Abstract
Chlamydomonas reinhardtii, a unicellular green alga, has been exploited as a reference organism for identifying proteins and activities associated with the photosynthetic apparatus and the functioning of chloroplasts. Recently, the full genome sequence of Chlamydomonas was generated and a set of gene models, representing all genes on the genome, was developed. Using these gene models, and gene models developed for the genomes of other organisms, a phylogenomic, comparative analysis was performed to identify proteins encoded on the Chlamydomonas genome which were likely involved in chloroplast functions (or specifically associated with the green algal lineage); this set of proteins has been designated the GreenCut. Further analyses of those GreenCut proteins with uncharacterized functions and the generation of mutant strains aberrant for these proteins are beginning to unmask new layers of functionality/regulation that are integrated into the workings of the photosynthetic apparatus.
Keywords: Chlamydomonas, GreenCut, Chloroplast, Phylogenomics, Regulation
Introduction
Chlamydomonas reinhardtii as a reference organism for the study of photosynthesis
The most well-characterized photosynthetic organisms that can be probed with powerful genetic and molecular tools include Synechocystis sp. PCC6803, Chlamydomonas reinhardtii (Chlamydomonas throughout) and Arabidopsis thaliana (Arabidopsis throughout). Complementary attributes of these organisms provide a synergistic view of basic biological and regulatory processes that occur in photosynthetic lineages. In this article, we emphasize the ways in which Chlamydomonas has been used to elucidate photosynthesis, especially with the aid of bioinformatic analyses to generate a set of proteins designated the “GreenCut” (Merchant et al. 2007).
Over the last half century, experimentation with Chlamydomonas has addressed numerous biological issues and elucidated mechanisms that underlie a variety of cellular activities. Recently, the state of Chlamydomonas biology has been described in the Chlamydomonas Sourcebook (Harris 2009), an invaluable, up-to-date resource on most aspects of Chlamydomonas biology. Those processes and analyses relevant to the focus of this article include characterization of the chloroplast genome (Higgs 2009) and chloroplast structure and function (de Vitry and Kuras 2009; Finazzi et al. 2009; Gokhale and Sayre 2009; Minagawa 2009; Niyogi 2009; Redding 2009; Rochaix 2009), post-translation regulation of chloroplast biogenesis (Rochaix 2001; Bollenbach et al. 2004; Drapier et al. 2007; Raynaud et al. 2007; Eberhard et al. 2008; Choquet and Wollman 2009; Goldschmidt-Clermont 2009; Herrin 2009; Klein 2009; Zerges and Hauser 2009; Zimmer et al. 2009), and elucidation of activities and regulatory circuits that control uptake and assimilation of various macronutrients (Camargo et al. 2007; Fernandez and Galvan 2007; Fernández and Galván 2008; González-Ballester et al. 2008; Fernández et al. 2009; González-Ballester and Grossman 2009; Moseley et al. 2009; Moseley and Grossman 2009; González-Ballester et al. 2010) and micronutrients (Merchant et al. 2006; Tejada-Jimenez et al. 2007; Kohinata et al. 2008; Long et al. 2008). Chlamydomonas also represents an important model for studies of light-driven H2 production (Ghirardi et al. 2007; Melis 2007; Posewitz et al. 2009).
The physiological, metabolic, and genetic characteristics of Chlamydomonas make it an ideal organism for dissecting the structure, function, and regulation of the photosynthetic apparatus. In early studies, Levine and colleagues exploited the haploid (single copy of the nuclear genome in each cell) genetics of Chlamydomonas, which was first highlighted by Sager (Sager 1960), to elucidate genes/proteins involved in photosynthetic electron transport and carbon fixation (Gorman and Levine 1966, 1967; Givan and Levine 1967; Lavorel and Levine 1968; Levine 1969; Levine and Goodenough 1970; Moll and Levine 1970; Sato et al. 1971). A significant advantage of working with an organism that displays haploid genetics is that the phenotype caused by a genetic lesion is manifest almost immediately after the generation of that lesion; this affords researchers the opportunity to select or screen for mutants with specific phenotypes without having to first generate diploid strains that are homozygous for the lesion. Another extremely important feature of this alga is that it exhibits robust growth under heterotrophic conditions in the dark, with acetate as a sole source of fixed carbon. This feature of the physiology of Chlamydomonas allows for the identification and maintenance of mutants that are completely blocked for photosynthetic function, as long as they are grown on medium supplemented with acetate. Furthermore, dark-grown, wild-type Chlamydomonas cells remain green, retain normal chloroplast structure, and resume photosynthesis immediately following their transfer to the light (Harris 1989). Hence, even mutants that are extremely sensitive to light (e.g., in some photosynthetic mutants, low light triggers photo-oxidative reactions that can cause peroxidation of membranes and oxidation of proteins) survive when maintained in the dark or near-dark conditions. Many other photosynthetic organisms are either unable to use exogenous reduced carbon, or use it to some extent, but show diminished growth rates and/or retarded developmental processes. Overall, the various biological features of Chlamydomonas make it an important, genetically tractable eukaryote in which lesions that eliminate photosynthesis are conditional rather than lethal or severely debilitating.
While Arabidopsis does not show optimal growth or completely normal development when maintained on a fixed source of carbon, studies of this organism are also important to our understanding of photosynthesis. For example, mutations of Arabidopsis in genes encoding proteins critical for photosynthetic function can be maintained in seeds as heterozygotes; these seeds can survive for years when stored under appropriate conditions. This feature of vascular plants also allows recessive mutations that are lethal in the homozygous diploid state to be maintained as heterozygous seeds; only when the homozygote strain is generated through crosses would the mutant plant die as photosynthetic function is lost in the developing seedlings. Furthermore, it is only in multicellular organisms that one can analyze the uptake, assimilation, and movement of nutrients between different tissues and organs, and elucidate various organ-specific developmental and regulatory processes associated with distinct plastid classes. Such processes might include temporal analyses of chromoplast and leucoplast development and the greening of etioplasts. Also, since different organisms thrive in different environments and occupy specific environmental niches, the exact mechanisms associated with photosynthetic function may be tailored to a specific environment. For example, while the PSBS protein, a member of the light harvesting family of proteins, may be critical for non-photochemical quenching of excess absorbed light energy in plants (Li et al. 2000), other light-harvesting family proteins, such as the LHCSRs, appear to be important for non-photochemical quenching in Chlamydomonas (Peers et al. 2009), while the orange carotenoid protein (OCP) is critical for non-photochemical quenching in cyanobacteria (Wilson et al. 2006). Organisms adapted to different environments may also exploit various electron outlets or valves to control the increased excitation pressure that can occur when the photosynthetic apparatus absorbs more light energy than it can use in downstream anabolic processes. For example, the flow of electrons to O2 via the Mehler reaction (oxidation of ferredoxin) may be significant in generating a specific redox poise that modulates cyclic electron flow around photosystem (PS) I and the formation of ATP, the activity of PSII, state transitions, non-photochemical quenching, and even aspects of chloroplast biogenesis (Asada 1999; Heber 2002; Makino et al. 2002; Forti 2008). A plastoquinone terminal oxidase may also significantly participate in at least some of these regulatory processes in certain organisms (Rumeau et al. 2007; Bailey et al. 2008; Stepien and Johnson 2009).
Mutant generation
In previous reports, photosynthetic mutants in Chlamydomonas were identified based on their inability to assimilate 14CO2 (Levine 1960). Photosynthetic mutants have been isolated based on their inability to grow in the absence of acetate (Eversole 1956), their resistance to metronidazole (Schmidt et al. 1977), or their chlorophyll fluorescence characteristics (Bennoun and Delepelaire 1982). Indeed, many fundamental discoveries leading to present-day knowledge of photosynthesis, including sequences of carriers critical for electron transfer, polypeptides involved in light harvesting and reaction center function, and enzymes of the Calvin–Benson–Bassham Cycle, have been elucidated through the generation and characterization of mutants (especially Chlamydomonas mutants) with lesions in components of the photosynthetic apparatus. Some processes critical for the dynamics of photosynthetic function have also been elucidated; these include state transitions and non-photochemical quenching. While the discoveries relating to photosynthetic structure and function are too numerous to detail here, many are summarized in various chapters of the new Chlamydomonas Sourcebook (Choquet and Wollman 2009; de Vitry and Kuras 2009; Finazzi et al. 2009; Gokhale and Sayre 2009; Goldschmidt-Clermont 2009; Herrin 2009; Higgs 2009; Klein 2009; Minagawa 2009; Niyogi 2009; Redding 2009; Rochaix 2009; Zerges and Hauser 2009), as well as in a number of recent review articles (Eberhard et al. 2008; Li et al. 2009; Grossman et al. 2010).
