Abstract
The signature for human immunodeficiency virus type 1 (HIV-1) neurovirulence remains a subject of intense debate. Macrophage viral tropism is one prerequisite but others, including virus-induced alterations in innate and adaptive immunity, remain under investigation. HIV-1–infected mononuclear phagocytes (MPs; perivascular macrophages and microglia) secrete toxins that affect neurons. The authors hypothesize that neurovirulent HIV-1 variants affect the MP proteome by inducing a signature of neurotoxic proteins and thus affect cognitive function. To test this hypothesis, HIV-1 isolates obtained from peripheral blood of women with normal cognition (NC) were compared to isolates obtained from women with cognitive impairment (CI) and to the laboratory adapted SF162, a spinal fluid R5 isolate from a patient with HIV-1–associated dementia. HIV-1 isolates were used to infect monocyte-derived macrophages (MDMs) and infection monitored by secreted HIV-1 p24 by enzyme-linked immunosorbent assay (ELISA). Cell lysates of uninfected and HIV-1–infected MDMs at 14 days post infection were fractionated by cationic exchange chromatography and analyzed by surface enhanced laser desorption ionization time of flight (SELDI-TOF) using generalized estimating equations statistics. Proteins were separated by one-dimensional sodium dodecyl sulfate–polyacrylamide gel electrophoresis (1D SDS-PAGE) and identified by tandem mass spectrometry. Levels of viral replication were similar amongst the HIV-1 isolates, although higher levels were obtained from one viral strain obtained from a patient with CI. Significant differences were found in protein profiles between virus-infected MDMs with NC, CI, and SF162 isolates (adjusted P value after multiple testing corrections, or q value < .10). The authors identified 6 unique proteins in NC, 7 in SF162, and 20 in CI. Three proteins were common to SF162 and CI strains. The MDM proteins linked to infection with CI strains were related to apoptosis, chemotaxis, inflammation, and redox metabolism. These findings support the hypothesis that the macrophage proteome differ when infected with viral isolates of women with and without CI.
Keywords: HIV-1, HIV-1, associated cognitive impairment, macrophages, proteomics
Introduction
Mononuclear phagocytes (MPs; peripheral blood derived macrophages, perivascular macrophages, and microglia) are cellular reservoirs for human immunodefiency virus type 1 (HIV-1) in the central nervous system (CNS). After HIV-1 enters the CNS, either as a cell free virus or in infected macrophages, it infects perivascular macrophages, eventually causing the cognitive and motor abnormalities characteristics of HIV-1–associated neurocognitive disorders (HAND) and in its most severe form, HIV-1–associated dementia (HAD). HAD is associated with immune activation and trafficking of peripheral HIV-1–infected monocytes across a disrupted blood-brain barrier (BBB). Upon differentiation, infected macrophages release a variety of postinflammatory factors into the brain parenchyma causing neuronal damage (Holguin et al, 2007). This process results in psychomotor slowing, memory impairment, and brain atrophy (Anderson et al, 2002; McArthur et al, 1999). Although the advent of antiviral therapy has diminished the severity of HAD, the disease is still common and presents with variable progression patterns (McArthur, 2004). This poses a challenge for diagnosis. In view of these changes the National Institutes of Health (NIH) and two of its institutes, National Institute of Mental Health (NIMH) and Neurological Disorders and Stroke (NINDS) charged a working group to critically review the definition of HAD (AAN AIDS Task Force 1991). This working group suggested a research nosology for HAND to include three conditions: asymptomatic neurocognitive impairment (ANI), mild neurocognitive disorder (MND), and HAD (Antinori et al, 2007).
During HAD, signaling pathways are activated with the release of inflammatory products, chemokines, and neurotoxins (cellular and viral) that attract other cells to the site of infection, resulting in an increase of intraneuronal calcium and subsequently producing neuronal death (Gendelman et al, 1997; Li et al, 2005; Persidsky et al, 1997). In fact, apoptosis of neurons and astrocytes has been detected in autopsy brain tissues from acquired immunodeficiency syndrome (AIDS) patients with HAD (Chen et al, 2005; Self et al, 2004). The communication between the nervous and the peripheral immune systems is an important pathway for activation of macrophages during HAD (Ballabh et al, 2004). The response of the CNS to systemic immune challenge results in brain inflammation caused by infected monocytes trafficking into the brain from the periphery, resulting in the disruption of the BBB (Gartner, 2000; Gonzalez-Scarano and Martin-Garcia, 2005; Reynolds et al, 2007).
The HIV-1 replication in monocytes is restricted, but viral replication increases after cell differentiation (Zybarth et al, 1999). After HIV-1 infects macrophages, several signals are activated causing morphological and functional changes to the cell (Chertova et al, 2006; Ciborowski et al, 2007; Kadiu et al, 2007; Lee et al, 2003). These changes induced by HIV-1 can drive the cell to a permissive state for viral replication (in favor of the virus) or can enhance phagocytosis and intracellular microbial killing. Despite all the confirmed information about the interactions between HIV-1 and macrophages, the exact pathways of macrophage activation after HIV-1 infection remain incompletely understood. We, as well as others, have demonstrated differences in protein profiles of macrophages from HIV-1–infected individuals with cognitive impairment (CI) by using surface enhanced laser desorption ionization time of flight (SELDI-TOF) (Luo et al, 2003; Sun et al, 2004). We have also found differences in the macrophage proteome (58 proteins up- or down-regulated) after infection with HIV-1 ADA (Carlson et al, 2004). This study identified -actin, annexin 5, and L-plastin among others in HIV-1 ADA–infected monocyte-derived macrophages (MDMs). Most recently, we have found that the macrophage secretome is affected by HIV infection (Ciborowski et al, 2007) where cystatins B and C, L-plastin, superoxide dismutase, and α-enolase were identified preferentially from infected cells. Taken together, these studies clearly demonstrate the effects of HIV-1 on macrophage activation, structure, and function (Carlson et al, 2004; Ciborowski et al, 2007).
Our goal in the current study was to determine the effects of primary HIV-1 isolates (Nieves et al, 2007) on the macrophage proteome. The viral isolates were derived from 21 women with or without CI. MDMs from normal donors were infected with the HIV-1 isolates and the cultures were followed for 2 weeks. The macrophage proteome was analyzed by SELDI-TOF protein chip assays to determine the differences in protein profiles among the groups. Following fractionation by cationic exchange chromatography, low abundant proteins were separated by one-dimensional sodium dodecyl sulfate–poly-acrylamide gel electrophoresis (1D SDS-PAGE) and sequenced by liquid chromatography tandem mass spectometer (LC-MS)/MS. We found proteins unique to macrophages infected with CI viruses. This study demonstrates that HIV-1 variants can affect the macrophage proteome differently and that CI isolates can induce a neurotoxic phenotype in MDMs with decreased antioxidant and more proapoptotic proteins.
Results
Viral replication of primary HIV-1 isolates in MDMs
We compared p24 antigen concentration as a measure of HIV-1 replication in MDMs infected with the HIV-1 isolate from NC (isolate 3), two isolates from patients with CI (isolates 11 and 20), and the laboratory-adapted spinal fluid isolate SF162 derived from a patient with HAD at different time points (days 0, 4, 7, 11, and 14 p.i.). All isolates showed increased infectivity over time in culture (Figure 1A). Significant differences in HIV-1 p24 antigen levels between isolates were found at day 14 p.i. Isolate 20 showed significantly higher HIV-1 p24 antigen levels (mean 223.07 ng/ml) than did the other viruses (SF162 mean 4.61 ng/ml; isolate 11 mean 31.65 ng/ml; Isolate 3 mean 50.37 ng/ml) at day 14 p.i. (P < .001) (Figure 1B).
Figure 1.
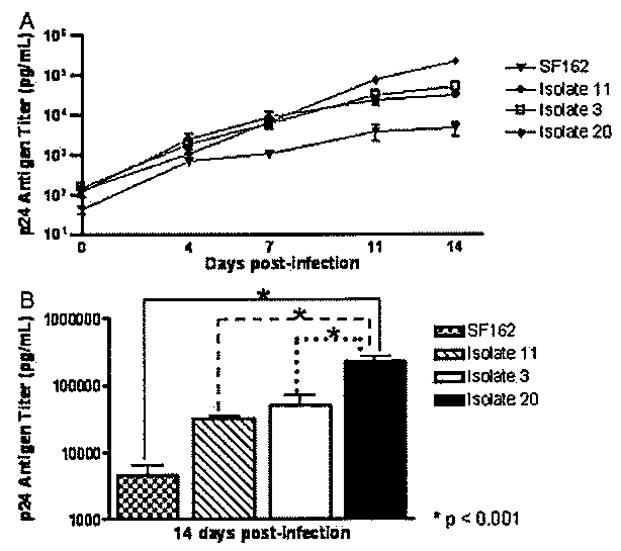
Replication of primary HIV-1 isolates in MDMs. Monocytes were cultured for 7 days for macrophage differentiation and in vitro infections. HIV-1 SF162 and primary isolates from Hispanic women with normal cognition (isolate 3) and cognitive impairment (isolates 11 and 20) were inoculated on day 7 and followed for 2 weeks. Culture supernatants were collected and tested for HIV-1 p24 antigen concentration at days 0, 4, 7, 11, and 14 p.i. to monitor virus replication (A). Significant differences (*P < .001) in HIV-1 p24 titer at day 14 p.i. were observed between isolate 20 as compared with all the others by analysis of variance (B).
