesis of each conserved amino acid to alanine revealed that multiple residues contributed to NSF interaction (Fig. 7diii,iv).
Conversely, to explore the binding site for Plk2 on NSF, we used Y2H analysis of individual domains of NSF (N-domain and two homologous ATP binding domains D1 and D2) and discovered no single domain was sufficient for PBind interaction (Fig. 7e). Similarly, all NSF domains are required for binding to GluA23, suggesting the overall structure of NSF may be important for both Plk2 and GluA2 recognition. Additionally, we found that Plk2 interaction was abolished by an ATP-binding mutation of NSF (K266A) but unaffected by an ATPase-deficient mutation (E329A) (Fig. 7e). These results suggest that ATP binding, but not hydrolysis, is necessary for NSF to recruit Plk2.
Plk2 binding NSF is sufficient to remove synaptic AMPARs
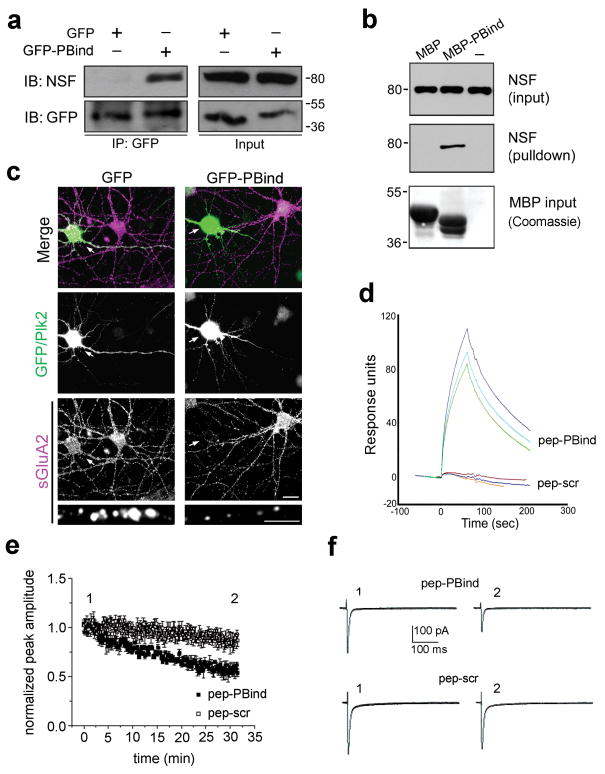
To confirm interaction of the minimal PBind motif with NSF, we showed that PBind fused to GFP could coimmunoprecipitate with NSF when coexpressed in COS-7 cells (Fig. 8a) and that a purified fusion protein of PBind linked to maltose binding protein (MBP-PBind) could pull-down native NSF from rat brain lysate (Fig. 8b). Furthermore, expression of GFP-PBind in cultured hippocampal neurons, but not GFP alone, fully mimicked the loss of sGluA2 immunofluorescence seen with intact Plk2 (Fig. 8c).
Figure 8. Plk2 binding NSF is sufficient for decreased surface GluA2.
(a) Lysates of COS-7 cells expressing NSF and GFP or GFP-PBind were subjected to immunoprecipitation (IP) with GFP antibodies and immunoblotted (IB) as indicated. Input, 5% of lysate for IP. Full-length blots are presented in Supplementary Fig. 9. (b) Pulldown of brain lysates with MBP or MBP-PBind and analyzed by IB for NSF and by Coomassie stain for MBP fusion proteins. Note MBP contains a 73aa polylinker C-terminal tail and therefore runs larger than MBP-PBind. (c) Cultured hippocampal neurons expressing GFP or GFP-PBind as indicated were immunostained for sGluA2 (bottom, violet) and GFP or Plk2 (middle, green). Arrows indicate transfected neurons. Colocalization appears white in merged images (top). Magnified views of representative dendrites at bottom. Scale bars, 10 μm (wide view), 5 μm (magnified). Surface GluA2 immunofluorescence intensities (in arbitrary units) were 82.8±7.3 for GFP, 18.6±3.3 for GFP-PBind; p=4.7×10-10; N=15–20 neurons. (d) Pep-PBind or scrambled control peptide (pep-scr) were analyzed for direct binding to His6-NSF by surface plasmon resonance. Averages of peak values were 89.4±7.6 response units for pep-PBind; -1.3±1.1 for pep-scr (negative value due to slight rundown during the experiment); p=0.0003, N=3). (e) Normalized AMPAR EPSCs peak amplitude vs time (pep-PBind, N=9, point 1 vs. point 2: p=0.0001; pep-scr, N=6, point 1 vs. point 2: p=0.17). (f) Representative EPSCs from individual neurons recorded at indicated times in the peak amplitude vs. time plot in the presence of intracellular pep-PBind (top) or pep-scr (bottom).
Finally, we generated a synthetic peptide comprising the PBind site (pep-pBind) to investigate the physiological effect of Plk2 binding NSF on synaptic AMPARs. We first verified via surface plasmon resonance that pep-PBind and purified His6-NSF bound directly, whereas a peptide with scrambled PBind sequence (pep-scr) showed no interaction (Fig. 8d). We then recorded synaptic AMPAR EPSCs from CA1 pyramidal neurons in acute hippocampal slices while electrically stimulating the Schaffer collateral input. We dialyzed pyramidal neurons with pep-PBind in the intracellular solution and observed a time-dependent decrease in EPSCs to ∼57% of its initial amplitude, while pep-scr caused no significant change in EPSC amplitude (Fig. 8e,f). These results strongly suggest that binding of NSF by the PBind site of Plk2 is sufficient to disrupt NSF-GluA2 interaction and cause profound and rapid loss of synaptic AMPARs.