Photoacclimation and the regulation of photosynthesis
The regulation of photosynthetic processes as a consequence of adaptation and acclimation is an area of research that several laboratories have approached, for which there are still large gaps in our knowledge remaining to be filled. Environmental signals impact chloroplast biogenesis and photosynthetic function, provoking marked changes in photosynthetic electron transport (PET) (Eberhard et al. 2008; Li et al. 2009). High light acclimation, for example, helps balance the harvesting of light energy by the two photosystems, and coordinates PET with the activity of the Calvin–Benson–Bassham Cycle; this type of modulation minimizes photodamage. Low light, in contrast, can elicit an increase in the cross section of the PSII antenna, which makes the capture of excitation energy more efficient. Furthermore, certain organisms respond dramatically to changes in the quality of the light that they are absorbing. For example, some cyanobacteria display a regulatory phenomenon called complementary chromatic adaptation. In this process, the polypeptide and pigment composition of the phycobilisome (the major light-harvesting complex in many cyanobacteria) can physically and functionally be tuned to light quality. When cyanobacteria experience light enriched in red wavelengths, the cells appear bluish because of elevated levels of phycocyanin, a blue-pigmented biliprotein associated with the phycobilisome. In contrast, when cells experience light enriched in green wavelengths, they appear red because of elevated levels of phycoerythrin, a red-pigmented biliprotein associated with the phycobilisome (Grossman et al. 2003; Kehoe and Gutu 2006). In addition, light triggers complex changes in thylakoid composition and cellular structure that may involve post-translational modifications as well as the synthesis of new polypeptide and pigment components (Bordowitz and Montgomery 2008; Eberhard et al. 2008; Whitaker et al. 2009). Despite considerable phenomenological and biochemical knowledge, little is known of underlying mechanisms that control photoacclimation (Eberhard et al. 2008). Although some evidence indicates that the cellular redox state may be a key regulatory signal (Huner et al. 1998), it is still not clear whether/how photoreceptors are integrated into the control networks. With respect to redox control (Eberhard et al. 2008; Pfannschmidt et al. 2009), increases in irradiance often act via an elevated redox state of the plastoquinone (PQ) pool, providing a signal that can develop very rapidly and elicit a multitude of downstream acclimation responses. This model of regulation predicts self-modulating control because as acclimation responses begin to alter the physiology of the cell, the PQ pool becomes reoxidized, which in turn would diminish or terminate aspects of the acclimation response; the cells would ultimately achieve a new steady state. Other aspects of redox control involve changes in the redox state of specific thioredoxins, the generation of reactive oxygen species, the flux of electrons through the cytochrome b 6 f complex, the extent of the ΔpH across the thylakoid membranes, and numerous aggregate metabolic signals that could include levels of ATP, NADPH, CO2, and various Calvin–Benson–Bassham Cycle metabolites. Hence, even though still not well understood, linear and cyclic electron flow appear to be precisely controlled and tightly integrated with the capacity of the cells to fix CO2. Furthermore, light-induced signals must be transduced to the chloroplast and nucleus/cytoplasm, influencing both transcriptional and post-transcriptional processes in the different subcellular compartments. Degradation of plastid components must also be tightly coordinated with de novo synthesis, the recycling of pigment molecules and the integration of polypeptides into photosynthetic complexes. Our understanding of most aspects of these processes is still at a relatively preliminary stage (Walters 2005). Indeed, there are still even structural proteins associated with the photosynthetic apparatus, which have only recently been identified. For example, examination of the crystal structure of PSI has revealed the presence of a previously unidentified protein, designated PsaR, which appears to be loosely associated with the PSI core and is positioned between the PsaK and Lhca3 subunits; this protein is potentially involved in the stabilization of PSI light-harvesting complexes (Amunts et al. 2010).
Photosynthesis in the era of genomics
The explosion of genomic information over the last decade is being used to identify the full complement of genes present on the nuclear, chloroplast, and mitochondrial genomes, elucidate relationships between gene content/expression patterns and ecological differences among related organisms, determine ways in which gene content has been arranged and modified by evolutionary processes, define the extent to which genes are transferred between organisms and the features of the transfer process, and uncover mechanisms critical for modulating gene expression in response to developmental processes and fluctuating environmental conditions. With the massive influx of genomic information and comparative genomic tools, it is becoming clear just how much is not understood about many biological processes, including those that are integral to global productivity, biogeochemical cycling, the structure and composition of ecological habitats, and the ways in which biological processes impact the geochemistry and geophysics of the Earth. Many researchers are beginning to mine fully characterized algal and cyanobacterial genomic information (Rocap et al. 2003; Armbrust et al. 2004; Matsuzaki et al. 2004; Barbier et al. 2005; Misumi et al. 2005; Mulkidjanian et al. 2006; Palenik et al. 2007; Bowler et al. 2008; Vardi et al. 2008; Maheswari et al. 2009; Worden et al. 2009), and there are now many newly generated sequences of algal nuclear genomes that either have been completed or are near completion; these include the sequences of Coccomyxa sp. C-169, Chlorella NC64A, Aureococcus anophagefferens, Emiliania huxleyi CCMP1516, Bathycoccus sp. (BAN7), Chondrus crispus, Porphyra umbilicalis, Ectocarpus siliculosus, Micromonas pusilla CCMP1545, Micromonas sp. RCC299, and Volvox carteri (see http://genome.jgi-psf.org and http://www.genoscope.cns.fr/spip/Plants-sequenced-at-Genoscope.html). It is likely that this list will rapidly expand over the next several years.
We and other researchers have been exploring the genomics of Chlamydomonas (Grossman et al. 2003, 2007; Gutman and Niyogi 2004; Ledford et al. 2004, 2007; Dent et al. 2005; Merchant et al. 2007; González-Ballester and Grossman 2009; Moseley et al. 2009; González-Ballester et al. 2010) in the context of a number of other algae, photosynthetic microbes, and plants. The Chlamydomonas genomic sequence was generated by the Joint Genome Institute (JGI) from the cell wall-deficient strain CC-503 cw92 mt+. A BAC library has been constructed from genomic DNA of this strain (https://www.genome.clemson.edu/cgi-bin/orders?&page=productGroup&service=bacrc&productGroup=162). Chlamydomonas EST libraries have also been generated and characterized; one (isolated by researchers at the Carnegie Institution) was constructed with RNA isolated from strain CC-1690 21 gr mt+ (Shrager et al. 2003), while cDNA libraries analyzed in Japan were constructed from C-9 mt (Asamizu et al. 1999, 2000). Both of the strains used for constructing the cDNA libraries are related to CC-503; they were derived from the same field isolate collected in Massachusetts in 1945. The mating partner used for mapping genetic loci in Chlamydomonas is designated S1D2, a field isolate (collected in Minnesota in the 1980s) for which significant EST information has also been generated. The EST sequences from S1D2 have been used to generate physical markers for fine scale map-based cloning of mutant alleles (Rymarquis et al. 2005). More recently, researchers have used the Chlamydomonas nuclear genome sequence and the gene models generated from that sequence for comparative analyses focused on identifying genes of unknown function that are potentially important for the regulation and/or activity of the various photosynthetic complexes.
An initial analysis of the Chlamydomonas genome (Merchant et al. 2007) used the version 3.0 assembly. This assembly represents ~13X coverage of the genome, which is ~121 Mb. The use of ab initio and homology-based algorithms resulted in the generation of 15,143 gene models. The version 4.0 assembly of the Chlamydomonas genome was released in March 2009 (http://genome.jgi-psf.org/Chlre4/Chlre4.home.html). This assembly is composed of 88 scaffolds with 112 Mb of genomic sequence information. The genome scaffolds have numerous gaps that comprise ~7.5% of the total scaffold lengths. After using both cDNA/EST and homology-based support to improve the gene models, manual annotation of many genes was completed, and the genome now has a total of 16,709 gene models.
There are presently over 300,000 publicly available ESTs that were generated from cDNAs constructed from RNA isolated from cultures of Chlamydomonas exposed to a variety of physiological conditions (Asamizu et al. 1999, 2000; Shrager et al. 2003; Jain et al. 2007). Although in some cases the libraries were normalized to increase the representation of lower abundance transcripts in the EST database, the existing data set covers a little over half of the predicted protein-coding gene models, with only about half of those covering full-length (or nearly full-length) transcripts. Hence, only ~25% of the protein-coding gene models are accurately computed and verified by transcript maps. Comparisons of the Chlamydomonas gene models to those of the close relative Volvox (shown on the Vista track of the JGI browser) and to available cDNA information, suggest that many JGI models are missing either the entire or part of the 5′ and 3′ UTRs, with several also under-predicted for the number of exons. Since in-depth sequencing of cDNA libraries may still not capture genes encoding low abundance transcripts and maximizing sequence information from cDNA libraries is neither time-efficient nor cost-effective, present efforts are directed toward the use of next generation transcript re-sequencing technologies (in which cDNA fragments derived from RNAs isolated from various conditions are sequenced without cloning) to generate new gene models and to correct those that have been previously constructed.
The rapid expansion of genomic sequence information for Chlamydomonas has also stimulated the establishment of strong proteomic initiatives (Stauber and Hippler 2004; Wagner et al. 2004, 2008, 2009; Keller et al. 2005; Schmidt et al. 2006; Naumann et al. 2007; Ozawa et al. 2009; Rolland et al. 2009) and integrative systems databases (May et al. 2008, 2009). Much of our attention has been focused on mechanisms of photosynthetic electron transport and its regulation and identification of specific genes/proteins associated with functional and regulatory aspects of photosynthesis, with an emphasis on acclimation of the photosynthetic apparatus to environmental change. With the genomic sequence information collected for Chlamydomonas and other photosynthetic and non-photosynthetic organisms, we are now in a position to perform comparative genomic analyses to link genes/proteins that have no assigned functions to specific biological processes.
The Greencut
The photosynthetic eukaryotic lineage comprising the Plantae is thought to have a single evolutionary origin that was initiated with the engulfment of a cyanobacterium by a non-photosynthetic protist. Following this primary endosymbiosis, the Plantae diverged into three lineages; the Rhodophyta, Glaucophyta, and Viridiplantae. The Viridiplantae then branched into the Chlorophyta or green algae, which include the Volvocales (e.g., Chlamydomonas and Volvox) and Prasinophytes (e.g., Ostreococcus and Micromonas), and the lineage that gave rise to the Spermatophyta (angiosperms, gymnosperms, bryophytes); this divergence occurred over 1 billion years ago. Genes common to the genomes of the Chlorophyta and Spermatophyta can be traced to the Viridiplantae ancestor of these lineages; a subset of genes in this category would be involved in photosynthesis and chloroplast function. This subset could potentially be identified by comparative genomic analyses.
Mining Chlamydomonas genomic sequence information
A comparative analysis was performed in which all predicted Chlamydomonas proteins (predicted from gene models) were compared against both Arabidopsis and human protein sequences using BLAST, and the best hit scores for each Chlamydomonas protein relative to the two genomes was shown in the analysis presented in Fig. 4 in the manuscript by Merchant et al. (2007). Some subsets of Chlamydomonas proteins were more similar to those of Arabidopsis, while others were more similar to those of humans. For example, Chlamydomonas thylakoid and stromal proteins, many of which are associated with photosynthetic function, were significantly more similar to polypeptides in Arabidopsis than to those in humans, as expected. Hence, some specific processes, including photosynthesis, have been preserved in Chlamydomonas and Arabidopsis but not in humans (animal lineage). In contrast, genes encoding proteins associated with the structure and function of Chlamydomonas flagella have been preserved in humans and other mammals, but not in seed plants. These observations indicate that the common ancestor to Chlamydomonas and Spermatophyta was ciliated, like animal cells. However, the cilia and the genes associated with their structure and assembly were lost during the evolution of the seed plants (Merchant et al. 2007).
