Abstract
Nephronophthisis-related ciliopathies (NPHP-RC) are recessive disorders featuring dysplasia or degeneration preferentially in kidney, retina, and cerebellum. Here we combine homozygosity mapping with candidate gene analysis by performing “ciliopathy candidate exome capture” followed by massively-parallel sequencing. We detect 12 different truncating mutations of SDCCAG8 in 10 NPHP-RC families. We demonstrate that SDCCAG8 is localized at both centrioles and directly interacts with NPHP-RC-associated OFD1. Depletion of sdccag8 causes kidney cysts and a body axis defect in zebrafish and induces cell polarity defects in 3D renal cell cultures. This work identifies SDCCAG8 loss of function as a novel cause of a retinal-renal ciliopathy and validates exome capture analysis for broadly heterogeneous single-gene disorders.
INTRODUCTION
Nephronophthisis (NPHP) is a recessive cystic kidney disease that represents the most frequent genetic cause of end-stage kidney disease in the first three decades of life. NPHP-related ciliopathies (NPHP-RC) are single-gene recessive disorders, which cause retinal-renal ciliopathies that affect kidney, retina, brain and liver by prenatal-onset dysplasia or by childhood-onset tissue degeneration. So far 9 different NPHP genes have been identified as causing NPHP-RC (NPHP1-NPHP9)1-11. The finding that proteins mutated in cystic kidney diseases are located at the primary cilium-centrosome complex provided a unifying disease mechanism for NPHP-RC characterizing them as ciliopathies. Many of the NPHP-RC gene products interact at the cilium-centrosome complex throughout the cell cycle. We recently demonstrated in a large cohort of 1,167 families with NPHP-RC that in more than 70% of cases the causative gene is unknown12. Furthermore, in families with severe forms of NPHP-RC we did not detect a mutation in any of the nine known NPHP-RC causing genes in 90 of 120 families (75%) (Otto et al., unpublished). Upon homozygosity mapping, 50 of these families exhibited putative homozygous disease loci, of which more than 30 showed no positional overlap, strongly indicating that many additional recessive NPHP-RC genes must exist.
RESULTS
Homozygosity mapping focuses exome capture
The finding that recently discovered NPHP-RC genes caused the disorder only in a small number of cases (<1%)9 necessitated the ability to map and identify disease genes in single families. We therefore developed a strategy that combines homozygosity mapping in single families13 with exon capture and consecutive massively parallel (MP) sequencing.14 Using the NimbleGen™ 385k platform we designed a ciliopathy candidate exon capture (CEC) array”, which contains oligonucleotides that interrogate ~13,000 exons from the “UCSC Gene” annotation (genome.ucsc.edu) of 828 NPHP-RC candidate genes. Candidates were derived from ciliopathy animal models, from the photoreceptor sensory cilia proteome15 and other candidate sources16 (Supplementary Table 1 Online).
Because exon capture with consecutive MP sequencing bears the problem of yielding too many variants from normal reference sequence (VRS) for making a safe call regarding the disease-causing mutation14, we devised a strategy of a priori reduction of VRS (Supplementary Table 1). These a priori restriction criteria consisted of, i) capturing only ~13,000 ciliopathy candidate exons instead of all ~180,000 CCDS exons (~15-fold reduction) (Supplementary Table 1), ii) evaluation of coding SNPs, splice variants and indels only (as other variants will be difficult to interpret), iii) absence of VRSs from a database of innocuous single nucleotide polymorphisms (dbSNP130) (2.3-fold reduction), iv) evaluation only within the mapped homozygous candidate region of an individual or family (~20-fold reduction), and v) preferential evaluation of truncating mutations (~4-fold reduction). This approach allowed a mean reduction of VRS by ~2,760-fold and led to the identification of the disease-causing gene in 3 out of 5 attempts (Supplementary Table 1). Homozygous mutations were discovered in the known NPHP-RC genes AHI1 (family A2045) and INVS (family A128) (Supplementary Table 1 Online). More importantly, a homozygous mutation was discovered in SDCCAG8 as a novel cause of NPHP-RC.
Null-mutations of SDCCAG8 cause retinal-renal degeneration
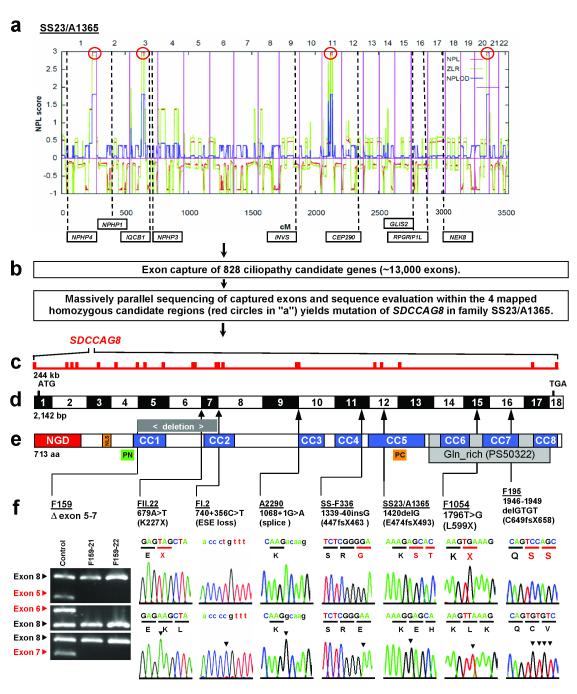
Specifically, in consanguineous family SS23/A1365 two siblings had Senior-Loken syndrome (SLSN), the association of nephronophthisis with retinal degeneration. Homozygosity mapping using the Affymetrix 250k StyI SNP platform yielded four segments of homozygosity (Fig. 1a).13 Because none of the 4 homozygous segments (red circles in Fig. 1a) coincided with any of the 9 known gene loci for NPHP-RC (dashed lines in Fig. 1a) we hypothesized that SLSN in SS23/A1365 must be caused by recessive mutations of a novel NPHP-RC causing gene. Exon capture and MP sequencing (Fig. 1b) led to the identification of a homozygous truncation mutation (p.E474fsX493) in SDCCAG8 (Fig. 1c-f, Table 1).
Figure 1. Homozygosity mapping, exon capture, and massively parallel sequencing identifies SDCCAG8 mutations as causing nephronophthisis with retinal degeneration.
(a) Non-parametric LOD (NPL) scores across the human genome in 2 sibs with nephronophthisis and retinal degeneration of consanguineous family SS23/A1365. X-axis gives Affymetrix 250k StyI array SNP positions on human chromosomes concatenated from p-ter (left) to q-ter (right). Genetic distance is given in cM. Four maximum NPL peaks (red circles) indicate candidate regions of homozygosity by descent.
(b) Exon capture of 828 ciliopathy candidate genes with consecutive massively parallel sequencing and sequence evaluation within the 4 mapped homozygous candidate regions (red circles in “a”) yields mutation of SDCCAG8 in SS23/A1365.
(c) The SDCCAG8 gene extends over 244 kb and contains 18 exons (vertical hatches).
(d) Exon structure of human SDCCAG8 cDNA. Positions of start codon (ATG) and of stop codon (TGA) are indicated. For mutations detected (see f ) arrows indicate positions relative to exons and protein domains (see e).
(e) Domain structure of the SDCCAG8 protein. N-terminal globular domain (NGD), nuclear localization (NLS) domain, coiled-coil domains (CC), and glutamine-rich region (Gln_rich). PN and PC denote peptides used for antibody generation.
(f) Eight homozygous SDCCAG8 mutations detected in 8 families with nephronophthisis and retinal degeneration. Family number, mutation and predicted translational changes are indicated (see Table 1). A homozygous deletion covering exons 5-7 is demonstrated by agarose gel electrophoresis. Sequence traces are shown for mutations above normal controls. Mutated nucleotides are indicated by arrow heads in traces of normal controls.
Table 1.
Twelve different truncating mutations of SDCCAG8 in 10 families with NPHP-RC.