MDM protein profiling
To determine the effect of HIV-1 infection with primary viral isolates and SF162 in the macrophage proteome, we devised the following experimental approach (Figure 2). Briefly, MDM lysates from day 15 p.i. were fractionated by cationic exchange chromatography. Differential protein fingerprints between uninfected and HIV-1–infected macrophages reported previously (Carlson et al, 2004) prompted us to further examine the effects of HIV-1 variants from NC and CI on macrophage protein expression. SELDI-TOF protein profiles from pH 9 fraction were chosen for further studies on protein identification based on higher protein concentration and increased number of differences among the groups (data not shown). Spectra from uninfected and infected MDMs with NC (isolate 3), CI (isolates 11 and 20), and HIV-1 SF162 were compared to determine differences in protein profiles (Table 1). When the spectra from MDMs infected with HIV-1 NC were compared to those infected with HIV-1 CI, we found a significant (q ≤ .10) increase in the mean intensity of the 6617-Da peak in MDMs infected with the viruses from CI (Figure 3A and B). Intensities of four peaks with m/z of 5300, 5468, 5482, and 8639 were decreased (q ≤ .10) in MDMs infected with NC virus when compared with MDMs infected with SF162 (Figure 4A to D). The intensity of these four peaks was similar in MDMs infected with NC and CI isolates.
Figure 2.
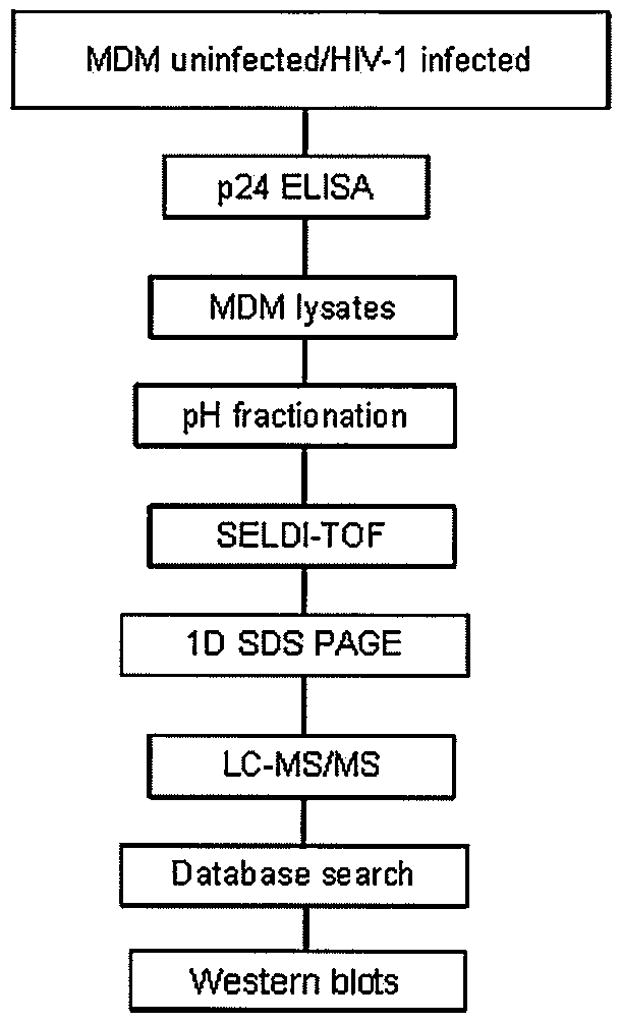
An overview of the experimental design and MDM proteome analyses used in this study.
Table 1.
Summary of differential peak expression between uninfected and HIV-1-infected MDMs
| Median of Intensity (natural scale) |
|||||
|---|---|---|---|---|---|
| Median m/z | q-value | Controla | NC | CI | SF162 |
| 5300 | 3.56E-02 | 1.343* | 0.79* | 1.31 | 1.16 |
| 5300 | 6.66E-14 | 1.343 | 0.79* | 1.31 | 1.16* |
| 5468 | 9.60E-03 | 6.659* | 4.244* | 5.41 | 6.41 |
| 5468 | 9.14E-03 | 6.659 | 4.244* | 5.41 | 6.41* |
| 5482 | 5.30E-02 | 11.585* | 11.278 | 12.43 | 13.38* |
| 5482 | 5.30E-02 | 11.585 | 11.278* | 12.43 | 13.38* |
| 5642 | 1.39E-03 | 7.464* | 6.053 | 5.91* | 6.5 |
| 6617 | 1.04E-01 | 2.585 | 1.982* | 2.81* | 2.58 |
| 8639 | 6.82E-02 | 1.127 | 0.967* | 1.16 | 1.36* |
| 9956 | 1.09E-01 | 14.048* | 16.761 | 16.7* | 16.11 |
Uninfected controls.
Level of significance determined by adjusted P value (q ≤ 0.1) between selected groups (Storey, 2003).
Figure 3.
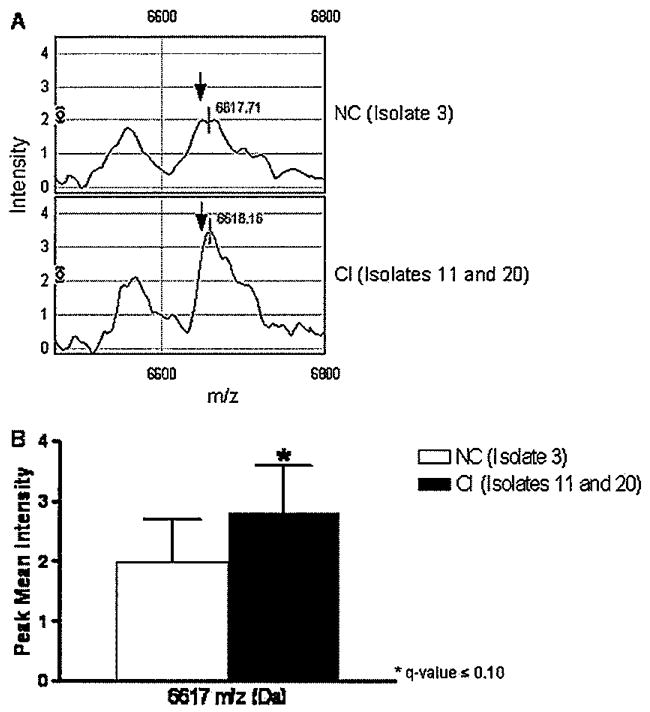
Differential expression of the 6617 m/z peak between MDMs infected with HIV-1 from normal cognition and cognitive impairment. Protein profiles from MDMs after 14 days of infection were compared by SELDI-TOF. Spectra from MDMs infected with a virus isolate from a patient with normal cognition (isolate 3) were compared with spectra from MDMs infected with viruses from patients with cognitive impairment (isolates 11 and 20) for intensity changes (A). The bar graph represents the difference between the two groups and the error bars represent one standard deviation for each group, respectively. Following generalized estimating equations with adjusted multiple comparisons, protein peaks with an adjusted P value (or a q-value) <.1 were considered significantly different among the groups (B). Data are representative of three independent experiments.
Figure 4.
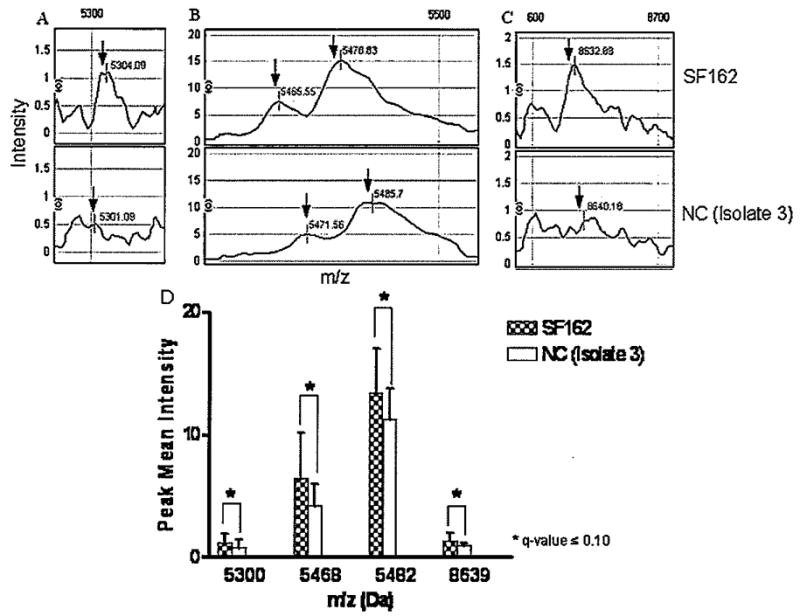
Differential protein profiles between MDMs infected with SF162 and primary isolate from NC. Protein profiles from MDMs after 14 days of infection were compared by SELDI-TOF. Spectra from MDMs infected with SF162 were compared with spectra from MDMs infected with NC (isolate 3) virus for differential protein peak intensities (A–C). The bar graph represents the comparison between NC and SF162, and the error bars represent one standard deviation for each group, respectively. Following generalized estimating equations with adjusted multiple comparisons, protein peaks with an adjusted P value (q-value <.1) were considered significantly different among the groups (D). Data are representative of three independent experiments.