Discussion
Homeostatic synaptic plasticity is thought to be important for stabilization of neuronal firing within an optimal operational range despite fluctuations in network activity27, but the mechanisms involved are not fully understood. Here we report that direct Plk2 binding to NSF was sufficient to cause its dissociation from GluA2. As Plk2 did not accelerate ATP hydrolysis by NSF, and NSF binding to Plk2 and GluA2 appeared comparable (Fig. 1c), we favor a model in which Plk2 exerts an allosteric conformational change in NSF that dislodges GluA2. The observed potency of such a noncatalytic mechanism may be explained in at least two ways: first, by the increase in Plk2 affinity for NSF bound to GluA2 (Fig. 2b). Second, stabilization of Plk2 binding to NSF by ATPγS, reminiscent of the GluA2-NSF interaction, suggests ATP-dependent cycling of Plk2 attachment to NSF could allow Plk2 to act processively on multiple NSF-GluA2 complexes.
A consequence of disrupting NSF-GluA2 interaction was the striking loss of surface GluA2 but not GluA1. Such preference mirrors the binding specificity of NSF for these subunits3-6,25, and is also consistent with the specific requirement for GluA2 in homeostatic synaptic scaling28. Furthermore, we found that all surface GluA2, dendritic as well as somatic, was subject to removal by Plk2. These results differ from previous reports blocking NSF-GluA2 interaction11,25. However, our study is the first to exploit an endogenous mechanism to perturb NSF-GluA2, which may exhibit different properties than nonphysiological competitive peptides.
Because the majority of AMPARs in hippocampus consists of GluA1/2 heteromers29,30, nearly complete depletion of sGluA2 with minimal effect on sGluA1 could be achieved by delivery of GluA1/3 heteromers or homomeric GluA1 to partly compensate for lost GluA2. Homomeric GluA1 insertion has been previously observed in various forms of synaptic plasticity31-33, in response to brain injury34, and in GluA2 knockout mice29. Extensive loss of sGluA2 by Plk2, as detected during homeostatic plasticity, suggests that GluA2-independent AMPAR trafficking35-37 may represent physiological mechanisms, rather than compensatory ones. Reduced GluA2 levels could contribute to decreased synaptic strength due to fewer synaptic receptors, but also increased calcium permeability, a property of GluA2-lacking receptors8,29,38. Both functions may be important for adaptation to hyperactivity; for instance, GluA2-lacking neurons undergo AMPAR internalization with more extensive spatial distribution37.
Plk2 caused rapid depletion of sGluA2 via heightened endocytosis and impaired receptor recycling, rather than degradation, since total GluA2 levels were insensitive to Plk2. These results unite previous reports that NSF constrains internalization7,25 but can promote receptor insertion39-43. NSF disassembles GluA2 from PICK144, a transport molecule that has also been implicated in both AMPAR endocytosis10,23,45,46 and surface delivery21. Our data support this dual role of NSF and PICK1 in AMPAR endocytosis/exocytosis; deprived of access to NSF by Plk2, GluA2 became trapped in greater complex with PICK1, which simultaneously promoted its endocytosis and hindered its release from intracellular retention. Plk2 also increased GluA2 phosphorylation at ser880, a PKC-dependent modification promoted by PICK110,23. Our results suggest ser880 phosphorylation correlates with removal of sGluA2, at least during homeostatic adaptation to overactivity, in contrast to a previous study that utilized primarily exogenous receptors and PKC stimulation24; such technical differences may explain these inconsistencies.
Interestingly, Plk2 dramatically increased GluA2 association with GRIP1 as well. GRIP/ABP family scaffolds have been proposed to tether AMPARs at synapses47 and intracellularly7,21. However, our data suggest the primary role of GRIP1 lies mainly at intracellular sites, although other GRIP/ABP family members may be more synaptically localized, possibly depending on palmitoylation state48. Because GluA2 ser880 phosphorylation blocks GRIP binding49, our data support the existence of two pools of intracellular GluA2, one tethered by GRIP1 and another associated with PICK1, which may reflect anchored and mobile pools, respectively21. The negligible amount of GluA2 associated with GRIP1 or PICK1 under basal conditions may explain the low level of AMPAR interaction of these adapters reported by some researchers50, and also suggests that GRIP1 and PICK1 do not constitutively retain large reserves of intracellular AMPARs until recruited by activity.
Another major finding of this work is the identification of a novel mode of polo-like kinase function. Plk2 has previously been implicated in homeostatic downscaling following overactivity via a mechanism critically dependent on PBD recognition and phosphorylation-induced degradation of SPAR17. In contrast, removal of sGluA2 by Plk2 was independent of both kinase activity and the classical PBD targeting domain of polo kinases. Disruption of NSF-GluA2 interaction, loss of sGluA2 and rapid rundown of AMPAR currents were caused by simple binding of NSF by the minimal 24-residue PBind region of Plk2. These findings uncover an additional regulatory mode that expands the repertoire by which polo-like kinases may exert influence over binding partners.