| Family -Individual |
Ethnic origin |
Nucleotide changea,b |
Deduced protein change |
Exon (state) |
Parental consan- guinity |
Kidney (age at ESKF) |
Eye (RD at age) |
Otherc |
|---|---|---|---|---|---|---|---|---|
| F159 -21 -22 |
Europe | c.421-?_ 740+?del |
p.E141_R247 del107fs |
Δ exon 5-7 (hom) |
Yes | NPHP, Bx -21: (11 yr) -22: (7 yr) |
RD -21: (7 yr) -22: (normal at 6 yr) |
-22: generalized seizures |
| FII.22 -21 -22 |
ND | c.679A>T | p.K227X | 7 (hom) | Yes | NPHP -21: (13 yr) -22: (13 yr) |
RD -21: (14 yr) -22: (1 yr) |
Mild MR, HG, OB |
| AR37 -05 -07 -02 |
Northern European |
c.696T>G c.740+1delG |
p.Y232X p.R247fsX250 |
7 (het) IVS7 (het) |
No | NPHP, Bx -05: (22 yr) -07: no ESKF -02: (28 yr) |
RD -05: (13 yr) -07: (11 yr), SNB -02: RD (2 yr) |
-05: OB, HG, PN -07: MR -02: OB, CD |
| FI.2 -21 -22 -23 -24 |
Gypsy | c.740+356C>T | loss of ESE site (aberrant ins IVS7) |
IVS 7 (hom) |
Yes | NPHP -21: (23 yr) -22: (13 yr) -23: (10 yr) -24: (6 yr) |
RD -21: (29 yr) -22: (13 yr) -23: (10 yr) -24: (6 yr) |
Mild MR, OB Mild MR, OB Mild MR |
| A2290 | Turkey | c.1068+1G>A | obligatory splice site |
IVS 9 (hom) |
Yes | NPHP, Bx (14 yr) |
RD | Brain: arachnoid cyst, mild MR |
| SS-F336 -21 -22 |
Algeria | c.1339- 1340insG |
p.E447fsX463 | 11 (hom) | Yes | NPHP, Bx -21: (7 yr) -22: (4 yr) |
RD -21: flat ERG (13 yr) -22: flat ERG (6 yr) |
-21: mild MR, No liver or bone disease |
| SS23/A1365 -21 -22 |
La Réunion | c.1420delG | p.E474fsX493 | 12 (hom) | Yes | NPHP, Bx -21: (4 yr) -22: (14 yr) |
RD -21: 30% vision (14 yr) -22: blind (7 yr) |
-21: Brain scan normal |
| NK-F1063 | India | c.1444delA c.1627_1630del GATA |
p.T482fsX493 p.D543fsX566 |
12 (het) 14 (het) |
No | NPHP, ESKF | RD | OB, mild MR |
| F1054 -21 -22 |
Pakistan | c.1796T>G | p.L599X | 15 (hom) | Yes | NPHP -21: (<10 yr) -22: (<9 yr) |
ND | -21: Polycystic ovary syndrome |
| F195 | Germany | c.1946-1949 delGTGT |
p.C649fsX658 | 16 (hom) | ND | NPHP, Bx (22 yr) |
RD | ND |
cDNA mutation numbering is based on human reference sequence NM_006642.2 for SDCCAG8, where +1 corresponds to the A of ATG start translation codon.
All mutations were absent from >270 healthy control individuals. Bx, Kidney biopsy demonstrates nephronophthisis.
Polydactyly was absent from all individuals.
CD, clinodactyly of 5th finger bilaterally; ERG, electroretinogram; ESKF, end-stage kidney failure; het, heterozygous; HG, hypogenitalism; hom, homozygous; IVS, intervening sequence; MR, mental retardation; ND, no data; NPHP, nephronophthisis; OB, obesity; PN, peripheral neuropathy; RD, retinal degeneration; SNB, stationary night blindness; yr, year(s
When investigating 11 families with NPHP-RC, in whom we had demonstrated homozygosity at the SDCCAG8 locus13 we detected four additional homozygous mutations (Fig. 1f). These were, (i) a deletion of exons 5-7 in F159, (ii) an obligatory splice site mutation (c.1068+1G>A) in A2290, (iii) a homozygous 1-bp insertion (resulting in p.E447fsX463) in SS-F336, and (iv) a homozygous nonsense mutation (p.L599X) in F1054. Direct exon sequencing of 118 families with SLSN yielded a 4-bp deletion resulting in a reading frame shift of the deduced amino acid sequence (p.C649fsX658) in F195 (Fig. 1f and Table 1). No mutations were detected by direct exon sequencing in 54 families with Joubert syndrome who had renal involvement with NPHP. All mutations were absent from >270 healthy control individuals and from healthy controls of the “1,000 genomes project” (http://www.1000genomes.org/page.php) (Table 1).
Furthermore, an independent homozygosity mapping study (using the Affymetrix 6.0 SNP) was carried out on 22 consanguineous families diagnosed as having Bardet-Biedl syndrome and for which no mutation was detected on genomic sequencing of 12 known BBS genes. A unique segment of homozygosity was identified for a large family (FI.2) on chromosome 1 (240 - 242.38 Mb) encompassing 5 genes (PLD5, CEP170, SDCCAG8, AKT3, ZNF238), for which sequencing of all coding exons was normal. Subsequently, RT-PCR with overlapping primer sets for the five genes was performed on RNA from fibroblast cell lines derived from the four affected family members. Abnormal fragments were obtained when amplifying exons 7 and 8 of SDCCAG8 (Supplementary Fig. 1 Online A,B). Sequencing of the products revealed various complex aberrant intron 7 insertions with a homozygous deep intronic mutation, c.740+356c>t, predicted to cause loss of an Exonic Splice Enhancer (ESE) site with the result of aberrant splicing introducing an in-frame stop codon (Supplementary Fig. 1 Online C-E).17 This leads to almost complete absence of the full-length SDCCAG8 product as confirmed by RT-PCR and immunoblotting (Supplementary Fig. 1 Online F-H). The finding that some residual full-length splice product and protein product remains (Supplementary Fig. 1 Online A,B and F-H) may explain the relatively late onset of renal failure and retinal degeneration observed in this kindred (Table 1).
Quantitative PCR analysis performed with primers bordering exons 8 and 9 amplified a significantly lower level of RNA compared to the control and Western blot confirmed trace amounts of protein (data not shown). The mutation was absent from >270 healthy control individuals and segregated with the disease in the family. In the same study, another consanguineous family (FII.22) with two siblings diagnosed with BBS (on the basis of obesity and mild mental retardation) showed overlapping homozygosity for the same chromosome 1 region. Exon sequencing of SDCCAG8 revealed a homozygous nonsense mutation (p.K227X). Because of these data, we initiated an unbiased (with regard to mutations at other loci) sequencing of SDCCAG8 in 96 unrelated BBS patients and found two families in which compound heterozygous loss of function mutations appeared sufficient to cause the disorder. Family NK-F1063 carried two deletions, causing frame shifts (p.T482fsX493 and p.D543fsX566), whereas in AR37, we found a nonsense allele and a frame shifting deletion (p.Y232X and p.R247fsX250) (Table 1). Both families had some of the six primary features of BBS (retinal degeneration, postaxial polydactyly, truncal obesity, learning disabilities, hypogonadism and renal anomalies).18 However, none of the patients had polydactyly (Table 1). We also detected two unique heterozygous alleles in two additional families, a p.E520G change in Northern European family AR605, and an p.E531K in middle-Eastern family KK11 (data not shown). No SDCCAG8 mutation was identified in 40 other BBS families, who did not carry a known BBS gene mutation. Likewise, no mutations were detected upon direct exon sequencing of 90 unrelated famlies with isolated juvenile-onset retinitis pigmentosa. All individuals from 10 families with 12 different homozygous or compound heterozygous truncating mutations in SDCCAG8 had nephronophthisis and retinal degeneration (Senior-Loken syndrome) (Table 1). Some had mild mental retardation, and two had signs of Bardet-Biedl syndrome (obesity and hypogenitalism).19 We thus identified recessive mutations of SDCCAG8 as a new cause of SLSN with some additional BBS-related features. Interestingly, all 10 families had two truncating mutations and exhibited a degenerative phenotype of kidney and eye only. Recessive mutations in SDCCAG8 explain 3.3 % (6/182) of cases in our worldwide SLSN cohort. To indicate the phenotypic relation of SDCCAG8 loss of function to other NPHP-RC we gave the related protein the aliases “SLSN7” and “NPHP10.
SDCCAG8 spans 244 kb on chromosome 1q44 (Fig. 1c). Up to 22 different putative alternatively spliced SDCCAG8 transcripts encoding different proteins have been proposed (www.aceview.org). To identify the relevant transcripts we designed primers complementary to the different potential splice variants and confirmed by sequencing the existence of 3 SDCCAG8 isoforms in retinal hTERT-RPE1 cells (Supplementary Fig. 3A). The full-length cDNA of SDCCAG8 (isoform-a; NM_006642) encodes an 82.7-kDa protein (713 amino acids) (Fig. 1d). Isoform-b lacks exon 6 compared to isoform-a, and isoform-e is C-terminally truncated beyond exon 9 (Supplementary Fig. 2I). Northern blot analysis had described a single 3.3 kb band expressed at low levels in mouse liver, spleen, kidney, brain, heart and muscle, consistent with the full-length (isoform-a) of 3,267 nt.20 The full-length cDNA of SDCCAG8 encodes an 82.7-kDa protein (713 amino acids) (Fig. 1e). Northern blot data and the finding that homozygous truncating mutations are found in most exons of full-length isoform-a (Fig. 1d-f) support the notion that this full-length isoform is the relevant one for the retinal-renal phenotype of this disease. Analysis of the deduced SDCCAG8 amino acid sequence yielded an N-terminal globular domain, a nuclear localization signal, and eight putative coiled-coil domains (Fig. 1e).20 Coiled coil domains represent a feature that is shared by most proteins mutated in NPHP-RC. Nuclear localization signals are also found in INVS and CEP290.21 In addition to being part of the photoreceptor sensory cilium proteome, SDCCAG8 (“serologically defined colon cancer antigen 8”; also known as CCCAP) is part of the human centrosomal proteome.22 The predicted SDCCAG8 protein sequence is highly conserved including Ciona intestinalis (sea squirt) (XP_002120666.1, 20% identity), and D. melanogaster (NP_732086.1; 16% aa identity), suggesting a conserved function of the domain assembly (Data available from the authors). By yeast-2-hybrid interaction the SDCCAG8 protein had been shown to homodimerize.20
SDCCAG8 colocalizes with other NPHP-RC proteins at centrosomes
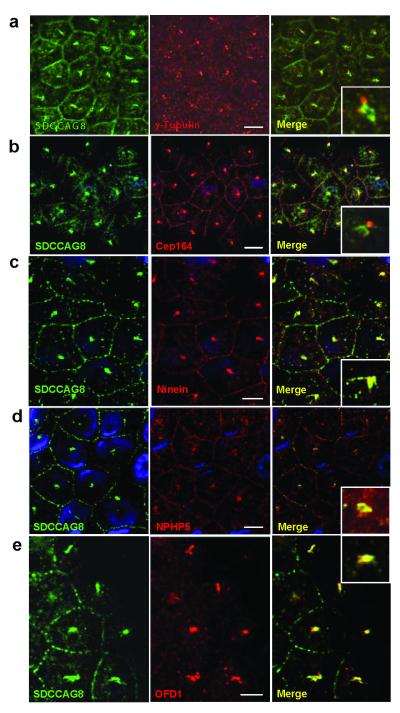
Because SDCCAG8 has been described as part of the human centrosomal proteome20,23, and since a defining feature of NPHP-RC proteins is their expression at centrosomes12,24, we evaluated SDCCAG8 subcellular localization using immunofluorescence studies in the renal epithelial cell line MDCK-II (Fig. 2).
