Abstract
A range of phenotypes including Greig cephalopolysyndactyly and Pallister-Hall syndromes (GCPS, PHS) are caused by pathogenic mutation of the GLI3 gene. To characterize the clinical variability of GLI3 mutations, we present a subset of a cohort of 174 probands referred for GLI3 analysis. Eighty-one probands with typical GCPS or PHS were previously reported, and we report the remaining ninety-three probands here. This includes nineteen probands (twelve mutations) who fulfilled clinical criteria for GCPS or PHS, forty-eight probands (sixteen mutations) with features of GCPS or PHS but who did not meet the clinical criteria (sub-GCPS and sub-PHS), twenty-one probands (six mutations) with features of PHS or GCPS and oral-facial-digital syndrome and five probands (one mutation) with non-syndromic polydactyly. These data support previously identified genotype-phenotype correlations and demonstrate a more variable degree of severity than previously recognized. The finding of GLI3 mutations in patients with features of oral-facial-digital syndrome supports the observation that GLI3 interacts with cilia. We conclude that the phenotypic spectrum of GLI3 mutations is broader than that encompassed by the clinical diagnostic criteria, but the phenotype-genotype correlation persists. Individuals with features of either GCPS or PHS should be screened for mutations in GLI3 even if they do not fulfill clinical criteria.
Keywords: GLI3, Greig syndrome, Pallister-Hall syndrome, Oral-facial-digital syndrome
Introduction
Mutations in the zinc finger transcription factor encoding gene GLI3 (MIM# 165240) on chromosome 7p14.1 cause Greig cephalopolysyndactyly syndrome (GCPS; MIM# 175700, (Vortkamp, et al., 1991)), Pallister-Hall syndrome (PHS, MIM# 146510 (Kang, et al., 1997)) and, less frequently, other phenotypes such as acrocallosal syndrome (MIM# 200990 (Elson, et al., 2002)) and non-syndromic polydactyly (MIM# 174700 (Radhakrishna, et al., 1999), 174200 (Radhakrishna, et al., 1997)). The GCPS and PHS phenotypes are clinically distinct and there is a robust genotype-phenotype correlation for truncating mutations in GLI3 for these two phenotypes (Johnston, et al., 2005). Truncating mutations in the middle third of the gene generally cause PHS whereas large deletions or truncating mutations elsewhere in the gene (amino terminal-encoding or carboxy terminal-encoding thirds of the gene) cause GCPS. There are important biologic correlates for this genotype-phenotype correlation. The mutations that predict truncations in the amino-terminal third of the gene are predicted to be null mutations, caused by loss of the zinc finger DNA binding domain. In contrast, the truncations in the middle third of the protein are predicted to generate a constitutive repressor protein that skews the balance of activator and repressor forms of GLI3, which is a key downstream modulator of SHH signaling. The mutations that predict truncations in the carboxy-terminal third of the gene are predicted to cause the loss of a transactivation domain of GLI3 (Shin, et al., 1999). To date, genotype-phenotype studies have been predominantly based on mutations found in patients with typical forms of GCPS and PHS and it therefore remains unclear whether there are variant phenotypes that are caused by mutations in GLI3 and if so, whether the same correlations hold for these other phenotypes. To address these questions, we have continued to analyze a large cohort of 174 probands with a wide spectrum of phenotypic manifestations that include features of GCPS or PHS. Of these 174 probands, we present data on ninety-three patients not previously reported representing a wide range of phenotypes. We have analyzed GLI3 in these patients to determine the frequency and type of mutations and assessed whether the mutation positions correlated with the phenotypes.
Methods
Patients
This study was reviewed and approved by the Institutional Review Board of the National Human Genome Research Institute. The overall GLI3 project included 174 probands with features of PHS or GCPS. Ninety-three probands were the focus of this report and they were subdivided into the following groups according to inclusion criteria in Table 1. Eighty-one probands (174-93) have been reported previously (Galasso, et al., 2001; Johnston, et al., 2005; Killoran, et al., 2000; Kos, et al., 2008; Ng, et al., 2004; Turner, et al., 2003) and details on these probands are not included in this report.
Table 1.
Inclusion criteria for patient groups
| Column A All required | Column B Minimum of one required | Column C Confirming features1 | |
|---|---|---|---|
| OFD-overlap | Polydactyly | Oral frenulae Oral hamartoma Clef lip/palate Cerebellar vermis hypoplasia Tibial hypoplasia |
|
| PHS | Mesoaxial polydactyly Hypothalamic hamartoma |
||
| GCPS | Preaxial polydactyly | Syndactyly Macrocephaly Hypertelorism Postaxial polydactyly |
|
| Sub-PHS | Mesoaxial polydactyly Hypothalamic hamartoma Oligodactyly OR Postaxial polydactyly plus one feature from column C |
Bifid epiglottis Imperforate anus Small nails Hypopituitarism Growth hormone deficiency Genital hypoplasia |
|
| Sub-GCPS | Preaxial polydactyly Broad thumbs or great toes Syndactyly Macrocephaly Hypertelorism OR Postaxial polydactyly plus one feature from column C |
Hypoplasia of the corpus callosum |
Confirming features were used to place individuals with into sub-PHS or sub-GCPS groups when their only feature from column B was postaxial polydactyly.
Probands were evaluated sequentially for inclusion in the OFD-overlap group, then the PHS or GCPS groups and lastly the sub-PHS or sub-GCPS groups. Probands were placed into the first group where they fulfilled the inclusion criteria. Individuals who fulfilled the criteria for both sub-PHS and sub-GCPS were placed based upon the number of features they demonstrated for each group.
Probands with features of GCPS or PHS insufficient to meet clinical criteria
These probands had one or more features of GCPS or PHS but did not meet clinical criteria for either disorder. Detailed clinical data are reported for these latter fifty-three probands (plus nine relatives) who did not fulfill clinical criteria for either GCPS or PHS. Anomalies were defined according to the recently published standard terminology (Biesecker, et al., 2009; Hall, et al., 2009). This pool of fifty-three probands was subdivided into three groups based on phenotypic manifestations. The first group (twenty-eight probands and six affected family members, Tables 2–3) was designated as sub-GCPS and comprised patients with one or more features of GCPS, including preaxial polydactyly, cutaneous syndactyly, widely spaced eyes, or macrocephaly, but who did not meet the suggested clinical criteria for GCPS. The second group comprised patients who had one or more features of PHS, polydactyly, bifid epiglottis and/or hypothalamic hamartoma, but who did not meet the published criteria. We refer to this group as sub-PHS patients (twenty probands and three affected family members, Tables 4–5). We placed individuals with isolated postaxial polydactyly (PAP-A) into a separate group which could overlap with PHS or GCPS as PAP-A is a manifestation of both GCPS and PHS (five probands).
Table 2.