Researchers can now integrate the power of full genome sequence analyses with the wealth of information amassed over the past several decades on photosynthetic and acclimation processes. The genomic information can be used to identify those genes present on the Chlamydomonas genome that encode proteins specifically associated with the green plant lineage; such proteins have been placed into an assemblage designated the “GreenCut” (Merchant et al. 2007; Grossman et al. 2010). Various analyses of GreenCut proteins and levels of transcripts encoding those proteins are providing new insights into their potential functions. Specific informatic tools have helped determine whether individual GreenCut proteins have a presequence that predicts their subcellular location. Furthermore, information on the abundance of transcripts encoding GreenCut proteins in the different tissue types of Arabidopsis, and as environmental conditions change, can be assembled from numerous previous experiments (Schmid et al. 2005). However, it is with the use of reverse genetic approaches for isolating strains harboring lesions in GreenCut proteins (both in Chlamydomonas and Arabidopsis) that researchers are most likely to be effective in deciphering the function(s) of these proteins. Mutant strains generated by insertional mutagenesis using a drug resistant marker gene (paromomycin or bleomycin resistance) can be identified by PCR-based screening of mutant libraries (Krysan et al. 1996) or by phenotypic analyses followed by identification of sequences flanking the insertion site (Dent et al. 2005). Given that the photosynthetic phenotype of the mutant co-segregates with the inserted marker gene, the consequences of the gene disruption can be further analyzed with powerful biophysical, biochemical, and molecular technologies. Such analyses are likely to result in the identification of proteins and activities, previously either never or minimally characterized, that influence the function or regulation of photosynthetic processes.
Generation of the GreenCut
The specific way in which the GreenCut was generated is described in Merchant et al. (Merchant et al. 2007). In brief, all protein sequences deduced from the gene models of the Chlamydomonas genome version 3.1 were compared by BLAST to all protein sequences in several phylogenetically diverse organisms including algae, land plants, cyanobacteria, respiring bacteria, archaea, oomycetes, amoebae, fungi, metazoans, and diatoms. Initially, all possible orthologous protein pairs, with one member of the pair a Chlamydomonas protein, were generated; orthologous proteins were defined as those proteins from the various organisms that exhibit a mutual best BLAST hit with a Chlamydomonas protein. However, the identification of orthologs is more complex in organisms where a gene may have duplicated after speciation, and even more complex when considering distantly related organisms where there may have been multiple occurrences of both pre- and post-speciation gene duplications as well as gene losses. For the GreenCut, the assignment of homologs into different or the same group of orthologs was based on sequence relatedness. The parameters were chosen empirically so that known gene families (such as LHCs) could be recovered and sets of orthologs distinguished (such as LHCAs vs. LHCBs). The application of this procedure resulted in the generation of 6,968 individual protein families, each containing one or more Chlamydomonas paralog(s), all mutual best BLAST hits to proteins of other species (orthologs), and all associated paralogs from those other species. However, it should be kept in mind that the GreenCut is under-represented for proteins encoded by large gene families since gene duplications and divergence of individuals within such families can make it difficult to generate precise orthology/paralogy assignments (e.g., there may not be any mutual best BLAST hit). This situation may also result in the introduction of non-homologous proteins into some protein families. An example of these difficulties is apparent when analyzing the light-harvesting protein family. Only two of the ~20 Chlamydomonas LHC proteins were retrieved in the initial GreenCut analysis; the paralogs were not similar enough to the orthologous sequences to be drawn into protein family clusters despite our attempt to do so.
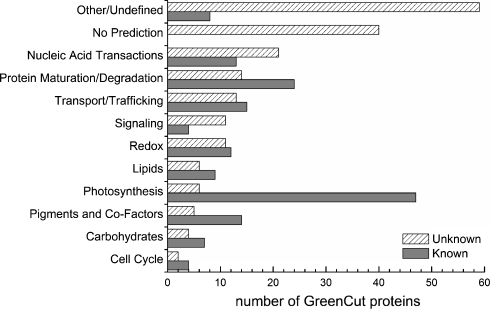
The families of proteins generated by the procedures described above were used for comparative analyses to identify those proteins that are specifically present in the green algal and plant lineages, and that in many cases may be associated with chloroplast/photosynthetic function. More specifically, families of homologous proteins for which all members were in the green lineage of the Plantae, which in this comparison included Chlamydomonas, Ostreococcus spp., Arabidopsis, and Physcomitrella, but were not present in the genomes of non-photosynthetic eukaryotes and prokaryotes, were identified. Based on the criteria outlined above, a set of 349 polypeptides of Chlamydomonas were grouped into the GreenCut (Merchant et al. 2007). Of these 349 polypeptides, 135 were previously known proteins with well-characterized functions. This set also included proteins whose function was known by inference based on comparisons with proteins from other organisms. Surprisingly, there was no specific functional information for 214 of these conserved proteins, although several did have a sequence motif (e.g., pfam domains for DNA binding, RNA binding, kinase activity etc.) that suggested a generalized biochemical function. Hints concerning protein functionality can also be inferred from co-expression profiles (e.g., tissue-specific expression in plants or expression based on different environmental conditions) and determination of potential subcellular location of the protein, based either on the presence/absence of a recognizable transit peptide, which targets polypeptides to the chloroplast, or subproteome analyses (Baginsky et al. 2007; Kleffmann et al. 2007; Rolland et al. 2009; Zybailov et al. 2008). The most recent groupings of the proteins of known and unknown functions of the GreenCut are shown in Fig. 1. As this figure indicates, there are many unknowns in the categories “Signaling,” which are mostly sensing proteins, and “Nucleic Acid Transactions,” which include many putative transcription factors and RNA-binding proteins. This emphasizes the point that most processes that regulate the biogenesis and function of the photosynthetic apparatus are still not defined. Furthermore, numerous hypothetical proteins are present in the categories “Other/Undefined,” and “No Prediction”; together, those categories contain nearly 100 proteins for which no function has been determined. GreenCut polypeptides were further categorized based on whether or not they were present in diatoms and cyanobacteria; 91 of the 349 GreenCut proteins are present in cyanobacteria. Of course, many unknowns will be moved to the various known categories as we learn more about their specific functions. Indeed, ever since the generation of the initial GreenCut (11/06), a number of unknowns have been characterized and attributed a function; these newly defined proteins are given in Table 1. Some of these proteins are involved in the biogenesis of chloroplast cytochromes (CPLD51, CPLD43, CPLD23) (Kuras et al. 2007; Lezhneva et al. 2008), others are involved in the breakdown of chlorophyll (Kusaba et al. 2007; Ren et al. 2007; Sato et al. 2009), and yet others are potentially involved in regulating photosynthetic functions (Lu et al. 2006; Lee et al. 2007; Schult et al. 2007; Zhu et al. 2007; DalCorso et al. 2008; Duan et al. 2008). The assignment of function to these unknowns validates the predictive power of the GreenCut.
Fig. 1.
Classification of GreenCut proteins. The bar graph presents the functional classification of GreenCut proteins (on y axis) and the number of proteins placed into each class (x axis). The solid bars represent proteins with known functions while the hatched bars represent proteins of unknown functions. The proteins of unknown function have been placed in generalized functional categories based on domains or motifs within the proteins
Table 1.