Figure 2. Indirect immunofluorescence detects SDCCAG8 at centrosomes together with other proteins mutated in NPHP-RC.
(a) Centrosomal SDCCAG8 is at centrosomes but slightly set apart from the signal of γ-tubulin, which marks the centrioles and from CEP164 (b), which marks distal centrosomal appendages22. (c) In contrast, there is tight colocalization with ninein, a marker of centrosomal appendages22. There is also colocalization at centrosomes with NPHP5 (d) and OFD1 (e) previously found to be mutated in NPHP-RC6,26. Size bars represent 5 μm. Insets show enlargement of representative results at 5-fold higher magnification. Nuclei are stained with DAPI. Antibody α-SDCCAG8-CG was used to detect SDCCAG8.
For this purpose we generated antibodies against N-terminal and C-terminal peptides of human SDCCAG8 (Fig. 1e). Antibodies α-SDCCAG8-NR, α-SDCCAG8-NG (raised against N-terminal peptides in rabbit and guinea pig, respectively) and α-SDCCAG8-CG (raised against C-terminal peptide in guinea pig) recognized upon immunoblotting endogenous and stably overexpressed SDCCAG8 full-length isoform-a of human and mouse SDCCAG8 (Supplementary Fig. 2A). α-SDCCAG8-NR also recognized isoform-a of zebrafish (Supplementary Fig. 2B). Upon coimmunoprecipitation of human retinal epithelial cell (hTERT-RPE) lysates with antibody α-SDCCAG8-PR (Proteintech, rabbit) immunoblotting recognized the SDCCAG8 full-length isoform-a (83 kDa) and isoform-b (78 kDa) when antibodies α-SDCCAG8-PR, α-SDCCAG8-NR, α-SDCCAG8-NG or α-SDCCAG8-CG were used. In contrast to antibodies α-SDCCAG8-PR and α-SDCCAG8-NR, α-SDCCAG8-CG did not recognize the short isoform-e at 41 kDa (Supplementary Fig. 2C-F), as the respective peptide is not part of short isoform-e (Fig. 1e). Immunofluorescence studies confirmed that these antibodies recognize overexpressed SDCCAG8 upon indirect immunofluorescence (Supplementary Fig. 3A-C) and that α-SDCCAG8-PR and α-SDCCAG8-NG colocalize in connecting cilia of mouse photoreceptors (Supplementary Fig. 3C). Taken together, these data demonstrate that these antibodies are specific for the full-length SDCCAG8 isoform-a and antibodies -PR and -NR also recognize C-terminal short isoform-e. We confirmed the presence of SDCCAG8 isoforms-a, -b and -e (Supplementary Fig. 3A) by RT-PCR in hTERT-RPE cells and direct sequencing (data not shown).
When we evaluated SDCCAG8 subcellular localization by immunofluorescence studies in the MDCK-II renal epithelial cell line, we found that SDCCAG8 localizes to centrosomes, however in a location set apart from the γ-tubulin signal that marks centrioles (Fig. 2a), and set apart from the signal of Cep164 that marks distal centrosomal appendages (Fig. 2b)22. In contrast, there was tight colocalization with ninein, a marker of centrosomal appendages (Fig. 2c)22. Colocalization was found between SDCCAG8 and NPHP5/IQCB1 (Fig. 2d) as well as OFD1 (Fig. 2e), which are mutated in NPHP-RC. By expression of human SDCCAG8 in mIMCD3 mouse renal epithelial cells we demonstrated expression of full-length isoform-a at centrosomes, whereas the N-terminal short isoform-e was primarily located in the cytoplasm (Supplementary Figure 2D). In addition, there was localization of SDCCAG8 at cell-cell junctions (Fig. 2a-e)25 similar to what has been described for NPHP1 and NPHP4.
SDCCAG8 occurs at centrosomes throughout the cell cycle
Because proteins mutated in NPHP-RC localize to cilia and centrosomes, which become the mitotic spindle poles bodies24, we examined subcellular SDCCAG8 localization in relation to cell cycle in MDCK-II cells using γ-tubulin as a centrosomal marker (Supplementary Fig. 4) and α-tubulin as a cytoplasmic microtubule marker (Supplementary Fig. 5). We demonstrate in cells stably transfected with C-terminally GFP-labelled SDCCAG8, that SDCCAG8 localizes to the centrosomes throughout the cell cycle, as shown by its co-localization with γ-tubulin (Supplementary Fig. 4) and α-tubulin (Supplementary Fig. 5), respectively. Additionaly, in both hTERT-RPE1 (ciliated; Supplementary Fig. 6A-B) and U2OS osteosarcoma cells (non ciliated; Supplementary Fig. 6C-D), SDCCAG8 localizes to each centriole on a centrosome. Similar to what has been described for OFD1,25 SDCCAG8 is present in all cell cycle phases at the distal part of both centrioles as shown by its co-localization with centrin, a distal centriolar marker (Supplementary Fig. 6A and C). The finding that SDCCAG8 is present even in non-ciliated cells and occurs at both centrioles may indicate a more general role in centrosomal function. We conclude that SDCCAG8 is located at the distal ends of both centrioles and that it colocalizes to centrosomes throughout the cell cycle together with other proteins that are mutated in NPHP-RC.
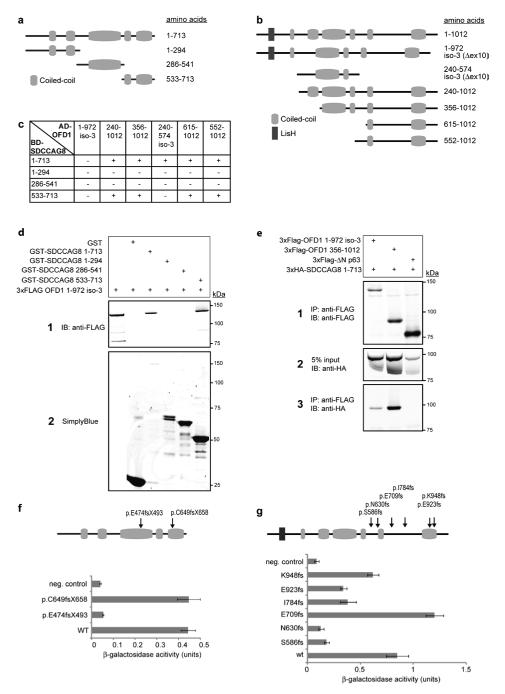
SDCCAG8 directly interacts with OFD1
To further characterize the protein complex in which SDCCAG8 participates, we performed yeast two-hybrid screening of a human retinal cDNA library, using SDCCAG8 as bait. Both, with full-length SDCCAG8 as well as with a C-terminal fragment (aa 533-713) (Fig. 3 a and b), we identified OFD1 as a direct interaction partner of SDCCAG8. The C-terminal OFD1 region containing the last two out of six predicted coiled-coil motifs (aa 615-1012) was found to bind to the C-terminal region of SDCCAG8 (aa 533-713) that also carried two predicted coiled-coil motifs (Fig. 3c). The identified interaction was validated by GST pull-down experiments (Fig. 3d) and co-immunoprecipitation (Fig. 3e).
Figure 3. SDCCAG8 Interaction with OFD1.
(a, b) Schematic of SDCCAG8 and OFD1 clones and protein domains. Full-length SDCCAG8 (Q86SQ7) and three cloned fragments are shown (a) as well as OFD1 fragments identified by yeast two-hybrid screening and isoforms O75665-1 and O75665-3.
(c) Yeast two-hybrid interaction assays confirmed binding of full-length SDCCAG8 and C-terminal fragment (533-713) to all cloned OFD1 fragments, except isoform-3. HIS3 and ADE2 reporter genes activation is indicated.
(d) Both GST-SDCCAG8 full-length and the GST-SDCCAG8 C-terminal fragment efficiently pulled down 3xFLAG-tagged OFD1 isoform-3, contrary to GST alone.
(e) 3xHA-tagged full-length SDDCAG8 coimmunoprecipitated with 3xFLAG-tagged OFD1 isoform 3, and fragment 356-1012, contrary to unrelated 3xFLAG-tagged ΔN-p63. Panel 2 shows 5% of cell lysate input.
(f) Interaction of wild-type (WT) and SDCCAG8 mutants with ODF1 isoform-1 in beta-galactosidase assay. Constructs encoding SDCCAG8 as GAL4-BD fusion protein, WT or indicated mutants, were cotransformed with pAD-OFD1 isoform-3 constructs in yeast. Cotransformation of pBD-SDCCAG8 and pAD-MUT vector served as negative control. Remaining LacZ reporter gene activity, corrected for background activity, is indicated.