Sub-GCPS patients with mutations
| Individual | Mutation | Findings and Symptoms | Additional Findings | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Mesoaxial Polydactyly |
Postaxial Polydactyly |
Preaxial Polydactyly |
Cutaneous Syndactyly |
Macrocephaly | Wide spaced eyes |
MRI Findings | |||
| G1-1 | c.1497+1G>C, IVS10 | HB | HB3, FB3 | + | + | ||||
| G1-2 | c.1497+1G>C, IVS10 | HB | HB3, FB3 | FB | + | Autism | |||
| G1-3 | c.1497+1G>C, IVS10 | HB | HB3, FB3 | 2,3 toe | + | Speech delay | |||
| G2 | c.2708C>T, p.S903L | - | HB3, FB | Hypospadias, nasal dermoid | |||||
| G3-1 | c.3474delG, p.I1160FfsX46 | HB | HB | Wilms tumor | |||||
| G3-2 | c.3474delG, p.I1160FfsX46 | HB | |||||||
| G4-1 | c.4240C>T, p.Q1414X | HR, FB | 2,3 toe | + | Normal | Trigonocephalic skull shape, - frontal bossing, depressed nasal bridge, double hair whorl, absence of kidney, DD | |||
| G4-2 | c.4240C>T, p.Q1414X | HR, FB | 2,3 toe | High anterior hairline, prominent metopic sutures, broad nasal root and tip, hypospadias, DD | |||||
| G5-1 | c.4430_4431delCT, p.S1477X | HB, FB | + | + | Normal | ||||
| G5-2 | c.4430_4431delCT, p.S1477X | HB, FB | + | Partial empty sella | Craniosynostosis of metopic sutures, bifid epiglottis, hypopituitarism, anosmia, Asperger’s | ||||
| G5-3 | c.4430_4431delCT, p.S1477X | FB | Normal | ||||||
| G6 | c.4432G>T, p.E1478X | FL | HB | FB3 | HR, FB | + | Hypoplasia of the CC | Fine motor delay | |
| G7 | c.4594_4596delTCCinsA, p.S1532TfsX2 | HB | HB3, FB | HB, FB | Enlarged ventricles | Gingival overgrowth, estropia, high, narrow palate, hypotonia, DD | |||
| G8 | Chr7:del41.7-44.9 Mb | HB | HB3, FB3 | HL, FL | + | + | CCM | Strabismus, RSV pneumonitis/asthma, Umbilical hernia, SZ/DD | |
HB, hands bilateral; HR, hand right; HL, hand left; HB3, wide thumbs; FB, foot bilateral; FL, foot left; FB3, wider great toes; CC, corpus collosum; CCM, cerebral cavernous malformation; SZ, seizures; DD, developmental delay; +, presence of finding. Nucleotide numbering reflects cDNA numbering with +1 corresponding to the A of the ATG translation initiation codon in the reference sequence, according to journal guidelines (www.hgvs.org/mutnomen). The initiation codon is codon 1.
Table 3.
Sub-GCPS patients without mutations
| Individual | Deletion Analysis |
Findings and Symptoms | Additional Findings | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Mesoaxial Polydactyly |
Postaxial Polydactyly |
Preaxial Polydactyly |
Cutaneous Syndactyly |
Macrocephaly | Wide spaced eyes |
MRI Findings | |||
| G9 | Array | HB | FL | HB | microcephaly | + | Normal | Craniosynostosis, hernia, hypospadias, SZ, DD | |
| G10 | HB, FB | + | Hypoplasia of the CC, frontal polymicrogyria, cerebellar grey matter heterotopia | Umbilical hernia, transverse vaginal membrane, DD | |||||
| G11 | FR | HB | + | + | Dental cysts | ||||
| G12 | QPCR | Oligodactyly HR | HL, FR | HB, 2,3 toe | + | + | Enlarged ventricles | Coarse face, ears low set, with increased posterior angulation, hypodontia, GH deficient, DD | |
| G13 | QPCR | HB | HB3 | + | DD | ||||
| G14 | QPCR | HB, FB | + | Normal | Short distal phalanges | ||||
| G15 | QPCR | HL, FB | FL | + | + | Mild colpocephaly | Skull asymmetry, high palate, dental crowding, long neck, pectus excavatum, prominent fetal pads, SZ, mild DD | ||
| G16 | FISH | HB, FB | + | Prominent ventricles | Broad forehead, hydronephrosis, inguinal hernias, DD | ||||
| G17 | Array | HB | HB | microcephaly | + | Agenesis of CC | Frontal bossing, infantile spasms, extra rib, DD, hearing loss, constipation, contractures | ||
| G18 | Array | HB | HB3 | Broad hands with unusual creases | + | + | Hypoplasia of the CC | Prominent forehead, depressed nasal bridge, down slanted palpebral fissures, distinctive ears, mild 6th cranial nerve palsy, tracheomalacia with one narrow bronchus, shawl scrotum, elbow dimples, low tone | |
| G19 | QPCR | HL | 2,3 toe | Hypoplasia of the CC | Bilateral club feet, VSD/ASD, iguinal hernia | ||||
| G20 | Array | FB | Agenesis of CC, cerebellar and brain stem hypoplasia, schizencephaly | High palate, 5th finger clinodactyly, left tibial bowing, deceased at 8 days | |||||
| G21 | QPCR | FB | |||||||
| G22 | Array | HB, FL | Short distal phalanges, absent finger nails, short humeri | ||||||
| G23 | Array | HR | Increased CSF space on ultrasound | Hemivertebrae, 10 ribs bilaterally, mild plagiocephaly, depressed nasal bridge, frontal | |||||
| G24 | Array | Bifid second toe FL | FB3 | Normal | Complex cardiac anatomy, bell shaped rib cage, hip dysplasia, small penis, retinal dysplasia, foveal hypoplasia, preauricular skin tag, diaphramatic hernia, supernumerary nipple, short metacarpals and metatarsals, cryptorchidism, scoliois | ||||
| G25 | Array | Complete 2,3 toe | + | + | Normal | Anteverted nares, short nose, hyperextensible joints, leg length discrepancy, DD | |||
| G26 | Array | HL | Agenesis of CC, cerebellar and brainstem hypoplasia, midline cyst | Oral frenula, hydronephrosis, DD | |||||
| G27 | QPCR | + | + | ||||||
| G28 | HB, FB | Pontocerebellar hypoplasia, hypoplasia of the CC | Facial dysmorphism, redundant tongue tissue, ruffled gums, horseshoe kidney, deceased at 5 months, 2 affected siblings | ||||||
HB, hands bilateral; HR, hand right; HL, hand left; HB3, wide thumbs; FB, foot bilateral; FR, foot right; FL, foot left; FB3, wide great toes; CC, corpus collosum; CSF, cerebral spinal fluid; GH, growth hormone; SZ, seizures; DD, developmental delay; VSD/ASD, ventricular/atrial septal defect; +, presence of finding.
Table 4.
Sub-PHS patients with mutations
| Individual | Mutation | Findings and Symptoms | Additional Findings | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Mesoaxial Polydactyly |
Postaxial Polydactyly |
Preaxial Polydactyly |
Cutaneous Syndactyly |
Craniofacial Features |
Bifid Epiglottis |
MRI Findings |
|||
| PH1 | c.2149C>T, p.Q717X | HB | 2,3 toe FB | Deep set eyes, small nose, diastema, small ears | + | HH with multicystic extension | Cloaca (many surgeries), unilateral renal agenesis, decreased renal function, thrombocytopenia, SZ, severe MR | ||
| PH2 | c.2437C>T, p.Q813X | HB, FB | NA | HH | Small nails, PDA, ASD, tricuspid regurgitation, absent pituitary and adrenal glands, pulmonary hypertension, abnormal lung lobulation, imperforate anus, genital hypoplasia, deceased at 1 day | ||||
| PH3 | c.2466delG, p.M824X | oligodactyly HL; fusion of metacarpals | FL | HR; 2,3 toe FL; 2,3,4 FR | NA | HH | Small nails, deceased at 3 months | ||
| PH4 | c.2542delG, p.D848TfsX12 | HR | Macrocephaly, small teeth | + | HH | Visual problems, hearing problems, ectopic right kidney, GH deficiency, obesity, SZ, DD | |||
| PH5 | c.2621_2624del, p.R874PfsX15 | Bilateral | NA | HH | Small nails, pulmonary hypoplasia, absent pituitary gland, adrenal hypoplasia, thyroid hypoplasia, vaginal atresia, vesicovaginal fistula, hydrocolpos, bilateral renal hypoplasia, deceased antepartum at 41 weeks | ||||
| PH6 | c.3004delG, p.V1002X | oligodactyly HR | NA | HH | Osseous syndactyly of metacarpals and metatarsals, short stature, growth hormone deficient, laughing spells | ||||
| PH7 | c.3302dupA, p.N1101KfsX28 | HB | + | HH | Small nails, hypoplastic toes, pointed teeth, midline frenula, laryngeal cleft, GH deficient, genital hypoplasia, neurosensory hearing loss, gelastic SZ | ||||
| PH8-1 | c.3887_3894del, p.L1297SfsX4 | HB | FB | + | Enlarged cerebellar tonsils | Growth hormone deficient, 13:17 translocation | |||
| PH8-2 | c.3887_3894del, p.L1297SfsX4 | HB, FB | + | Normal | |||||
| PH8-3 | c.3887_3894del, p.L1297SfsX4 | HB | + | Thoracic scoliosis, nystagmus, DD | |||||
| PH8-4 | c.3887_3894del, p.L1297SfsX4 | HB, FB | Broad forehead | + | Sphenoid sinus | Extra bone in right foot, chronic sinus problems, 13:17 translocation | |||
HB, hands bilateral; HR, hand right; HL, hand left; FB, foot bilateral; FR, foot right; FL, foot left; HH, hypothalamic hamartoma; SZ, seizures; MR, mental retardation; PDA, patent ductus arteriosus; ASD, atrial septal defect; GH, growth hormone; DD, developmental delay; +, presence of finding. Nucleotide numbering reflects cDNA numbering with +1 corresponding to the A of the ATG translation initiation codon in the reference sequence, according to journal guidelines (www.hgvs.org/mutnomen). The initiation codon is codon 1.