Genes of the GreenCut encoding proteins that have recently been assigned a function
| Cre gene name | Other gene name | Cre v4 protein ID | AT identifier | Functional annotation | Functional category |
|---|---|---|---|---|---|
| PGRL1 | AthPGRL1A/AthPGRL1B | 162423 | At4g22890/At4g11960 | Modulation of photosynthetic electron transport | Photosynthesis |
| CCB1 | AthCCB1 | 185128 | At3g26710 | Heme attachment to cyt b6 of the b 6 f complex | Photosynthesis |
| CCB3 | AthCCB3 | 205830 | At5g36120 | Heme attachment to cyt b6 of the b6f complex | Photosynthesis |
| CCB4 | AthCCB4 | 205823 | At1g59840 | Heme attachment to cyt b 6 of the b 6 f complex | Photosynthesis |
| NYC1 | AthNYC1 | 205572 | At4g13250 | Chlorophyll b reductase | Pigments and co-factors |
| CGLD28 | AthABA4 | 187252 | At1g67080 | Neoxanthin synthase | Pigments and co-factors |
| CPL6 | BolOR | 205873 | At5g61670 | DnaJ-domain protein involved in differentiation of proplastids into chromoplasts | Pigments and co-factors |
| CGL61 | AthNYE1 | 205911 | At4g22920 | Regulates chlorophyll degradation | Pigments and co-factors |
| CPLD10 | AthHCF173 | 205878 | At1g16720 | Initiation of translation of the psbA mRNA | Protein maturation/degradation |
| CGL13 | AthTPST | 206702 | At1g08030 | Tyrosylprotein sulfotransferase | Protein maturation/degradation |
| RNB2 | AthRNR1 | 188559 | At5g02250 | rRNA processing exoribonuclease | Nucleic acid transactions |
| CPLD46 | AthISE2 | 116679 | At1g70070 | DEVH box RNA helicase involved in plasmodesmata function | Nucleic acid transactions |
| CGLD18 | AthCOL2 | 159133 | At3g02380 | Constans-like zinc finger transcription factor influencing flowering-time | Nucleic acid transactions |
| CGL19 | AthCDF2 | 205894 | At5g39660 | Photoperiodic flowering response transcription factor | Nucleic acid transactions |
| CGL15 | AthPHT4 | 205891 | At5g20380 | Phosphate transporter 4;5 | Transport/Trafficking |
| CPLD23 | AthGONST3 | 149680 | At1g76340 | Golgi nucleotide-sugar transporter | Transport/trafficking |
| TRXL1 | AthACHT1 | 205754 | At4g26160 | Atypical Cys His rich thioredoxin 1 | Redox |
| CPLD30 | AthENH1 | 183051 | At5g17170 | Rubredoxin domain protein involved in ROS detoxification | Redox |
| GRX6 | AthCXIP2 | 195615 | At2g38270 | Activates calcium transporter CAX4 | Redox |
| CPLD41 | SynDsbAB | 178821 | At4g35760 | Disulfide bond formation | Redox |
| CGLD21 | AthEX2 | 205932 | At1g27510 | Plastid protein signaling 1O2-dependent nuclear gene expression changes | Signaling |
| CGL8 | AthSPX2 | 205638 | At2g26660 | Transcriptional control of gene expression during phosphate starvation/cold stress | Signaling |
| CPLD33 | AthRPH1 | 184621 | At2g48070 | Positive regulator of oxidative burst in plant immune response | Signaling |
| CPLD27 | AthTNMT | 119132 | At4g33110 | S-Adenosyl-L-methionine:tetrahydroprotoberberine cis-N-methyltransferase | Other/undefined |
Note: Cre is used as an abbreviation of Chlamydomonas reinhardtii
Advantageous features of the GreenCut
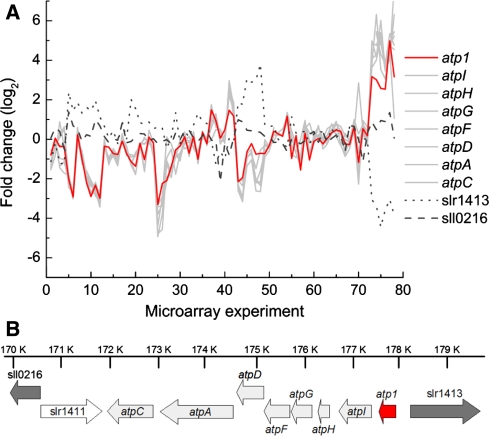
There are several advantages in using informatics to determine sets of proteins, such as the GreenCut, likely to be associated with specific cellular functions. The relatively small number of proteins grouped in such sets helps to establish a focus for constructing an indexed library of mutant organisms. For example, lesions in genes encoding GreenCut proteins could be profitably generated in any oxygenic phototroph for which genomic and molecular tools are sufficiently advanced. The logistics of generating and managing genome-scale numbers of mutants is not required, and the relatively small mutant library can be subjected to in-depth phenotypic analyses. Individual mutants can be analyzed using a barrage of techniques that allows for measurements of almost all aspects of photosynthetic function and thylakoid composition, under a range of environmental conditions. In this way, any mutant aberrant in processes critical for modulating the structure and composition of the photosynthetic apparatus in response to environmental conditions, or involved in the biogenesis and repair of photosynthetic complexes, has little chance of slipping through the “phenotyping net.” Furthermore, informatic analyses of the set of proteins in the GreenCut may help address questions concerning the evolution of the photosynthetic apparatus. For example, the genomic location of cyanobacterial GreenCut genes and their potential inclusion within operons may provide hints as to the biological process in which the encoded proteins participate. An analysis of this type was recently performed for the unknowns of the GreenCut. As shown in Table 2, specific GreenCut unknowns are present in putative cyanobacterial operons that are associated with isoprenoid biosynthesis, ribosome biogenesis, and photosynthetic function, including ATP synthesis and the management of reducing equivalents, which suggests a function for the unknowns in those pathways. The assignment of a gene to an operon (with a potential function that is related to the other genes in that operon) is further supported by co-expression analyses, as depicted in Fig. 2 for CGLD22 (corresponding gene in Synechocystis sp. PCC6803 is sll1321); this gene appears to be coordinately expressed with seven other genes that are likely in the same operon (sll1322 to sll1327 plus ssl2615), all of which encode ATP synthase subunits. Co-expression was examined under 38 different conditions (from past studies); which included studies relating to osmotic activity, UV irradiation, heavy metal toxicity, H2O2 treatment, and iron depletion. Gene expression data are also helpful for the the analysis of CGLD14, a GreenCut protein that is conserved in the green lineage and diatoms. Transcripts encoding CGLD14 are elevated in green organs (stems and leaves) with little accumulation in root and floral organs. Very similar expression patterns have been observed for the photosynthetic proteins CYN38, a cyclophilin involved in assembly and maintenance of a PSII supercomplex (Fu et al. 2007), and PSBY, a PSII thylakoid membrane protein that has not been attributed a specific function (Gau et al. 1998). These results suggest a role for CGLD14 in photosynthetic function (Grossman et al. 2010).
Table 2.
Genes encoding GreenCut proteins of unknown physiological function that are present in cyanobacterial operons
| Cre gene name | AT identifier | Locus in Synechocystis sp. PCC6803 | Functional annotation | Number of cyanobacteria with similar gene arrangementa | Linked gene(s) in cyanobacterial operons |
|---|---|---|---|---|---|
| CPLD47 | At4g19100 | sll0933 | Conserved expressed membrane protein | 33 | Ribosomal protein S15 |
| CPLD38 | At3g17930 | slr0815 | Conserved expressed protein | 26 | NADH dehydrogenase subunit NdhL |
| CGLD22 | At2g31040 | sll1321 | Conserved expressed protein; some similarity to ATP synthase I protein | 32 | ATP synthase chain a |
| CGLD27 | At5g67370 | sll0584 | Conserved expressed protein of unknown function (DUF1230). This family consists of several hypothetical plant and photosynthetic bacterial proteins of around 160 residues in length. | 25 | Iojap-related protein |
| CGL68 | At1g67600 | slr1394 | Acid phosphatase/vanadium-dependent haloperoxidase related, DUF212 | 31 | Geranylgeranyl pyrophosphate synthase |
| CGL83 | At3g61770 | slr1394 | Conserved expressed protein of unknown function | 33 | Geranylgeranyl pyrophosphate synthase |
Note: Cre is used as an abbreviation of Chlamydomonas reinhardtii
aThe total number of cyanobacterial genomes used in this analysis was 36 (those present in CyanoBase) and the syntenic associations are only given when the contiguous gene has a functional annotation; other associations with hypothetical conserved genes, not shown, have also been noted
Fig. 2.
Co-expression of genes of the ATP synthase operon with CGLD22 (sll1321) in Synechocystis sp. PCC 6803. a The microarray data used to generate the expression curves were obtained from the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/). The atp1 gene is the putative ortholog of CGLD22; the curve showing the expression profile of atp1 is in red. The curves showing the expression profiles of slr1413 and sll0216, genes that are not part of the ATP synthase operon and were used as a control for these analyses, are a dotted and broken line, respectively. The curves showing expression profiles of all other genes of the ATP synthase operon are in gray. Microarray values were background-corrected, normalized against the median of the ratio of each sample against the reference, and log-transformed. The plotted data include microarray replicates of 38 biological experiments. b The arrangement of genes of the ATP synthase operon. The genes are depicted as arrows, with the orientation indicated by the direction of the arrow. The location of the genes on the chromosome relative to the origin is indicated. This information was obtained from CyanoBase (http://genome.kazusa.or.jp/cyanobase/) (Nakao et al. 2010). The genes of the operon are atp1 (sll1321), atpI (sll1322), atpH (ssl2615), atpG (sll1323), atpF (sll1324), atpD (sll1325), atpA (sll1326), and atpC (sll1327). slr1413 is upstream, and slr1411 and sll0216 are downstream of the ATP synthase operon, respectively, and neither is co-expressed with atp1. All of the genes of the ATP synthase operon are depicted as light gray-filled arrows, except for atp1; this arrow is red-filled. Arrows representing genes outside the operon, slr1411, slr1413, and sll0216, are unfilled and dark gray-filled
Phenotypic analysis of GreenCut mutants
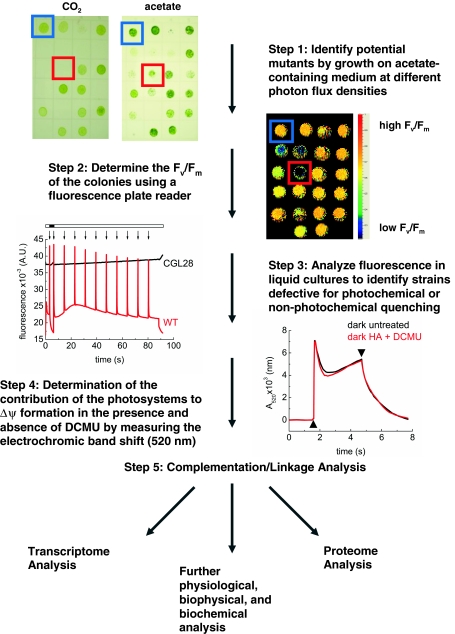
Identification of numerous proteins potentially involved in photosynthetic function allows for the exploitation of reverse genetic approaches to generate specific strains that are null or suppressed for a specific targeted gene. Strategies that have been successfully used to generate such strains include RNAi (Rohr et al. 2004; Im et al. 2006) and amiRNA approaches (Molnar et al. 2009; Zhao et al. 2009), as well as PCR identification of strains harboring specific mutations (Pootakham et al. 2010). Thus far, approximately 30 strains of Chlamydomonas and well over 100 strains of Arabidopsis have been identified with insertions in genes encoding GreenCut proteins of unknown function. Both sets of mutants are being analyzed using a specific set of assays that are relatively rapid. An example of a specific Chlamydomonas mutant strain that has gone through the primary assays of the characterization platform potentially harbors a lesion in the gene encoding CGL28, which has a motif that may allow it to bind RNA. Initially, the cells are grown on both minimal medium (no fixed carbon source) supplemented with bicarbonate and medium containing acetate. As shown in Fig. 3, a Chlamydomonas strain with a lesion in CGL28 (colony within red box, step 1) appears to be unable to grow on minimal medium, although it can grow on medium supplemented with acetate. The colonies that grew on acetate-containing medium were examined for fluorescence to determine the quantum yield of PSII. The fluorescence image shown in Fig. 3, step 2, suggests that the strain with an insertion in CGL28 has an extremely low quantum yield for PSII. The mutant strain was further analyzed with respect to fluorescence kinetics. The fluorescence curve demonstrates that the fluorescence yield is constant and equal to FM (step 3); the results suggest that the mutant exhibits essentially no photochemical or non-photochemical quenching. Furthermore, analysis of the carotenoid electrochromic shift (a measure of the electrochemical gradient generated from electron flow through PSI and PSII; step 4) indicates that DCMU has no effect on the membrane potential. Considering the overall information, the results suggest that PSII activity in the cgl28 mutant is severely compromised, although further spectroscopic and biochemical analyses are required.