(g) Interaction of WT isoform-3 and OFD1 mutants with SDCCAG8 in yeast two-hybrid assay. Constructs encoding WT SDCCAG8 as GAL4-BD fusion protein were cotransformed in PJ694a with pAD-OFD1 isoform-3 constructs, WT or indicated mutations. Interaction of SDCCAG8 with OFD1 mutant p.E709fs and WT was detected as described above. Beta-galactosidase assays revealed decreased LacZ reporter gene activity for OFD1 all mutations except E709fs, indicating significantly reduced interaction with SDCCAG8. Error bars represent standard error of the mean in f and g.
Dominant mutations of OFD1 cause oral-facial-digital syndrome type 1, a male-lethal X-linked dominant condition, and recessive OFD1 mutations cause the NPHP-RC Joubert syndrome and X-linked recessive Simpson-Golabi-Behmel syndrome type 2 (SGBS2)26. Whereas dominant mutations in OFD1 abolish its interaction and its pericentriolar localization, recessive mutations only weaken OFD1 interaction with lebercilin (LCA5)26. To explore the hypothesis that a loss of binding to OFD1 as result of mutations in SDCCAG8 underlies the disease mechanism for SLSN in patients carrying SDCCAG8 mutations, we tested whether the SDCCAG8 mutations, p.C649fsX658 or p.E474fsX493, affect interaction with OFD1. In a liquid β-galactosidase assay, demonstrating activation of the LacZ reporter, we identified that only the SDCCAG8 p.E474fsX493 mutation that affects the OFD1 interacting domain completely disrupts the interaction, whereas no difference in binding affinity for OFD1 was observed for the C-terminal p.C649fsX658 mutation (Fig. 3f). Similarly, we also assessed the effects on SDCCAG8 binding affinity of six previously published OFD1 mutations, p.K948fs and p.E923fs26, p.I784fs27, p.E709fs28, p.N630fs29, and p.S586fs30. All dominant OFD1 mutations disrupted the binding, while only the p.E709fs mutation did not affect the affinity for SDCCAG8 (Fig. 3g).
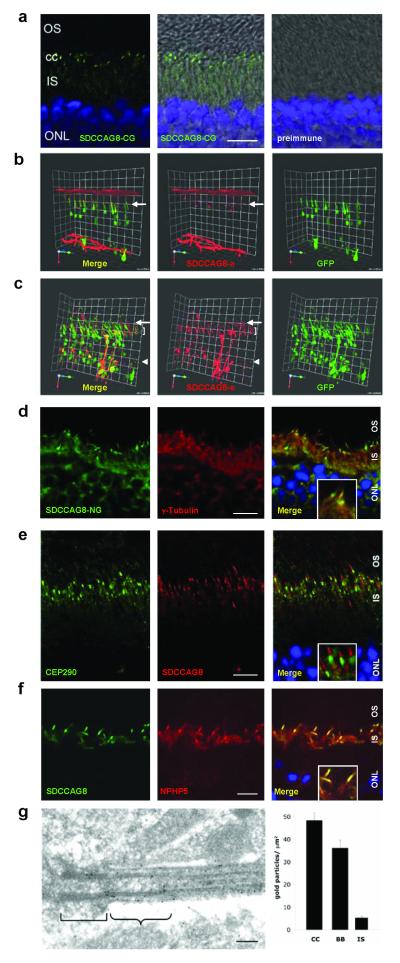
SDCCAG8 colocalizes with other NPHP-RC proteins to photoreceptors
To elucidate the role of SDCCAG8 in the pathogenesis of NPHP-RC we examined expression of SDCCAG8 and other proteins mutated in NPHP-RC for potential colocalization in mouse retina using indirect immunofluorescence studies (Fig. 4). We detect a strong signal for SDCCAG8 with antibody α-SDCCAG8-CG in transition zone and a weaker signal in inner segments of mouse photoreceptors (Fig. 4a). This localization was confirmed at high resolution-3D fluorescence imaging following electroporation of two different human SDCCAG8 constructs into rat retina (Fig. 4b-c). Specifically, human full-length SDCCAG8 isoform-a localizes to transition zone of photoreceptor cells (Fig. 4b), whereas the short N-terminal isoform-e exhibits expression in connecting cilia, inner segments, and the cytoplasm (Fig. 4c). This confirms the finding in mIMCD3 renal epithelial cells that full-length SDCCAG8 isoform-a localizes to the vicinity of centrosomes whereas the short N-terminal isoform-e shows diffuse cytoplasmic expression (Supplementary Fig. 3D).
Figure 4. SDCCAG8 is located in mouse photoreceptor basal bodies and connecting cilia transition zone.
(a) Immunofluorescence using the α-SDCCAG8-CG antibody demonstrates strong SDCCAG8 expression in transition zone of connecting cilia (cc) and weak expression in inner segments (IS) of mouse photoreceptors.
(b-c) Upon in vivo electroporation into rat retinae, human full-length SDCCAG8 isoform-a (red) (b) localizes to transition zone of photoreceptor cells (level of arrow), whereas the short N-terminal isoform-e (red) (c) exhibits expression in transition zone (level of arrow), inner segments (level of bracket) and cytoplasm (level of arrow head). Photoreceptor cytoplasms are counterstained by GFP overexpression (green).
(d) In mouse photoreceptors SDCCAG8 is located in transition zone of connecting cilia, distal to basal body marker γ-tubulin in a contiguous but non-overlapping location.
(e) SDCCAG8 is located in the transition zone, distal to, and clearly set apart from the pericentriolar marker CEP290.
(f) SDCCAG8 tightly colocalizes with NPHP5. Scale bars are 10 μm. Insets in d-f show enlargement of representative results at 3-fold higher magnification.
(g) Upon ultrathin sectioning of mouse retina SDCCAG8 expression is particularly prominent at distal basal body (bracket) and the transition zone (winged bracket) connecting cilium of mouse photoreceptors (antibody α-SDCCAG8-PR). The bar graph represents the density of immunogold labeling over the photoreceptor transition zone (TZ), basal bodies (BB) and inner segments (IS) after subtracting background labeling. Scale bar = 100 nm.
We then examined SDCCAG8 localization in mouse photoreceptors at high resolution. We demonstrate that SDCCAG8 is located in the transition zone, distal to basal body marker γ-tubulin in a contiguous but not overlapping position (Fig. 4d), and distal to, but clearly set apart from the pericentriolar marker CEP290 (Fig. 4e). This subcellular localization at fine resolution mirrors the findings of centrosomal localization in MDCK-II cell lines, where SDCCAG8 located just distal to the centrosomal marker γ-tubulin (Fig. 2a). We tested for colocalization with NPHP5, because NPHP5 is located in photoreceptor transition zone6 and shares a similar genotype-phenotype correlation with SDCCAG8, in that homozygous truncating mutations cause the retinal-renal ciliopathy Senior-Loken syndrome. As demonstrated in MDCK-II cells (Fig. 2a-d) we detect tight colocalization of SDCCAG8 with NPHP5 in photoreceptor transition zone (Fig. 4f), compatible with the notion of a shared function of both proteins in photoreceptors. A similar result was obtained with antibody α-SDCCAG8-PR (data not shown). Immunogold labeling of antibody α-SDCCAG8-PR confirmed upon ultrathin sectioning of mouse retina that SDCCAG8 expression is particularly prominent at the distal basal body and the transition zone of mouse photoreceptors (Fig. 4g). Immunofluorescence studies also demonstrated colocalization of SDCCAG8 with the retinitis pigmentosa proteins RPGRIP and RP1 (Supplementary Fig. 7).
Sdccag8 knockdown causes multiple developmental defects in zebrafish
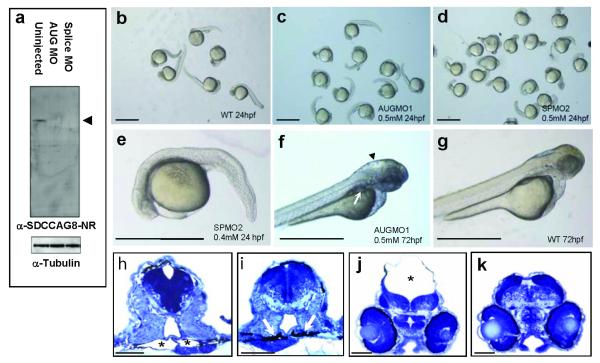
Morpholino oligonucleotide (MO)-mediated gene knockdown in zebrafish has been successfully employed in disease models of human NPHP-RC, yielding phenotypes of kidney cysts, brain defects and phenotypes of body axis development.2,7 To utilize the zebrafish model we demonstrated cross-reactivity of the antibody α-SDCCAG8-NR against zebrafish SDCCAG8 (Supplementary Fig. 2B). We examined for sdccag8 knockdown zebrafish phenotypes using an MO directed against the start codon (AUGMO1) as well as against the donor splice site of exon 6 (SPMO2), both of which efficiently abrogated expression of SDCCAG8 protein (Fig. 5a). Compared to standard control morpholino (Fig. 5b), sdccag8 knockdown using both MOs resulted in a developmental phenotype of body axis curvature and shortened and broadened tails (Fig. 5c-e). In addition, AUGMO knockdown caused kidney cysts at 72 hpf in 34% (n=243) of zebrafish (Fig. 5f,h) compared to standard MO negative control (3%, n=158) (Fig. 5g,i). Furthermore, hydrocephalus resulted in 36% (n=128) of knockdown zebrafish (Fig. 5f,j) compared to standard MO negative control (7%, n=158) (Fig. 5g,k). These findings demonstrate loss of sdccag8 function results in multiple developmental phenotypes that have been described in related NPHP-RC genes including invs, cep290, and nek8 pointing to possible shared disease mechanisms.
