Abstract
Aniline-catalyzed hydrazone ligation between surface immobilized hydrazines and aldehyde-modified antibodies is shown to be an efficient method for attaching protein capture agents to model oxide-coated biosensor substrates. Silicon photonic microring resonators are used to directly evaluate the efficiency of this surface bioconjugate reaction at various pHs and in the presence or absence of aniline as a nucleophilic catalyst. It is found that aniline significantly increases the net antibody loading for surfaces functionalized over a pH range from 4.5 to 7.4, allowing derivatization of substrates with reduced incubation time and sample consumption. This increase in antibody loading directly results in more sensitive antigen detection when functionalized microrings are employed in a label-free immunoassay. Furthermore, these experiments also reveal an interesting pH dependent non-covalent binding trend that plays an important role in dictating the amount of antibody attached onto the substrate, highlighting the competing contributions of the bioconjugate reaction rate and the dynamic interactions that control opportunities for a solution-phase biomolecule to react with a substrate-bound reagent.
Introduction
Many biomolecular analysis methods rely upon surface-bound capture probes, including well-established techniques such as microarrays,1, 2 enzyme-linked immunosorbent assays (ELISAs), and surface plasmon resonance,3 as well as a multitude of emerging optical, electronic, and gravimetric analysis technologies.4 In all cases and regardless of detection modality, the performance of each of these biosensing schemes is impacted by the underlying chemistry that links the probe to the sensor surface. Immobilization is particularly critical for proteomic applications, where concerns with reagent consumption and capture agent stability are common.5
In general, all bioconjugate schemes for functionalizing biosensor surfaces can be broken down into two classes: covalent and non-covalent.5-8 Covalent linkages between the capture agent and surface are often preferred over non-covalent approaches based upon electrostatic or van der Waals interactions on the basis of sensor stability, i.e., non-covalently attached proteins can be removed from the surface during a sensing experiment giving an inconsistent response. Non-covalent attachment methods that take advantage of high affinity interactions such as biotin-avidin often have stability comparable to covalent linkages but require addition of non-native chemical functionalities that can affect target recognition.
Covalent functionalization schemes can be further subdivided into two distinct groups based upon their requirement for chemical reactive groups that are either native or non-native to the protein to be immobilized.5 Reactions between appropriately-modified surfaces with free amines (from lysines) or sulfhydryls (from cysteines) have been the most widely exploited on account of their generality and single-step functionalization. However, these methods encounter challenges in that the most common reactive surface groups, succinimidyl esters and maleimides for amines and sulfhydryls, respectively, decompose via hydrolysis under conditions optimal for biocoinjugation,6 meaning that it is difficult to create defined surfaces wherein each immobilized capture agent is attached to the surface to a similar extent. In recent years, chemoselective ligation chemistries based on the use of bioorthogonal moieties,9 such as Staudinger ligation,10 Cu(I)-catalyzed Huisgen cycloaddition,11 Diels-Alder cycloaddition,12 and native chemical ligation, 13, 14 have been widely explored as immobilization methods, offering exquisite control over the extent of chemical ligation between the protein and underlying surface. However, for these reactions a non-native chemical functionality must be incorporated into the capture agent of interest, often involving recombinant expression of non-native proteins or solid phase synthesis of modified peptides.15
As a compromise between the generality of reactions at functionalities natively available on proteins and the control afforded by chemoselective approaches, our group, as well as others, have employed imine ligation16-24—i.e., the reaction between α-effect amines (hydrazine or aminooxy groups) and aldehydes or ketones. More specifically, we have used the commercially-available reagent, S-4FB, to covalently incorporate an aryl aldehyde group onto antibody capture agents via a general reaction of a succinimidyl ester with lysine groups. The antibody is then coupled to a surface immobilized 6-hydrazinopyridine installed onto the sensor surface via a single-step silanization using a second commercially-available reagent, HyNic silane. An advantage of this approach for sensor derivatization is that the succinimide-amine reaction is performed in solution and therefore the distribution in number of incorporated aryl aldehydes can be regulated and optimized by controlling the excess reagent concentration and solution pH. In contrast, succinimide-amine reaction on surfaces, where the succinimide is presented on the surface, is more difficult to control due to the competing hydrolysis of the underlying surface, which can then become the limiting reagent. However, the rate of imine bond formation and thus surface bioconjugation is relatively slow and, in some cases, the capture agent concentration required to efficiently derivatize the sensor surface can detract from the advantages of many chip-based measurement schemes that typically feature reduced reagent consumption.