Fig. 3.
Analyses of mutants defective for genes encoding GreenCut proteins. Step 1: Mutants are grown at varying light intensities on medium containing acetate or in minimal medium supplemented with CO2. In this example, a strain with a mutation in the CGL28 gene (red box, step 1) grew slower than wild-type cells (blue box) on acetate-containing medium, and did not grow at all on minimal medium supplemented with CO2. Step 2: Fv/Fm values, shown as a false color image, are determined for colonies grown on solid medium containing acetate. In this case, the cgl28 mutant (red box) was determined to have a markedly reduced Fv/Fm relative to wild-type cells (blue box). Step 3: The mutants are further analyzed after growth in the dark in liquid medium containing acetate for photochemical and non-photochemical quenching using fluorescence assays. This strain (blue curve) has no variable fluorescence (which can be observed in the pink curve of wild-type [WT] cells). When the horizontal bar at the top of the image is unfilled (white, outlined in black), the sample is being exposed to actinic light, while the black-filled region of the bar indicates that the sample is in the dark. All downward arrows are the times at which the sample is exposed to a pulse of saturating light, which allows for the determination of maximal fluorescence yield. Step 4: Samples are further analyzed for the contribution of each of the reaction centers to the generation of the electrochemical gradient across the thylakoid membranes by measuring the electrochromic band shift (carotenoid band shift at 520 nm) induced by illumination in the presence and the absence of the PSII inhibitors DCMU and hydroxylamine (HA). The upward arrow indicates light on, while the downward arrow indicates light off. PSII inhibitors have no effect on the electrochemical gradient generated in the cgl28 mutant in the presence of illumination, indicating that PSII cannot perform a charge separation. Step 5: In order to verify that the mutation is linked to the observed phenotype, the mutant is backcrossed with wild-type cells to determine whether the mutant phenotype is linked to the insertion (drug-resistant marker gene). A wild-type copy of the gene altered in the mutant strain is introduced into that strain (using the pSL18 plasmid and under the control of the PSAD promotor) to determine whether it rescues the mutant phenotype. If the mutation is indeed linked to the phenotype, then the mutant is further studied by additional transcriptomic, proteomic, physiological, biochemical, and biophysical analyses. Preliminary studies in this case suggest that the cgl28 mutation is not linked to the photosynthetic phenotype
Before we can be certain that the insertion in CGL28 is responsible for the mutant phenotype, it is critical that genetic crosses be done to demonstrate that the CGL28 gene is linked to the mutant phenotype (Zeocin or paromomycin resistance, depending on the marker gene used in the screen, always segregates with the photosynthetic phenotype) and ultimately that the phenotype can be rescued by introducing a wild-type copy of the CGL28 gene into the mutant strain (step 5); not all phenotypes identified by reverse genetic screening are actually caused by the inserted DNA. In most cases, the linkage and complementation analyses would be performed either before or at the same time that the physiological and biophysical characterizations are being performed. Additional analyses of the mutant strains, such as detailed studies of light sensitivity, sensitivity to compounds that facilitate the generation of reactive oxygen compounds, and analyses of the polypeptides present in the individual complexes associated with photosynthetic activities would add new perspectives to our view of photosynthesis and its regulation.
Concluding remarks
Numerous studies over the last half century have defined activities associated with photosynthetic function and identified proteins critical for the harvesting and utilization of excitation energy, electron transport reactions, ATP formation, and CO2 fixation. However, with more in-depth analyses of photosynthetic function, it is becoming clear that photosynthetic activities are exquisitely sensitive to environmental change (and developmental stage) and that various regulatory mechanisms interact to yield a final output from the system. Rapid responses of photosynthetic activities to fluctuations in the environment help to coordinate the products of photosynthesis with the metabolic demands of the cell and minimize damage associated with reactive oxygen species that may be formed as a consequence of excitation of pigment molecules and the generation of reactive intermediates. These short-term responses may reflect changes in protonation, phosphorylation, and the association of various pigment and protein components of the photosynthetic complexes. Longer-term responses may result in changes in subunit stoichiometries, pigment composition, and the insertion of novel proteins into individual complexes. The acquisition of a broad genomic view of various biological processes, which can be coupled with results from high throughput, sophisticated molecular and genetic technologies, will allow us to unravel novel and critical aspects of photosynthetic control and the impact that the various control mechanisms exert on overall physiological and metabolic processes in the cell.
Acknowledgments
This collaborative project has received multiple sources of support. ARG was supported by NSF grants MCB 0824469 and MCB 0235878, and BH was supported by funds from Stanford University, Department of Biology. SJK was supported in part by a Ruth L. Kirschstein National Research Service Award GM07185. SM and HL were supported in part by the Office of Science (BER), U.S. Department of Energy, Cooperative Agreement No. DE-FC02-02ER63421. RD and KKN were supported by NSF grant MCB 0235878 and the Simon Family Fund. XJ, JA, and FAW were supported by CNRS UMR7141.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Amunts A, Toporik H, Borovikova A, Nelson N. Structure determination and improved model of plant photosystem I. J Biol Chem. 2010;285:3478–3486. doi: 10.1074/jbc.M109.072645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armbrust EV, Berges JA, Bowler C, Green BR, Martinez D, Putnam NH, et al. The genome of the diatom Thalassiosira pseudonana: ecology, evolution, and metabolism. Science. 2004;306:79–86. doi: 10.1126/science.1101156. [DOI] [PubMed] [Google Scholar]
- Asada K. The water–water cycle in chloroplasts: scavenging of active oxygens and dissipation of excess photons. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:601–639. doi: 10.1146/annurev.arplant.50.1.601. [DOI] [PubMed] [Google Scholar]
- Asamizu E, Nakamura Y, Sato S, Fukuzawa H, Tabata S. A large scale structural analysis of cDNAs in a unicellular green alga Chlamydomonas reinhardtii. Generation of 3, 433 non-redundant expressed sequence tags. DNA Res. 1999;6:369–373. doi: 10.1093/dnares/6.6.369. [DOI] [PubMed] [Google Scholar]
- Asamizu E, Miura K, Kucho K, Inoue Y, Fukuzawa H, Ohyama K, et al. Generation of expressed sequence tags from low-CO2 and high-CO2 adapted cells of Chlamydomonas reinhardtii. DNA Res. 2000;7:305–307. doi: 10.1093/dnares/7.5.305. [DOI] [PubMed] [Google Scholar]
- Baginsky S, Grossmann J, Gruissem W. Proteome analysis of chloroplast mRNA processing and degradation. J Proteome Res. 2007;6:808–820. doi: 10.1021/pr060473q. [DOI] [PubMed] [Google Scholar]
- Bailey S, Melis A, Mackey KR, Cardol P, Finazzi G, van Dijken G, et al. Alternative photosynthetic electron flow to oxygen in marine Synechococcus. Biochim Biophys Acta. 2008;1777:269–276. doi: 10.1016/j.bbabio.2008.01.002. [DOI] [PubMed] [Google Scholar]
- Barbier G, Oesterhelt C, Larson MD, Halgren RG, Wilkerson C, Garavito RM, et al. Comparative genomics of two closely related unicellular thermo-acidophilic red algae, Galdieria sulphuraria and Cyanidioschyzon merolae, reveals the molecular basis of the metabolic flexibility of Galdieria sulphuraria and significant differences in carbohydrate metabolism of both algae. Plant Physiol. 2005;137:460–474. doi: 10.1104/pp.104.051169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennoun P, Delepelaire P. Isolation of photosynthesis mutants in Chlamydomonas. In: Edelman M, Hallick RB, Chua NH, editors. Methods in chloroplast molecular biology. Amsterdam: Elsevier Biomedical Press; 1982. pp. 25–38. [Google Scholar]
- Bollenbach TJ, Schuster G, Stern DB. Cooperation of endo- and exoribonucleases in chloroplast mRNA turnover. Prog Nucleic Acid Res Mol Biol. 2004;78:305–337. doi: 10.1016/S0079-6603(04)78008-3. [DOI] [PubMed] [Google Scholar]
- Bordowitz JR, Montgomery BL. Photoregulation of cellular morphology during complementary chromatic adaptation requires sensor-kinase-class protein RcaE in Fremyelladiplosiphon. J Bacteriol. 2008;190:4069–4074. doi: 10.1128/JB.00018-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowler C, Allen AE, Badger JH, Grimwood J, Jabbari K, Kuo A, et al. The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature. 2008;456:239–244. doi: 10.1038/nature07410. [DOI] [PubMed] [Google Scholar]