Although Plk2 kinase activity results in loss of proximal dendritic excitatory synapses15, the kinase-independent mechanism described here affected sGluA2 in secondary dendrites where postsynapse loss was not evident. It is currently unclear what mechanisms restrict synapse loss mainly to proximal dendrites; we speculate that competing phosphatase activity may exist that is overcome only in proximal dendrites where Plk2 expression is highest. Regardless, it is clear that two distinct homeostatic mechanisms are governed by Plk2 which may support complementary functions.
We propose NSF acts as a one-way “homeostatic gate” (Supplementary Fig. 8) that corrals sGluA2 by continual detachment of GluA2 from PICK1, dynamically retrieving GluA2 from endocytic zones as well as promoting its reinsertion from intracellular mobile pools. Activity-induced Plk2 engages and reverses this gating mechanism to allow graded endocytosis of sGluA2 while slowing down recycling, providing continuous homeostatic negative feedback in direct proportion to the level of activity recently experienced. Plk2-NSF interactions observed under basal conditions of synaptic activity in cultured hippocampal neurons and in brain further suggest this kinase-independent noncatalytic mechanism may operate over a fairly wide dynamic range and not only during conditions of overstimulation.
In the event of sustained hyperactivity, the Plk2 kinase-dependent pathway may be invoked to target postsynaptic substrates for degradation, leading to all-or-none loss of synapses. We envision that such a tiered response system, perhaps in concert with other described activity-inducible factors27, enables homeostatic negative feedback mechanisms to compensate for a continuum of synaptic input levels ranging from normal ongoing activity to excitotoxic insults.
Methods
DNA constructs
Myc or HA epitope-tagged Plk2, hyperactivating Plk2 (T236E), kinase-dead Plk2 (K108M), C-terminal Plk2 (aa395–682), polo domain mutant Plk2 (W504F), NSF, GluA2 and SPAR were expressed from pGW1-CMV. The following oligonucleotide (5′ to 3′) was inserted into pLL3.7 vector for Plk2-RNAi: GCATAGGGATCTCAAGCTA. For Plk2 rescue the following silent mutations were made (in bold) in full-length rat Plk2: GCATAGAGACCTCAAGCTA. For yeast-two-hybrid analysis, Plk2 fragments were cloned into pBHA and NSF into pGAD. Plk2 PBind was cloned into pEGFPC1 for GFP fusion protein and pMAL-c2x for MBP fusion. Site-directed mutagenesis was performed using QuikChange Kit (Stratagene) and confirmed by DNA sequencing.
Antibodies
Rabbit Plk215 and SPAR antibodies14 have been described. The following antibodies were purchased commercially: mouse NSF (Calbiochem), mouse GluA2 (BD Pharmingen), mouse and rabbit GFP (Qbiogene and Invitrogen), goat Plk2 (SNK C-18, Santa Cruz Biotechnology), rabbit GluA1 (Calbiochem and Millipore), mouse PSD-95 clone K28/43 (NeuroMab), rabbit GRIP1 (Millipore), mouse PICK1 clone L20/8 (NeuroMab), rabbit phospho-Ser880 GluA2 (Millipore). AlexaFluor-488, -555 and -647 (Invitrogen) were used as secondary antibodies.
Affinity purification
Glutathione-S-transferase (GST) fused to Plk2c (aa352–682) was purified from E. coli and coupled to glutathione sepharose resin. Adult rat brain lysates were applied to a GST-Plk2c-resin column, washed extensively, and eluted. Eluates were separated on SDS-PAGE gels and visualized by Coomassie blue. Bands were excised in the ∼50–120kD range that contained most of the visible proteins, cut into slices, and digested in-gel by trypsin. Peptides were identified by liquid chromatography-tandem mass spectrometry.
GST-pulldown
Cells were lysed in IP buffer (50mM Tris pH 8.0, 150mM NaCl, 1% NP-40, plus pepstatin, leupeptin, PMSF), centrifuged to remove cell debris, and incubated 3–4 h, 4°C with 5 μg GST-Plk2c (or GST alone). Following wash with GST-lysis buffer (20mM Tris-Cl, pH 8.0, 200mM NaCl, 1mM EDTA, 0.5% Nonidet P-40, 2 μg/ml aprotinin, 1 μg/ml leupeptin, 0.7 μg/ml pepstatin and 25 μg/ml PMSF), bound proteins were analyzed by SDS-PAGE and immunoblotting. For SDS wash, GST-Plk2c coupled resin was incubated in 1% SDS for 1 min, then quenched by 10-fold dilution into IP buffer.
In vitro binding and ATPase Assay
His6-NSF was purified from E. coli by disruption in lysis buffer (100mM Hepes/KOH, pH 7.4, 500mM KCl, 5mM MgCl2, 1mM DTT, and 0.5mM ATP plus protease inhibitors), incubation with Ni-NTA resin, and elution by 50–500mM imidizaole gradient in binding buffer (20mM Hepes/KOH pH 7, 200mM KCl, 2mM β-mercaptoethanol, 10% glycerol). His6-NSF was then incubated with GST-Plk2c or GST-GluA2c (aa834–883), with ATP, ADP or ATP-γ-S (all at 2mM) in binding buffer. For ATPase assay, 1 μg His6-NSF was incubated in ATPase buffer (25mM Tris-HCl, pH 8.8, 100mM KCl, 0.5mM DTT, 2mM MgCl2, 0.6mM purified ATP, 10% glycerol) at 37°C with 10 μg of GST, GST-GluA2c, or GST-Plk2c. An ATPase colorimetric assay (Innova Biosciences) was used according to manufacturer's directions to detect formation of free phosphate and values read at 595nm.