Table 5.
Sub-PHS patients without mutations
| Individual | Deletion Analysis |
Findings and Symptoms | Additional Findings | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Mesoaxial Polydactyly |
Postaxial Polydactyly |
Preaxial Polydactyly |
Cutaneous Syndactyly |
Craniofacial Features |
Bifid Epiglottis |
MRI Findings | |||
| PH9 | HB | Fine scalp hair | + | Disruption between hypothalamus and | Nuchal folds, hypotonia, panhypopituitarism, DD | ||||
| PH10 | HB | Microcephaly, pointed | Normal | Small nails, pituitary problem, microphallus, SZ, DD | |||||
| PH11 | HB | Depressed nasal | + | Normal | Small nails, panhypopituitarism, vaginal tag, hydronephrosis, | ||||
| PH12 | HB, FB | HB, FB | Large anterior | + | NA | Microphallus, unilateral undescended testes | |||
| PH13 | HB | HB | Small mouth and | + | HH | Atrioventricular canal defect, deceased at 5 months | |||
| PH14 | Array | HB, FB | Microcephaly, frontal bossing, mild dolichocephaly, high | Ventriculomegaly, periventricular leukomalacia | Slightly hypoplastic left 5th metacarpal, hypotonia, postnatal growth failure, pseudostrabismus, mild right estropia, bilateral accessory nipples, multiple bladder infection, urethral | ||||
| PH15 | HB | Plagiocephaly | short epiglottis | Possible HH, absent anterior pituitary | Laryngeal web, laryngeal cleft, ASD, mitral valve cleft, urethral reflux, hypopituitarism | ||||
| PH16 | HB3, FB3 | Hypotelorism, left microphthamia, right anophthalm | NA | HH | Panhypopituitarism, choanal atresia, diaphragmatic hernia, severe DD | ||||
| PH17 | NA | HH | SZ, DD | ||||||
| PH18 | Microcephaly, cleft lip | NA | HH | Microphallus, DD | |||||
| PH191 | Oral frenulae | NA | HH | Hypoplastic fifth finger | |||||
| PH201 | Oral frenulae | NA | HH | Hypoplastic middle phalanx of fifth digit, endocrine deficiency | |||||
HB, hands bilateral; HB3, wide thumbs; FB, foot bilateral; FB3, wide great toes; NA, not assessed; HH, hypothalamic hamartoma; DD, developmental delay; SZ, seizures; ASD, atrial septal defect; +, presence of finding.
Probands with features that overlapped with the oral-facial-digital syndromes
Key features of the oral-facial-digital syndromes (OFDS) include tongue and other oral hamartomas, multiple buccal-oral frenula, cleft lip and/or cleft palate, polydactyly, tibial hypoplasia, or cerebellar vermis hypoplasia (Gurrieri, et al., 2007). There are thirteen clinical types of OFDS but only OFDS type 1 has a known molecular etiology (Ferrante, et al., 2001). We delineated this group because there have been reports of patients with manifestations that overlapped PHS, oral-facial-digital syndrome, and other disorders (Muenke, et al., 1991). We identified twenty-one probands that had polydactyly and one or more features of an OFDS (Tables 6–7).
Table 6.
OFD-overlap patients with mutations
| Individual | Mutation | Findings and Symptoms | Other Findings |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mesoaxial Polydactyly |
Postaxial Polydactyly |
Preaxial Polydactyly |
Oral frenula |
Oral Hamartoma |
Cleft Lip/ Palate |
Cerebellar Vermis Hypoplasia |
Tibial hypoplasia |
Cutaneous Syndactyly |
MRI Findings |
|||
| OFD11 | c.2077A>T, p.K693X | HB | FR | + | + | HH | Esotropia, amblyopia, optic nerve hypoplasia, precocious puberty, supernumerary maxillary incisor, gelastic seizures, DD | |||||
| OFD2 | c.2977C>T, p.Q993X | HL | HL | + | Short palpebral fissures, short fingers, small nails, imperforate anus, ASD, deceased at 5 days | |||||||
| OFD3 | c.3002delG, p.G1001AfsX2 | HB | Palate | + | HH, Agenesis of the CC | Bilateral choanal atresia, small wide spaced eyes, small mouth, syngnathia, dysplastic kidney, short limbs, imperforate anus, short fingers, small nails, pregnancy terminated at 22 weeks | ||||||
| OFD4 | c.3040G>T, p.E1014X | Oligodactyly HL | HR | + | HR | Hypothalamic mass | Absent left kidney, imperforate anus, deceased at 1 week | |||||
| OFD52 | c.3370dupC, p.H1124PfsX5 | HB, FB | + | + | R | HB, FB | HH, left cerebral atrophy | Short left ulna, small fibulae, hydrometrocol pos with a vagino-cystic fistula, precocious puberty, MR | ||||
| OFD6 | Chr7:del33.2-47.2 Mb | FB | + | + | + | Dilated ventricles, dural dermoid cyst | Macrocephaly, hypertelorism, bifid epiglottis, bilateral choanal hypoplasia, patent foramen ovale, horseshoe kidney, accessory spleen, bilateral undescended testes, kyphosis, deceased at 2.5 years | |||||
HB, hands bilateral; HR, hand right; HL, hand left; FB, foot bilateral; FR, foot right; FB3, wide great toes; HH, hypothalamic hamartoma; CC, corpus callosum; ASD, atrial septal defect; DD, developmental delay; +, presence of finding. Nucleotide numbering reflects cDNA numbering with +1 corresponding to the A of the ATG translation initiation codon in the reference sequence, according to journal guidelines (www.hgvs.org/mutnomen). The initiation codon is codon 1.
Table 7.