- Camargo A, Llamas A, Schnell RA, Higuera JJ, González-Ballester D, Lefebvre PA, et al. Nitrate signaling by the regulatory gene NIT2 in Chlamydomonas. Plant Cell. 2007;19:3491–3503. doi: 10.1105/tpc.106.045922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choquet Y, Wollman FA. The CES process. In: Stern D, Witman GB, Harris EH, editors. The Chlamydomonas sourcebook. Amsterdam: Elsevier; 2009. pp. 1027–1063. [Google Scholar]
- DalCorso G, Pesaresi P, Masiero S, Aseeva E, Schunemann D, Finazzi G, et al. A complex containing PGRL1 and PGR5 is involved in the switch between linear and cyclic electron flow in Arabidopsis. Cell. 2008;132:273–285. doi: 10.1016/j.cell.2007.12.028. [DOI] [PubMed] [Google Scholar]
- de Vitry C, Kuras R. The cytochrome b6f complex. In: Harris EH, Stern D, Witman GB, editors. The Chlamydomonas sourcebook. Amsterdam: Elsevier; 2009. pp. 603–638. [Google Scholar]
- Dent RM, Haglund CM, Chin BL, Kobayashi MC, Niyogi KK. Functional genomics of eukaryotic photosynthesis using insertional mutagenesis of Chlamydomonas reinhardtii. Plant Physiol. 2005;137:545–556. doi: 10.1104/pp.104.055244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drapier D, Rimbault B, Vallon O, Wollman FA, Choquet Y. Intertwined translational regulations set uneven stoichiometry of chloroplast ATP synthase subunits. EMBO J. 2007;26:3581–3591. doi: 10.1038/sj.emboj.7601802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan K, Yi K, Dang L, Huang H, Wu W, Wu P. Characterization of a sub-family of Arabidopsis genes with the SPX domain reveals their diverse functions in plant tolerance to phosphorus starvation. Plant J. 2008;54:965–975. doi: 10.1111/j.1365-313X.2008.03460.x. [DOI] [PubMed] [Google Scholar]
- Eberhard S, Finazzi G, Wollman FA. The dynamics of photosynthesis. Annu Rev Genet. 2008;42:463–515. doi: 10.1146/annurev.genet.42.110807.091452. [DOI] [PubMed] [Google Scholar]
- Eversole RA. Biochemical mutants of Chlamydomonas reinhardtii. Am J Bot. 1956;43:404–407. [Google Scholar]
- Fernandez E, Galvan A. Inorganic nitrogen assimilation in Chlamydomonas. J Exp Bot. 2007;58:2279–2287. doi: 10.1093/jxb/erm106. [DOI] [PubMed] [Google Scholar]
- Fernández E, Galván Á. Nitrate assimilation in Chlamydomonas. Eukaryot Cell. 2008;7:555–559. doi: 10.1128/EC.00431-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández E, Llamas A, Galván Á. Nitrogen assimilation and its regulation. In: Harris EH, Stern D, Witman GB, editors. The Chlamydomonas sourcebook. Amsterdam: Elsevier; 2009. pp. 69–114. [Google Scholar]
- Finazzi G, Drapier D, Rappaport F. The CFoF1 ATP-synthase complex of photosynthesis. In: Stern D, Witman GB, Harris EH, editors. The Chlamydomonas sourcebook. Amsterdam: Elsevier; 2009. pp. 639–670. [Google Scholar]
- Forti G. The role of respiration in the activation of photosynthesis upon illumination of dark adapted Chlamydomonas reinhardtii. Biochim Biophys Acta. 2008;1777:1449–1454. doi: 10.1016/j.bbabio.2008.08.011. [DOI] [PubMed] [Google Scholar]
- Fu A, He Z, Cho HS, Lima A, Buchanan BB, Luan S. A chloroplast cyclophilin functions in the assembly and maintenance of photosystem II in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2007;104:15947–15952. doi: 10.1073/pnas.0707851104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gau AE, Thole HH, Sokolenko A, Altschmied L, Hermann RG, Pistorius EK. PsbY, a novel manganese-binding, low-molecular-mass protein associated with photosystem II. Mol Gen Genet. 1998;260:56–68. doi: 10.1007/s004380050870. [DOI] [PubMed] [Google Scholar]
- Ghirardi ML, Posewitz MC, Maness PC, Dubini A, Yu J, Seibert M. Hydrogenases and hydrogen photoproduction in oxygenic photosynthetic organisms. Annu Rev Plant Biol. 2007;58:71–91. doi: 10.1146/annurev.arplant.58.032806.103848. [DOI] [PubMed] [Google Scholar]
- Givan AL, Levine RP. The photosynthetic electron transport chain of Chlamydomonas reinhardtii. VII. Photosynthetic phosphorylation by a mutant strain of Chlamydomonas reinhardtii deficient in active P700. Plant Physiol. 1967;42:1264–1268. doi: 10.1104/pp.42.9.1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokhale X, Sayre RT. Photosystem II, a structural perspective. In: Stern D, Witman GB, Harris EH, editors. The Chlamydomonas sourcebook. Amsterdam: Elsevier; 2009. pp. 573–602. [Google Scholar]
- Goldschmidt-Clermont M. Chloroplast RNA splicing. In: Stern D, Witman GB, Harris EH, editors. The Chlamydomonas sourcebook. Amsterdam: Elsevier; 2009. pp. 915–936. [Google Scholar]
- González-Ballester D, Grossman AR. Sulfur: from acquisition to assimilation. In: Harris EH, Witman GB, Stern D, editors. The Chlamydomonas sourcebook. Amsterdam: Elsevier; 2009. pp. 159–188. [Google Scholar]
- González-Ballester D, Pollock SV, Pootakham W, Grossman AR. The central role of a SNRK2 kinase in sulfur deprivation responses. Plant Physiol. 2008;147:216–227. doi: 10.1104/pp.108.116137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Ballester D, Cassero, D, Pellegrini M, Merchant S, Grossman AR (2010) Insights into sulfur deprivation responses of Chlamydomonas from RNA seq. Plant Cell (in press) [DOI] [PMC free article] [PubMed]
- Gorman DS, Levine RP. Photosynthetic electron transport chain of Chlamydomonas reinharditi. VI. Electron transport in mutant strains lacking either cytochrome 553 or plastocyanin. Plant Physiol. 1966;41:1648–1656. doi: 10.1104/pp.41.10.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman AR, Harris EE, Hauser C, Lefebvre PA, Martinez D, Rokhsar D, et al. Chlamydomonas reinhardtii at the crossroads of genomics. Eukaryot Cell. 2003;2:1137–1150. doi: 10.1128/EC.2.6.1137-1150.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman AR, Croft M, Gladyshev VN, Merchant SS, Posewitz MC, Prochnik S, Spalding MH. Novel metabolism in Chlamydomonas through the lens of genomics. Curr Opin Plant Biol. 2007;10:190–198. doi: 10.1016/j.pbi.2007.01.012. [DOI] [PubMed] [Google Scholar]
- Grossman, AR, Bailey S, González-Ballester D, Karpowicz SJ, and Merchant SS (2010) Understanding photosynthetic electron transport using Chlamydomonas: the path from classical genetics to high throughput genomics. In: Govindjee (ed) Advances in photosynthesis and respiration: functional genomics and evolution of photosynthetic systems. Kluwer Academic Publishers, Dordrecht (in submission)
- Gutman BL, Niyogi KK. Chlamydomonas and Arabidopsis. A dynamic duo. Plant Physiol. 2004;135:607–610. doi: 10.1104/pp.104.041491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris EH. The Chlamydomonas sourcebook. A comprehensive guide to biology and laboratory use. San Diego: Academic Press; 1989. [DOI] [PubMed] [Google Scholar]
- Harris EH. The Chalmydomonas sourcebook. San Diego, CA: Academic Press; 2009. [Google Scholar]
- Heber U. Irrungen, Wirrungen? The Mehler reaction in relation to cyclic electron transport in C3 plants. Photosynth Res. 2002;73:223–231. doi: 10.1023/A:1020459416987. [DOI] [PubMed] [Google Scholar]
- Herrin DL. Chloroplast RNA processing and stability. In: Stern D, Witman GB, Harris EH, editors. The Chlamydomonas sourcebook. Amsterdam: Elsevier; 2009. pp. 937–966. [Google Scholar]
- Higgs D. The chloroplast genome. In: Stern D, Witman GB, Harris EH, editors. The Chlamydomonas sourcebook. Amsterdam: Elsevier; 2009. pp. 871–892. [Google Scholar]
- Huner NPA, Öquist G, Sarhan F. Energy balance and acclimation to light and cold. Trends Plant Sci. 1998;3:224–235. [Google Scholar]
- Im C-S, Eberhard S, Huang K, Beck C, Grossman AR. Phototropin involvement in expression of genes encoding chlorophyll and carotenoid biosynthesis enzymes and LHC apoproteins in Chlamydomonas reinhardtii. Plant J. 2006;48:1–16. doi: 10.1111/j.1365-313X.2006.02852.x. [DOI] [PubMed] [Google Scholar]
- Jain M, Shrager J, Harris EH, Halbrook R, Grossman AR, Hauser C, Vallon O. EST assembly supported by a draft genome sequence: an analysis of the Chlamydomonas reinhardtii transcriptome. Nucleic Acids Res. 2007;35:2074–2083. doi: 10.1093/nar/gkm081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehoe DM, Gutu A. Responding to color: the regulation of complementary chromatic adaptation. Annu Rev Plant Biol. 2006;57:127–150. doi: 10.1146/annurev.arplant.57.032905.105215. [DOI] [PubMed] [Google Scholar]
- Keller LC, Romijn EP, Zamora I, Yates JR, 3rd, Marshall WF. Proteomic analysis of isolated Chlamydomonas centrioles reveals orthologs of ciliary-disease genes. Curr Biol. 2005;15:1090–1098. doi: 10.1016/j.cub.2005.05.024. [DOI] [PubMed] [Google Scholar]
- Kleffmann T, von Zychlinski A, Russenberger D, Hirsch-Hoffmann M, Gehrig P, Gruissem W, Baginsky S. Proteome dynamics during plastid differentiation in rice. Plant Physiol. 2007;143:912–923. doi: 10.1104/pp.106.090738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein U. Chloroplast transcription. In: Stern D, Witman GB, Harris EH, editors. The Chlamydomonas sourcebook. Amsterdam: Elsevier; 2009. pp. 893–914. [Google Scholar]