MDM protein identification
After SELDI-TOF protein profiling, pH 9 protein fractions from three individual experiments were selected based on significant spectra differences between groups and separated by 1D SDS-PAGE gels. Bands corresponding to regions of SELDI-TOF spectra were digested with trypsin and the resulting peptides were sequenced by LC-MS/MS. Sixteen (16) proteins were identified with high confidence as common to all groups. The three proteins with higher number of peptides were SH3 domain-binding glutamic acid rich-like protein (seven peptides), Cu/Zn superoxide dismutase (five peptides), and thioredoxin (five peptides). These three proteins have antioxidant activity.
When we compared HIV-1–infected MDMs with the different isolates, three proteins (A-kinase anchor protein 9, annexin V, and ubiquitin) were common to MDMs infected with SF162, CI, and NC viruses (Table 2). Two of these, A-kinase anchor protein 9 and ubiquitin, are important in HIV-1 trafficking, whereas annexin 5 is important in prevention of HIV infection and apoptosis (Munoz et al, 2007). Three proteins (myotrophin, protein kinase C inhibitor protein 1, and thymosin beta-4) were identified only in MDMs infected with SF162 and CI viruses (Table 2). Myotrophin is a cystoplasmic protein related to muscle differentiation whereas protein kinase C inhibitor protein 1 and thymosin beta-4 have a common function in macrophage migration and inflammation. Seven proteins were identified only in MDMs infected with SF162: alpha-1 protease inhibitor, aspartylglucosaminidase, DNA-directed RNA polymerase III subunit, genome polyprotein, heparan sulfate proteoglycan 2, mannose 6 phosphate receptor binding protein 1, and triosephosphate isomerase (Table 2). Most of these are important enzymes involved in cellular metabolism, cell adhesion, and transport of cellular receptors. Six proteins related to cell structure, nucleic acids, and signaling were identified only in MDMs infected with NC virus: chromosome-associated protein E, coactosin-like protein, GTP-binding protein 3, putative methyltransferase NSUN5, teneurin-3, and uncharacterized protein C7orf24 (Table 2).
Table 2.
Identified proteins from macrophages infected with HIV-1 isolates
| Proteins | MWa | NCBIb | SwissProtc | Function |
|---|---|---|---|---|
| Proteins in NC | ||||
| Chromosome-associated protein E | 135781 | 30173225 | O95347 | Central component of the condensin complex, a complex required for conversion of interphase chromatin into mitotic-like condense chromosomes. |
| Coactosin-like protein | 15945 | 21624607 | Q14019 | Actin and enzyme binding in a calcium-independent manner. |
| GTP-binding protein 3 | 52030 | 74731069 | Q969Y2 | Intracellular protein with GTPase activity and involved in tRNA modification. |
| Putative methyltransferase NSUN5 | 46692 | 146289861 | Q96P11 | May have S-adenosyl-L-methionine-dependent methyl-transferase activity. |
| Teneurin-3 | 300950 | 118573058 | Q9P273 | May function as a cellular signal transducer. |
| Uncharacterized protein C7orf24 | 21008 | 34222503 | O75223 | Unknown |
| Proteins in CI | ||||
| Adenine phosphoribosyltransferase | 19608 | 4502171 | P07741 | Catalyzes a salvage reaction resulting in the formation of AMP, which is energetically less costly than de novo synthesis. |
| Adenosylhomocysteinase | 47716 | 20141702 | P23526 | Involved in the control of methylations via regulation of the intracellular concentration of adenosylhomocysteine. |
| ADP-ribosylation factor GTPase-activating protein 3 (ARF3) | 56928 | 21263420 | Q9NP61 | Involved in protein secretion, Golgi vesicle transport and promotes the hydrolysis of the ARF1-bound GTP. |
| Apoptosis-inducing factor 3 (AIF3 or AIFL) | 66791 | 74732608 | Q96NN9 | Induces apoptosis through a caspase dependent pathway. Reduces mitochondrial membrane potential. |
| Beta-actin** | 41737 | 46397333 | P60709 | Cytoskeletal protein wihich binds to ATP and is involved in cell motility (structural molecule activity). |
| Cathepsin A | 54466 | 20178316 | P10619 | Protective lysosomal protein apparently essential for the activity of beta-galactosidase and neuraminidase. Component of the endoplasmic reticulum; Caiboxypeptidase, enzyme activator, and intracellular protein transporter activity. |
| Conserved oligomeric Golgi complex component 4 | 89095 | 48429262 | Q9H9E3 | Required for normal Golgi function. |
| DNA fragmentation factor (DFF-40 or CAD) | 39110 | 4758150 | O76075 | Nuclease that induces DNA fragmentation and chromatin condensation during apoptosis. Caspase-activated deoxyribonuclease activity involved in intracellular signaling cascade. |
| Ferritin heavy chain | 21226 | 56682959 | P02794 | Plasma membrane protein involved in iron homeostasis. Kinase binding and ferroxidase activity. Involved in the negative regulation of cell proliferation (immune response). Protects DNA from damage. |
| Flavin reductase (FAD) | 22119 | 4502419 | P30043 | Role in protecting cells from oxidative damage or in regulating iron metabolism. |
| L-Lactate dehydrogenase A chain | 36689 | 13786849 | P00338 | Cytosolic protein involved in the final step of anaerobic glycolysis. |
| L-plastin | 70289 | 4504965 | P13796 | Cytosolic actin-binding protein expressed in neutrophils, monocytes, B lymphocytes, and myeloid cells; Calcium ion binding. |
| Myosin RLC | 19794 | 15809016 | P19105 | Involved in the regulation of smooth muscle and nonmuscle cell contractile activity. |
| Pyrin | 86444 | 8928170 | O15553 | Nuclear or cytoplasmic protein involved in controlling the inflammatory response in myelomonocytic cells at the level of the cytoskeleton organization. |
| Ras-related protein Rab-18 | 18023 | 10880989 | Q9NP72 | Involved in vesicular trafficking and neurotransmitter release. Plays a role in apical endocytosis/recycling. |
| Ras-related protein Rab-7 | 23490 | 1709999 | P51149 | Involved in late endocytic transport. Contributes to the maturation of phagosomes. |
| Ras-related protein Rap-1A | 20987 | 51338607 | P62834 | Membranal protein with GTPase activity involved in signal transduction. |
| Ribosome biogenesius protein | 145807 | 113420775 | Q14692 | May act as a molecular switch during maturation of the 40S ribosomal subunit in the nucleolus. |
| BMS1 | ||||
| homolog | ||||
| RING finger protein 165 | 39526 | 57165361 | Q6ZSG1 | Unknown. |
| Signal recognition particle 54 (SRP54) | 42779 | 46577650 | P61011 | Membranal protein probably involved in the reception and insertion of a subset of proteins at the cytoplasmic membrane. |
| Proteins in SF162 | ||||
| Alpha-1 protease inhibitor | 46737 | 50363217 | P01009 | Angiotensinogen; Serine-type endopeptidase inhibitor activity. |
| Aspartylglucosaminidase | 37194 | 262567 | P20933 | Lysosomal protein that cleaves the GlcNAc-Asn bond that joins oligosaccharides to the peptide of asparagine-linked glycoproteins. |
| DNA-directed RNA polymerase III subunit | 127785 | 29428029 | Q9NW08 | Catalyzes the transcription of DNA into RNA using the four ribonucleoside triphosphates as substrates. |
| Genome polyprotein | 244392 | 31076989 | Q9YLG5 | Polyprotein containing capsid proteins VP1, VP2, VP3, and VP4 that forms a closed capsid enclosing the viral positive strand RNA genome. Other subunits (P2A, P3C-cysteine proteases; P2C-associated with viral RNA synthesis of poliovirus and picornavirus). |
| Heparan sulfate proteoglycan 2 (perlecan) | 468830 | 55665909 | Q5VU27 | Extracellular matrix component involved in cell adhesion. |
| Mannose 6 phosphate receptor binding | 47047 | 20127486 | O60664 | Cytoplasmic protein required for the transport of mannose 6-phosphate receptors (MPR) from endosomes to the trans-Golgi network. |
| protein 1 | ||||
| Triosephosphate isomerase | 26669 | 4507645 | P60174 | Involved in carbohydrate biosynthesis (gluconeogenesis) and carbohydrate degradation (glycolysis). |
| Proteins shared by SF162 and CI | ||||
| Myotrophin | 12895 | 21956645 | P58546 | Cytoplasmic protein with a potential role in neuron differentiation. Also, associated to regulation of translation and development of striated muscle. |
| Protein kinase C inhibitor protein 1 | 27745 | 4507953 | P63104 | Adapter protein implicated in the regulation of general and specialized signaling pathways. |
| Thymosin beta-4 | 5053 | 78103211 | P62328 | Cytoplasmic protein that plays a role in the organization of the cytoskeleton. Inhibits actin polymerization and the entry of hematopoeitic pluripotent stem cells into the S-phase. |
| Proteins shared by NC and CI | ||||
| Fatty acid-binding protein 5 | 15164 | 4557581 | Q01469 | Cytoplasmic protein involved in fatty acids and protein binding. |
| HERV-K[III] Pol protein | 165184 | 3702894 | P63135 | Protein synthesized as Gag-Pro and Gag-Pro-Pal polyprotein precursors. Involved in converting the viral RNA genome into double-stranded viral DNA, early post infection. |
| Proteins shared by SF162, NC, and CI | ||||
| A-kinase anchor protein 9 | 453667 | 22538387 | Q99996 | Binds to type II regulatory subunits of protein kinase A. Scaffolding protein that assembles protein kinases and phosphatases on the centrosome and Golgi apparatus. |
| Annexin V** | 35937 | 4502107 | P08758 | Anticoagulant protein that inhibits the thromboplastin-specific complex, which is involved in the blood coagulation cascade. |
| Ubiquitin | 8565 | 4507761 | P62988 | Protein modifier which can be attached to target lysines as a monomer or as a lysine-linked polymer (this form leads to their degradation by proteasome). Involved in maintaining chromatin structure, regulation of gene expression, stress response, ribosome biogenesis and DNA repair. |
Molecular weight (Da);
NCBI accession number;
SwissProt accession number.