Cell culture, Transfection, Sindbis virus infection and Picrotoxin induction
COS-7 cells were grown in DMEM (Gibco) supplemented with 10% fetal bovine serum and 0.1% gentamicin (Invitrogen). COS-7 cells were transiently transfected with 1μg of plasmid DNA using Lipofectamine 2000 (Invitrogen), expressed for 24 h and harvested in IP buffer (see above). Primary hippocampal neurons were prepared from day 18 rat embryos and maintained 18–24 DIV. All animals were used in accordance with guidelines of the Georgetown University Animal Care and Use Committee. Cells were plated at medium density (∼150 cells/mm2) on coverslips coated with poly-D-lysine (Sigma) and laminin (2μg/mL, Roche). Cultures were grown in Neurobasal medium (Invitrogen) supplemented with B27 (Invitrogen), 0.5mM glutamine and 12.5 μM glutamate. Neurons were transfected using calcium phosphate as described14 or Lipofectamine 2000 (Invitrogen). Transfected neurons were incubated for 1–2 days, 3 days for RNAi experiments. For some experiments picrotoxin (100μM, Tocris Bioscience) was applied to neurons for 24 h. Plk2, K108M, or GFP were cloned into the pSinRep5 Sindbis virus vector (Invitrogen) and replication-defective pseudovirions produced according to the manufacturer's directions. Neurons were DIV18–21 at time of infection and duration of infection limited to 18–24 h, during which no significant toxicity could be observed.
Immunoprecipitation and immunoblotting
For immunoprecipitation, cells were lysed in IP buffer (see above), centrifuged to remove cell debris, and incubated ∼18 h at 4°C with 1 μg antibody. Complexes were precipitated with Protein A-sepharose or glutathione sepharose, as appropriate. Bound proteins were eluted by boiling in Laemmli sample buffer, centrifuged and the supernatants separated by SDS-PAGE, transferred to nitrocellulose and imaged with enhanced chemiluminescence (Western Lightning (Perkin Elmer) or SuperSignal West Femto (Pierce)) following HRP-linked secondary antibody.
Immunocytochemistry
For immunolabeling of cell surface receptors, infected primary hippocampal cultured neurons DIV19–22 were treated with primary N-terminal specific antibody under nonpermeabilizing conditions, fixed, washed and treated with saturating concentrations of fluorescently-tagged secondary antibody. Cells were permeabilized and immunostained for Plk2 (or GFP) to visualize infected cells. Antibodies for immunostaining were diluted in conditioned media or ADB buffer (3% normal goat serum, 0.1% BSA, PBS) prior to permeabilization and GDB buffer (0.1% gelatin, 0.3% Triton X-100, 16mM sodium phosphate pH 7.4, 450mM NaCl), thereafter. For experiments with NEM, drug was applied for 10min at 10μM immediately before immunolabeling.
Cell Surface Biotinylation
COS-7 cells were transfected with GluA2 and combinations of NSF and Plk2 (WT or KN) and expressed for 24 h. Cells were washed twice with PBS, and surface proteins labeled with Sulfo-NHS-SS-Biotin (Pierce) (10 μl at 500 μg/ml in PBS) with gentle shaking at 4°C for 30 min. Cells were bathed in quenching solution (50μL, Pierce), washed twice with Tris-buffered saline, lysed in 500 μl of IP buffer, collected and disrupted by sonication on ice. After 30-min incubation on ice, cell lysates were clarified by centrifugation (10,000×g, 2 min). To isolate biotin-labeled proteins, lysates were added to immobilized NeutrAvidin TM Gel (50 μl) and incubated for 1 h at room temperature. Gels were washed 5 times with wash buffer and incubated 1 h with SDS-PAGE sample buffer including 50mM dithiothreitol. Surface proteins were then analyzed by immunoblotting for GluA2. For neurons, labeling was performed in ice-cold ACSF for 60 min, and lysis was performed in RIPA buffer. Intracellular receptor pool was calculated as [(total – surface)/total receptors].
Fluorescence Lifetime Microscopy
Proximity of NSF and Plk2 was assessed by FLIM, a fluorescence resonance energy transfer-based technique using multiphoton microscopy. Myc-tagged NSF and HA-tagged Plk2 were transfected into COS-7 cells and labeled by indirect immunocytochemistry using mouse myc and rabbit HA antibodies, and then labeled with secondary Alexa488 anti-mouse (donor fluorophore) and Cy3-anti-rabbit secondary as the acceptor fluorophore. Lifetime of the Alexa488 donor fluorophore attached to NSF decreases due to nonradiative energy transfer if Cy3-Plk2 (acceptor fluorophore) is positioned within 1–10 nm of the donor. Thus, a decreased donor lifetime indicates proximity of the two fluorophores (and the proteins labeled with these fluorophores). Images of Alexa488 fluorescence lifetimes on a pixel-by-pixel basis were acquired using a Bio-Rad Radiance 2000 multiphoton microscope, Ti:sapphire laser (Spectra Physics), a high-speed detector MCP R3809 (Hamamatsu), and a time-correlated single-photon counting acquisition board (SPC 830, Becker and Hickl GmbH). Images were analyzed using SPC-Image V2.8 (Becker and Hickl GmbH). Donor fluorophore lifetimes were fit using this software to two exponential decay curves to determine a fraction of the donor interacting with an acceptor. Results are presented in pseudocolored images representing color-coded donor lifetime on a pixel-by-pixel basis.