- Kohinata T, Nishino H, Fukuzawa H. Significance of zinc in a regulatory protein, CCM1, which regulates the carbon-concentrating mechanism in Chlamydomonas reinhardtii. Plant Cell Physiol. 2008;49:273–283. doi: 10.1093/pcp/pcn003. [DOI] [PubMed] [Google Scholar]
- Krysan PJ, Young JE, Tax F, Sussman MR. Identification of transferred DNA insertions within Arabidopsis genes involved in signal transduction and ion transport. Proc Natl Acad Sci USA. 1996;93:8145–8150. doi: 10.1073/pnas.93.15.8145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuras R, Saint-Marcoux D, Wollman FA, de Vitry C. A specific c-type cytochrome maturation system is required for oxygenic photosynthesis. Proc Natl Acad Sci USA. 2007;104:9906–9910. doi: 10.1073/pnas.0702340104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusaba M, Ito H, Morita R, Iida S, Sato Y, Fujimoto M, et al. Rice NON-YELLOW COLORING1 is involved in light-harvesting complex II and grana degradation during leaf senescence. Plant Cell. 2007;19:1362–1375. doi: 10.1105/tpc.106.042911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavorel J, Levine RP. Fluorescence properties of wild-type Chlamydomonas reinhardtii and three mutant strains having impaired photosynthesis. Plant Physiol. 1968;43:1049–1055. doi: 10.1104/pp.43.7.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledford HK, Baroli I, Shin JW, Fischer BB, Eggen RI, Niyogi KK. Comparative profiling of lipid-soluble antioxidants and transcripts reveals two phases of photo-oxidative stress in a xanthophyll-deficient mutant of Chlamydomonas reinhardtii. Mol Genet Genomics. 2004;272:470–479. doi: 10.1007/s00438-004-1078-5. [DOI] [PubMed] [Google Scholar]
- Ledford HK, Chin BL, Niyogi KK. Acclimation to singlet oxygen stress in Chlamydomonas reinhardtii. Eukaryot Cell. 2007;6:919–930. doi: 10.1128/EC.00207-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KP, Kim C, Landgraf F, Apel K. EXECUTER1- and EXECUTER2-dependent transfer of stress-related signals from the plastid to the nucleus of Arabidopsis thaliana. Proc Natl Acad Sci USA. 2007;104:10270–10275. doi: 10.1073/pnas.0702061104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine RP. Genetic control of photosynthesis in Chlamydomonas reinhardtii. Science. 1960;162:768–771. doi: 10.1073/pnas.46.7.972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine RP. The analysis of photosynthesis using mutant strains of algae and higher plants. Annu Rev Plant Physiol. 1969;20:523–540. [Google Scholar]
- Levine RP, Goodenough UW. The genetics of photosynthesis and of the chloroplast in Chlamydomonas reinhardii. Annu Rev Genet. 1970;4:397–408. doi: 10.1146/annurev.ge.04.120170.002145. [DOI] [PubMed] [Google Scholar]
- Lezhneva L, Kuras R, Ephritikhine G, de Vitry C. A novel pathway of cytochrome c biogenesis is involved in the assembly of the cytochrome b6f complex in Arabidopsis chloroplasts. J Biol Chem. 2008;283:24608–24616. doi: 10.1074/jbc.M803869200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Bjorkman O, Shih C, Grossman A, Rosenquist M, Jansson C, Niyogi KK. A pigment-binding protein essential for regulation of photosynthetic light harvesting. Nature. 2000;403:391–395. doi: 10.1038/35000131. [DOI] [PubMed] [Google Scholar]
- Li Z, Wakao S, Fischer BB, Niyogi KK. Sensing and responding to excess light. Annu Rev Plant Biol. 2009;60:239–260. doi: 10.1146/annurev.arplant.58.032806.103844. [DOI] [PubMed] [Google Scholar]
- Long JC, Sommer F, Allen MD, Lu SF, Merchant SS. FER1 and FER2 encoding two ferritin complexes in Chlamydomonas reinhardtii chloroplasts are regulated by iron. Genetics. 2008;179:137–147. doi: 10.1534/genetics.107.083824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S, Van Eck J, Zhou X, Lopez AB, O’Halloran DM, Cosman KM, et al. The cauliflower Or gene encodes a DnaJ cysteine-rich domain-containing protein that mediates high levels of beta-carotene accumulation. Plant Cell. 2006;18:3594–3605. doi: 10.1105/tpc.106.046417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maheswari U, Mock T, Armbrust EV, Bowler C. Update of the Diatom EST Database: a new tool for digital transcriptomics. Nucleic Acids Res. 2009;37:D1001–D1005. doi: 10.1093/nar/gkn905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino A, Miyake C, Yokota A. Physiological functions of the water-water cycle (Mehler reaction) and the cyclic electron flow around PSI in rice leaves. Plant Cell Physiol. 2002;43:1017–1026. doi: 10.1093/pcp/pcf124. [DOI] [PubMed] [Google Scholar]
- Matsuzaki M, Misumi O, Shin IT, Maruyama S, Takahara M, Miyagishima SY, et al. Genome sequence of the ultrasmall unicellular red alga Cyanidioschyzon merolae 10D. Nature. 2004;428:653–657. doi: 10.1038/nature02398. [DOI] [PubMed] [Google Scholar]
- May P, Wienkoop S, Kempa S, Usadel B, Christian N, Rupprecht J, et al. Metabolomics- and proteomics-assisted genome annotation and analysis of the draft metabolic network of Chlamydomonas reinhardtii. Genetics. 2008;179:157–166. doi: 10.1534/genetics.108.088336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May P, Christian JO, Kempa S, Walther D. ChlamyCyc: an integrative systems biology database and web-portal for Chlamydomonas reinhardtii. BMC Genomics. 2009;10:209. doi: 10.1186/1471-2164-10-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melis A. Photosynthetic H2 metabolism in Chlamydomonas reinhardtii (unicellular green algae) Planta. 2007;226:1075–1086. doi: 10.1007/s00425-007-0609-9. [DOI] [PubMed] [Google Scholar]
- Merchant SS, Allen MD, Kropat J, Moseley JL, Long JC, Tottey S, Terauchi AM. Between a rock and a hard place: trace element nutrition in Chlamydomonas. Biochim Biophys Acta. 2006;1763:578–594. doi: 10.1016/j.bbamcr.2006.04.007. [DOI] [PubMed] [Google Scholar]
- Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, et al. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science. 2007;318:245–250. doi: 10.1126/science.1143609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minagawa J. Light-harvesting proteins. In: Stern D, Witman GB, Harris EH, editors. The ‘Chlamydomonas sourcebook’. Amsterdam: Elsevier; 2009. pp. 503–540. [Google Scholar]
- Misumi O, Matsuzaki M, Nozaki H, Miyagishima SY, Mori T, Nishida K, et al. Cyanidioschyzon merolae genome. A tool for facilitating comparable studies on organelle biogenesis in photosynthetic eukaryotes. Plant Physiol. 2005;137:567–585. doi: 10.1104/pp.104.053991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll B, Levine RP. Characterization of a photosynthetic mutant strain of Chlamydomonas reinhardi deficient in phosphoribulokinase activity. Plant Physiol. 1970;46:576–580. doi: 10.1104/pp.46.4.576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnar A, Bassett A, Thuenemann E, Schwach F, Karkare S, Ossowski S et al (2009) Highly specific gene silencing by artificial microRNAs in the unicellular alga Chlamydomonas reinhardtii. Plant J (Epub ahead of print) [DOI] [PubMed]
- Moseley J, Grossman AR (2009) Phosphorus limitation from the physiological to the genomic. In: Harris EH, Stern D, Witman GB (eds) ‘The Chlamydomonas sourcebook’, pp 189–216
- Moseley J, González-Ballester D, Pootakham W, Bailey S, Grossman AR. Genetic interactions between regulators of Chlamydomonas phosphorus and sulfur deprivation responses. Genetics. 2009;181:889–905. doi: 10.1534/genetics.108.099382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulkidjanian AY, Koonin EV, Makarova KS, Mekhedov SL, Sorokin A, Wolf YI, et al. The cyanobacterial genome core and the origin of photosynthesis. Proc Natl Acad Sci USA. 2006;103:13126–13131. doi: 10.1073/pnas.0605709103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakao M, Okamoto S, Kohara M, Fujishiro T, Fujisawa T, Sato S, Tabata S, Kaneko T, Nakamura Y. CyanoBase: the cyanobacteria genome database update 2010. Nucleic Acids Res. 2010;38:D379–D381. doi: 10.1093/nar/gkp915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumann B, Busch A, Allmer J, Ostendorf E, Zeller M, Kirchhoff H, Hippler M. Comparative quantitative proteomics to investigate the remodeling of bioenergetic pathways under iron deficiency in Chlamydomonas reinhardtii. Proteomics. 2007;7:3964–3979. doi: 10.1002/pmic.200700407. [DOI] [PubMed] [Google Scholar]
- Niyogi K. Photoprotection and high light responses. In: Stern D, Witman GB, Harris EH, editors. The Chlamydomonas sourcebook. Amsterdam: Elsevier; 2009. pp. 847–870. [Google Scholar]
- Ozawa S, Nield J, Terao A, Stauber EJ, Hippler M, Koike H, et al. Biochemical and structural studies of the large Ycf4-photosystem I assembly complex of the green alga Chlamydomonas reinhardtii. Plant Cell. 2009;21:2424–2442. doi: 10.1105/tpc.108.063313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palenik B, Grimwood J, Aerts A, Rouze P, Salamov A, Putnam N, et al. The tiny eukaryote Ostreococcus provides genomic insights into the paradox of plankton speciation. Proc Natl Acad Sci USA. 2007;104:7705–7710. doi: 10.1073/pnas.0611046104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peers G, Truong TB, Ostendorf E, Busch A, Elrad D, Grossman AR, et al. An ancient light-harvesting protein is critical for the regulation of algal photosynthesis. Nature. 2009;462:518–521. doi: 10.1038/nature08587. [DOI] [PubMed] [Google Scholar]