These proteins were identified with two peptides, all the other proteins were identified with one peptide.
Twenty proteins were identified in MDMs infected with viral strains related to CI. These included proteins with molecular weights ranging from 5000 to 300,000 Da, many of in a mass range higher than SELDI-TOF m/z values (5000 to 20,000). Identified proteins were categorized into the following four main functions: (1) structural and chemotaxis; (2) protein trafficking; (3) apoptosis; and (4) redox. The first group, structural and chemotaxis, included: L-plastin, myosin RLC, adenine phosphoribosyltransferase, adenosylhomocysteinase, and ADP-ribosylation factor GTPase-activating protein 3. The second group, protein trafficking, included -actin, cathepsin A, conserved oligomeric Golgi complex component 4, Ras-related proteins Rab-7, Rab-18, and Rap-1A, ribosome biogenesis protein BMS1 homolog, RING finger protein 165, and signal recognition particle 54. The third group, apoptosis, included apoptosis-inducing factor 3, DNA fragmentation factor, and pyrin. The fourth group, redox, included ferritin, flavin reductase, and 1-lactate dehydrogenase A chain (Table 2). All of the proteins identified in macrophages infected with primary isolates (NC and CI viruses) and SF162 matched with only one peptide with the exceptions of annexin V and β-actin, which matched with two peptides. Although most of the proteins were identified with only one peptide, they met all five criteria and were accepted as “true positives” in accordance with the results published by the Plasma Proteome Project performed by the Human Proteome Organization (Omenn et al, 2005), which shows “true positive” identifications of 25% of the proteins based on the sequence of one peptide.
Protein expression
Western Blots were performed to validate the expression of eight proteins that were identified in MDMs infected with HIV-1 CI virus and selected following the criteria outlined at the Methods. Proteins in gel bands corresponding to molecular weight regions 5 to 20 kDa as indicated by SELDI-TOF experiments were sequenced. As expected, besides high confidence identification, we found proteins represented by one sequenced peptide. Such identifications were considered as low confidence based on the guidelines published by the Plasma Proteome Project (Omenn et al, 2005). For all the proteins tested for the different groups, we were only able to determine their relative abundance because Western Blot is only a qualitative method.
There was no difference in L-plastin, cathepsin A (data not shown), or Rap 1A expression between NC, CI, and SF162 viruses, whereas apoptosis-inducing factor like (AIF-L; data not shown) and DNA fragmentation factor (DFF-40; CAD) showed a trend to increase in isolate 11 (CI). Ferritin heavy chain presented a relative increase in expression with both CI viruses, whereas lactate dehydrogenase (LDH) did not show any significant difference. Representative Western blots for some of these proteins are included in Figure 5A. To summarize, although various monoclonal and polyclonal antibodies were tested against these proteins, their relative abundance was low. Due to the limited amount of sample obtained from all three experiments, Western Blot was not the most suitable method to measure the differential expression of these low-abundance proteins. Other methods such as ELISA could be more suitable, because they require a smaller amount of sample; however, no ELISAs were available for most of the proteins under study. We assayed the expression of superoxide dismutase (Cu/Zn SOD; 16 kDa) by Western Blots because it was identified with five peptides in all groups, and found abundant in the CSF of women with CI (Laspiur et al, 2007). Surprisingly, we found that Cu/Zn SOD protein expression decreased in MDMs infected with HIV-1 from CI as compared with that from NC (Figure 5B). Following densitometry analysis and normalization against β-tubulin, we confirmed decreased abundance of SOD in MDMs infected with CI (P < .05, Kruskal-Wallis chi-square) (Figure 5C).
Figure 5.
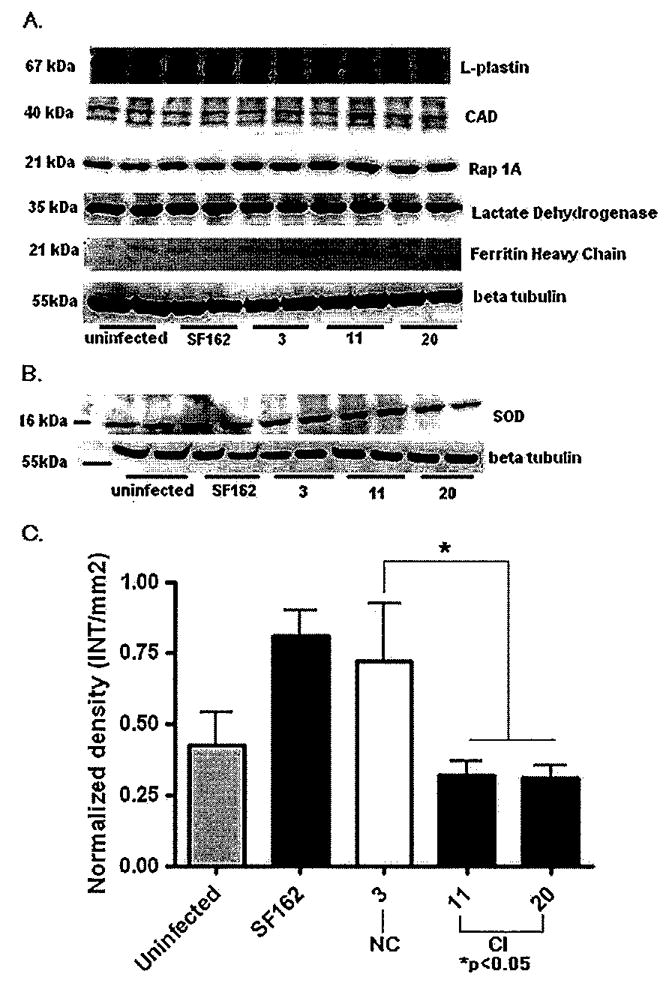
Protein expression of MDMs infected with HIV-1 SF162, NC, and CI isolates by Western blots. Representative Western blots of uninfected controls and MDMs infected with HIV-1 SF162, NC, and CI isolates (A). SOD-1 expression in uninfected MDMs and MDMs infected with HIV-1 SF162, CI, and NC (B). Densities of protein bands were normalized to beta-tubulin expression on the same blot (C). MDMs infected with primary isolates from CI showed significant decrease in SOD expression when compared with NC (P <.05). Data analyzed correspond to the densitometry of three different experiments.
Discussion
In this study, we identified proteins expressed in macrophages after in vitro infection with HIV-1 strains derived from patients with CI. Primary viral isolates induced significant changes in 10 protein peaks between groups: uninfected MDMs and MDMs infected with HIV-1 recovered from NC and CI patients as well as the laboratory adapted SF162 strain. One of these peaks (m/z = 6617) was found to be significantly different between strains isolated from NC and CI subjects. Interestingly, we have found peaks in similar mass range in macrophages (m/z =6900) and CSF (m/z = 6436, 6695, and 6974) from patients with CI (Wojna et al, 2004; Laspiur et al, 2007). These findings could suggest similar proteins or disease mechanisms that need to be studied further.
After protein separation by gel electrophoresis and sequencing by tandem mass spectrometry, we identified 16 common proteins among all groups, 20 proteins in MDMs infected with HIV-1 isolates from CI that were not found in cells infected with the NC and SF162 isolates, and only 3 that were shared by SF162 and CI among other comparison groups. These results demonstrate that more proteins were identified at higher molecular weight range by tandem mass spectrometry than peaks observed in SELDI-TOF spectra. Proteins identified by tandem mass spectrometry did not matched with the SELDI-TOF data. This could be due to the fact that in SELDI-TOF experiments we investigated proteins within an m/z range of 5000 to 20,000 Da under non-reducing conditions. Aggregation of proteins thus having total mass higher than 20,000 m/z cannot be detected. Tandem mass spectrometry is done from gel-tryptic digested proteins. In SDS-PAGE electrophoresis disruption of protein complexes is facilitated by heat denaturation, therefore, proteins may not migrate at the same mass owing to different charges or conformational states.