Figure 5. sdccag8 knockdown results in multiple developmental defects.
(a) Injection of both sdccag8-targeting MOs abrogates sdccag8 protein from MO injected embryo morphant lysates. sdccag8 protein is detected by the α-SDCCAG8-NR antibody as a single band in uninjected embryos, compatible with the sdccag8 full-length product at ~83 kDa (arrow head). Anti-α-tubulin antibody was used to demonstrate equal loading.
(b-d) In comparison to embryos injected at the 2-cell stage with standard negative control MO (b) embryos injected with AUG-targeting MO (c) or splice site targeting MO (d,e) exhibit a dose-dependent phenotype of body axis curvature and shortened and broadened tails at 24 hpf.
(f-k) Knockdown of sdccag8 in zebrafish embryos at 72 hpf caused pronephric cysts as evidenced by a rounded structure (arrow in f) and hollow spaces (asterisk in h) compared to control morpholino injected control (g,j) , which shows slender pronephric tubular lumina (arrows in i). It also caused hydrocephalus (asterisk in j) compared to control morpholino injected control (g,k). Scale bars represent 1 mm in b-g and 100 μm in h-k.
Loss of Sdccag8 function disturbs renal epithelial lumen formation
Formation of lumen-containing spheroids by renal epithelial cells in 3D culture is an established test for cell polarity defects31,32, which are central to the pathogenesis of NPHP-RC33. For the proteins NPHP1 and NPHP4 it was shown that their knockdown leads to abnormal lumen formation in the 3D culture spheroid assay.25 To investigate the role of SDCCAG8 in 3D kidney architecture, we transfected mIMCD3 cells with siRNA oligonucleotides against murine Sdccag8, random sequences (negative control), or murine Ift88, which causes a very severe NPHP-RC phenotype if mutated (positive control).34 Using quantitative RT-PCR, we determined that Sdccag8 transcript levels were depleted by 80% and Ift88 was depleted by 89% (data not shown).
We next examined whether Sdccag8 depletion would affect 3D spheroid growth in matrigel. After 3 days of culture, cells transfected with control random oligonucleotides formed spheroid structures with a clear lumen, apical cilia, defined tight junctions and clear basolateral structure. Cells transfected with siIft88 developed virtually no spheroids, or grew as clumps of cells with few cilia evident. Cells transfected with siSdccag8 developed spheroids with architectural defects characterized by disturbed localization of β-catenin at the basolateral membrane, fewer tight junctions, and an irregular lumen. Abnormal lumina were clearly seen upon siSdccag8 knockdown (in 75.8% spheroids with siSdccag8 knockdown, versus 19.5% for control siRNA; p=0.0055). To exclude the possibility that the effect observed was due to off-target effects, we stably transfected mIMCD3 cells with full-length human SDCCAG8. We confirmed that these stable lines are still capable of forming spheroids with only occasional irregular lumina (16.7%; data not shown). We then repeated the siRNA of the endogenous murine Sdccag8 ortholog and observed no significant change in lumen irregularities (16.7%) compared to negative control siRNA (19.5%). The phenotypic rescue of Sdccag8 knock-down by co-expression of human SDCCAG8, supports the conclusion that lack of SDCCAG8 induces a defect in cell polarity and lumen formation which may reflect renal tubular defects present in individuals with a congenital SDCCAG8 mutation.
SDCCAG8 abandons cell-cell junctions in response to increased intracellular cAMP
Increased levels of intracellular cAMP have been strongly associated with the renal cystic phenotype of NPHP-RC35. Pharmacologic reduction of intracellular cAMP levels by use of the vasopressin-2-receptor antagonist tolvaptan or the somatostatin analog octreotide reduced renal cystic disease in mouse models of NPHP-RC including the mouse model (Pcy) of Nphp3 loss of function36. These drugs are currently tested in phase II trials in humans with polycystic kidney disease37. We therefore studied whether localization of SDCCAG8 at cell-cell junctions (Fig. 2a-e) would be altered upon changes of intracellular cAMP.
We demonstrate that upon increasing doses of the analog 8Br-cAMP there is dose-dependent loss of SDCCAG8 from cell-cell junctions in relation to three controls, i.e. peripheral E-cadherin, centrosomal γ-tubulin, and centrosomal SDCCAG8 (Fig. 6) (see also Supplementary Fig. 8 Online). The same effect was seen upon treatment of serum starved cells with forskolin and IBMX, which increase endogenous intracellular cAMP via stimulation of adenylyl cyclase and inhibition of phosphodiesterase, respectively, and it was reversible with the adenylyl cyclase inhibitor tolvaptan (Supplementary Fig. 9 Online). We conclude that SDCCAG8 translocates away from cell-cell junctions in response to increased intracellular cAMP. Treatment of spheroids with 8Br-cAMP did not result in an effect similar to SDCCAG8 knockdown in spheroids, suggesting that redistribution of SDCCAG8 from the cell periphery in response to 8Br-cAMP and disruption of lumen formation in spheroids may be independent phenomena (data not shown).
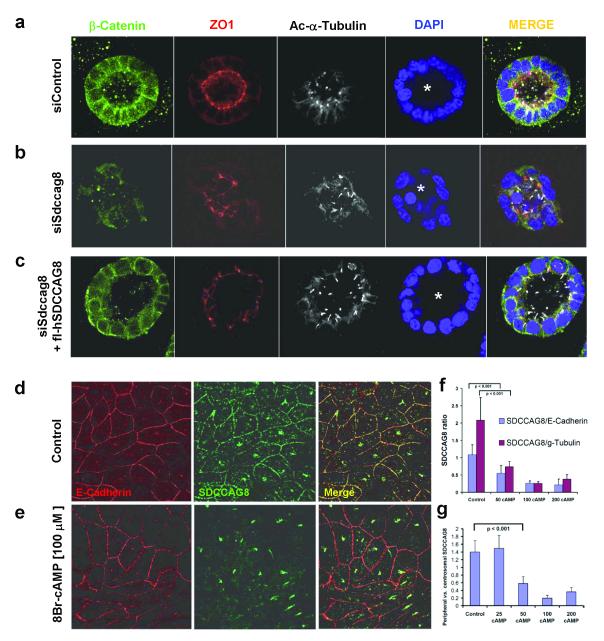
Figure 6. siRNA knockdown of siSdccag8 perturbs lumen formation of renal epithelial cells in 3D spheroid culture.
(a) IMCD3 murine renal epithelial cells, when grown in 3D matrigel culture for 3 days, polarize as demonstrated by staining for β-catenin (green) and tight junction marker ZO1 (red). They ciliate apically as shown by acetylated α-tubulin (white) and form a spheroid containing a central lumen (asterisk). Nuclei are stained blue with DAPI.
(b) Upon siSdccag8 knockdown cells develop spheroids that have architectural defects characterized by disturbed localization of β-catenin (green) away from the basolateral membrane, fewer tight junctions (red), and an irregular lumen (asterisk). Abnormal lumina were seen in 19.5% of siRNA controls versus 75.8% in siSdccag8 knockdown (p=0.0055).
(c) When siSdccag8 knockdown was performed in mIMCD3 cells that were stably transfected with human full-length SDCCAG8, the knockdown phenotype was fully rescued leading to no significant reduction in irregular lumina (16.7%) compared to negative control siRNA (19.5%).
(d-g) SDCCAG8 abandons cell-cell junctions in response to increased intracellular cAMP.
(d) In the renal epithelial cell line MCDK-II SDCCAG8 stained with antibody α-SDCCAG8-CG (green) is located at centrosomes and cell-cell junctions, which are marked with an α-E-cadherin antibody (red). (e) Upon treatment with 8-Br-cAMP [100 μM], there is dose-dependent loss of SDCCAG8 signal from cell-cell junctions relative to E-cadherin (f-g). When 100 linear segments of the pentagonal cell-cell junctions of non-neighboring cells were evaluated for the ratio of relative fluorescence signals for SDCCAG8 versus E-cadherin there was a redistribution depending on cAMP concentration [μM] away from cell junction staining for SDCCAG8 relative to peripheral E-cadherin, centrosomal γ-tubulin (f) and relative to centrosomal SDCCAG8 (g) (see also Supplementary Figure 6 ). Error bars represent standard error of the mean.
DISCUSSION
The discovery of single-gene causes of retinal-renal ciliopathies has generated novel insights into the role of the cilia/centrosome/mitotic spindle complex (CCC) for disease mechanisms of dysplasia and degeneration of kidney, retina, liver, cerebellum and many other organs.12 In addition, it revealed that multiple signaling mechanisms including Wnt signaling and sonic hedgehog signaling are closely connected to ciliary and centrosomal function. However, dozens of essential CCC components within this pathogenesis are still unknown.18 Their identification has been limited by the fact that additional monogenic causes of ciliopathies are very rare, often occurring only in single families of large international cohorts9. This necessitates the ability of identifying ciliopathy-causing genes in single families. The utility of the emerging techniques of exon capture and MP sequencing, however, is limited by the high number of variants form normal reference sequence that they generate. We here demonstrate the efficiency of an a priori approach to restricting the yield of not disease-causing variants by combining homozygosity mapping and a candidate gene approach with exon capture and MP sequencing. This approach has the potential of greatly advancing disease gene identification in a multitude of single-gene disorders, thereby allowing assembly of the related pathogenic pathways.