Several recent reports by Dirksen and Dawson describing solution phase reactions have illustrated that the rates of imine ligations can be significantly enhanced via the addition of aniline, which serves as a nucleophilic catalyst.19-21 Under mild conditions the rate constant of the reaction in solution can be increased by as much as three orders of magnitude.18 In this paper we employ the aniline-catalyzed hydrazone ligation reaction for the bioconjugation of a representative protein capture agent (antibody) to a silicon oxide biosensor surface using commercially available reagents in a simple two-step scheme. We show that this approach is an efficient and generally applicable method for immobilizing antibodies onto surfaces for immuno-based biosensing platforms and highlight an interesting pH dependence on surface conjugation. Moreover, we find that the amount of immobilized capture agent depends not exclusively on the rate of chemoselective ligation, but also the amount of non-specific interaction between the protein capture agent and underlying surface. That is to say that increased non-specific interactions between the proteins and underlying surface, which are pH dependent, provide a greater opportunity for the bioconjugate reaction to occur. As a result, we find that for our system bioconjugation can be accomplished with relatively high efficiency even at pHs as high as 7.4 and with reduced sample consumption when aniline is added, compared to our previous report of non-catalyzed imine ligation.25 We also compare, under identical pH and antibody concentration conditions, the catalyzed hydrazone conjugation scheme to a more conventional reaction between the lysine groups of an antibody and surface-bound succinimide esters and find that imine ligation represents a superior method for substrate modification. Finally, we compare the performance of biosensors functionalized identically with the same capture agent and show that the addition of aniline as a nucleophilic catalyst leads to improved sensor performance.
In this report, chemical and biomolecular functionalization is monitored using silicon photonic microring resonators as a model silicon oxide sensor surface. Silicon photonic microring resonators are an emerging surface-sensitive analysis method that our group has recently utilized for monitoring chemical deposition and biomolecular binding and detection in an array-based, label-free biosensor format.25-28 This detection platform uses optical interferences within ring-shaped silicon waveguide cavities that are extremely sensitive to changes in the local refractive index on account of the chemical or biochemical reactions and/or interactions at the sensor surface. The addition of biomolecules to the sensor surface leads to changes in microring optical properties that are observed as shifts in the resonance frequency of the device, measured as an increase in the relative resonance wavelength shift and plotted with units of picometer shift (Δpm). For applications in analysis of surface bioconjugate reactions it is most important to note that the silicon waveguide is natively passivated with a silicon oxide overlayer and therefore the reported results are relevant to many common biosensor substrates, including glass, quartz, and many of the other oxide surfaces utilized by micro-electro-mechanical systems (MEMS) technologies.
Experimental
Materials
3-N-((6-(N′-Isopropylidene-hydrazino))nicotinamide)propyltriethyoxysilane (HyNic silane) and succinimidyl 4-formylbenzoate (S-4FB) were purchased from SoluLink (San Diego, CA). The 3-aminopropyltriethoxysilane (APTES), was purchased from Gelest (Morrisville, PA). Mouse monoclonal anti-(human thrombin) (Cat. # AHT-5020) and human α-thrombin (Cat. # HCT-0020) were purchased from Haematologic Technologies Inc. (Essex Junction, VT). Vivaspin 500 columns (5 kDa MW cutoff) were purchased from Sartorius (Aubugne, France). All other chemicals were obtained from Sigma-Aldrich (St. Louis, MO) and used as received.
All buffers were made with purified water (ELGA PURELAB filtration system; Lane End, UK), and the pH adjusted using 1 M HCl or 1 M NaOH. PBS was reconstituted from Dulbecco's phosphate buffered saline packets and adjusted to either pH 6.0 or 7.4. Acetate buffer was prepared from 50 mM sodium acetate and 150 mM sodium chloride adjusted to pH 4.5. Glycine buffer consists of 10 mM glycine and 160 mM NaCl adjusted to pH 2.2. A blocking solution containing 2 % (w/v) BSA was prepared by dissolving bovine serum albumin in PBS (pH 7.4); BSA-PBS buffer was prepared from this solution by dilution with PBS (pH 7.4) to a final concentration of 0.1 mg/mL BSA. BSA solutions were degassed under vacuum before being flowed across the sensor surface.
Sensor Substrates and Instrumentation
Microring resonator sensor chips were designed in collaboration with Genalyte, Inc. (San Diego, CA)25, 29 and fabricated on 8” silicon-on-insulator (SOI, 200 nm thick top-layer Si) wafers by the silicon foundry at LETI (Grenoble, France). The entire 8” wafer was spin coated with a fluoropolymer cladding material and the cladding removed from individual microrings via photolithography and reactive ion etching. The 6 × 6 mm chips with an array of 64 microrings measuring 30 μm in diameter were diced from the 8” wafers by Grinding and Dicing Services, Inc. (San Jose, CA). Linear waveguides having input and output diffractive grating couplers at either end were located next to the microrings, allowing for the optical properties of each microring to be interrogated independently. Light from the linear waveguide is coupled into the microring cavity through the evanescent field, and resonance is observed when the following condition is satisfied:
where m is an integer, λ is the wavelength of light, r is the radius of the cavity, and neff is the effective refractive index of the optical mode. Binding events on the surface lead to changes in neff and cause shifts in resonance wavelength, λ, which provides the detection modality.