Among the 16 common proteins found in uninfected MDMs and MDMs infected with isolates NC, CI, and HIV-1 SF162, the most abundant proteins belonged to the antioxidants group. These included SH3 domain–binding glutamic acid protein (identified with seven peptides), Cu/Zn superoxide dismutase (SOD; identified with five peptides), and thioredoxin (identified with five peptides). These results reflect the importance of antioxidants in cell viability and function. Interestingly, the 20 proteins identified in MDMs infected with CI HIV-1 isolates included functions related to structure, chemotaxis, trafficking, apoptosis, and redox. All of these, except two structural proteins, were identified with a single peptide, In order to determine the relative abundance of these proteins in MDMs infected with CI viruses and given the limited concentration of proteins available from these experiments, we chose to study eight proteins among these four groups. The criteria for selecting these proteins included number of peptides, antibody availability, and functional group as described in the Materials and Methods section, Among the functional groups, for structural we tested L-plastin; for trafficking we tested cathepsin A and Rap 1A; for apoptosis we tested apoptosis inducing factor like (AIF-L) and DNA fragmentation factor (DFF-40; CAD); and for redox we tested ferritin heavy chain (FHC) and lactate dehydrogenase (LDH). From our results we were able to detect expression of these enzymes in uninfected MDMs and MDMs infected with isolates NC, CI, and HIV-1 SF162. We found a relatively higher expression of apoptotic inducing factor like (AIF-L), CAD, and the antioxidant protein FHC in MDMs infected with CI viruses after normalization. The AIF-L is a homologous protein of the apoptosis-inducing factor protein (AIF) that induces apoptosis through the caspase 3 activation pathway (Xie et al, 2005). Even though HIV-1–infected macrophages live for prolonged time in tissue compartments, acting as viral reservoirs and promoting virus replication, signal transduction of apoptotic genes has been reported (Vazquez et al, 2005; Wahl et al, 2003). These include the apoptotic protein TRAIL and TRAIL-over-expressing macrophages in the brain of HAD patients (Zhang et al, 2001). Apoptosis-inducing factor-like and DNA fragmentation factor could be inducing apoptotic pathways in the macrophage and this may be due to HIV-1 infection. Ferritin heavy chain, which was found increased in MDMs infected with CI, sequesters the excess of free iron and mediates the recently discovered “antioxidant activities” of nuclear factor (NF)-κB, and by these means suppresses the generation of reactive oxygen species (Bubici et al, 2006; Tsuji, 2005).
We selected to determine expression of another antioxidant protein, Cu/Zn SOD, by Western blots. Although, this protein was common to all the samples, we wanted to test differences between NC and CI groups because it was identified with a high number of peptides. Recently, we found that this protein is decreased in monocytes from Hispanic women with CI (Velásquez et al, 2007). Interestingly, after densitometry and statistical analyses, we found a significant decrease of this protein in MDMs infected with CI viruses. These results suggest that HIV-1 variants from CI could induce oxidative stress by reducing SOD-1 expression in infected MPs.
Because we could not isolate viruses from the CSF of HIV-1–infected women due to HAART (Nieves et al, 2007), the HIV-1 SF162 was chosen to compare with the HIV-1 isolates from NC and CI. We obtained common and unique protein profiles in MDMs infected with SF162 and the viruses from NC and CI. Protein identification by LC-MS/MS also revealed common and unique proteins in MDMs infected with NC, CI, and SF162, demonstrating that there are common and distinct changes in the MDM proteome induced by viral isolates from CI.
Taking these observations together, our findings suggest that the HIV-1 variants isolated from patients with NC and CI can activate different pathways in macrophage protein trafficking, apoptosis, and oxidative stress. The differences in protein expression found in macrophages infected with primary isolates from NC and CI suggest a specific virus-cell interaction that activates diverse signaling events and cellular pathways that affect cellular function. It is important to emphasize that the presence of these proteins was not related to HIV-1 replication levels. All the cultures were inoculated with at the same MOI and these proteins were not detected in macrophages infected with HIV-1 BaL, a laboratory-adapted macrophage-tropic virus with high replication level (data not shown). There are several limitations of this study, which include the small number of HIV-1 primary macrophage-tropic isolates obtained from NC and CI groups and the limited concentration of protein obtained from virus-infected MDMs for Western blots. Because most of the study patients were on HAART, the viral isolation and growth proved to be a challenging task.
Our study is novel in the field of NeuroAIDS and our findings are very significant as they show for the first time that different HIV-1 viral strains provoke changes in the macrophage’s proteome that most likely lead to differences in function during infection. This finding could define a neurotoxic role for the macrophage determined in part by HIV-1 variants that could influence the development of neurotoxicity and cognitive impairment.
Materials and methods
Patient cohort
This study was conducted as part of the NeuroAIDS Specialized Neuroscience Research Program (SNRP) at the University of Puerto Rico, Medical Sciences Campus (UPR, MSC). HIV-1–seropositive women were recruited from primary HIV-1 clinics at UPR, MSC. The study had the approval of the Institutional Review Board (number 0720102) and was conducted with the informed consent of all participants. The inclusion and exclusion criteria, recruitment, and evaluation have been described previously (Wojna et al, 2006). Plasma and cerebrospinal fluid (CSF) viral loads were determined with use of an Ultrasensitive RNA Roche Amplicor at an AIDS Clinical Trial Group (ACTG)-certified laboratory with a detection range of 50 to 75,000 copies of RNA/ml, Cognitive impairment was determined in accordance with the American Academy of Neurology HIV-1 associated dementia criteria (1991 (1996) (AAN criteria) (American Academy of Neurology AIDS Task Force, 1991, 1996), modified (m-AAN criteria) to include an asymptomatic cognitively impaired group (Wojna et al, 2006). The asymptomatic cognitively impaired group is denned as patients with abnormal neuropsychological tests (1 SD in two or more tests or 2 SD in one or more tests below the normal control group) but who presented neither functional/emotional nor neurological findings. According to the m-AAN criteria, patients were classified as having normal cognition (NC), asymptomatic cognitive impairment, minor cognitive motor disturbance (MCMD), or HAD. For our study, patients with, asymptomatic cognitive impairment, MCMD, or HAD were grouped together under the term cognitively impaired (CI) and were compared with patients having normal cognition (NC) as described (Nieves et al, 2007). All patients included in this study were negative for other infectious diseases and toxicology.
Monocyte isolation
Peripheral blood mononuclear cells were isolated from HIV-1–, HIV-2–, and hepatitis-seronegative healthy donors by leukopheresis and purified by countercurrent centrifugal elutriation (Gendelman et al, 1988). Monocytes were >95% pure as determined by CD14-FITC (fluorescein isothiocyanate) staining and flow cytometry analysis. Cells were seeded at a concentration of 1 × 106 cells/ml on monocyte medium (Dulbecco’s modified Eagle’s Medium [DMEM], 1000 U macrophage colony-stimulating factor [M-CSF; a generous gift from Wyeth, Cambridge, MA], 10% human serum, 1% l-glutamine, 50 μg/ml gentamicin, and 10 μg/ml ciprofloxacin) and seeded in 6-well plates for 7 days. Medium was half-exchanged every 2 to 3 days. At day 7, the supernatant was removed and cells were washed twice with phosphate-buffered saline (PBS) at room temperature prior to HIV-1 infection.
Preparation of viral stocks
Primary isolates were obtained from the peripheral blood of 62 HIV-1–seropositive women characterized for cognitive function who were using highly active antiretroviral therapy (HAART) as described (Nieves et al, 2007). HIV-1–infected peripheral blood mononuclear cells (PBMCs) from these women were cocultured with mitogen-activated PBMCs from normal donors for 2 weeks and supernatants recovered for detection of virus by HIV-1 p24 antigen enzyme-linked immunosorbent assay (ELISA). Out of 62 viral cultures, 21 HIV-1 isolates were recovered and characterized for tropism in lymphocytes and macrophages. From these, three viruses were identified that replicated in macrophages and used the CXCR4 and CCR5 coreceptors (isolates 3, 11, and 20). HIV-1 isolates 11 and 20 were derived from women with CI and isolate 3 was derived from one woman with NC. We compared the peripheral blood primary isolates with HIV-1 SF162, a spinal fluid isolate derived from a patient with HAD and acquired from the NIH-AIDS Reagent Program, National Institutes for Allergy and Infectious Diseases. Virus stocks were prepared from these four viruses (3, 11, 20, and SF162) by high-speed centrifugation of supernatants at 19,000 rpm, −10°C, for 2 h, resuspended in monocyte medium containing 20% of human serum, and titrated in MDMs by limited dilution analyses using HIV-1 p24 ELISA as previously described (Arroyo et al, 1996; Renta et al, 1997).
HIV-1 infection of MDMs
MDMs from eight different donors (2 × 105) were inoculated with 25 ng of HIV-p24 antigen of each viral isolate per well in 6-well plates in triplicate cultures. This was equivalent to a multiplicity of infection (MOI) of 0.1. Cells were incubated with virus for 4 h at 37°C, and thereafter each well was washed twice with monocyte medium at room temperature to remove unbound virus. After three medium washes, cells were resuspended in monocyte medium without M-CSF and cultured at the same concentration. Supernatants were collected on days 0, 4, 7, 11, and 14 post infection (p.i.), and viral infection was monitored by HIV-1 p24 antigen ELISA (Coulter) according to the manufacturer’s instructions. On day 14 p.i., cells were washed and resuspended in serum-free medium for 24 h prior to cell lysis for proteomic analyses. The three experiments with productive HIV-1 infection were selected for proteomics studies.