Particularly in the degenerative variants of NPHP-RC there is a theoretical time window for prevention or treatment before degeneration of renal and retinal tissue develops in late childhood. However, no treatment is currently available for any of these disorders. In the “Pcy” mouse model of Nphp3 loss of function, reduction of the intracellular cAMP level using a vasopressin-2-receptor antagonist resulted in successful delay or even abrogation of renal cystic disease36. It will therefore be important to delineate the role of intracellular cAMP for disease mechanisms of NPHP-RC. In this context, both the spheroid assay and the cell culture assay described here provide useful models for the study of the relation between intracellular cAMP levels and the function of genes mutated in NPHP-RC. These in vitro models as well as the zebrafish model of sdccag8 loss of function lend themselves to high-throughput testing of small molecules that may halt disease mechanisms of NPHP-RC.
ONLINE METHODS
Research subjects
We obtained blood samples and pedigrees following informed consent from individuals with NPHP-RC and/or their parents. Approval for human subjects research was obtained from the University of Michigan Institutional Review Board, McGill University Health Centre and the other institutions involved. The diagnosis of NPHP-RC was based on published clinical criteria.
Linkage analysis
For genome-wide homozygosity mapping the GeneChip® Human Mapping 250k StyI Array from Affymetrix was used. Non-parametric LOD scores were calculated using a modified version of the program GENEHUNTER 2.138,39 through stepwise use of a sliding window with sets of 110 SNPs and the program ALLEGRO40 in order to identify regions of homozygosity as described7,13 using a disease allele frequency of 0.0001 and Caucasian marker allele frequencies.
Exon capture array design
We custom designed an exon sequence capture hybridization 385K array (NimbleGen™) to capture 13,221 exons of 828 selected ciliopathy candidate genes. Candidate genes were derived from ciliopathy animal models, from the photoreceptor sensory cilia proteome15, the human centrosomal proteome23, and from the cilia proteome database16 (Supplementary Table 3). DNA capture resulted in an average 200-fold enrichment of targeted exons. Captured DNA fragment size was reduced by exo- and endonuclease treatment to be suitable for high throughput sequencing on a Solexa/Illumina™ GA2 platform. In order to generate random start position for MP sequencing, ablate NimbleGen linker sequences and to reduce fragment sizes, we digested the captured DNA fragments with exonuclease Bal-31 (New England Biolabs) and endonuclease DNase I (Roche). 5 μg DNA was digested with 2 units of Bal-31 exonuclease for 5 min at 30°C in a 200 μl reaction. The reaction was stopped by adding EGTA to a final concentration of 20 mM and immediate heat inactivation at 65°C for 10 min, followed by Qiaquick PCR column purification (Qiagen). DNA (about 2 μg) was further digested by incubating with 1 unit DNAse I (Roche) and freshly prepared reaction buffer [2× reaction buffer contains 20 mM Tris-Cl (pH 7.5), 2 mM CaCl2, and 20 mM MnCl2] for 3 min at 16°C. The reaction was stopped by adding 2 μl EDTA (500 mM) and immediate heat inactivation at 65°C for 10 min.
Massively parallel sequencing
Library construction (adapter ligation) of the modified captured fragments was performed using “Genomic DNA Sample Prep Kit” according to the manufactures instructions (Illumina, San Diego, CA) and fragments separated on a 1.5% agarose gel and excised in the 150-200 bp range. Fragments were purified and subjected to 10 rounds of PCR amplification using complementary linker specific primers. Amount and size distribution of each sample was analyzed on a Bioanalyzer 2100 (Agilent Technologies, Inc). Single-stranded DNA fragments were annealed to a flow cell surface in a cluster station (Illumina) and 46 cycles of bridge amplification were applied. Fragments were run on a single lane of a Solexa/Illumina Genome Analyzer II platform, generating about 6-14 million single-end sequence reads of 36-39 bases each. Image analysis and base calling was generated by the Genome Analyzer Pipeline 1.5 with default parameters. Illumina specific FASTQ file containing sequence information and quality scores for each base call were exported for further analysis. Sequence evaluation was performed with the CLC™ Genomics Workbench software. On average we obtained about 10 million single short reads (5.8-14 million) per lane with 68% of reads on target (range 51%-81%). The median sequencing depth per coding nucleotide was 49 with 99% of the targeted exons covered at least 5-fold.
Bioinformatics
Genetic location is according to the March 2006 Human Genome Browser data (http://www.genome.ucsc.edu).
cDNA cloning
To generate inducible SDCCAG8-EGFP the SDCCAG8 open reading frame was subcloned by PCR from full length human ESTs (SDCCAG8 human EST Clone ID: 5174799) into the vector pRetroX-Tight-Puro (Clontech). RT-PCR was performed to identify SDCCAG8 isoforms in hTERT-RPE1 cell line. For in vivo photoreceptor localization studies, the wild-type and mutant human SDCCAG8 cDNAs were cloned into a custom-made pCAG-V5-cDNA-IRES-EGFP-pA Gateway destination expression vector using LR clonase II (Invitrogen) mediated recombination.
Yeast two-hybrid assays
The GAL4-based yeast two-hybrid system (HybriZAP, Stratagene, La Jolla, USA) was used for identifying protein-interaction partners of SDCCAG8. The SDCCAG8 full-length and fragments (amino acids 1-294, 286-541, 533-713) fused to a DNA-binding domain (GAL4-BD), were used as baits for screening a human oligo-dT primed retinal cDNA library. The yeast strain PJ69-4A, which carried the HIS3 (histidine), ADE2 (adenine), MEL1 (alpha-galactosidase) and LacZ (beta-galactosidase) reporter genes, was used as a host. Interactions were analyzed by assessment of reporter gene activation via growth on selective media (HIS3 and ADE2 reporter genes), alpha-galactosidase colorimetric plate assays (MEL1 reporter gene), and beta-galactosidase colorimetric filter lift assays (LacZ reporter gene). For analysis of the binding capacities of SDCCAG8 mutant proteins to OFD1, expression constructs encoding SDCCAG8 as a GAL4-BD fusion protein, either wild-type or containing the p.C649fsX658 or p.E474fsX493 mutation were cotransformed in PJ694a with a construct pAD-OFD1 isoform-3, encoding OFD1 as a GAL4-AD fusion protein. For analysis of the binding capacities of OFD1 mutant proteins to SDCCCAG8, expression constructs encoding amino acids 356–1012 of OFD1 as a GAL4-AD-fusion protein, either wild-type or containing the p.K948fs, p.K923fs, p.I784fs, p.E709fs, p.N630fs, or p.S586fs mutation, were cotransformed with a construct encoding the full-length SDCCAG8 fused to GAL4-BD (pBD-SDCCAG8) in PJ69-4a. As a negative control, the empty pAD vector was cotransformed with pBD-SDCCAG8. Detailed protocols for evaluation of the protein interactions are available from the authors.
GST Pull-Down Assay
SDCCAG8 full-length and fragments (amino acids 1-294, 286-541, and 533-713) were cloned into pDEST15 (Gateway cloning system, Invitrogen). For the creation of GST-fusion proteins, BL21-DE3 cells were transformed with pDEST15 constructs. Detailed protocols for GST pull down are available from the authors.
Coimmunoprecipitation
HA-SDCCAG8 (full-length) and 3xFLAG-OFD1 (both splice variants and the fragments encompassing amino acids 356–1012) were coexpressed in HEK293T cells. As a negative control, HA-SDCCAG8 was coexpressed with the functionally unrelated 3xFLAG-dN-p63 protein. After 48 hr of expression, cells were lysed on ice in lysis buffer (50 mM Tris–HCl [pH 7.5], 150 mM NaCl, 0.5% Triton X-100) supplemented with complete protease inhibitor cocktail. Lysates were incubated with anti- FLAG M2-agarose from mouse (Sigma-Aldrich), for 5 hr at 4 degrees Celsius. After incubation, beads with bound protein complexes were washed in lysis buffer. Then beads were taken up in NuPage Sample Buffer and heated for 10 min at 70 degrees Celsius. Beads were precipitated by centrifugation, and supernatant was run on a NuPAGE Novex 4%–12% Bis-Tris SDS-PAGE gel. The interaction of HA-SDCCAG8 with 3xFLAG-OFD1 was assessed by immunoblotting, followed by staining with either monoclonal mouse anti-FLAG or monoclonal mouse anti-HA (Sigma-Aldrich) as a primary antibody and goat anti-mouse RDye800 as a secondary antibody. Fluorescence was analyzed on a Li-Cor Odyssey 2.1 infrared scanner.