Internalization and recycling assay
Cultured hippocampal neurons were live labeled with 1° antibody recognizing GluA2 N-terminus. After incubation with antibody, cells were returned to conditioned media at 37°C, 5% CO2 for 0–30 min to allow receptor endocytosis. Cells were then fixed and remaining surface GluA2 labeled using saturating concentrations of 2° antibody. Following permeabilization, internalized receptors were labeled using a different 2° antibody. For recycling assays, the same procedure as above was followed except that cells were returned to conditioned media at 37°C, 5% CO2 for 30 min to allow for receptor internalization. Remaining surface-bound primary antibody was then acid stripped (0.5M NaCl and 0.2M acetic acid, at 4°C, 2 min). Neurons were incubated at 37°C, 5% CO2 for 1 hour with 2° antibody to allow for and label internalized receptors recycling back to the cell surface, or immediately fixed and incubated with 2° antibody to verify complete acid stripping. After washing, neurons were subsequently fixed and permeabilized and immunolabeled to show transfected cells (GFP or Plk2).
Yeast two-hybrid assay
Yeast two-hybrid analysis was conducted as described15. NSF was cloned into the prey vector, pGAD, and fragments of Plk2c cloned into the bait vector, pBHA. L40 yeast strain and beta-galactosidase reporter were used to assay for interaction of NSF and Plk2 fragments.
Maltose binding protein fusion purification and pull-down
Plk2 PBind was cloned into the vector pMAL-c2× (New England Biolabs), expressed in E.coli, and purified according to the manufacturer's directions with amylose resin and maltose elution. Purified MBP-PBind or MBP alone (10 μg) was added to binding buffer (20mM Tris pH 7.4, 150mM NaCl) with 200 μg of whole brain homogenate (extracted with 1% deoxycholate, clarified by centrifugation at 100,000×g, and dialyzed against PBS) and 10 μl amylose beads for 2 h at 4°C with gentle agitation. Pellets were washed 3× with 20mM Tris pH 7.4, 500mM NaCl and analyzed by immunoblotting against NSF as well as Coomassie staining for fusion proteins.
Quantification and image analysis
Images were acquired using an Axiovert 200M epifluorescnece inverted microscope (Zeiss) using consistent laser intensity or camera exposure levels for each fluorescent marker in each experiment. For image analysis and quantification, measurements were made using MetaMorph software (Molecular Devices). Average intensity was calculated from integrated intensity and area for each selected area. Secondary dendritic branches (>20μm from the cell body) were used for quantification except where noted.
Surface plasmon resonance (SPR)
Biacore T100 instrument was used to measure direct interaction between recombinant NSF protein and synthetic Plk2 peptides, pep-PBind (sequence SAVENKQQIGDAIRMIVRGTLGSC) and a scrambled version, pep-scr (sequence SGDISVKINARITVCARLQGEMQG). Biacore series S NTA sensor chip was charged with 3 min injection of 100mM Ni2SO4. Injection of 122nM NSF protein for 4 min captured 5842 RU ligand on flow cell 2. Ni+ charged flow cell 1 was used as reference surface. Peptides were injected at 1 μM concentration for 1 min. Injections were repeated 3 times. All injections were made at 10μl/min flow rate. HBS-P (10mM Hepes pH 7.4, 150mM NaCl, 0.05% P-20) buffer supplemented with 2mM ATP was used as running and dilution buffers.
Electrophysiology
Wistar rats 14–18 days old were anesthetized with isoflurane and decapitated in accordance with NIH animal care and use guidelines. Transverse hippocampal slices (400 μm) were cut in ice-cold artificial cerebrospinal fluid (ACSF) containing (mM): 119 NaCl, 2.5 KCl, 2.5 CaCl2, 9 MgSO4, 1 NaH2PO4, 26.2 NaHCO3, 11 glucose equilibrated with 95% O2 and 5% CO2. Slices were allowed to recover for at least 1 hour in ACSF at room temperature (composition as above except 1.3mM MgSO4). Whole cell patch clamp somatic recordings were made from visually identified CA1 pyramidal neurons in the presence of 50 μM picrotoxin at room temperature. The whole cell solution contained (mM): 135 CsMeSO4; 8 NaCl; 10 HEPES; 0.5 EGTA; 4 Mg-ATP; 0.3 Na-GTP; 5 QX-314 (pH 7.25), 285 mOsm. In addition, 100 μM of peptide was included together with 100 μM of each of the following protease inhibitors: bestatin, leupeptin, and pepstatin-A. EPSCs (at a holding potential of −70 mV) were evoked by afferent stimulation (0.1Hz) of fibers in the stratum radiatum. All recordings were made using a Multiclamp 700B amplifier (Axon Instruments), signals were filtered at 4 kHz, digitized at 10 kHz, and analyzed on-line using pClamp 9.2 (Axon Instruments). EPSCs were averaged during the first and last 2 minutes of recording for statistical analysis.
Statistical analysis
All values were expressed as means ± SEM, unless otherwise stated. Experiments were performed at least in triplicate and Student's t-test used for pairwise tests of significance, and ANOVA with Tukey's post-hoc test for group comparisons.