Instrumentation enabling measurement of resonance frequency shifts for the microring resonators was purchased from Genalyte, Inc. and has been previously discussed in detail.25, 29 Briefly, a tunable beam from a diode laser with a center frequency of 1560 nm is rapidly rastered across the sensor chip, and the back reflection used to register the location of the grating couplers. The beam is then focused onto a single input coupler, and the light projected off the output coupler is measured as the input beam is swept through a suitable spectral window. Resonance is detected as a minimum in intensity of the output at a given wavelength, since under this condition light is strongly coupled into the cavity, leading to a strong negative attenuation in transmission down the linear waveguide past the microring. This configuration allows up to 32 microring resonators to be individually addressed, eight of which are not exposed to test solutions (i.e., under the cladding layer) and serve as controls for thermal drift. Solutions are introduced through two inlets in a custom Teflon lid that delivers liquids across to the sensor chip through two microfluidic channels defined by a laser cut Mylar gasket (fabricated by RMS Laser; El Cajon, CA) via negative pressure applied by syringe pumps (11 Plus syringe pump, Harvard Apparatus; Holliston, MA).
Surface Functionalization and Antibody Attachment
Sensor chips were cleaned with piranha solution (3:1 H2SO4:30%H2O2)30 for 3 minutes to remove any organic contaminants, rinsed with copious amounts of distilled (DI) water, and dried under nitrogen. Microring substrates were then loaded into the described Teflon cell and Mylar gasket microfluidic device and functionalized by exposure to 1 mg/mL HyNic silane solution in 95% ethanol and 5% DMF at 5 μL/min for 30 min, followed by rinsing with 100% ethanol.
A solution containing 100 μg of anti-(human thrombin) was buffer-exchanged into PBS (pH 7.4) using Vivaspin 500 spin columns. A 5-fold molar excess of S-4FB solution (2 mg/mL stock) was added to the antibody solution (~0.3 mg/mL) and the reaction mixture allowed to incubate for 3 h. Following the incubation, unreacted S-4FB was removed by buffer-exchanging into the appropriate working pH buffer (e.g., 4.5, 6.0, or 7.4) using Vivaspin 500 spin columns. Conjugation solutions were prepared with 5 μg/mL (33 nM) 4FB-modified antibody both with and without 100 mM aniline as a nucleophilic catalyst at each of the three different pHs.
Sensor functionalization was monitored by flowing the S-4FB-modified antibody solution over HyNic-modified microring resonators at a rate of 30 μL/min, 150 μL/min, and 6 μL/min for 30 min and recording the shift in resonance wavelength as a function of time. A two-channel gasket was employed to observe surface conjugation simultaneously in the presence or absence of 100 mM aniline, with one channel presenting aniline- and antibody-containing solution to the substrate and the other channel filled with a catalyst-free antibody solution. The buffer used for obtaining background measurements and rinsing was adjusted to mimic the presence or absence of aniline in the respective conjugation solutions to eliminate bulk refractive index differences upon solution switching. By doing this, observed shifts in resonance wavelength can be attributed solely to surface bioconjugation. After a 30 min reaction time for hydrazone bond formation between HyNic presented on the surface and S-4FB moiety on the antibody, the flow chamber and microrings were rinsed with PBS buffer for 20 min, followed by a 2 min exposure to a low pH glycine buffer (pH = 2.2) in order to remove any non-covalently attached antibody.
For succinimidyl ester attachment of antibody onto microring surfaces, the sensor surface was first exposed to a 2 % (v/v) solution of APTES in 95 % ethanol at 5 μL/min for 60 min, followed by rinsing with 95 % ethanol to remove any residual silane. Anhydrous DMF containing 100 mM N,N-diisopropylethylamine and 100 mM N,N′-disuccinimidyl carbonate was then flowed over the amine modified microring surface at 5 μL/min for 3 h, and rinsed with anhydrous DMF containing 100 mM N,N-diisopropylethylamine for 20-30 min, followed by 95% ethanol for 10-20 min. Shortly after switching to PBS (pH 7.4), the microring surface was exposed to 5 μg/mL anti-(human thrombin) in PBS (pH 7.4) for antibody functionalization. After 30 min incubation at 30 μL/min, the sensor surface was rinsed with PBS buffer for 20 min, followed by a 2 min exposure to glycine buffer (pH = 2.2) to remove any non-covalently attached antibody.