OFD-overlap patients without mutations
| Individual | Deletion Analysis | Findings and Symptoms | Other Findings | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mesoaxial Polydactyly |
Postaxial Polydactyly |
Preaxial Polydactyly |
Oral frenula |
Oral Hamartoma |
Cleft Lip/Palate |
Cerebellar Vermis Hypoplasia |
Tibial hypoplasia |
Cutaneous Syndactyly |
MRI Findings | |||
| OFD7 | HB, FB | HB, FB | Lip and Palate | + | Agenesis of CC, fused basal ganglia, abnormal cortical gyral pattern | Short limbs, club feet, AV canal, cystic pancreas, deceased | ||||||
| OFD8 | FISH | HL | + | 2,3 toe (−) | High anterior hairline, pyloric stenosis, underdeveloped ribs, microphallus, mild DD | |||||||
| OFD9 | Array | FB | palate | Normal CT | Depressed nasal bridge, epilepsy, hydronephrosis, reflux, severe DD | |||||||
| OFD10 | Array | HB | FB | + | lip, tongue | + | HB, FB | Mild hearing loss, wide spaced eyes, clefted epiglottis, DD | ||||
| OFD12 | FL | + | HH | Mild hypospadias, SZ, DD | ||||||||
| OFD12 | HB, FB | + | + | Endocardial cushion defect, Dandy-Walker malformation | Small ears, bilateral polycystic kidney | |||||||
| OFD13 | HB, FB | Lip/palate | Absent premaxilla and midline frenulum, VSD/ASD, absent pituitary, panhypopituitarism, DD | |||||||||
| OFD14 | HB | + | + | + | HH, Agenesis of CC | Wide spaced eyes, hypoplastic left heart, imperforate anus, deceased at 4 months of age | ||||||
| OFD15 | FISH | HB | HB | palate | Normal | Wide spaced eyes, frontal bossing, depressed nasal bridge, micrognathia, retinal dysplasia, optic nerve hypoplasia, detached retina, severe hearing loss | ||||||
| OFD16 | Array | HL, FB | HB, FB | Lip/palate | + | FB | HH | Absent fibulae, short ribs, short long bones, small jaw, pregnancy terminated at 20 weeks | ||||
| OFD17 | + | HB | + | HB | Agenesis of CC | Unilateral radius hypoplasia, bilateral tibia hypoplasia, gingiva overgrowth, cystic kidneys | ||||||
| OFD18 | HB | FB | + | + | Lip/palate | + | Molar tooth sign, HH | Marked rhizomelic and mesomelic shortening with small hands and feet and brachydactyly, absent epiglottis, optic nerve colobomas with searching nystagmus and absent VERs, notched midline small jaw | ||||
| OFD19 | HL | + | + | Hamartoma | Macrocephaly, small finger nails, short 5th finger, second degree microtia, gelastic seizures, bifid tooth, absent tooth, supernumerary tooth | |||||||
| OFD20 | HB, FB | HB, FB | + | Lip/palate | HH | Tethered tongue, vaginal atresia, DD, deceased | ||||||
| OFD21 | + | + | HH, hypoplasia of cerebellum, dandy walker cyst with molar tooth sign | Macrocephaly, wide sutures, frontal bossing, broad depressed nasal bridge, deceased at 2 months | ||||||||
HB, hands bilateral; HL, hand left; FB, foot bilateral; FL, foot left; CC; corpus callosum; HH, hypothalamic hamartoma; NA, not assessed; DD, developmental delay; SZ, seizures; VSD/ASD, ventricular/atrial septal defect; +, presence of finding.
Probands with typical GCPS or PHS
Nineteen probands who fulfilled diagnostic criteria for GCPS (Johnston, et al., 2005) (seventeen probands) or PHS (Biesecker, et al., 1996) (two probands) were included in this report as they have not been reported previously. Detailed clinical data are reported for these nineteen probands (plus five relatives, Tables 8–11). The clinical diagnostic criteria for PHS require the presence of mesoaxial polydactyly and a hypothalamic hamartoma in the proband (Biesecker, et al., 1996). Suggested clinical criteria for GCPS include 1) preaxial polydactyly in at least one limb or broad great toes or thumbs, and 2) cutaneous syndactyly, macrocephaly, and wide spaced eyes (Biesecker, 2001). For this study we set GCPS eligibility criteria of pre-axial polydactyly and the presence of at least one additional feature (cutaneous syndactyly, macrocephaly, wide spaced eyes, postaxial polydactyly).
Table 8.
GCPS patients with mutations
| Individual | Mutation | Findings and Symptoms | Additional Findings | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Mesoaxial Polydactyly |
Postaxial Polydactyly |
Preaxial Polydactyly |
Cutaneous Syndactyly |
Macrocephaly | Wide spaced eyes | MRI Findings | |||
| G29 | c.1096C>T, p.R366X | HB | HB3, FB | HB, FB | + | + | Dental crowding, talipes equinovarus, undescended testis, right inguinal hernia | ||
| G30 | c.1561_1576del, p.S521PfsX9 | FB | FB | + | + | Hypoplasia of corpus callosum, calcified falx | Lipoma on forehead, delayed eruption of molars | ||
| G31 | c.1728C>A, p.Y576X | HB | FB | FB | + | + | Craniosynostosis, epicanthal folds, depressed nasal bridge | ||
| G32 | c.1748G>T, p.C583F | HB | FB | FB | Normal U/S | Umbilical hernia | |||
| G33-1 | c.2374C>T, p.R792X | HB | FB | FB | + | Normal | SZ | ||
| G33-2 | c.2374C>T, p.R792X | HB | FB | Broad nasal bridge | |||||
| G34-1 | c.2708C>T, p.S903L | FB | FB | Normal | Asthma | ||||
| G34-2 | c.2708C>T, p.S903L | HB, FB | FB | + | + | Agenesis of the CC, mild ventricular prominence | Dolichocephaly, sagittal craniosynostosis, bulbous nose, umbilical hernia with diastasis recti, DD | ||
| G34-3 | c.2708C>T, p.S903L | FB | FB | High anterior hairline, | |||||
| G35-1 | c.2741delG, p.G914AfsX38 | HB, FB | 2,3 toe | + | +/atrial sep | Family history of preaxial polydactyly | |||
| G35-2 | c.2741delG, p.G914AfsX38 | HB, FB | + | ||||||
| G36-1 | c.4072C>T, p.Q1358X | HB, FB | FB | ||||||
| G36-2 | c.4072C>T, p.Q1358X | HB, FB | FB | + | + | Umbilical hernia, SZ, DD | |||
| G37 | Chr7:del37.1-49.3 Mb | HB, FB | HB, FB | + | CCM, abnormal CC | Bilateral hydronephrosis, L-ureteral reflux, Course liver, Laryngomalacia, SZ/DD | |||
| G38 | Chr7:del39.7-45.8 Mb | HB3, FB | 2,3 toe | + | + | CCM, ventriculomegaly | Duane syndrome, VSD/ASD, SZ/DD | ||
| G39 | Chr7:del41.0-45.1 Mb | FB | HB | FB, HB | + | Subdural effusion | Cryptorchidism, horizontal earlobe creases, antihelix pit, single transverse palmar crease of the left hand, SZ, DD | ||
HB, hands bilateral; HR, hand right; FB, foot bilateral; CC, corpus collosum; CCM, cerebral cavernous malformation; SZ, seizures; DD, developmental delay; VSD/ASD, ventricular/atrial septal defect; +, presence of finding. Nucleotide numbering reflects cDNA numbering with +1 corresponding to the A of the ATG translation initiation codon in the reference sequence, according to journal guidelines (www.hgvs.org/mutnomen). The initiation codon is codon 1.
Table 11.
PHS patients without mutations
| Individual | Deletion Analysis |
Findings or Symptoms | Additional Findings | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Mesoaxial Polydactyly |
Postaxial Polydactyly |
Preaxial Polydactyly |
Cutaneous Syndactyly |
Craniofacial Features |
Bifid Epiglottis | MRI Findings | |||
| PH22 | Array | HB | + | HH | Small nails, pointed teeth, genital hypoplasia, microglossia, MR | ||||
HB, hands bilateral; HH, hypothalamic hamartoma; MR, mental retardation; +, presence of finding.
DNA Isolation, PCR and Sequencing
DNA was isolated from whole blood using the salting out method (Qiagen, Valencia, CA) following the manufacturer’s instructions. PCR of GLI3 exons and flanking intron sequences was performed using standard methods and primers as described (Johnston, et al., 2005). Sequencing of the GLI3 coding exons was performed with v3.1 BigDye terminator cycle sequencing kit (Applied Biosystems, Foster City, CA) and either the ABI 377 (Applied Biosystems) or ABI 3100 (Applied Biosystems) per the manufacturer’s protocol. Sequence data were compared with the published GLI3 sequence (GenBank reference number NM_000168.5) using Sequencher 4.9 (Gene Codes Corp., Ann Arbor, MI). Nucleotide numbering reflects cDNA numbering with +1 corresponding to the A of the ATG translation initiation codon in the reference sequence, according to journal guidelines (www.hgvs.org/mutnomen). The initiation codon is codon 1. The entire coding region was sequenced for all probands except OFD2 due to insufficient DNA.
DHPLC Analysis
For some probands, screening of exons 3 through 12 and the last third of exon 15 was performed using dHPLC as described in (Johnston, et al., 2005).
Classification of Sequence Variants
We classified sequence variants as causative mutations if they were:
a nonsense or frameshift variant or,
a missense variant that predicted a non-conservative amino acid change and segregated with the phenotype in multiple family members or was de novo in a patient with a GLI3-related phenotype and unaffected parents
qPCR Analysis
qPCR was performed in a subset of individuals to identify deletions and duplications of GLI3 exons. qPCR analysis of the GLI3 coding exons was performed with the Platinum SYBR Green qPCR SuperMix UDG kit (Invitrogen) and the ABI PRISM 7000 (PE Applied Biosystems) as described in Johnston et al. 2005 (Table 3).