Preparation and fractionation of MDM lysates
After overnight incubation with serum-free medium, on day 15 after infection, cells were washed twice with cold PBS. Adherent MDMs were incubated for 25 min on ice with cold lysis buffer (5 mM Tris-HCl buffer at pH 8.0, 0.1% Triton X-100, and 0.05% of protease inhibitor solution) at a ratio of 100 μl of buffer/1 × 105 cells. Cell lysates were collected, vortexed, and centrifuged at 4°C for 10 min at 500 × g (1500 rpm). The protein concentration of resultant MDM lysates was determined using the BioRad DC protein assay (Richmond, CA) following the manufacturer’s instructions. Cell lysates from triplicate wells from each individual experiment were pooled and stored in aliquots at − 80°C for proteomics analyses. Fractionation of MDM lysates was performed to positively detect low abundant proteins by cationic exchange chromatography using Ciphergen CM Ceramic HyperD spin columns (C540-0026; Ciphergen Biosystems), These columns isolated positively charged proteins that bound to the resin from the unbound negatively charged proteins that were eluted. In brief, columns were washed three times with 200 μl of equilibration buffer (0.1 M ammonium acetate pH 4.0) and centrifuged at 80 × g for 30 s to remove the buffer. From each sample, 300 μg of protein was added to the spin columns and incubated for 35 min with shaking. Two washes were performed with 200 μl of equilibration buffer followed by vortex and centrifugation (30 s) of the columns. Subsequent washes were performed with 200 μl of buffer with pH 9, 7, 5, 4, and 3, and with an organic buffer consisting of 33.3% isopropanol/16.7% acetonitrile/0.1% trifluoroacetic acid. These washes were followed by incubation for 10 minutes with shaking. After columns were centrifuged for 30 s, fractions were collected and stored at −80°C for subsequent experiments.
SELDI-TOF
Proteomics analysis was performed by SELDI-TOF protein chip assay using the weak cationic exchange chip, CM10 (BioRad, Inc. former Ciphergen Biosystems, CA). A total protein concentration of 0.05 μg/ml from samples derived from each one of the three experiments was applied to the chips in quadruplicates. Briefly, each protein sample was diluted in binding buffer (100 mM ammonium acetate pH 4 with 0.1% Triton-X) to a final volume of 250 μl. Before sample loading, each CM10 spot was equilibrated with binding buffer. An aliquot of 50 μl of the protein sample was loaded on each spot and incubated using a Bioprocessor (Ciphergen) for 30 min at room temperature with shaking. Spots were rinsed twice with binding buffer followed by a wash with high-performance liquid chromatography (HPLC)-grade water to remove unbound proteins. Spots were air-dried and the matrix sinapinic acid (SPA) diluted to 50% in the solvent (30% acetronitrile, 15% isopropanol, 0.5% trifluoroacetic acid [TFA] 1%, 0.05% Triton-X100, and HPLC-grade water) was applied (1 μl) twice. The molecular mass/charge (m/z) ratios of proteins were detected with a ProteinChip reader (PBS IIc series; Ciphergen). The ProteinChip reader was externally calibrated for each analysis by mass using a mixture of five standard proteins: bovine ubiquitin (5,733.6 Da), bovine cytochrome c (12,230.9 Da), bovine superoxide dismutase (SOD) (15,591.4 Da), equine cardiac myoglobin (16,951.5 Da), and beta-lactoglobulin (18,363.3 Da). The spectra obtained from each sample were analyzed using the Biomarker Wizard ProteinChip software 3.2 (Ciphergen). The following parameters for peak detection were used: first pass signal/noise (S/N) ratio =5, second pass S/N ratio = 2, mass tolerance = 0.5%.
Protein separation by 1D SDS-PAGE
Samples from pH 9 fraction were selected based on the most abundant protein differences and 20 μg of each protein sample were dehydrated using Speed Vac, rehydrated with NuPAGE LDS buffer (Invitrogen, Carlsbud, CA) and separated by ID electrophoresis on a NuPAGE Novex 10% Bis-Tris (Invitrogen) gel. Proteins obtained from HIV-1–infected macrophages derived from three different donors were isolated in three different gels and stained with Coomassie Brilliant Blue (BioRad, Hercules, CA). Protein bands from each one of the three gels (n = 34), for a total of 102 bands corresponding to the molecular weights observed in SELDI-TOF (m/z of 5 to 20 kDa), were excised using a sterile razor blade. Gel pieces were destained with 50% acetonitrile (ACN), 50 mM NH4CO3/50% ACN, and 10 mM NH4HCO3/50%. After gel pieces were dried, proteins were digested with trypsin (Promega, Madison, WI) by overnight incubation at 37°C. Peptides were extracted with 0.1% trifluoroacetic acid/60% ACN, dried, and purified using Zip Tip (Millipore) before liquid chromatography–mass spectrometry (LC-MS)/MS protein analysis.
Protein identification
Resulting peptides were resuspended in 0.1% formic acid/HPLC-graded water, The ionized peptides were detected on a LTQ LC-MS/MS system (Thermo Electron, Waltham, MA) as previously described (Ciborowski et al, 2007; Laspiur et al, 2007). Data obtained from the LC-MS/MS analysis were searched against the human NCBI.fasta protein database to identify the matching protein using BioWorks 3.1SR (ThermoElectron). Protein identifications were accepted as true positives according with previously described parameters (Omenn et al, 2005). The criteria used for data mining were as follows: BioWorks unified score ≥3000, Xcorr for singly charged precursor ion ≥2, Xcorr for doubly charged precursor ion ≥ 2.5, Xcorr for triply charged precursor ion ≥3, DeltaCn ≥0.3, and more than 55% of fragment ions per sequenced peptide. Proteins with one peptide were considered candidates for identification if the peptide met all the parameters. These proteins were accepted as “true positive” identifications in accordance with the results published by the Plasma Proteome Project performed by the Human Proteome Organization (Omenn et al, 2005), Their results showed that as many as 25% of identifications based on the sequence of one peptide were confirmed as “true positive.”
Protein expression
Western Blot analysis was performed in order to determine the relative abundance of several proteins identified in MDMs infected with viruses from CI. Due to sample limitation, we selected eight proteins based on the following criteria: (1) number of peptides identified, (2) antibody availability, and (3) functional group in relation to CI. All proteins assayed were identified with only one peptide with the exception of Cu/Zn superoxide dismutase, which was identified with five peptides. The proteins were classified in four groups, according to their major function: structural (L-plastin), apoptotic (apoptosis inducing factor like [AIF-L] and DNA fragmentation factor subunit beta [DFF-40; CAD]), trafficking (cathepsin A and Rap 1A), and redox (1-lactate dehydrogenase A chain, ferritin heavy chain [FHC] and Cu/Zn superoxide dismutase). Protein lysates and controls were resolved by SDS-PAGE (in duplicates), transferred to nitrocellulose membranes, blocked with 3% bovine serum albumin (BSA), and incubated overnight at 4°C with the corresponding primary antibody. Immunoblot detection was performed for selected proteins from the different functional groups with the following antibodies: goat anti-human L-plastin (Santa Cruz Biotechnology, Santa Cruz, CA), rabbit anti-human apoptosis inducing factor like (AIF-L) was provided by Dr. Joseph Bonanno, Indiana University (Xie et al, 2005), mouse anti-DNA fragmentation factor subunit beta (DFF-40; CAD) (Novus Biologicals, Littleton, CO), mouse anti-cathepsin A (R&D Systems, Minneapolis, MN), mouse anti-Ras-related protein Rap 1A (BD Biosciences, Franklin Lakes, NJ), goat anti-1-lactate dehydrogenase A chain (LDH) (Santa Cruz Biotechnology), rabbit anti-ferritin heavy chain (FHC) (Novus Biologicals), and sheep anti-Cu/Zn superoxide dismutase (SOD-1) (Calbio-chem, San Diego, CA). All membranes were incubated for 1 h at room temperature with the corresponding secondary antibody conjugated to horseradish peroxidase enzyme (HRP), Expressed proteins were detected using the Super Signal West Femto Maximum Sensitivity Substrate (Thermo Scientific-Pierce Biotechnology, Rockford, IL), enhanced chemiluminescent substrate. Images were obtained using a Versa Doc Imaging System (BioRad Laboratories, Hercules, CA) and densitometric analysis of images was performed using the Quantity One software. All data obtained were normalized against a monoclonal anti β-tubulin antibody (Sigma Aldrich, St. Louis, MO). ELISA for SOD-1 (Calbiochem) was determined from a 1:200 dilution of uninfected and HIV-infected cell lysates.
Statistical analysis
For HIV-1 replication studies, we used a linear regression model to determine significant differences in HIV-1 replication in MDMs at day 14 p.i. Comparisons of the p24 antigen levels between the isolates at day 14 p.i. were analyzed using analysis of variance in SAS software version 9.1 (SAS Institute, Cary, NC). The significance level was determined using a type I error rate of 0.05. SELDI-TOF protein profiles were initially analyzed using the Biomarker Wizard software 3.2, All peaks in spectra were baseline subtracted, calibrated on mass accuracy, and normalized with the Biomarker Wizard program, Data were exported to Microsoft Excel for analysis using SAS software. Profiles from uninfected MDMs and MDMs infected with HIV-1 (SF162, CI, and NC primary isolates) were compared on the basis of normalized peak height. Generalized estimating equations (GEEs) with adjusted multiple comparisons were used to identify peaks with significant differences in the distribution of intensity scores among the groups. Specifically, the raw intensity values were found to be asymmetrical and were adjusted prior to analysis by means of the following transformation: Y=log2 (X+SQRT[X**2 + 1]), where ‘X’ is the observed intensity. This transformation has been used previously to stabilize the intensity variance and make the data more normally distributed, and it has the advantage over a log-transformation of being able to handle negative intensities (Beyer et al, 2006; Huber et al, 2002). An “adjusted P value after multiple testing corrections” (a ‘q-value’) was computed and we chose a cutoff (10%) that controlled the expected number of false positives (Storey, 2003). Therefore, there were no more than 10% false positives among the selected significant differences between groups. These comparisons were made using GEE statistics. Western blot experiments were then conducted to verify those differentially expressed proteins. An unpaired Student’s t test was used to analyze densitometry data from Western blots to determine statistically significant differences between the compared groups at a significance level of .05.