Antibodies
Antipeptide antibodies were raised against the following targets: α-SDCCAG8-NG (raised against human N-terminal peptide PN (Fig. 1e) in guinea pig, aa 119-138), α-SDCCAG8-CG (raised against C-terminal peptide PC (Fig. 1e) in guinea pig, aa 483-502) and α-SDCCAG8-NR (raised against N-terminal peptide PN (Fig. 1e) in rabbit, aa 119-138). Cystein was added to the N-terminus of peptides to facilitate the coupling to maleimide activated mcKLH (Thermo Fisher Scientific, Rockford, IL). Rabbits or Guinea Pigs were then immunized with KLH-coupled peptides (Cocalico Biologicals Inc, Reamstown, PA). To affinity purify peptide antibodies, appropriate peptides were coupled to Sulfolink resin according to manufacturers recommendation (Thermo Fisher Scientific).The following primary antibodies were used: mouse anti-GFP (Clontech); mouse anti-γ-tubulin (Sigma), mouse anti-acetylated-α-tubulin (Sigma), rabbit anti-γ-tubulin (Sigma), mouse anti-β actin (BD Biosciences), mouse-anti-glutamylated tubulin (Sigma), mouse anti-Centrin (M. Dias), mouse anti-GT335 (C. Janke); rabbit anti-ninein (Biolegend); rabbit anti-SDCCAG8 (Proteintech), rabbit anti-Nphp5 (E. Otto), rabbit anti-OFD1 (E. Nigg), anti-OFD1-GR (GenTex), rabbit anti-Cep164 (E. Nigg), rabbit ant-CEP290 (B. Chang), rabbit anti-γ-catenin (BD Biosciences), rabbit anti-ZO1 (P. Jackson), rabbit anti-Ecadherin (BD Biosciences), anti-FLAG (Sigma), anti-HA (Sigma). Alexa-488, Alexa-594 and Alexa-647 conjugated secondary antibodies were obtained from Invitrogen.
mIMCD3 spheroid growth assay
Mouse kidney collecting duct mIMCD3 cells were cultured in DMEM:F12 1:1 in 10% FBS. 25,000 cells were seeded per well in 24-well dishes (plastic and glass bottom) in triplicate 24 hours prior to being transfected with 50 nM Dharmacon OTP siRNA against mouse Sdccag8 (smartpool, L-045871-01-0005), mouse Ift88 (J-050417-06), or control (Ctrl; D001206-14-05, SiGenome) random oligos. One plate was harvested to determine knock-down efficiency by quantitative real-time PCR three days later, another was fixed in 4% PFA and immunostained for acetylated tubulin (T7451, Sigma, 1:2,000) and pericentrin (PRB-432C, Covance, 1:500), and scored for % nuclei with cilia (n=300-700 nuclei counted). The third plate was used to examine the effect of Sdccag8 on spheroid growth in 3D culture conditions. The detailed protocol is available from the authors.
For quantitative real-time PCR, total RNA was prepared using the RNeasy Mini kit (QIAGEN). 100 ng of the total RNA was added to a master mix with One-step RT-PCR reagents (Applied Biosystems 4309169) and Taqman Gene Expression Assays (Applied Biosystems) for each individual gene: Sdccag8 (Mm00659616_m1), Ift88 (Mm00493675_m1), Rpl19 (Mm02601633_g1). Triplicate reactions were run and analyzed on an ABI 7500 thermocycler. The mRNA levels of Ift88 and Sdccag8 were normalized to the endogenous Rpl19.
Immunofluorescence and confocal microscopy in cell lines and mouse retina
For immuno-staining MDCK-II cells, cells were seeded onto 0.4 μm Transwell filters (Corning, Lowell, MA) and grown for 7 days post-confluency. For immuno-staining centrosomes and basal body, filters were incubated in ice cold acetone:methanol (1:1) for 10 minutes. Filters were then rehydrated in PBS prior to blocking in 2% goat serum/PBS. For all other immuno-staining, filters were fixed for 20 minutes in 4%PFA/PBS followed by a 10 minutes permeabilization step in 0.1%Triton-X100/PBS. Filters were then blocked as above. Primary antibody incubations were performed overnight in 2% goat serum/PBS. Secondary antibody and DAPI incubations were performed for 1 hour. Filters were mounted in Prolong Gold antifade reagent (Invitrogen). For immunostaining hTERT-RPE cells, cells were grown on #1 coverslips and fixed and stained as above. Confocal imaging was performed using Leica SP5X system with an upright DM6000 compound microscope and images were processed with the Leica AF software suite. Immunofluorescence of mouse retina was performed essentially as described41. The dilutions of various antibodies used are as follows: SDCCAG8-PR (Proteintech, 1:1000), SDCCAG8-CG (guinea pig 1:800), Ninein (1:500), NPHP5 (1:1000), OFD1 (Nigg, 1:200), Cep164 (1:1000), Cep170 (1:500), CEP290 (Chang et al., 2006, 1:500), RPGRIP1L (Khanna et al., 2009; 1:1000), VHL, SCBT (1:1000), Acetylated α-tubulin (Sigma 1:1000).
Electronmicroscopy studies
For immunogold labeling studies of mouse retina mouse eyecups were fixed in 0.25% glutaraldehyde + 4% formaldehyde in 0.1 M cacodylate buffer, pH 7.4, and processed for embedment in LR White. Ultrathin sections (70 nm) were labeled with primary antibody, followed by secondary antibody conjugated to 12 nm gold.
In vivo electroporation of rat retinas
We used the in vivo electroporation technique to express human SDCCAG8 proteins in rat photoreceptor cells. Injected plasmids were electroporated into retinal cells using tweezer-type electrodes in the right eyes of neonatal rats (n=6-8 rats/injection) as described.42 Animals were sacrificed 4 weeks following transfection, and 60 μm thick sections of the transfected portions of retinas were then prepared from the portions of the eyecups with EGFP signal. Sections were stained with anti-V5 antibodies (Invitrogen), followed by Alexa-555 conjugated secondary antibodies (Invitrogen), and imaging using a Zeiss LSM510 confocal microscope. Three-dimensional reconstructions of the confocal image stacks were generated and analyzed using Volocity 3D imaging software (Improvision, Waltham, MA).
Zebrafish morpholino oligo-mediated knockdown
Morpholino oligonucleotides (MOs) were obtained from Gene Tools, LLC (Philomath, OR). The sequences of the morpholino oligonucleotides used are given in Supplementary Table 1 Online. Fertilized eggs were microinjected with the specified amount of MO dissolved in 0.1M KCl. JB4-embedded sections were prepared as described previously. 6 μm sections were stained with methylene-blue and imaged with a compound microscope. To examine the zebrafish sdccag8 protein, 24 hpf embryos were lysed with RIPA buffer containing protease inhibitor.
Statistical analysis
Student’s two-tailed nonpaired t-tests and normal distribution two-tailed z-tests were carried out using pooled standard error and s.d. values to determine the statistical significance of differences between samples. The significance level was set at P < 0.05.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to families and study individuals for their contribution and thank E. Nigg for providing the OFD1 antibody. This research was supported by grants from the National Institutes of Health to F.H. (DK1069274, DK1068306, DK064614), to H.K. (EY007961), to D.S.W. (EY13408), to N.K. (HD042601, DK075972, DK072301), and to E.A.P (EY12910) by grants from the Netherlands Organization for Scientific Research to K.L.M.C. (NWO Toptalent-021.001.014), to R.R. (NWO Vidi-91786396) and to R.H.G. (NWO Vidi-917.66.354 ), by the WellChild and Wellcome Trust to E.R.M., by the Avenir-INSERM program, the Agence Nationale pour la Recherche, the Union Nationale pour les Aveugles et Déficients Visuels, RETINA France, Programme Hospitalier de Recherche National 2007, and the Association Bardet-Biedl, France to H.D., C.S. and E.A.P by the Foundation Fighting Blindness, the Research to Prevent Blindness, the F.M. Kirby Foundation, and the Rosanne Silbermann Foundation to E.A.P., the Midwest Eye Banks and Transplantation Center and Rare Disease Initiative, University of Michigan to H.K., by Instituto Gulbenkian de Ciência and EMBO to M.B.D., by CIHR, FFB-Canada, FRSQ, and Reseau Vision to R.K.K., by the “Else Kröner-Fresenius-Stiftung“ (P66/09//A75/09) to H.P.H., and by EU FP7 Consortium “Syscilia” to R.H.G, R.R. and N.K.. F.H. is an Investigator of the Howard Hughes Medical Institute, a Doris Duke Distinguished Clinical Scientist, and a Frederick G. L. Huetwell Professor. D.S.W. is a Jules and Doris Stein RPB professor. N.K. is a George R. Brumley Professor. S.S. is a laureat of the Equipe FRM (DEQ20071210558) and the Agence National de la Recherche (R07089KS). The authors thank the families and the physicians who contributed to this study, Annick Toutain, Marie-Claire Gubler, Rémi Salomon, Marie-Alice Macher and Michel Fischbach for contribution of clinical data, Susan J. Allen, Alexei Saveliev, and Yun Liu for excellent technical assistance, Kàlmàn Tory and Céline Becker for linkage analysis and exon sequencing, Songhai Shi and Ryan Insolera for providing shRNA clones, Carsten Janke for providing GT335 antibody, Jeff Salisbury for providing centrin-2 antibody, Eric Nigg for providing Cep164 antibody, and Bo Chang for Cep290 antibody.
Footnotes
Competing interests statement The authors declare that they have no competing financial interests.
Accession numbers. SDCCAG8 full-length isoform-a is GenBank accession number NM_006642.2 and human isoform-e is accession number BC032454.