- Pfannschmidt T, Bräutigam K, Wagner R, Dietzel L, Schröter Y, Steiner S, Nykytenko A. Potential regulation of gene expression in photosynthetic cells by redox and energy state: approaches towards better understanding. Ann Bot. 2009;103:599–607. doi: 10.1093/aob/mcn081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pootakham W, González-Ballester D, Grossman AR (2010) The sulfate transporters of Chlamydononas reinhardtii and their regulation (in submission) [DOI] [PMC free article] [PubMed]
- Posewitz M, Dubini A, Meuser JE, Seibert M, Ghirardi ML. Hydrogenase, hydrogen production and anoxia. In: Stern D, Witman GB, Harris EH, editors. The Chlamydomonas sourcebook. Amsterdam: Elsevier; 2009. pp. 218–256. [Google Scholar]
- Raynaud C, Loiselay C, Wostrikoff K, Kuras R, Girard-Bascou J, Wollman FA, Choquet Y. Evidence for regulatory function of nucleus-encoded factors on mRNA stabilization and translation in the chloroplast. Proc Natl Acad Sci USA. 2007;104:9093–9098. doi: 10.1073/pnas.0703162104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redding K. Photosystem I. In: Stern D, Witman GB, Harris EH, editors. The Chlamydomonas sourcebook. Amsterdam: Elsevier; 2009. pp. 541–572. [Google Scholar]
- Ren G, An K, Liao Y, Zhou X, Cao Y, Zhao H, et al. Identification of a novel chloroplast protein AtNYE1 regulating chlorophyll degradation during leaf senescence in Arabidopsis. Plant Physiol. 2007;144:1429–1441. doi: 10.1104/pp.107.100172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocap G, Larimer FW, Lamerdin J, Malfatti S, Chain P, Ahlgren NA, et al. Genome divergence in two Prochlorococcus ecotypes reflects oceanic niche differentiation. Nature. 2003;424:1042–1047. doi: 10.1038/nature01947. [DOI] [PubMed] [Google Scholar]
- Rochaix JD. Posttranscriptional control of chloroplast gene expression. From RNA to photosynthetic complex. Plant Physiol. 2001;125:142–144. doi: 10.1104/pp.125.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochaix JD. State transitions. In: Stern D, Witman GB, Harris EH, editors. The Chlamydomonas sourcebook. Amsterdam: Elsevier; 2009. pp. 819–846. [Google Scholar]
- Rohr J, Sarkar N, Balenger S, Jeong BR, Cerutti H. Tandem inverted repeat system for selection of effective transgenic RNAi strains in Chlamydomonas. Plant J. 2004;40:611–621. doi: 10.1111/j.1365-313X.2004.02227.x. [DOI] [PubMed] [Google Scholar]
- Rolland N, Atteia A, Decottignies P, Garin J, Hippler M, Kreimer G, et al. Chlamydomonas proteomics. Curr Opin Microbiol. 2009;12:285–291. doi: 10.1016/j.mib.2009.04.001. [DOI] [PubMed] [Google Scholar]
- Rumeau D, Peltier G, Cournac L. Chlororespiration and cyclic electron flow around PSI during photosynthesis and plant stress response. Plant Cell Environ. 2007;30:1041–1051. doi: 10.1111/j.1365-3040.2007.01675.x. [DOI] [PubMed] [Google Scholar]
- Rymarquis LA, Handley JM, Thomas M, Stern DB. Beyond complementation. Map-based cloning in Chlamydomonas reinhardtii. Plant Physiol. 2005;137:557–566. doi: 10.1104/pp.104.054221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sager R. Genetic systems in Chlamydomonas. Science. 1960;132:1459–1465. doi: 10.1126/science.132.3438.1459. [DOI] [PubMed] [Google Scholar]
- Sato V, Levine RP, Neumann J. Photosynthetic phosphorylation in Chlamydomonas reinhardti. Effects of a mutation altering an ATP-synthesizing enzyme. Biochim Biophys Acta. 1971;253:437–448. doi: 10.1016/0005-2728(71)90047-8. [DOI] [PubMed] [Google Scholar]
- Sato Y, Morita R, Katsuma S, Nishimura M, Tanaka A, Kusaba M. Two short-chain dehydrogenase/reductases, NON-YELLOW COLORING 1 and NYC1-LIKE, are required for chlorophyll b and light-harvesting complex II degradation during senescence in rice. Plant J. 2009;57:120–131. doi: 10.1111/j.1365-313X.2008.03670.x. [DOI] [PubMed] [Google Scholar]
- Schmidt GW, Matlin KS, Chua NH. A rapid procedure for selective enrichment of photosynthetic electron transport mutants. Proc Natl Acad Sci USA. 1977;74:610–614. doi: 10.1073/pnas.74.2.610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Scholkopf B, Weigel D, Lohmann JU. A gene expression map of Arabidopsis thaliana development. Nat Genet. 2005;5:501–506. doi: 10.1038/ng1543. [DOI] [PubMed] [Google Scholar]
- Schmidt M, Gessner G, Luff M, Heiland I, Wagner V, Kaminski M, et al. Proteomic analysis of the eyespot of Chlamydomonas reinhardtii provides novel insights into its components and tactic movements. Plant Cell. 2006;18:1908–1930. doi: 10.1105/tpc.106.041749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schult K, Meierhoff K, Paradies S, Toller T, Wolff P, Westhoff P. The nuclear-encoded factor HCF173 is involved in the initiation of translation of the psbA mRNA in Arabidopsis thaliana. Plant Cell. 2007;19:1329–1346. doi: 10.1105/tpc.106.042895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrager J, Hauser C, Chang CW, Harris EH, Davies J, McDermott J, et al. Chlamydomonas reinhardtii genome project. A guide to the generation and use of the cDNA information. Plant Physiol. 2003;131:401–408. doi: 10.1104/pp.016899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stauber EJ, Hippler M. Chlamydomonas reinhardtii proteomics. Plant Physiol Biochem. 2004;42:989–1001. doi: 10.1016/j.plaphy.2004.09.008. [DOI] [PubMed] [Google Scholar]
- Stepien P, Johnson GN. Contrasting responses of photosynthesis to salt dtress in the glycophyte Arabidopsis and the halophyte Thellungiella: role of the plastid terminal oxidase as an alternative electron sink. Plant Physiol. 2009;149:1154–1165. doi: 10.1104/pp.108.132407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tejada-Jimenez M, Llamas A, Sanz-Luque E, Galván A, Fernández E. A high-affinity molybdate transporter in eukaryotes. Proc Natl Acad Sci USA. 2007;104:20126–20130. doi: 10.1073/pnas.0704646104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vardi A, Thamatrakoln K, Bidle KD, Falkowski PG. Diatom genomes come of age. Genome Biol. 2008;9:245. doi: 10.1186/gb-2008-9-12-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner V, Fiedler M, Markert C, Hippler M, Mittag M. Functional proteomics of circadian expressed proteins from Chlamydomonas reinhardtii. FEBS Lett. 2004;559:129–135. doi: 10.1016/S0014-5793(04)00051-1. [DOI] [PubMed] [Google Scholar]
- Wagner V, Kreimer G, Mittag M. The power of functional proteomics: components of the green algal eyespot and its light signaling pathway(s) Plant Signal Behav. 2008;3:433–435. doi: 10.4161/psb.3.7.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner V, Boesger J, Mittag M. Sub-proteome analysis in the green flagellate alga Chlamydomonas reinhardtii. J Basic Microbiol. 2009;49:32–41. doi: 10.1002/jobm.200800292. [DOI] [PubMed] [Google Scholar]
- Walters RG. Towards an understanding of photosynthetic acclimation. J Exp Bot. 2005;56:435–447. doi: 10.1093/jxb/eri060. [DOI] [PubMed] [Google Scholar]
- Whitaker MJ, Bordowitz JR, Montgomery BL. CpcF-dependent regulation of pigmentation and development in Fremyella diplosiphon. Biochem Biophys Res Commun. 2009;389:602–606. doi: 10.1016/j.bbrc.2009.09.030. [DOI] [PubMed] [Google Scholar]
- Wilson A, Ajlani G, Verbavatz JM, Vass I, Kerfeld CA, Kirilovsky D. A soluble carotenoid protein involved in phycobilisome-related energy dissipation in cyanobacteria. Plant Cell. 2006;18:992–1007. doi: 10.1105/tpc.105.040121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worden AZ, Lee JH, Mock T, Rouze P, Simmons MP, Aerts AL, et al. Green evolution and dynamic adaptations revealed by genomes of the marine picoeukaryotes Micromonas. Science. 2009;324:268–272. doi: 10.1126/science.1167222. [DOI] [PubMed] [Google Scholar]
- Zerges W, Hauser C. Protein synthesis in the chloroplast. In: Stern D, Witman GB, Harris EH, editors. The Chlamydomonas sourcebook. Amsterdam: Elsevier; 2009. pp. 967–1026. [Google Scholar]
- Zhao T, Wang W, Bai X, Qi Y. Gene silencing by artificial microRNAs in Chlamydomonas. Plant J. 2009;58:157–164. doi: 10.1111/j.1365-313X.2008.03758.x. [DOI] [PubMed] [Google Scholar]
- Zhu J, Fu X, Koo YD, Zhu JK, Jenney FE, Jr, Adams MW, et al. An enhancer mutant of Arabidopsis salt overly sensitive 3 mediates both ion homeostasis and the oxidative stress response. Mol Cell Biol. 2007;27:5214–5224. doi: 10.1128/MCB.01989-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer SL, Schein A, Zipor G, Stern DB, Schuster G. Polyadenylation in Arabidopsis and Chlamydomonas organelles: the input of nucleotidyltransferases, poly(A) polymerases and polynucleotide phosphorylase. Plant J. 2009;59:88–99. doi: 10.1111/j.1365-313X.2009.03853.x. [DOI] [PubMed] [Google Scholar]
- Zybailov B, Rutschow H, Friso G, Rudella A, Emanuelsson O, Sun Q, van Wijk KJ. Sorting signals, N-terminal modifications and abundance of the chloroplast proteome. PLoS One. 2008;3:21994. doi: 10.1371/journal.pone.0001994. [DOI] [PMC free article] [PubMed] [Google Scholar]