Array Hybridization
Zoom-in comparative genomic hybridization (CGH) for chromosome 7p14 was performed as described previously (Johnston, et al., 2007) in a subset of individuals to identify large deletions and duplications on chromosome 7 including GLI3 (Tables 2, 3, 5, 6, 7, 8, 9 and 11).
Table 9.
GCPS patients without mutations
| Individual | Deletion Analysis | Findings and Symptoms | Additional Findings | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Mesoaxial Polydactyly |
Postaxial Polydactyly |
Preaxial Polydactyly |
Cutaneous Syndactyly |
Macrocephaly | Wide spaced eyes |
MRI Findings | |||
| G41 | Array | FL | + | ||||||
| G42 | HB | FB, soft tissue | HB | Agenesis of CC, brain cyst, brain stem hypoplasia | |||||
| G43 | Array | HB | FB | FB | |||||
| G44 | Array | HB | HR | + | Agenesis of CC | Small nose, prominent forehead, deformed ear, MR | |||
| G45 | Array | FB | + | Porencephaly of left occipital and left temporal lobes, absence of septum pellucidum, hypoplastic optic nerves | Bilateral hernia, midline capillary vascular malformation, tetralogy of Fallot | ||||
| G46 | Array | HB3, FB | HB, FB | Trigonocephaly | |||||
HB, hands bilateral; HR, hand right; FB, foot bilateral; FL, foot left; HB3, wide thumbs; CC, corpus collosum; MR, mental retardation; +, presence of finding.
FISH Analysis
FISH analysis was performed in a subset of individuals to identify large deletions on chromosome 7 in the vicinity of GLI3. FISH analysis was performed as described in Johnston et al. 2003 (Tables 3 and 7).
Results
The cohort delineated in this study included ninety-three probands and was drawn from a pool of 174 probands who were referred to our research protocol because they had one or more manifestations consistent with either (or both) GCPS or PHS. In addition, some of these probands were from multiplex families and clinical data on some of those affected family members are included in this cohort. These ninety-three probands were divided into several groups (see inclusion criteria, table 1) and each group is described in turn.
GLI3 mutations in probands with features of GCPS or PHS insufficient to meet clinical criteria
The first group included fifty-three probands with features that overlapped with GCPS or PHS, but these probands did not have sufficient features to warrant a clinical diagnosis of either disorder. Of these fifty-three probands, twenty-eight were categorized in the sub-GCPS group and eight of them had mutations. Of these eight mutations, five were frameshift or nonsense mutations, one was a splice mutation, one was a missense mutation, and one was a large genomic deletion. Four of the truncation or termination mutations were in the predicted domains (either 5′ of position 1998 or 3′ of 3481); c.4240C>T, which predicts p.Q1414X; c.4430_4431delCT, which predicts p.S1477X; c.4432G>T, which predicts p.E1478X; and c.4594_4596delTCCinsA, which predicts p.S1532TfsX2. The fifth was at the 3′ border of the PHS region; c.3474delG, which predicts p.I1160FfsX46. The splice site alteration, c.1497+1G>C, IVS10, has been identified previously (Kalff-Suske, et al., 1999). The missense alteration, c.2708C>T, which predicts p.S903L, was also identified in this study in a proband who fulfilled the clinical criteria for GCPS.
We noted that all five of the frameshift or nonsense mutations in the sub-GCPS group were located in the 3′ region of the gene. Overall, the mutation yield for patients with typical GCPS was thirty-nine of fifty-seven (68%), as compared to eight of twenty-eight (29%) for the sub-GCPS group (p = 0.0006, Fisher’s exact test). The distribution of mutations for typical GCPS with frameshift or nonsense mutations is as follows; thirty-one were in the 5′ region, nine were in the PHS region, and fourteen were in the 3′ region (Fujioka, et al., 2005; Furniss, et al., 2009; Johnston, et al., 2005). Interestingly, all five of the patients with sub-GCPS who have frameshift or nonsense mutations have those mutations in the 3′ region (p = 0.0023 Fisher’s exact test). Of the twenty probands in the sub-PHS group, eight had mutations (40%). Of these eight mutations, all were nonsense or frameshift mutations and all but one of these mutations were in the previously defined PHS region (between cDNA positions 1998 and 3481). One proband had a c.3887_3894del mutation that predicts p.L1297SfsX4. As this mutation would be predicted to cause GCPS, some clinical details are provided here. The proband had bilateral mesoaxial polydactyly of the hand, isolated growth hormone deficiency without a hypothalamic hamartoma, and a bifid epiglottis. Her three affected family members have two to four limb postaxial polydactyly with a bifid epiglottis without a hypothalamic hamartoma. One family member had a broad forehead. The biologic mechanism of how this variant causes a sub-PHS phenotype requires further study.
Seven of the eight mutations in the sub-PHS group were novel. One mutation (c.2149C>T, p.Q717X) has been described previously in a patient with typical PHS (Johnston, et al., 2005). The overall mutation yield for the sub-PHS probands was eight of twenty (40%), which is significantly lower than for patients with typical PHS (twenty of twenty-two, 91%; p = 0.0008, Fisher’s exact test).
One of five patients (20%) in the isolated PAP-A group was found to have a mutation in GLI3, c.874C>T, p.R292C. This mutation is upstream of the zinc finger in a conserved region of the protein.
GLI3 mutations in probands with features of oral-facial-digital syndromes
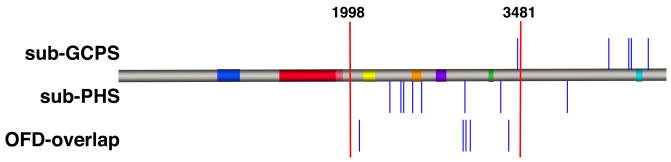
We identified twenty-one probands from our cohort who had one or more features of PHS or GCPS and in addition, one or more features of OFDS. Among these twenty-one probands we identified five frameshift or nonsense mutations that we concluded were pathologic and one large genomic deletion of 14.0 Mb. All five of the frameshift or nonsense mutations were similar in position within GLI3 to other mutations that have been reported to cause PHS (Figure 1). Indeed, several of the probands in this group met the clinical criteria for PHS (OFD1, c.2077A>T, p.K693X; OFD2, c.2977C>T, p.Q993X, OFD3, c.3002delG, p.G1001AfsX2). Patient OFD4 with the c.3040G>T, p.E1014X mutation did not meet clinical criteria for PHS but he had oligodactyly, which we have observed in affected relatives of probands with typical PHS (unpublished observations). Similarly, patient OFD5 with the c.3371dupC, p.H1124PfsX5 mutation had postaxial polydactyly and a hypothalamic hamartoma. Although not sufficient for a clinical diagnosis of PHS, this combination of features has been observed in affected relatives of probands with PHS. Six of twenty-one patients with features that overlap an OFDS had a GLI3 mutation for an overall yield of 29%. This yield of mutations is significantly below that for typical PHS (twenty of twenty-two, 91%, p < 0.0001, Fisher’s exact test).
Figure 1.
Diagram of the position within the gene of newly described nonsense and frameshift mutations in probands with sub-GCPS, sub-PHS and OFD-overlap. Some of the closely spaced mutations have been adjusted for increased visual clarity. Red bars denote the 5′ and 3′ limits of the PHS region at nucleotides 1998 and 3481 respectively. The colored bars on the protein show the conserved domains of GLI3 as defined elsewhere (Ruppert, et al., 1990).