Supplementary Material
Supplementary Figure 1: Validation of GST-Plk2c affinity reagent.
Supplementary Figure 2: NSF ATPase activity is not affected by Plk2 or GluA2 in vitro.
Supplementary Figure 3: High NSF overexpression causes loss of surface GluA2, but Plk2 does not affect NSF expression or localization.
Supplementary Figure 4: Plk2 causes loss of postsynaptic integrity in proximal but not secondary dendrites.
Supplementary Figure 5: RNAi knockdown prevents Plk2 induction by picrotoxin.
Supplementary Figure 6: NEM treatment occludes Plk2 effect on surface GluA2.
Supplementary Figure 7: Plk2 C-terminus alone does not affect PSD-95 in proximal or secondary dendrites.
Supplementary Figure 8: Homeostatic gating model for Plk2-NSF function.
Supplementary Figure 9: Uncropped blots for main Figures.
Supplementary Table 1: Plk2c-associated membrane trafficking proteins.
Supplementary Table 2: FLIM analysis of Plk2-NSF interaction.
Acknowledgments
D.M.E., H.S.H., J.A.M., D.Z. and D.T.S.P. performed experiments; D.M.E., S.H.L., J.T.I. and D.T.S.P. designed experiments, D.M.E. and D.T.S.P. wrote the manuscript, and D.T.S.P. supervised the project. This work was supported by NIH/NINDS grants NS048085 (D.T.S.P.), F31NS061467 (D.M.E.) and P30CA051008 (Biacore Molecular Interactions Shared Resource of Lombardi Comprehensive Cancer Center). We thank Leslie Bilello for technical assistance.
References
- 1.Sheng M, Lee SH. AMPA receptor trafficking and the control of synaptic transmission. Cell. 2001;105:825–8. doi: 10.1016/s0092-8674(01)00406-8. [DOI] [PubMed] [Google Scholar]
- 2.Zhao C, Slevin JT, Whiteheart SW. Cellular functions of NSF: Not just SNAPs and SNAREs. FEBS Lett. 2007 doi: 10.1016/j.febslet.2007.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nishimune A, et al. NSF binding to GluR2 regulates synaptic transmission. Neuron. 1998;21:87–97. doi: 10.1016/s0896-6273(00)80517-6. [DOI] [PubMed] [Google Scholar]
- 4.Song I, et al. Interaction of the N-ethylmaleimide-sensitive factor with AMPA receptors. Neuron. 1998;21:393–400. doi: 10.1016/s0896-6273(00)80548-6. [DOI] [PubMed] [Google Scholar]
- 5.Osten P, et al. The AMPA receptor GluR2 C terminus can mediate a reversible, ATP-dependent interaction with NSF and alpha- and beta-SNAPs. Neuron. 1998;21:99–110. doi: 10.1016/s0896-6273(00)80518-8. [DOI] [PubMed] [Google Scholar]
- 6.Luscher C, et al. Role of AMPA receptor cycling in synaptic transmission and plasticity. Neuron. 1999;24:649–58. doi: 10.1016/s0896-6273(00)81119-8. [DOI] [PubMed] [Google Scholar]
- 7.Braithwaite SP, Xia H, Malenka RC. Differential roles for NSF and GRIP/ABP in AMPA receptor cycling. Proc Natl Acad Sci U S A. 2002;99:7096–101. doi: 10.1073/pnas.102156099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shi S, Hayashi Y, Esteban JA, Malinow R. Subunit-specific rules governing AMPA receptor trafficking to synapses in hippocampal pyramidal neurons. Cell. 2001;105:331–43. doi: 10.1016/s0092-8674(01)00321-x. [DOI] [PubMed] [Google Scholar]
- 9.Luthi A, et al. Hippocampal LTD expression involves a pool of AMPARs regulated by the NSF-GluR2 interaction. Neuron. 1999;24:389–99. doi: 10.1016/s0896-6273(00)80852-1. [DOI] [PubMed] [Google Scholar]
- 10.Kim CH, Chung HJ, Lee HK, Huganir RL. Interaction of the AMPA receptor subunit GluR2/3 with PDZ domains regulates hippocampal long-term depression. Proc Natl Acad Sci U S A. 2001;98:11725–30. doi: 10.1073/pnas.211132798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Noel J, et al. Surface expression of AMPA receptors in hippocampal neurons is regulated by an NSF-dependent mechanism. Neuron. 1999;23:365–76. doi: 10.1016/s0896-6273(00)80786-2. [DOI] [PubMed] [Google Scholar]
- 12.Simmons DL, Neel BG, Stevens R, Evett G, Erikson RL. Identification of an early-growth-response gene encoding a novel putative protein kinase. Mol Cell Biol. 1992;12:4164–9. doi: 10.1128/mcb.12.9.4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kauselmann G, et al. The polo-like protein kinases Fnk and Snk associate with a Ca(2+)- and integrin-binding protein and are regulated dynamically with synaptic plasticity. Embo J. 1999;18:5528–39. doi: 10.1093/emboj/18.20.5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pak DT, Yang S, Rudolph-Correia S, Kim E, Sheng M. Regulation of dendritic spine morphology by SPAR, a PSD-95-associated RapGAP. Neuron. 2001;31:289–303. doi: 10.1016/s0896-6273(01)00355-5. [DOI] [PubMed] [Google Scholar]
- 15.Pak DT, Sheng M. Targeted protein degradation and synapse remodeling by an inducible protein kinase. Science. 2003;302:1368–73. doi: 10.1126/science.1082475. [DOI] [PubMed] [Google Scholar]