Acknowledgments
The authors would like to thank their patients for supporting this research. Tania Ginebra and Tania de la Torre supported patient outreach, and Dr. Rosa Hechavarría performed neuropsychological testing. The authors thank Drs. Carmen Zorrilla and Hermes García (their clinic directors) for referring patients. The RCMI–Clinical Research Center is thanked for providing staff and supplies for laboratory sample collections. Thanks to Claribel Luciano for support on the gel processing. The authors thank and acknowledge Richard Skolasky for statistical support. They thank Dr. Joseph Bonanno for providing them with the AIF-L antibody. They acknowledge Drs. Howard Gendelman for his critical review of the manuscript, and Edmundo Kraiselburd for his continuous support of this work as part of the Puerto Rico Specialized Neuroscience Program in NeuroAIDS, and Idalí Martínez for her helpful suggestions. This work was supported by U54NS430, MBRS-SCORE-SO6GMO822, RCMI-CRC P20RR11126, RCMI-12-RR03051, and MBRS-RISE GM061838. The NIH AIDS Research and Reference Reagent Program is acknowledged for providing the HIV-1 SF162 viral isolate.
Footnotes
Declaration of interest: The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- (AAN criteria). American Academy of Neurology AIDS Task Force. Nomenclature and research case definitions for neurologic manifestations of human immunodeficiency virus-type 1 (HIV-1) infection. Report of a working group of the American Academy of Neurology AIDS task force. Neurology. 1991;41:778–785. doi: 10.1212/wnl.41.6.778. [DOI] [PubMed] [Google Scholar]
- (AAN criteria). American Academy of Neurology AIDS Task Force. Clinical confirmation of the American Academy of Neurology algorithm for HIV-1-associated cognitive/motor disorder. The Dana Consortium on Therapy for HIV Dementia and Related Cognitive Disorders. Neurology. 1996;47:1247–1253. doi: 10.1212/wnl.47.5.1247. 1996. [DOI] [PubMed] [Google Scholar]
- Aksenov MY, Aksenova MV, Butterfield DA, Geddes JW, Markesbery WR. Protein oxidation in the brain in Alzheimer’s disease. Neuroscience. 2001;103:373–383. doi: 10.1016/s0306-4522(00)00580-7. [DOI] [PubMed] [Google Scholar]
- Anderson E, Zink W, Xiong H, Gendelman HE. HIV-1-associated dementia: a metabolic encephalopathy perpetrated by virus-infected and immune-competent mononuclear phagocytes. J Acquir Immune Defic Syndr. 2002;31(Suppl 2):S43–S54. doi: 10.1097/00126334-200210012-00004. [DOI] [PubMed] [Google Scholar]
- Antinori A, Arendt G, Becker JT, Brew BJ, Byrd DA, Cherner M, Clifford DB, Cinque P, Epstein LG, Goodkin K, Gisslen M, Grant I, Heaton RK, Joseph J, Marder K, Marra CM, McArthur JC, Nunn M, Price RW, Pulliam L, Robertson KR, Sacktor N, Valcour V, Wojna VE. Updated research nosology for HIV-associated neuro-cognitive disorders. Neurology. 2007;69:1789–1799. doi: 10.1212/01.WNL.0000287431.88658.8b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arroyo MA, Tien H, Pagán M, Swanstrom R, Hillyer GV, Cadilla CL, Meléndez-Guerrero LM. Virologic risk factors for vertical transmission of HIV type 1 in Puerto Rico. AIDS Res Hum Retroviruses. 2002;18:447–460. doi: 10.1089/088922202753614218. [DOI] [PubMed] [Google Scholar]
- Ballabh P, Braun A, Nedergaard M. The blood-brain barrier: an overview: structure, regulation, and clinical implications. Neurobiol Dis. 2004;16:1–13. doi: 10.1016/j.nbd.2003.12.016. [DOI] [PubMed] [Google Scholar]
- Beyer S, Walter Y, Hellmann J, Kramer PJ, Kopp-Schneider A, Kroeger M, Ittrich C. Comparison of software tools to improve the detection of carcinogen induced changes in the rat liver proteome by analyzing SELDI-TOF-MS spectra. J Proteome Res. 2006;5:254–261. doi: 10.1021/pr050279o. [DOI] [PubMed] [Google Scholar]
- Bubici C, Papa S, Pham CG, Zazzeroni F, Franzoso G. The NF-kappaB-mediated control of ROS and JNK signaling. Histol Histopathol. 2006;21:69–80. doi: 10.14670/HH-21.69. [DOI] [PubMed] [Google Scholar]
- Carlson KA, Ciborowski P, Schellpeper CN, Biskup TM, Shen RF, Luo X, Destache CJ, Gendelman HE. Proteomic fingerprinting of HIV-1-infected human monocyte-derived macrophages: a preliminary report. J Neuroimmunol. 2004;147:35–42. doi: 10.1016/j.jneuroim.2003.10.039. [DOI] [PubMed] [Google Scholar]
- Chen W, Tang Z, Fortina P, Patel P, Addya S, Surrey S, Acheampong EA, Mukhtar M, Pomerantz RJ. Ethanol potentiates HIV-1 gp120-induced apoptosis in human neurons via both the death receptor and NMDA receptor pathways. Virology. 2005;334:59–73. doi: 10.1016/j.virol.2005.01.014. [DOI] [PubMed] [Google Scholar]
- Chertova E, Chertov O, Coren LV, Roser JD, Trabey CM, Bess JW, Jr, Sowder RC, 2nd, Barsov E, Hood BL, Fisher RJ, Nagashima K, Conrads TP, Veenstra TD, Lifson JD, Ott DE. Proteomic and biochemical analysis of purified human immunodeficiency virus type 1 produced from infected monocyte-derived macrophages. J Virol. 2006;80:9039–9052. doi: 10.1128/JVI.01013-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciborowski P, Kadiu I, Rozek W, Smith L, Bernhardt K, Fladseth M, Ricardo-Dukelow M, Gendelman HE. Investigating the human immunodeficiency virus type 1-infected monocyte-derived macrophage secretome. Virology. 2007;363:198–209. doi: 10.1016/j.virol.2007.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartner S. HIV infection and dementia. Science. 2000;287:602–604. doi: 10.1126/science.287.5453.602. [DOI] [PubMed] [Google Scholar]
- Gendelman HE, Orenstein JM, Martin MA, Ferrua C, Mitra R, Phipps T, Wahl LA, Lane HC, Fauci AS, Burke DS, et al. Efficient isolation and propagation of human immunodeficiency virus on recombinant colony-stimulating factor 1-treated monocytes. J Exp Med. 1988;167:1428–1441. doi: 10.1084/jem.167.4.1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendelman HE, Persidsky Y, Ghorpade A, Limoges J, Stins M, Fiala M, Morrisett R. The neuropathogenesis of the AIDS dementia complex. AIDS. 1997;11 (Suppl A):S35–S45. [PubMed] [Google Scholar]
- Glanzer JG, Enose Y, Wang T, Kadiu I, Gong N, Rozek W, Liu J, Schlautman JD, Ciborowski PS, Thomas MP, Gendelman HE. Genomic and proteomic micro-glial profiling: pathways for neuroprotective inflammatory responses following nerve fragment clearance and activation. J Neurochem. 2007;102:622–645. doi: 10.1111/j.1471-4159.2007.04568.x. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Scarano F, Martin-Garcia J. The neuropathogenesis of AIDS. Nat Rev Immunol. 2005;5:69–81. doi: 10.1038/nri1527. [DOI] [PubMed] [Google Scholar]
- Holguin A, Frank MG, Biedenkapp JC, Nelson K, Lippert D, Watkins LR, Rudy JW, Maier SF. Characterization of the temporo-spatial effects of chronic bilateral intrahippocampal cannulae on interleukin-1beta. J Neurosci Methods. 2007;161:265–272. doi: 10.1016/j.jneumeth.2006.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber W, von Heydebreck A, Sultmann H, Poustka A, Vingron M. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics. 2002;18(Suppl 1):S96–S104. doi: 10.1093/bioinformatics/18.suppl_1.s96. [DOI] [PubMed] [Google Scholar]
- Kadiu I, Ricardo-Dukelow M, Ciborowski P, Gendelman HE. Cytoskeletal protein transformation in HIV-1-infected macrophage giant cells. J Immunol. 2007;178:6404–6415. doi: 10.4049/jimmunol.178.10.6404. [DOI] [PubMed] [Google Scholar]