Antigen Detection
Following antibody conjugation the microring surfaces were blocked with 2 % (w/v) BSA in PBS (pH 7.4) in order to reduce non-specific protein adsorption in subsequent antigen detection experiments. A 4-μL aliquot of 8.3 mg/mL solution of human α-thrombin was buffer exchanged to BSAPBS using Vivaspin 500 spin columns. Working solutions containing 20 nM, 5 nM, 2 nM, and 0.5 nM antigen in BSA-PBS were prepared by successive dilution of a 1 μM stock solution. These solutions were then flowed over the antibody-functionalized sensor chips at 30 μL/min for 10 min as antigen association was monitored, followed by a BSA-PBS rinse for 20 min to observe antigen dissociation. The surface was then regenerated by flowing glycine buffer for 2 minutes followed by 10 min in BSAPBS to re-establish baseline for subsequent detection experiments.
Data Processing
Sensor microring data was corrected for drift related to thermal and instrumental fluctuations by normalizing against the response of reference microrings—those (four microrings in each channel) that were kept under the fluoropolymer cladding, unexposed to solution. Offline, residual slope as well as offset of the baseline were corrected, and data aligned temporally at the point when antibody or antigen was introduced.
Results and Discussion
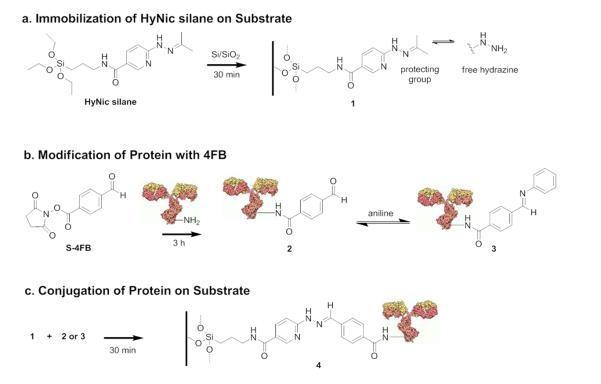
The general imine reaction used to couple a representative protein capture agent, a mouse monoclonal anti-(human thrombin) antibody, to the microring silicon oxide surface is shown in Figure 1. In parallel, the silicon oxide surface is modified with HyNic silane to install hydrazine moieties (Figure 1a), while aromatic aldehydes are incorporated onto antibodies (Figure 1b) in a controlled manner via reaction with succinimidyl 4-formylbenzoate (S-4FB). The modified-antibody is then flowed across the HyNic surface at various pHs and in the presence or absence of aniline to enable bioconjugation (Figure 1c). In the presence of aniline, a more reactive Schiff base, 3, is formed (Figure 1b) that is more easily protonated under mildly acidic conditions than the starting carbonyl; this results in the acceleration of hydrazone ligation via transimination.20, 21
Figure 1.
Surface reaction scheme for conjugating antibodies onto oxide-passivated silicon photonic microring resonators via hydrazone ligation. (a) 3-N-((6-(N′-Isopropylidenehydrazino))-nicotinamide)propyl triethyoxysilane (HyNic silane) is immobilized onto the silicon/silica surface. (Note that the free hydrazine is protected as an acetone hydrazone, 1.) (b) In parallel, proteins are modified with succinimidyl 4-formylbenzoate (S-4FB) via their primary amines, 2; and, if added, aniline forms a Schiff base, 3. (c) The 4FB-modified protein is conjugated to the HyNic-modified silicon/silica surface via the transimination of the Schiff base with the hydrazine to form the hydrazone linkage, 4.
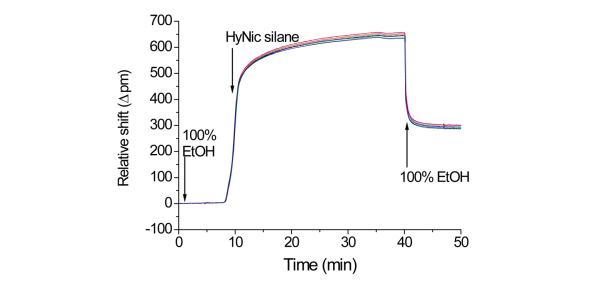
A key attribute of the microring resonator analysis platform is that it enables real time, label-free monitoring of chemical and biomolecular reactions and interactions that occur on the surface of the microring. This feature allows visualization of each step involved in surface conjugation on our model silicon oxide biosensor substrate. Figure 2 shows the real-time shift in resonance wavelength for ten microrings simultaneously exposed to a 1 mg/mL HyNic silane solution; the first surface reaction shown in Figure 1a. It is worth mentioning here that HyNic silane is an effective and more convenient method of installing hydrazine moieties on the silicon oxide microring surface, compared to our previous sequential two step functionalization involving addition first of APTES followed by S-HyNic (succinimidyl 6-hydrazinonicotinamide acetone hydrazone).25, 26 Specifically, the reaction time has been significantly reduced from several hours to 30 min and we have found sensor functionalization to be much more reproducible.
Figure 2.
Real-time monitoring of the shift in resonance frequency for ten microrings within the same sample flow chamber during organic modification via reaction with HyNic silane. The chamber was initially filled with 100% ethanol and a solution of 1 mg/mL HyNic silane in 95% EtOH and 5% DMF introduced under flow (5 μL/min) at t = 8 min. After 30 min, the HyNic silane was flushed from the chamber and microrings returned to 100% ethanol. The residual wavelength shift corresponds to immobilized HyNic silane.