GLI3 mutations in probands with typical GCPS or PHS
The final group included patients with typical manifestations of GCPS or PHS. These patients were similar in their clinical manifestations to patients described previously (Johnston, et al., 2005). Among the seventeen probands with GCPS we identified eleven mutations. Of these eleven mutations, six were frameshift or nonsense mutations, two were missense mutations and three were large genomic deletions. Of the six frameshift or nonsense mutations, three were in the 5′ segment of GLI3 (between the start codon and cDNA position 1998); c.1096C>T, which predicts p.R366X; c.1561_1576del, which predicts p.S521PfsX9, and c.1728C>A, which predicts p.Y576X. One nonsense mutation (c.4072C>T, p.Q1358X) was in the 3′ segment of GLI3 (between cDNA position 3481 and the normal stop codon). Two nonsense or frameshift mutations were in the middle region of GLI3 (between cDNA positions 1998 and 3481), which in most cases is associated with a phenotype of Pallister-Hall syndrome. Mutation (c.2374C>T; p.R792X) represents the eighth report of this variant associated with GCPS and this variant has been associated with nonsense-mediated mRNA decay (Furniss, et al., 2007). The second frameshift or nonsense mutation in the middle region of GLI3 is c.2741delG, which predicts p.G914AfsX38. Two missense mutations were identified, c.1748G>T, p.C583F, and c.2708C>T, p.S903L. Of these eight mutations, six are novel. There were three probands in this group with deletions that included GLI3 and ranged from 4.1 Mb to 12.2 Mb. All three of these deletions have novel breakpoints. These individuals were given a diagnosis of GCPS contiguous gene syndrome based on their molecular findings. This phenotype can include microcephaly or normocephaly, cognitive impairment, seizures, and other manifestations.
Of the two probands with PHS, one had a mutation in GLI3, c.2685C>G, p.Y895X. This mutation conforms to the previously described correlation that PHS mutations lie between cDNA positions 1998 and 3481 and is novel.
The overall yield of mutations was 65% for GCPS (eleven of seventeen) and 50% for PHS (one of two). We previously showed that among patients with typical GCPS, twenty-eight of forty patients had a GLI3 mutation (70%) and nineteen of twenty probands with PHS had GLI3 mutations (95%)(Johnston, et al., 2005). The results in the current study are similar for GCPS. Merging these data, the current estimates for GCPS would be thirty-nine of fifty-seven (68%) and for PHS would be twenty of twenty-two probands (91%).
Discussion
GLI3 mutations have been associated with several phenotypes including GCPS (Vortkamp, et al., 1991), PHS (Kang, et al., 1997) isolated polydactyly types A, A/B, and preaxial polydactyly type 4 (Radhakrishna, et al., 1999; Radhakrishna, et al., 1997), and a single case of acrocallosal syndrome (Elson, et al., 2002). By combining the data in this report with those of our prior work (Johnston, et al., 2005) we predict that when an individual manifests features sufficient for the clinical diagnostic criteria for PHS or GCPS, their chance of having a mutation in GLI3 is high; 91% and 68%, respectively. The data presented here extend these observations into several distinct groups of patients.
In the early phases of gene discovery efforts, it is important to maximize the likelihood of locus homogeneity by setting strict clinical eligibility criteria. This was done successfully for PHS, and was likely done for GCPS as well. As noted above, nearly all patients who met the clinical criteria for PHS had a truncating mutation in the middle third of GLI3. In this study we hypothesized that a relaxation of the clinical criteria would identify additional patients with GLI3 mutations. By relaxing the criteria to allow subjects with either mesoaxial polydactyly or hypothalamic hamartoma (but not requiring both), we show that a substantial proportion (50% or eight of sixteen) of patients have mutations in GLI3, a substantial and clinically useful yield that is slightly more than half the rate for patients who meet clinical criteria. When the criteria are relaxed even further to allow patients with syndromic postaxial polydactyly without mesoaxial polydactyly or hypothalamic hamartoma, no mutations were identified in four additional probands. Similar to the situation for PHS, the relaxation of the clinical criteria for GCPS allowed us to identify mutations in 29% of patients in the sub-GCPS category, again about half the yield for patients who meet the former criteria. We had a limited set of probands enrolled in the study who had non-syndromic polydactyly, which was mostly postaxial polydactyly. The yield in these patients was one of five or 20% but because this cohort is small, we believe that the implications of this finding are limited.
We also identified a cohort of patients who had one or more features of an oral-facial-digital syndrome. Other than OFDS type 1, there is no known molecular etiology for the many types that have been described (up to thirteen types have been proposed). We reasoned that some cases of OFDS could be caused by mutations in GLI3 because: (1) there were a number of clinical reports of patients whose findings overlapped OFDS and PHS; (2) OFDS type 1 is a ciliopathy (Ferrante, et al., 2006); and (3) GLI3 requires ciliary function for proper processing (Haycraft, et al., 2005). We selected twenty-one cases from our cohort with one or more features of an OFDS. Some of the patients had sufficient features to warrant a diagnosis of PHS or GCPS as well, but some of the patients have been accepted as examples of an OFDS as evidenced by their publication in the literature (Fujiwara, et al., 1999; Stephan, et al., 1994). Among these twenty-one probands, we identified six cases with causative mutations in GLI3, establishing molecular evidence that mutations of this gene can cause phenotypes within the OFDS spectrum. Taken together, these data suggest that clinicians and molecular diagnostic laboratories should encourage a relaxation of clinical criteria for GLI3 testing for patients with one or more features of GCPS or PHS. This would include patients with a feature of PHS or GCPS and one or more features of an OFDS. In this way additional patients will be diagnosed molecularly, which can be valuable for directing further clinical evaluations (endocrine and imaging studies), prognostic advice, molecular diagnostics in other family members, and family planning.
Beyond the clinical diagnostic utility, these data further the understanding of the biology of this gene and its pathway. The mutational spectra of typical GCPS and PHS are distinct; GCPS is caused by a wide range of mutations, but PHS is caused essentially only by truncating mutations. The data presented here, combined with published cases (Borg, et al., 2007; Fujioka, et al., 2005; Furniss, et al., 2009; Johnston, et al., 2005; Mendoza-Londono, et al., 2005; Roscioli, et al., 2005; Yilmaz, et al., 2008), describe 147 mutations in patients with typical GCPS or PHS. The mutation distribution in these two phenotypes is distinct. The GCPS mutations include large deletions/duplications (n=31) and translocations (n=5), and a variety of point mutations including missense (n=9), in frame deletions (n=1), splice (n=11), and frameshift or nonsense mutations (n=54). The distribution among patients with PHS is limited to one splice mutation and thirty-five frameshift or nonsense mutations. The difference in these mutation spectra is highly statistically significant (frameshift/nonsense vs. all other types; Fisher’s exact test < 0.0001). We have previously shown that among the patients with truncating or frameshift mutations, the position of the mutations in GLI3 robustly correlates with the phenotype; patients with PHS have mutations only in the middle portion of the gene (cDNA position 1998 to 3481), whereas patients with GCPS typically have mutations 5′ of position 1998 or 3′ of 3481. Again, the association between mutation position and phenotype is highly significant (3′ mutation vs. 5′ or middle segment, Fisher’s exact test <0.0001).
The data presented here not only strengthen the known association among those with typical GCPS and PHS but also show the same mutation trend in atypical forms of the disorders. Among eight probands with sub-PHS, all eight mutations are frameshift or nonsense, whereas this is the case for slightly more than half, five of eight, of the sub-GCPS probands. Seven of eight of the PHS truncation or nonsense mutations lie in the middle third of the gene, whereas this is the case for none of five frameshift or nonsense mutations among patients with sub-GCPS. These data support the notion that the anomalies of GCPS and PHS are specific to their mutational mechanism, whether those anomalies are typical (PHS and GCPS) or atypical (sub-PHS and sub-GCPS).
There was no apparent correlation for the type or position of frameshift or nonsense mutations within the sub-PHS group that explained or predicted that these mutations caused an atypical phenotype as distinct from typical PHS, as all or nearly all were in the middle third of the gene. However, we did find a correlation of mutations in sub-GCPS patients that distinguished them from GCPS. In probands with GCPS, the frameshift or nonsense mutations were distributed among the three segments of the gene; 5′ segment (n=31), middle segment (n=9), and 3′ segment (n=14). In contrast, five of five frameshift or nonsense mutations in probands with sub-GCPS were in the 3′ segment of the gene (3′ mutation vs. 5′ or middle segment, Fisher’s exact test = 0.0023). These data suggest that the frameshift and nonsense mutations in the 3′ segment of the gene cause distinct biologic and phenotypic consequences from those in the other two segments of the gene.