- 16.Ang XL, Seeburg DP, Sheng M, Harper JW. Regulation of postsynaptic RapGAP SPAR by Polo-like kinase 2 and the SCFbeta-TRCP ubiquitin ligase in hippocampal neurons. J Biol Chem. 2008;283:29424–32. doi: 10.1074/jbc.M802475200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seeburg DP, Feliu-Mojer M, Gaiottino J, Pak DT, Sheng M. Critical role of CDK5 and Polo-like kinase 2 in homeostatic synaptic plasticity during elevated activity. Neuron. 2008;58:571–83. doi: 10.1016/j.neuron.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seeburg DP, Sheng M. Activity-induced Polo-like kinase 2 is required for homeostatic plasticity of hippocampal neurons during epileptiform activity. J Neurosci. 2008;28:6583–91. doi: 10.1523/JNEUROSCI.1853-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elia AE, et al. The molecular basis for phosphodependent substrate targeting and regulation of Plks by the Polo-box domain. Cell. 2003;115:83–95. doi: 10.1016/s0092-8674(03)00725-6. [DOI] [PubMed] [Google Scholar]
- 20.Feng Y, et al. Association of polo-like kinase with alpha-, beta- and gamma-tubulins in a stable complex. Biochem J. 1999;339(Pt 2):435–42. [PMC free article] [PubMed] [Google Scholar]
- 21.Daw MI, et al. PDZ proteins interacting with C-terminal GluR2/3 are involved in a PKC-dependent regulation of AMPA receptors at hippocampal synapses. Neuron. 2000;28:873–86. doi: 10.1016/s0896-6273(00)00160-4. [DOI] [PubMed] [Google Scholar]
- 22.Dong H, Zhang P, Liao D, Huganir RL. Characterization, expression, and distribution of GRIP protein. Ann N Y Acad Sci. 1999;868:535–40. doi: 10.1111/j.1749-6632.1999.tb11323.x. [DOI] [PubMed] [Google Scholar]
- 23.Perez JL, et al. PICK1 targets activated protein kinase Calpha to AMPA receptor clusters in spines of hippocampal neurons and reduces surface levels of the AMPA-type glutamate receptor subunit 2. J Neurosci. 2001;21:5417–28. doi: 10.1523/JNEUROSCI.21-15-05417.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.States BA, Khatri L, Ziff EB. Stable synaptic retention of serine-880-phosphorylated GluR2 in hippocampal neurons. Mol Cell Neurosci. 2008;38:189–202. doi: 10.1016/j.mcn.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee SH, Liu L, Wang YT, Sheng M. Clathrin adaptor AP2 and NSF interact with overlapping sites of GluR2 and play distinct roles in AMPA receptor trafficking and hippocampal LTD. Neuron. 2002;36:661–74. doi: 10.1016/s0896-6273(02)01024-3. [DOI] [PubMed] [Google Scholar]
- 26.Lee KS, Grenfell TZ, Yarm FR, Erikson RL. Mutation of the polo-box disrupts localization and mitotic functions of the mammalian polo kinase Plk. Proc Natl Acad Sci U S A. 1998;95:9301–6. doi: 10.1073/pnas.95.16.9301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Turrigiano G. Homeostatic signaling: the positive side of negative feedback. Curr Opin Neurobiol. 2007;17:318–24. doi: 10.1016/j.conb.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 28.Gainey MA, Hurvitz-Wolff JR, Lambo ME, Turrigiano GG. Synaptic scaling requires the GluR2 subunit of the AMPA receptor. J Neurosci. 2009;29:6479–89. doi: 10.1523/JNEUROSCI.3753-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu W, et al. Subunit composition of synaptic AMPA receptors revealed by a single-cell genetic approach. Neuron. 2009;62:254–68. doi: 10.1016/j.neuron.2009.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wenthold RJ, Petralia RS, Blahos J, II, Niedzielski AS. Evidence for multiple AMPA receptor complexes in hippocampal CA1/CA2 neurons. J Neurosci. 1996;16:1982–9. doi: 10.1523/JNEUROSCI.16-06-01982.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shepherd JD, et al. Arc/Arg3.1 mediates homeostatic synaptic scaling of AMPA receptors. Neuron. 2006;52:475–84. doi: 10.1016/j.neuron.2006.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thiagarajan TC, Lindskog M, Malgaroli A, Tsien RW. LTP and adaptation to inactivity: overlapping mechanisms and implications for metaplasticity. Neuropharmacology. 2007;52:156–75. doi: 10.1016/j.neuropharm.2006.07.030. [DOI] [PubMed] [Google Scholar]
- 33.Plant K, et al. Transient incorporation of native GluR2-lacking AMPA receptors during hippocampal long-term potentiation. Nat Neurosci. 2006;9:602–4. doi: 10.1038/nn1678. [DOI] [PubMed] [Google Scholar]
- 34.Liu B, et al. Ischemic insults direct glutamate receptor subunit 2-lacking AMPA receptors to synaptic sites. J Neurosci. 2006;26:5309–19. doi: 10.1523/JNEUROSCI.0567-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jia Z, et al. Enhanced LTP in mice deficient in the AMPA receptor GluR2. Neuron. 1996;17:945–56. doi: 10.1016/s0896-6273(00)80225-1. [DOI] [PubMed] [Google Scholar]