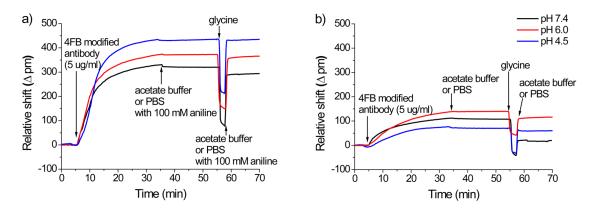
Following silanization, the 4FB-modified antibody is flowed across the HyNic-functionalized microring surface and the amount of antibody localized at the sensor surface is monitored in real time as a shift in resonance wavelength. Figure 3 shows real-time wavelength shifts as 4FB-modified anti-(human thrombin) antibody is introduced over HyNic-modified surfaces at three different pHs either in the presence (Figure 3a) or absence (Figure 3b) of aniline. To simplify the graphical display of the data, the response of single ring, which is representative to the average response of all the microrings (n = 10-12 for all measurements), was chosen for inclusion in Figure 3. The first key observation in comparing these bioconjugate reactions in the presence or absence of the nucleophilic catalyst (Figure 3a vs. Figure 3b) is that for all pHs the total amount of immobilized antibody is significantly increased in the presence of aniline, as indicated by larger resonance wavelength shifts. By comparing the net peak shifts measured after the glycine rinse to remove non-covalently bound protein (Table S1), average antibody loading is improved with the addition of aniline by a factor of 7.2 at pH 4.5, 3.2 at pH 6.0, and 14.8 at pH 7.4. Furthermore, for the aniline-catalyzed reactions (Figure 3a) the amount of covalently bound antibody increases with decreasing pH, consistent with previous reports of enhanced solution reaction rates at lower pHs.31
Figure 3.
Real-time shifts in resonance wavelength from representative microrings upon covalent immobilization of anti-(human thrombin) antibody onto the sensor surfaces at three different pHs and in the presence (a) and absence (b) of 100 mM aniline. In each trace the sensors were initially incubated in buffer (with or without aniline, respectively) at a flow rate of 30 μL/min and a 5 μg/mL solution of 4FB-modified antibody with or without 100 mM aniline was flowed for 30 min. After switching back to the original buffer for 20 minutes, non-covalently attached antibody was removed with a low pH (pH 2.2) glycine buffer rinse. The sensors were then returned to the respective buffer at t = 57 min to determine the residual net shift corresponding to the amount of covalently immobilized antibody.
Recently, we empirically determined the relationship between the amount of bound mass, determined via radioimmunoassay, and resonance wavelength shift measured from our microring resonators.32 Using the resultant conversion factor, 14.7 (pg/mm2)/Δpm, we are able to convert the observed antibody loadings into relative surface coverage in terms of both mass per unit area and area per antibody. We find that when aniline is used as a catalyst, antibody loadings up to 6.4 ± 0.3 ng/mm2 (39 nm2/antibody) can be achieved, while the best non-catalyzed loading is only 1.6 ± 0.5 ng/mm2 (150 nm2/antibody). For reference, the theoretical maximum coverage for antibodies arranged in the most space efficient edge-on orientation is 34 nm2/antibody,33 meaning that the aniline-catalyzed method described in this paper is able to achieve 87% of the maximal loading. However, for optimal biomolecular recognition, the edge-on orientation would not be ideal and thus a compromise must be struck between loading density and antibody activity—a balance that will likely vary for each antibody and depend on the eventual application of the modified substrate.34
Returning to the non-catalyzed reaction (Figure 3b), the pH-dependence does not appear to be as straightforward as it is in the case of the catalyzed reaction, as pH 6.0 gives the largest resonance wavelength shift after glycine rinse followed by 4.5 and 7.4, respectively. To understand this discrepancy we focus on the difference in resonance wavelength shift before and after the low pH glycine rinse, which is used to remove proteins that are non-covalently bound to the surface. Interestingly, the amount of antibody removed by the glycine rinse increases dramatically with increasing pH—relative resonance wavelength shift decreases of 16.5 ± 2.6 % (n=12), 21.8 ± 6.0 % (n=12), and 82.8 ± 12.3 % (n=11) for pH 4.5, 6.0, and 7.4, respectively—revealing a significant pH-dependence of non-specific adsorption. Furthermore the pH-dependence of non-specific adsorption is opposite that of the hydrazone bond formation reaction, which leads to the complex relationship between pH and amount of covalently bound antibody. While the reaction is more efficient at lower pH, proteins have less of an opportunity to react with the surface due to reduced residence time at the sensor surface. This is in contrast to the purely solution phase situation where yield is predominantly dictated by reaction rate. When one of the reactants is bound to a substrate then the other reactant is required to interact with the surface, a process that for biomolecules is often accomplished via non-specific and initially non-covalent adsorption. This