The transition at nucleotide 1998 relates to the position of these mutations with respect to the zinc finger domain-encoding region and the normal proteolytic processing site of the GLI3 protein (Kalff-Suske, et al., 1999). The transition at nucleotide 3481 may relate to the presence of the transactivation domain (Ruppert, et al., 1990; Shin, et al., 1999). There are known exceptions to these correlations. There is a recurrent c.2374C>T, p.R792X mutation, which lies within the PHS region of the gene, but in eight of eight families (including one proband in this report) is associated with a typical GCPS phenotype (Debeer, et al., 2003; Furniss, et al., 2009; Johnston, et al., 2005; Kalff-Suske, et al., 1999). A similar mutation, c.2741delG, p.G914AfsX38, has been identified in a single family with a typical GCPS phenotype in this report. The proband in this case manifested postaxial polydactyly with macrocephaly and hypertelorism and had a family history of preaxial polydactyly. A third exception is a single family with PHS that has a splice mutation instead of a frameshift or nonsense mutation, although that mutation likely produces a truncated gene product (Johnston, et al., 2005). These data show that the clinical spectrum of phenotypes caused by mutations in GLI3 is wider than previously appreciated. Further, they demonstrate that some mutant alleles of GLI3 can cause malformations that are milder than the typical, clinically defined pleiotropic picture of these disorders, in that they do not demonstrate all of the features required for a clinical diagnosis. The previously reported association of mutation type and phenotype (PHS vs. GCPS) is strengthened by this report and it is extended into milder phenotypes as well. In addition, the distribution of frameshift and nonsense mutations in patients with sub-GCPS is distinct from that in those with typical GCPS, which suggests that these mutations are pathogenetically distinct. The data presented here should encourage molecular diagnostic laboratories to test a wider array of patients and the data should be useful to further understand the pathogenesis of these distinct pleiotropic developmental anomalies.
Table 10.
PHS patients with mutations
| Individual | Mutation | Findings and Symptoms | Additional Findings | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Mesoaxial Polydactyly |
Postaxial Polydactyly |
Preaxial Polydactyly |
Cutaneous Syndactyly |
Craniofacial Features |
Bifid Epiglottis | MRI Findings | |||
| PH21 | c.2685C>G, p.Y895X | HB | HB | HB, FB | HH | Bilateral renal hypoplasia | |||
HB, hands bilateral; FB, foot bilateral; HH, hypothalamic hamartoma. Nucleotide numbering reflects cDNA numbering with +1 corresponding to the A of the ATG translation initiation codon in the reference sequence, according to journal guidelines (www.hgvs.org/mutnomen). The initiation codon is codon 1.
Acknowledgments
The authors thank the following genetic professionals for referring patients to our study: William P. Allen, David J. Aughton, Christopher Cunniff, Sally Davies, William B. Dobyns, Linda Genen, Daniel Gruskin, Ketil Heimdal, Gail Herman, Jodi Hoffman, Helen Hughes, LaDonna Immken, Jeffrey Innis, Ian Krantz, David Manchester, Elizabeth McPherson, Thomas Morgan, Maximilian Muenke, Tracy Oh, Melissa Parisi, Betsy Peach, Lynda Pollack, Nazneen Rahman, Miranda Splitt and LuAnn Weik. JMG is supported by SHARE’s Childhood Disability Center, the Steven Spielberg Pediatric Research Center, the NIH/NICHD Program Project Grant (HD22657), and the Medical Genetics NIH/NIGMS Training Program Grant (5-T32-GM08243). We also acknowledge the Manchester NIHR Biomedical research Centre. This research was supported by funding from the Intramural Research Program of the National Human Genome Research Institute of the National Institutes of Health.
Footnotes
DISCLAIMER: The opinions and assertions contained herein are the views of the authors and are not to be construed as official or as reflecting the views of the United States Department of Defense.
References
- Biesecker LG. GeneReviews at GeneTests: Medical genetics information resource (database online) Copyright, University of Washington; Seattle: 2001. Greig cephalopolysyndactyly syndrome; pp. 1997–2004. [PubMed] [Google Scholar]
- Biesecker LG, Aase JM, Clericuzio C, Gurrieri F, Temple IK, Toriello H. Elements of morphology: standard terminology for the hands and feet. Am J Med Genet A. 2009;149A(1):93–127. doi: 10.1002/ajmg.a.32596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biesecker LG, Abbott M, Allen J, Clericuzio C, Feuillan P, Graham JM, Jr, Hall J, Kang S, Olney AH, Lefton D, et al. Report from the workshop on Pallister-Hall syndrome and related phenotypes. Am J Med Genet. 1996;65(1):76–81. doi: 10.1002/(SICI)1096-8628(19961002)65:1<76::AID-AJMG12>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Bonnemann CG, Drishnamoorthy KS, Johnston JJ, Lee MM, Fowler DJ, Bsiesecker LG, Holmes LB. Clinical and Molecular Heterogeneity of Syndromic Hypothalamic Hamartoma: Delineation of a Recognizable Mild Phenotype. Am J Med Genet. doi: 10.1002/ajmg.a.63306. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borg K, Nowakowska B, Obersztyn E, Cheung SW, Brycz-Witkowska J, Korniszewski L, Mazurczak T, Stankiewicz P, Bocian E. Complex balanced translocation t(1;5;7)(p32.1;q14.3;p21.3) and two microdeletions del(1)(p31.1p31.1) and del(7)(p14.1p14.1) in a patient with features of Greig cephalopolysyndactyly and mental retardation. Am J Med Genet A. 2007;143A(22):2738–43. doi: 10.1002/ajmg.a.32017. [DOI] [PubMed] [Google Scholar]
- Debeer P, Peeters H, Driess S, De Smet L, Freese K, Matthijs G, Bornholdt D, Devriendt K, Grzeschik KH, Fryns JP, et al. Variable phenotype in Greig cephalopolysyndactyly syndrome: Clinical and radiological findings in 4 independent families and 3 sporadic cases with identified GLI3 mutations. Am J Med Genet. 2003;120A(1):49–58. doi: 10.1002/ajmg.a.20018. [DOI] [PubMed] [Google Scholar]
- Elson E, Perveen R, Donnai D, Wall S, Black GC. De novo GLI3 mutation in acrocallosal syndrome: Broadening the phenotypic spectrum of GLI3 defects and overlap with murine models. J Med Genet. 2002;39(11):804–6. doi: 10.1136/jmg.39.11.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrante MI, Giorgio G, Feather SA, Bulfone A, Wright V, Ghiani M, Selicorni A, Gammaro L, Scolari F, Woolf AS, et al. Identification of the gene for oral-facial-digital type I syndrome. Am J Hum Genet. 2001;68(3):569–76. doi: 10.1086/318802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrante MI, Zullo A, Barra A, Bimonte S, Messaddeq N, Studer M, Dolle P, Franco B. Oral-facial-digital type I protein is required for primary cilia formation and left-right axis specification. Nat Genet. 2006;38(1):112–7. doi: 10.1038/ng1684. [DOI] [PubMed] [Google Scholar]