- 36.Meng Y, Zhang Y, Jia Z. Synaptic transmission and plasticity in the absence of AMPA glutamate receptor GluR2 and GluR3. Neuron. 2003;39:163–76. doi: 10.1016/s0896-6273(03)00368-4. [DOI] [PubMed] [Google Scholar]
- 37.Biou V, Bhattacharyya S, Malenka RC. Endocytosis and recycling of AMPA receptors lacking GluR2/3. Proc Natl Acad Sci U S A. 2008;105:1038–43. doi: 10.1073/pnas.0711412105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gardner SM, et al. Calcium-permeable AMPA receptor plasticity is mediated by subunit-specific interactions with PICK1 and NSF. Neuron. 2005;45:903–15. doi: 10.1016/j.neuron.2005.02.026. [DOI] [PubMed] [Google Scholar]
- 39.Beretta F, et al. NSF interaction is important for direct insertion of GluR2 at synaptic sites. Mol Cell Neurosci. 2005;28:650–60. doi: 10.1016/j.mcn.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 40.Narisawa-Saito M, et al. Brain-derived neurotrophic factor regulates surface expression of alpha-amino-3-hydroxy-5-methyl-4-isoxazoleproprionic acid receptors by enhancing the N-ethylmaleimide-sensitive factor/GluR2 interaction in developing neocortical neurons. J Biol Chem. 2002;277:40901–10. doi: 10.1074/jbc.M202158200. [DOI] [PubMed] [Google Scholar]
- 41.Lin DT, Huganir RL. PICK1 and phosphorylation of the glutamate receptor 2 (GluR2) AMPA receptor subunit regulates GluR2 recycling after NMDA receptor-induced internalization. J Neurosci. 2007;27:13903–8. doi: 10.1523/JNEUROSCI.1750-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yao Y, et al. PKM zeta maintains late long-term potentiation by N-ethylmaleimide-sensitive factor/GluR2-dependent trafficking of postsynaptic AMPA receptors. J Neurosci. 2008;28:7820–7. doi: 10.1523/JNEUROSCI.0223-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang Y, et al. S-nitrosylation of N-ethylmaleimide sensitive factor mediates surface expression of AMPA receptors. Neuron. 2005;46:533–40. doi: 10.1016/j.neuron.2005.03.028. [DOI] [PubMed] [Google Scholar]
- 44.Hanley JG, Khatri L, Hanson PI, Ziff EB. NSF ATPase and alpha-/beta-SNAPs disassemble the AMPA receptor-PICK1 complex. Neuron. 2002;34:53–67. doi: 10.1016/s0896-6273(02)00638-4. [DOI] [PubMed] [Google Scholar]
- 45.Xia J, Chung HJ, Wihler C, Huganir RL, Linden DJ. Cerebellar long-term depression requires PKC-regulated interactions between GluR2/3 and PDZ domain-containing proteins. Neuron. 2000;28:499–510. doi: 10.1016/s0896-6273(00)00128-8. [DOI] [PubMed] [Google Scholar]
- 46.Terashima A, et al. Regulation of synaptic strength and AMPA receptor subunit composition by PICK1. J Neurosci. 2004;24:5381–90. doi: 10.1523/JNEUROSCI.4378-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Osten P, et al. Mutagenesis reveals a role for ABP/GRIP binding to GluR2 in synaptic surface accumulation of the AMPA receptor. Neuron. 2000;27:313–25. doi: 10.1016/s0896-6273(00)00039-8. [DOI] [PubMed] [Google Scholar]
- 48.DeSouza S, Fu J, States BA, Ziff EB. Differential palmitoylation directs the AMPA receptor-binding protein ABP to spines or to intracellular clusters. J Neurosci. 2002;22:3493–503. doi: 10.1523/JNEUROSCI.22-09-03493.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chung HJ, Xia J, Scannevin RH, Zhang X, Huganir RL. Phosphorylation of the AMPA receptor subunit GluR2 differentially regulates its interaction with PDZ domain-containing proteins. J Neurosci. 2000;20:7258–67. doi: 10.1523/JNEUROSCI.20-19-07258.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fukata Y, et al. Molecular constituents of neuronal AMPA receptors. J Cell Biol. 2005;169:399–404. doi: 10.1083/jcb.200501121. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1: Validation of GST-Plk2c affinity reagent.
Supplementary Figure 2: NSF ATPase activity is not affected by Plk2 or GluA2 in vitro.
Supplementary Figure 3: High NSF overexpression causes loss of surface GluA2, but Plk2 does not affect NSF expression or localization.
Supplementary Figure 4: Plk2 causes loss of postsynaptic integrity in proximal but not secondary dendrites.
Supplementary Figure 5: RNAi knockdown prevents Plk2 induction by picrotoxin.
Supplementary Figure 6: NEM treatment occludes Plk2 effect on surface GluA2.
Supplementary Figure 7: Plk2 C-terminus alone does not affect PSD-95 in proximal or secondary dendrites.
Supplementary Figure 8: Homeostatic gating model for Plk2-NSF function.
Supplementary Figure 9: Uncropped blots for main Figures.
Supplementary Table 1: Plk2c-associated membrane trafficking proteins.
Supplementary Table 2: FLIM analysis of Plk2-NSF interaction.