preadsorption helps increase the amount of protein on the surface that is available to react.35-37 Once bound, the rate of the bioconjugate reaction helps dictate the overall amount of antibody loading, hence the reduction in antibody desorption after glycine rinse at lower pHs, which follows the same pH dependence observed in free solution.19-21 Returning to the data in Figure 3b, at pH 7.4 a large amount of antibody initially non-covalently binds to the substrate, but the reaction is so inefficient that it is almost entirely removed with the glycine rinse. By contrast, a reduced amount of protein initially binds at pH 4.5, but the enhanced reaction rate (even in the absence of aniline) leads to a greater amount of covalently attached antibody and a larger net resonance wavelength shift after glycine. This model, which involves a competition between reaction rate and non-specific binding, is also observed for the aniline-catalyzed hydrazone ligations (Figure 3a), with bound antibody losses after glycine rinse of 1.3 ± 0.4 % (n=10), 3.4 ± 0.4 % (n=12), and 8.0 ± 2.3 % (n=11) observed for pH 4.5, 6.0 and 7.4, respectively. Again, more non-covalently adsorbed antibody is removed from surfaces conjugated at higher pHs where the reaction rate is reduced; however, the addition of aniline significantly increases the rate of reaction at all pHs such that net losses of bound protein are significantly reduced, as compared to the non-catalyzed reaction. Furthermore, aniline catalyzed reactions at 150 μL/min and 6 μL/min were measured to investigate the effect flow rate during antibody immobilization (Figure S1). For these additional flow rates, the pH dependent trend in antibody loading and the percent loss of bound antibody after glycine rinse is consistent, but the overall responses are, in both cases, smaller than those observed for 30 μL/min.
Beyond just comparing catalyzed versus non-catalyzed hydrazone ligation, we also investigated the efficiency of the often used succinimidyl ester/amine (lysine) conjugation chemistry. Unmodified antibody was flowed across succinimidyl ester presenting microrings after the surface was modified with APTES followed by N,N′-disuccinimidyl carbonate. While this conjugation reaction is similar in efficiency to that of the non-conjugated hydrazone ligation at the optimal condition (pH = 6.0), aniline catalyzed imine ligation is superior at all pHs, showing at least a 3-fold improvement in antibody loading (see supporting information Figure S2a). Using the conversion factor between resonance wavelength shift and bound mass, we can determine that the succinimidyl ester scheme, under the described conditions, yields an antibody coverage of ~1.7 ng/mm2.
To demonstrate the improvement in sensor response resulting from the effective aniline-catalyzed functionalization of the microring with an antibody capture agent, we monitored the interactions between human α-thrombin and the sensor surface. These experiments not only verify that the immobilized antibodies are still functional but also facilitate direct comparison of sensor responses from microrings bioconjugated in the presence or absence of aniline.
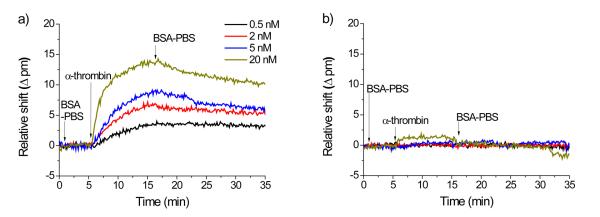
Microrings functionalized with antibody both in the presence and absence of aniline and at all three pHs were exposed to solutions of human α-thrombin at various concentrations ranging from 0.5 to 20 nM. Figure 4 shows the real time shifts in resonance wavelength upon the introduction of thrombin across microrings functionalized with anti-(human thrombin) antibodies immobilized at pH 7.4 in the presence (Figure 4a) and absence (Figure 4b) of aniline. Notably, a strong, concentration-dependent sensor response is observed for microrings functionalized in the presence of aniline. Conversely, the binding of thrombin is hardly detectable even at the highest concentrations when flowed across microrings functionalized in the absence of aniline at pH 7.4. Thrombin detection for sensors functionalized with antibody at pH 6.0 and 4.5 (Figures S3 and S4) show similar results. At all pHs microrings functionalized in the presence of aniline displayed stronger responses to the antigen; however, the relative difference in signal is most pronounced at pH 7.4, where the difference in antibody loading in the catalyzed versus non-catalyzed reactions is largest.
Figure 4.
Time-resolved response from human α-thrombin binding to microrings modified with anti-(human thrombin) antibody at pH 7.4 in the presence (a) and absence (b) of 100 mM aniline. The microrings are initially in BSA-PBS buffer and thrombin is introduced at t = 5 min. Thrombin is then introduced and incubated for 10 min, after which BSA-PBS buffer is flowed to observe antigen dissociation. The remaining antigen-antibody interactions on the surface are then disrupted via exposure to a low pH (pH 2.2) glycine buffer for 2 min (not shown) to regenerate the sensor for subsequent detection experiments.