REFERENCES
- 1.Hildebrandt F, et al. A novel gene encoding an SH3 domain protein is mutated in nephronophthisis type 1. Nat Genet. 1997;17:149–153. doi: 10.1038/ng1097-149. [DOI] [PubMed] [Google Scholar]
- 2.Otto EA, et al. Mutations in INVS encoding inversin cause nephronophthisis type 2, linking renal cystic disease to the function of primary cilia and left-right axis determination. Nat Genet. 2003;34:413–420. doi: 10.1038/ng1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Olbrich H, et al. Mutations in a novel gene, NPHP3, cause adolescent nephronophthisis, tapeto-retinal degeneration and hepatic fibrosis. Nat Genet. 2003;34:455–459. doi: 10.1038/ng1216. [DOI] [PubMed] [Google Scholar]
- 4.Otto E, et al. A gene mutated in nephronophthisis and retinitis pigmentosa encodes a novel protein, nephroretinin, conserved in evolution. Am J Hum Genet. 2002;71:1167–1171. doi: 10.1086/344395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mollet G, et al. The gene mutated in juvenile nephronophthisis type 4 encodes a novel protein that interacts with nephrocystin. Nat Genet. 2002;32:300–305. doi: 10.1038/ng996. [DOI] [PubMed] [Google Scholar]
- 6.Otto E, Loeys B, Khanna H, Hellemans J, Sudbrak R, Fan S, Muerb U, O’Toole JF, Helou J, Attanasio M, Utsch B, Sayer JA, Lillo C, Jimeno D, Coucke P, De Paepe A, Reinhardt R, Klages S, Tsuda M, Kawakami I, Kusakabe T, Omran H, Imm A, Tippens M, Raymond PA, Hill J, Beales P, He S, Kispert A, Margolis B, Williams DS, Swaroop A, Hildebrandt F. A novel ciliary IQ domain protein, NPHP5, is mutated in Senior-Loken syndrome (nephronophthisis with retinitis pigmentosa), and interacts with RPGR and calmodulin. Nat Genet. 2005;37:282–288. doi: 10.1038/ng1520. [DOI] [PubMed] [Google Scholar]
- 7.Sayer JA, et al. The centrosomal protein nephrocystin-6 is mutated in Joubert syndrome and activates transcription factor ATF4. Nat Genet. 2006;38:674–681. doi: 10.1038/ng1786. [DOI] [PubMed] [Google Scholar]
- 8.Valente EM, et al. Mutations in CEP290, which encodes a centrosomal protein, cause pleiotropic forms of Joubert syndrome. Nat Genet. 2006;38:623–625. doi: 10.1038/ng1805. [DOI] [PubMed] [Google Scholar]
- 9.Attanasio M, et al. Loss of GLIS2 causes nephronophthisis in humans and mice by increased apoptosis and fibrosis. Nat Genet. 2007;39:1018–1024. doi: 10.1038/ng2072. [DOI] [PubMed] [Google Scholar]
- 10.Delous M, et al. The ciliary gene RPGRIP1L is mutated in cerebello-oculo-renal syndrome (Joubert syndrome type B) and Meckel syndrome. Nat Genet. 2007;39:875–881. doi: 10.1038/ng2039. [DOI] [PubMed] [Google Scholar]
- 11.Otto EA, et al. NEK8 mutations affect ciliary and centrosomal localization and may cause nephronophthisis. J Am Soc Nephrol. 2008;19:587–592. doi: 10.1681/ASN.2007040490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hildebrandt F, Zhou W. Nephronophthisis-associated ciliopathies. J Am Soc Nephrol. 2007;18:1855–1871. doi: 10.1681/ASN.2006121344. [DOI] [PubMed] [Google Scholar]
- 13.Hildebrandt F, et al. A Systematic Approach to Mapping Recessive Disease Genes in Individuals from Outbred Populations. PloS Genetics. 2009;5:31000353. doi: 10.1371/journal.pgen.1000353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ng SB, et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461:272–276. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Q, et al. The proteome of the mouse photoreceptor sensory cilium complex. Mol Cell Proteomics. 2007;6:1299–1317. doi: 10.1074/mcp.M700054-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gherman A, Davis EE, Katsanis N. The ciliary proteome database: an integrated community resource for the genetic and functional dissection of cilia. Nat Genet. 2006;38:961–962. doi: 10.1038/ng0906-961. [DOI] [PubMed] [Google Scholar]
- 17.Zhang XH, Kangsamaksin T, Chao MS, Banerjee JK, Chasin LA. Exon inclusion is dependent on predictable exonic splicing enhancers. Mol Cell Biol. 2005;25:7323–7332. doi: 10.1128/MCB.25.16.7323-7332.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baker K, Beales PL. Making sense of cilia in disease: the human ciliopathies. Am J Med Genet C Semin Med Genet. 2009;151C:281–295. doi: 10.1002/ajmg.c.30231. [DOI] [PubMed] [Google Scholar]
- 19.Beales PL, Elcioglu N, Woolf AS, Parker D, Flinter FA. New criteria for improved diagnosis of Bardet-Biedl syndrome: results of a population survey. J Med Genet. 1999;36:437–446. [PMC free article] [PubMed] [Google Scholar]
- 20.Kenedy AA, Cohen KJ, Loveys DA, Kato GJ, Dang CV. Identification and characterization of the novel centrosome-associated protein CCCAP. Gene. 2003;303:35–46. doi: 10.1016/s0378-1119(02)01141-1. [DOI] [PubMed] [Google Scholar]
- 21.Simms RJ, Eley L, Sayer JA. Nephronophthisis. Eur J Hum Genet. 2009;17:406–416. doi: 10.1038/ejhg.2008.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Graser S, et al. Cep164, a novel centriole appendage protein required for primary cilium formation. J Cell Biol. 2007;179:321–330. doi: 10.1083/jcb.200707181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Andersen JS, et al. Proteomic characterization of the human centrosome by protein correlation profiling. Nature. 2003;426:570–574. doi: 10.1038/nature02166. [DOI] [PubMed] [Google Scholar]
- 24.Watnick T, Germino G. From cilia to cyst. Nat Genet. 2003;34:355–356. doi: 10.1038/ng0803-355. [DOI] [PubMed] [Google Scholar]
- 25.Delous M, et al. Nephrocystin-1 and nephrocystin-4 are required for epithelial morphogenesis and associate with PALS1/PATJ and Par6. Hum Mol Genet. 2009;18:4711–4723. doi: 10.1093/hmg/ddp434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coene KL, et al. OFD1 is mutated in X-linked Joubert syndrome and interacts with LCA5-encoded lebercilin. Am J Hum Genet. 2009;85:465–481. doi: 10.1016/j.ajhg.2009.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thauvin-Robinet C, et al. Clinical, molecular, and genotype-phenotype correlation studies from 25 cases of oral-facial-digital syndrome type 1: a French and Belgian collaborative study. J Med Genet. 2006;43:54–61. doi: 10.1136/jmg.2004.027672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Budny B, et al. A novel X-linked recessive mental retardation syndrome comprising macrocephaly and ciliary dysfunction is allelic to oral-facial-digital type I syndrome. Hum Genet. 2006;120:171–178. doi: 10.1007/s00439-006-0210-5. [DOI] [PubMed] [Google Scholar]
- 29.Rakkolainen A, Ala-Mello S, Kristo P, Orpana A, Jarvela I. Four novel mutations in the OFD1 (Cxorf5) gene in Finnish patients with oral-facial-digital syndrome 1. J Med Genet. 2002;39:292–296. doi: 10.1136/jmg.39.4.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ferrante MI, et al. Identification of the gene for oral-facial-digital type I syndrome. Am J Hum Genet. 2001;68:569–576. doi: 10.1086/318802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schluter MA, Margolis B. Apical lumen formation in renal epithelia. J Am Soc Nephrol. 2009;20:1444–1452. doi: 10.1681/ASN.2008090949. [DOI] [PubMed] [Google Scholar]
- 32.Elia N, Lippincott-Schwartz J. Culturing MDCK cells in three dimensions for analyzing intracellular dynamics. Curr Protoc Cell Biol. 2009:22. doi: 10.1002/0471143030.cb0422s43. Chapter 4, Unit 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim E, Walz G. Sensitive cilia set up the kidney. Nat Med. 2007;13:1409–1411. doi: 10.1038/nm1207-1409. [DOI] [PubMed] [Google Scholar]
- 34.Moyer JH, et al. Candidate gene associated with a mutation causing recessive polycystic kidney disease in mice. Science. 1994;264:1329–1333. doi: 10.1126/science.8191288. [DOI] [PubMed] [Google Scholar]
- 35.Torres VE. Role of vasopressin antagonists. Clin J Am Soc Nephrol. 2008;3:1212–1218. doi: 10.2215/CJN.05281107. [DOI] [PubMed] [Google Scholar]
- 36.Gattone VH, 2nd, Wang X, Harris PC, Torres VE. Inhibition of renal cystic disease development and progression by a vasopressin V2 receptor antagonist. Nat Med. 2003;9:1323–1326. doi: 10.1038/nm935. [DOI] [PubMed] [Google Scholar]
- 37.Patel V, Chowdhury R, Igarashi P. Advances in the pathogenesis and treatment of polycystic kidney disease. Curr Opin Nephrol Hypertens. 2009;18:99–106. doi: 10.1097/MNH.0b013e3283262ab0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kruglyak L, Daly MJ, Reeve-Daly MP, Lander ES. Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet. 1996;58:1347–1363. [PMC free article] [PubMed] [Google Scholar]
- 39.Strauch K, et al. Parametric and nonparametric multipoint linkage analysis with imprinting and two-locus-trait models: application to mite sensitization. Am J Hum Genet. 2000;66:1945–1957. doi: 10.1086/302911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gudbjartsson DF, Jonasson K, Frigge ML, Kong A. Allegro, a new computer program for multipoint linkage analysis. Nat Genet. 2000;25:12–13. doi: 10.1038/75514. [DOI] [PubMed] [Google Scholar]
- 41.Khanna H, et al. A common allele in RPGRIP1L is a modifier of retinal degeneration in ciliopathies. Nat Genet. 2009;41:739–745. doi: 10.1038/ng.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Matsuda M, et al. The administration of retinoic acid down-regulates cAMP-responsive element modulator (CREM) mRNA in vitamin A-deficient testes. Biosci Biotechnol Biochem. 2005;69:261–266. doi: 10.1271/bbb.69.261. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.