- Kanmogne GD, Schall K, Leibhart J, Knipe B, Gendelman HE, Persidsky Y. HIV-1 gp120 compromises blood-brain barrier integrity and enhances monocyte migration across blood-brain barrier: implication for viral neuropathogenesis. J Cereb Blood Flow Metab. 2007;27:123–134. doi: 10.1038/sj.jcbfm.9600330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laspiur JP, Anderson ER, Ciborowski P, Wojna V, Rozek W, Duan F, Mayo R, Rodriguez E, Plaud-Valentin M, Rodriguez-Orengo J, Gendelman HE, Meléndez LM. CSF proteomic fingerprints for HIV-associated cognitive impairment. J Neuroimmunol. 2007;192:157–170. doi: 10.1016/j.jneuroim.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Liu QH, Tomkowicz B, Yi Y, Freedman BD, Collman RG. Macrophage activation through CCR5- and CXCR4-mediated gp120-elicited signaling pathways. J Leukoc Biol. 2003;74:676–682. doi: 10.1189/jlb.0503206. [DOI] [PubMed] [Google Scholar]
- Li W, Galey D, Mattson MP, Nath A. Molecular and cellular mechanisms of neuronal cell death in HIV dementia. Neurotox Res. 2005;8:119–134. doi: 10.1007/BF03033824. [DOI] [PubMed] [Google Scholar]
- Luo X, Carlson KA, Wojna V, Mayo R, Biskup TM, Stoner J, Anderson J, Gendelman HE, Melendez LM. Macrophage proteomic fingerprinting predicts HIV-1-associated cognitive impairment. Neurology. 2003;60:1931–1937. doi: 10.1212/01.wnl.0000064396.54554.26. [DOI] [PubMed] [Google Scholar]
- McArthur JC. HIV dementia: an evolving disease. J Neuroimmunol. 2004;157:3–10. doi: 10.1016/j.jneuroim.2004.08.042. [DOI] [PubMed] [Google Scholar]
- McArthur JC, Sacktor N, Seines O. Human immunodeficiency virus-associated dementia. Semin Neurol. 1999;19:129–150. doi: 10.1055/s-2008-1040831. [DOI] [PubMed] [Google Scholar]
- Munoz LE, Franz S, Pausch F, Furnrohr B, Sheriff A, Vogt B, Kern PM, Baum W, Stach C, von Laer D, Brachvogel B, Poschl E, Herrmann M, Gaipl US. The influence on the immunomodulatory effects of dying and dead cells of annexin V. J Leukoc Biol. 2007a;81:6–14. doi: 10.1189/jlb.0306166. [DOI] [PubMed] [Google Scholar]
- Nieves DM, Plaud M, Wojna V, Skolasky R, Melendez L. Characterization of peripheral blood human immunodeficiency virus isolates from Hispanic women with cognitive impairment. J NeuroVirol. 2007;13:315–327. doi: 10.1080/13550280701361508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omenn GS, States DJ, Adamski M, Blackwell TW, Menon R, Hermjakob H, Apweiler R, Haab BB, Simpson RJ, Eddes JS, Kapp EA, Moritz RL, Chan DW, Rai AJ, Admon A, Aebersold R, Eng J, Hancock WS, Hefta SA, Meyer H, Paik YK, Yoo JS, Ping P, Pounds J, Adkins J, Qian X, Wang R, Wasinger V, Wu CY, Zhao X, Zeng R, Archakov A, Tsugita A, Beer I, Pandey A, Pisano M, Andrews P, Tammen H, Speicher DW, Hanash SM. Overview of the HUPO Plasma Proteome Project: results from the pilot phase with 35 collaborating laboratories and multiple analytical groups, generating a core dataset of 3020 proteins and a publicly-available database. Proteomics. 2005;5:3226–3245. doi: 10.1002/pmic.200500358. [DOI] [PubMed] [Google Scholar]
- Persidsky Y, Stins M, Way D, Witte MH, Weinand M, Kim KS, Bock P, Gendelman HE, Fiala M. A model for monocyte migration through the blood-brain barrier during HIV-1 encephalitis. J Immunol. 1997;158:3499–3510. [PubMed] [Google Scholar]
- Renta JY, Cadilla CL, Vega ME, Hillyer GV, Estrada C, Jiménez E, Abreu E, Méndez I, Gandia J, Meléndez-Guerrero LM. Longitudinal studies on maternal HIV-1 variants by biological phenotyping, sequence analysis and viral load. Cell Mol Biol (Noisy-le-grand) 1997;43:1097–1114. [PubMed] [Google Scholar]
- Reynolds A, Laurie C, Mosley RL, Gendelman HE. Oxidative stress and the pathogenesis of neurodegenerative disorders. Int Rev Neurobiol. 2007;82:297–325. doi: 10.1016/S0074-7742(07)82016-2. [DOI] [PubMed] [Google Scholar]
- Schafer U, Seibold S, Schneider A, Neugebauer E. Isolation and characterisation of the human rab18 gene after stimulation of endothelial cells with histamine. FEBS Lett. 2000;466:148–154. doi: 10.1016/s0014-5793(99)01778-0. [DOI] [PubMed] [Google Scholar]
- Self RL, Mulholland PJ, Nath A, Harris BR, Prendergast MA. The human immunodeficiency virus type-1 transcription factor Tat produces elevations in ultra-cellular Ca2+ that require function of an N-methyl-D-aspartate receptor polyamine-sensitive site. Brain Res. 2004;995:39–45. doi: 10.1016/j.brainres.2003.09.052. [DOI] [PubMed] [Google Scholar]
- Stehlik C, Fiorentino L, Dorfleutner A, Bruey JM, Ariza EM, Sagara J, Reed JC. The PAAD/PYRIN-family protein ASC is a dual regulator of a conserved step in nuclear factor kappaB activation pathways. J Exp Med. 2002;196:1605–1615. doi: 10.1084/jem.20021552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD. The positive false discovery rate: a Bayesian interpretation and the q-value. Ann Stat. 2003;31:2013–2035. [Google Scholar]
- Sun B, Rempel HC, Pulliam L. Loss of macrophage-secreted lysozyme in HIV-1 associated dementia detected by SELDI-TOF mass spectrometry. AIDS. 2004;18:1009–1012. doi: 10.1097/00002030-200404300-00008. [DOI] [PubMed] [Google Scholar]
- Tsuji Y. JunD activates transcription of the human ferritin H gene through an antioxidant response element during oxidative stress. Oncogene. 2005;24:7567–7578. doi: 10.1038/sj.onc.1208901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez N, Greenwell-Wild T, Marines NJ, Swaim WD, Nares S, Ott DE, Schubert U, Henklein P, Orenstein JM, Sporn MB, Wahl SM. Human immunodeficiency virus type 1-induced macrophage gene expression includes the p21 gene, a target for viral regulation. J Virol. 2005;79:4479–4491. doi: 10.1128/JVI.79.7.4479-4491.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velásquez I, Plaud M, Perez Laspiur J, Wojna V, Skolasky R, Ciborowski P, Gendelman HE, Melendez LM. Dysregulation of Cu/Zn superoxide disrmutase-1 in the CSF and monocytes of Hispanic women with cognitive impairment. J NeuroVirol. 2007;13(Suppl 1):131. [Google Scholar]
- Wahl SM, Greenwell-Wild T, Peng G, Ma G, Orenstein JM, Vazquez N. Viral and host cofactors facilitate HIV-1 replication in macrophages. J Leukoc Biol. 2003;74:726–735. doi: 10.1189/jlb.0503220. [DOI] [PubMed] [Google Scholar]
- Wojna V, Carlson KA, Luo X, Mayo R, Meléndez LM, Kraiselburd E, Gendelman HE. Proteomic fingerprinting of human immunodeficiency virus type 1-associated dementia from patient monocyte-derived macrophages: a case study. J NeuroVirol. 2004;10(Suppl 1):74–81. doi: 10.1080/753312756. [DOI] [PubMed] [Google Scholar]
- Wojna V, Skolasky RL, Hechavarria R, Mayo R, Selnes O, McArthur JC, Melendez LM, Maldonado E, Zorrilla CD, Garcia H, Kraiselburd E, Nath A. Prevalence of human immunodeficiency virus-associated cognitive impairment in a group of Hispanic women at risk for neurological impairment. J NeuroVirol. 2006;12:356–364. doi: 10.1080/13550280600964576. [DOI] [PubMed] [Google Scholar]
- Xie Q, Tianxin L, Zhang Y, Zheng J, Bonanno JA. Molecular cloning and characterization of a human AIF-like gene with ability to induce apoptosis. J Biol Chem. 2005;280:19673–19681. doi: 10.1074/jbc.M409517200. [DOI] [PubMed] [Google Scholar]
- Zhang M, Li X, Pang X, Ding L, Wood O, Clouse K, Hewlett I, Dayton AI. Identification of a potential HIV-induced source of bystander-mediated apoptosis in T cells: upregulation of trail in primary human macrophages by HIV-1 tat. J Biomed Sci. 2001;8:290–296. doi: 10.1007/BF02256603. [DOI] [PubMed] [Google Scholar]
- Zybarth G, Reiling N, Schmidtmayerova H, Sherry B, Bukrinsky M. Activation-induced resistance of human macrophages to HIV-1 infection in vitro. J Immunol. 1999;162:400–406. [PubMed] [Google Scholar]