For our application, the enhanced efficiency of aniline-catalyzed bioconjugation coupled with the interesting pH dependence of non-specific interaction allows us to effectively functionalize biosensor surfaces for shorter times and with less antibody consumption (~30 pmoles). It also permits bioconjugation at neutral pHs, which is advantageous for some proteins over to the lower pHs where the solution-phase hydrazone ligation is most efficient. The relative extents of the competing effects of non-covalent adsorption and bioconjugation will clearly vary as a function of many factors, including underlying surface charge (electrostatics) and relative hydrophobicity (van der Waals interactions), but this paper highlights an interesting interplay between reaction rate and opportunity for surface interaction, perhaps by non-specific adsorption, as both play important roles in dictating the efficiency of surface bioconjugation and therefore should be considered in concert for process optimization.
Conclusion
Aniline-catalyzed hydrazone ligation is shown to be an efficient method for attaching protein capture agents to a model silicon dioxide surface. Using silicon photonic microring resonators as a label-free, real-time surface analysis tool we demonstrate that substrate functionalization is improved across a broad pH range when aniline is added as a nucleophilic catalyst. Importantly, we show an important pH-dependent role that non-covalent protein adsorption plays in dictating the overall antibody loading. A role that in our system allows efficient functionalization of biosensor surfaces with reduced sample consumption, incubation time, and at neutral pH. We anticipate that aniline catalyzed hydrazone ligation will become a useful tool in a wide range of surface biofunctionalization applications which include, amongst others, quantitative antigen detection and target-specific immunocapture/depletion.
Supplementary Material
Acknowledgements
We acknowledge financial support from the NIH Director's New Innovator Award Program, part of the NIH Roadmap for Medical Research, through grant number 1-DP2-OD002190-01, the U.S. National Science Foundation, through the Science and Technology Center of Advanced Materials for the Purification of Water with Systems (WaterCAMPWS, CTS-0120978), the Roy J. Carver Charitable Trust, and the Camille and Henry Dreyfus Foundation.
Footnotes
Supporting Information Available: Aniline-catalyzed antibody immobilization at different flow rates, succinimidyl ester antibody immobilization, and thrombin binding results at pH 6.0 and 4.5 are presented in the supporting information. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Oh SJ, Hong BJ, Choi KY, Park JW. OMICS. 2006;10:327–343. doi: 10.1089/omi.2006.10.327. [DOI] [PubMed] [Google Scholar]
- 2.MacBeath G, Schreiber SL. Science. 2000;289:1760–1763. doi: 10.1126/science.289.5485.1760. [DOI] [PubMed] [Google Scholar]
- 3.Homola J. Chem. Rev. 2008;108:462–493. doi: 10.1021/cr068107d. [DOI] [PubMed] [Google Scholar]
- 4.Qavi AJ, Washburn AL, Byeon JY, Bailey RC. Anal. Bioanal. Chem. 2009;394:121–135. doi: 10.1007/s00216-009-2637-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jonkheijm P, Weinrich D, Schroder H, Niemeyer CM, Waldmann H. Angew. Chem. Int. Ed. 2008;47:9618–9647. doi: 10.1002/anie.200801711. [DOI] [PubMed] [Google Scholar]
- 6.Wong LS, Khan F, Micklefield J. Chem. Rev. 2009;109:4025–4053. doi: 10.1021/cr8004668. [DOI] [PubMed] [Google Scholar]
- 7.Tomizaki KY, Usui K, Mihara H. Chembiochem. 2005;6:783–799. doi: 10.1002/cbic.200400232. [DOI] [PubMed] [Google Scholar]
- 8.Rusmini F, Zhong ZY, Feijen J. Biomacromolecules. 2007;8:1775–1789. doi: 10.1021/bm061197b. [DOI] [PubMed] [Google Scholar]
- 9.Prescher JA, Bertozzi CR. Nat. Chem. Biol. 2005;1:13–21. doi: 10.1038/nchembio0605-13. [DOI] [PubMed] [Google Scholar]
- 10.Saxon E, Bertozzi CR. Science. 2000;287:2007–2010. doi: 10.1126/science.287.5460.2007. [DOI] [PubMed] [Google Scholar]