- Fujioka H, Ariga T, Horiuchi K, Otsu M, Igawa H, Kawashima K, Yamamoto Y, Sugihara T, Sakiyama Y. Molecular analysis of non-syndromic preaxial polydactyly: preaxial polydactyly type-IV and preaxial polydactyly type-I. Clin Genet. 2005;67(5):429–33. doi: 10.1111/j.1399-0004.2005.00431.x. [DOI] [PubMed] [Google Scholar]
- Fujiwara I, Kondo Y, Iinuma K. Oral-facial-digital syndrome with hypothalamic hamartoma, postaxial ray hypoplasia of the limbs, and vagino-cystic communication: a new variant? Am J Med Genet. 1999;83(2):77–81. [PubMed] [Google Scholar]
- Furniss D, Critchley P, Giele H, Wilkie AO. Nonsense-mediated decay and the molecular pathogenesis of mutations in SALL1 and GLI3. Am J Med Genet A. 2007;143A(24):3150–60. doi: 10.1002/ajmg.a.32097. [DOI] [PubMed] [Google Scholar]
- Furniss D, Kan SH, Taylor IB, Johnson D, Critchley PS, Giele HP, Wilkie AO. Genetic screening of 202 individuals with congenital limb malformations and requiring reconstructive surgery. J Med Genet. 2009;46(11):730–5. doi: 10.1136/jmg.2009.066027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galasso C, Scire G, Fabbri F, Spadoni GL, Killoran CE, Biesecker LG, Boscherini B. Long-term treatment with growth hormone improves final height in a patient with Pallister-Hall syndrome. Am J Med Genet. 2001;99(2):128–31. doi: 10.1002/1096-8628(2001)9999:9999<::aid-ajmg1128>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- Gurrieri F, Franco B, Toriello H, Neri G. Oral-facial-digital syndromes: review and diagnostic guidelines. Am J Med Genet A. 2007;143A(24):3314–23. doi: 10.1002/ajmg.a.32032. [DOI] [PubMed] [Google Scholar]
- Hall BD, Graham JM, Jr, Cassidy SB, Opitz JM. Elements of morphology: standard terminology for the periorbital region. Am J Med Genet A. 2009;149A(1):29–39. doi: 10.1002/ajmg.a.32597. [DOI] [PubMed] [Google Scholar]
- Haycraft CJ, Banizs B, Aydin-Son Y, Zhang Q, Michaud EJ, Yoder BK. Gli2 and Gli3 localize to cilia and require the intraflagellar transport protein polaris for processing and function. PLoS Genet. 2005;1(4):e53. doi: 10.1371/journal.pgen.0010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston JJ, Olivos-Glander I, Killoran C, Elson E, Turner JT, Peters KF, Abbott MH, Aughton DJ, Aylsworth AS, Bamshad MJ, et al. Molecular and clinical analyses of Greig cephalopolysyndactyly and Pallister-Hall syndromes: robust phenotype prediction from the type and position of GLI3 mutations. Am J Hum Genet. 2005;76(4):609–22. doi: 10.1086/429346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston JJ, Walker RL, Davis S, Facio F, Turner JT, Bick DP, Daentl DL, Ellison JW, Meltzer PS, Biesecker LG. Zoom-in comparative genomic hybridisation arrays for the characterisation of variable breakpoint contiguous gene syndromes. J Med Genet. 2007;44(1):e59. doi: 10.1136/jmg.2006.042473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalff-Suske M, Wild A, Topp J, Wessling M, Jacobsen EM, Bornholdt D, Engel H, Heuer H, Aalfs CM, Ausems MG, et al. Point mutations throughout the GLI3 gene cause Greig cephalopolysyndactyly syndrome. Hum Mol Genet. 1999;8(9):1769–77. doi: 10.1093/hmg/8.9.1769. [DOI] [PubMed] [Google Scholar]
- Kang S, Graham JM, Jr, Olney AH, Biesecker LG. GLI3 frameshift mutations cause autosomal dominant Pallister-Hall syndrome. Nat Genet. 1997;15(3):266–8. doi: 10.1038/ng0397-266. [DOI] [PubMed] [Google Scholar]
- Killoran CE, Abbott M, McKusick VA, Biesecker LG. Overlap of PIV syndrome, VACTERL and Pallister-Hall syndrome: Clinical and molecular analysis. Clin Genet. 2000;58(1):28–30. doi: 10.1034/j.1399-0004.2000.580105.x. [DOI] [PubMed] [Google Scholar]
- Kos S, Roth K, Korinth D, Zeilinger G, Eich G. Hydrometrocolpos, postaxial polydactyly, and hypothalamic hamartoma in a patient with confirmed Pallister-Hall syndrome: a clinical overlap with McKusick-Kaufman syndrome. Pediatr Radiol. 2008;38(8):902–6. doi: 10.1007/s00247-008-0870-5. [DOI] [PubMed] [Google Scholar]
- Mendoza-Londono R, Kashork CD, Shaffer LG, Krance R, Plon SE. Acute lymphoblastic leukemia in a patient with Greig cephalopolysyndactyly and interstitial deletion of chromosome 7 del(7)(p11.2 p14) involving the GLI3 and ZNFN1A1 genes. Genes Chromosomes Cancer. 2005;42(1):82–6. doi: 10.1002/gcc.20100. [DOI] [PubMed] [Google Scholar]
- Muenke M, Ruchelli ED, Rorke LB, McDonald-McGinn DM, Orlow MK, Isaacs A, Craparo FJ, Dunn LK, Zackai EH. On lumping and splitting: a fetus with clinical findings of the oral-facial-digital syndrome type VI, the hydrolethalus syndrome, and the Pallister-Hall syndrome. Am J Med Genet. 1991;41(4):548–56. doi: 10.1002/ajmg.1320410436. [DOI] [PubMed] [Google Scholar]
- Ng D, Johnston JJ, Turner JT, Boudreau EA, Wiggs EA, Theodore WH, Biesecker LG. Gonadal mosaicism in severe Pallister-Hall syndrome. Am J Med Genet. 2004;124A(3):296–302. doi: 10.1002/ajmg.a.20338. [DOI] [PubMed] [Google Scholar]
- Radhakrishna U, Bornholdt D, Scott HS, Patel UC, Rossier C, Engel H, Bottani A, Chandal D, Blouin JL, Solanki JV, et al. The phenotypic spectrum of GLI3 morphopathies includes autosomal dominant preaxial polydactyly type-IV and postaxial polydactyly type-A/B; No phenotype prediction from the position of GLI3 mutations. Am J Hum Genet. 1999;65(3):645–55. doi: 10.1086/302557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishna U, Wild A, Grzeschik KH, Antonarakis SE. Mutation in GLI3 in postaxial polydactyly type A. Nat Genet. 1997;17(3):269–71. doi: 10.1038/ng1197-269. [DOI] [PubMed] [Google Scholar]
- Roscioli T, Kennedy D, Cui J, Fonseca B, Watson GF, Pereira J, Xie YG, Mowat D. Pallister-Hall syndrome: unreported skeletal features of a GLI3 mutation. Am J Med Genet A. 2005;136A(4):390–4. doi: 10.1002/ajmg.a.30818. [DOI] [PubMed] [Google Scholar]
- Ruppert JM, Vogelstein B, Arheden K, Kinzler KW. GLI3 encodes a 190-kilodalton protein with multiple regions of GLI similarity. Mol Cell Biol. 1990;10(10):5408–15. doi: 10.1128/mcb.10.10.5408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin SH, Kogerman P, Lindstrom E, Toftgard R, Biesecker LG. GLI3 mutations in human disorders mimic Drosophila cubitus interruptus protein functions and localization. Proc Natl Acad Sci U S A. 1999;96(6):2880–4. doi: 10.1073/pnas.96.6.2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan MJ, Brooks KL, Moore DC, Coll EJ, Goho C. Hypothalamic hamartoma in oral-facial-digital syndrome type VI (Varadi syndrome) Am J Med Genet. 1994;51(2):131–6. doi: 10.1002/ajmg.1320510209. [DOI] [PubMed] [Google Scholar]
- Turner C, Killoran C, Thomas NS, Rosenberg M, Chuzhanova NA, Johnston J, Kemel Y, Cooper DN, Biesecker LG. Human genetic disease caused by de novo mitochondrial-nuclear DNA transfer. Hum Genet. 2003;112(3):303–9. doi: 10.1007/s00439-002-0892-2. [DOI] [PubMed] [Google Scholar]
- Vortkamp A, Gessler M, Grzeschik KH. GLI3 zinc-finger gene interrupted by translocations in Greig syndrome families. Nature. 1991;352(6335):539–40. doi: 10.1038/352539a0. [DOI] [PubMed] [Google Scholar]
- Yilmaz Z, Gokdemir M, Derbent M, Sahin FI. Greig syndrome based on a de novo translocation. Pediatr Int. 2008;50(2):248–50. doi: 10.1111/j.1442-200X.2008.02550.x. [DOI] [PubMed] [Google Scholar]