- 11.Rostovtsev VV, Green LG, Fokin VV, Sharpless KB. Angew. Chem. Int. Ed. 2002;41:2596–2599. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 12.Houseman BT, Huh JH, Kron SJ, Mrksich M. Nat. Biotechnol. 2002;20:270–274. doi: 10.1038/nbt0302-270. [DOI] [PubMed] [Google Scholar]
- 13.Dawson PE, Muir TW, Clarklewis I, Kent SBH. Science. 1994;266:776–779. doi: 10.1126/science.7973629. [DOI] [PubMed] [Google Scholar]
- 14.Camarero JA, Kwon Y, Coleman MA. J. Am. Chem. Soc. 2004;126:14730–14731. doi: 10.1021/ja0456611. [DOI] [PubMed] [Google Scholar]
- 15.Hackenberger CP, Schwarzer D. Angew. Chem. Int. Ed. Engl. 2008;47:10030–10074. doi: 10.1002/anie.200801313. [DOI] [PubMed] [Google Scholar]
- 16.Sander EG, Jencks WP. J. Am. Chem. Soc. 1968;90:6154–6162. [Google Scholar]
- 17.King TP, Zhao SW, Lam T. Biochemistry. 1986;25:5774–5779. doi: 10.1021/bi00367a064. [DOI] [PubMed] [Google Scholar]
- 18.Dirksen A, Dawson PE. Curr. Opin. Chem. Biol. 2008;12:760–766. doi: 10.1016/j.cbpa.2008.10.009. [DOI] [PubMed] [Google Scholar]
- 19.Dirksen A, Hackeng TM, Dawson PE. Angew. Chem. Int. Ed. 2006;45:7581–7584. doi: 10.1002/anie.200602877. [DOI] [PubMed] [Google Scholar]
- 20.Dirksen A, Dirksen S, Hackeng TM, Dawson PE. J. Am. Chem. Soc. 2006;128:15602–15603. doi: 10.1021/ja067189k. [DOI] [PubMed] [Google Scholar]
- 21.Dirksen A, Dawson PE. Bioconjug. Chem. 2008;19:2543–2548. doi: 10.1021/bc800310p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zeng Y, Ramya TNC, Dirksen A, Dawson PE, Paulson JC. Nat. Methods. 2009;6:207–209. doi: 10.1038/nmeth.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luo W, Chan EW, Yousaf MN. Journal of the American Chemical Society. 2010;132:2614–2621. doi: 10.1021/ja907187f. [DOI] [PubMed] [Google Scholar]
- 24.Lamb BM, Park S, Yousaf MN. Langmuir. 2010 doi: 10.1021/la1022642. [DOI] [PubMed] [Google Scholar]
- 25.Washburn AL, Gunn LC, Bailey RC. Anal. Chem. 2009;81:9499–9506. doi: 10.1021/ac902006p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Washburn AL, Luchansky MS, Bowman AL, Bailey RC. Anal. Chem. 2010;82:69–72. doi: 10.1021/ac902451b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qavi AJ, Bailey RC. Angew Chem Int Ed. 2010;49:1–5. doi: 10.1002/anie.201001712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luchansky MS, Bailey RC. Anal. Chem. 2010;82:1975–1981. doi: 10.1021/ac902725q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iqbal M, Gleeson M, Tybor F, Spaugh B, Gunn W, Hochberg M, Baehr-Jones T, Bailey RC, Gunn LC. IEEE J. Sel. Top. Quant. 2009 in press. [Google Scholar]
- 30.Caution! Pirahna solutions are extraordinarily dangerous, reacting explosively with trace quantities of organics.
- 31.Cordes EH, Jencks WP. J. Am. Chem. Soc. 1962;84:832–837. [Google Scholar]
- 32.Luchansky MS, Washburn AL, Martin TA, Iqbal M, Gunn LC, Bailey RC. Biosensors and Bioelectronics. 2010 doi: 10.1016/j.bios.2010.07.010. doi:10.1016/j.bios.2010.1007.1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koh I, Wang X, Varughese B, Isaacs L, Ehrman SH, English DS. J. Phys. Chem. B. 2006;110:1553–1558. doi: 10.1021/jp0556310. [DOI] [PubMed] [Google Scholar]
- 34.Xu H, Lu JR, Williams DE. J. Phys. Chem. B. 2006;110:1907–1914. doi: 10.1021/jp0538161. [DOI] [PubMed] [Google Scholar]
- 35.Kamruzzahan ASM, Ebner A, Wildling L, Kienberger F, Riener CK, Hahn CD, Pollheimer PD, Winklehner P, Holzl M, Lackner B, Schorkl DM, Hinterdorfer P, Gruber HJ. Bioconjugate Chemistry. 2006;17:1473–1481. doi: 10.1021/bc060252a. [DOI] [PubMed] [Google Scholar]
- 36.Hahn CD, Leitner C, Weinbrenner T, Schlapak R, Tinazli A, Tampe R, Lackner B, Steindl C, Hinterdorfer P, Gruber HJ, Holzl M. Bioconjugate Chemistry. 2007;18:247–253. doi: 10.1021/bc060292e. [DOI] [PubMed] [Google Scholar]
- 37.Holzl M, Tinazli A, Leitner C, Hahn CD, Lackner B, Tampe R, Gruber HJ. Langmuir. 2007;23:5571–5577. doi: 10.1021/la0627664. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.