Abstract
It is well documented that placental macrophages show lower levels of HIV-1 infection than monocyte-derived macrophages (MDM). We used proteomic methods to test the hypothesis that placental macrophages secrete different proteins as compared to MDM that may contribute to decreased HIV-1 replication. Placental macrophages and MDM were cultured for 12 days and supernatant was collected. To characterize supernatants, the protein profiles of placental macrophages and MDM were compared using the protein chip assay. Subsequently, proteins were separated by one-dimensional gel electrophoresis and identified by tandem mass spectrometry at the corresponding mass to charge (m/z) range of 5000–20,000. Significant differences were found between placental macrophages and MDM in seven protein peaks with m/z values of 6075, 6227, 11,662, 14,547, 6158, 7740, and 11,934 on the CM10 and IMAC chips. After sequencing and identification, five proteins were validated for differential expression in placental macrophages and MDM by Western blot analyses. Peroxiredoxin 5, found to be more abundant in placental macrophage supernatants, is important in the cellular antioxidant mechanisms, and other members of its family have shown antiviral activity. Cystatin B was less abundant in PM supernatant, and decreased intracellular levels have recently been shown to be associated with lower HIV-1 replication in placental macrophages than in MDM. This study elucidates for the first time the placental macrophage secretome corresponding to 5000–20,000 Da and advances our understanding of the proteins secreted in the placenta that can protect the fetus against HIV-1 and other viral infections.
Keywords: Placental, macrophages, Proteomics, Secretome
1. Introduction
The placenta is a natural barrier to numerous bacterial and viral infections. It is known that viruses like Hepatitis B, C, and Herpes Simplex have a very low rate of transmission across the placenta [1]. Even HIV-1 which is known to cross the placenta has a low rate of vertical transmission ranging from 10% to 25% without antiviral treatment [2]. Studies done on infants born to HIV-1 positive mothers suggest that 80% of transmissions occur at the moment of birth [3]. This demonstrates that the placenta limits viral replication including HIV-1.
Immune cells in the maternal–fetal interface are specialized for this environment, and have been shown to possess different phenotypes than their counterparts in the blood [4–6]. A number of placental products have been found to inhibit bacterial growth and viral replication [7–10], suggesting that factors produced by placental cells are important in the protection of the fetus against infection. Soluble suppressor factors are important innate defenses because they are able to inhibit viral replication without killing the cells. Soluble factors can inhibit different cells and viral strains simultaneously.
Inside the placenta the primary cells involved in the defense against infections are the placental macrophages (PM) [11]. Despite their protective role these cells are the primary targets for HIV-1 in the placenta. They can be productively infected with HIV-1 laboratory strains and primary isolates and express CCR5 and CXCR4 co-receptors [12–18]. However, PM are less susceptible than monocyte-derived macrophages (MDM), express lower levels of CD4 [12] and CCR5 co-receptors that may contribute to decreased susceptibility [19] When stimulated with LPS, PM produce fewer pro-inflammatory cytokines than do MDM. Despite these differences, the level of PM cytokines does not correlate with HIV-1 production [17]. However, PM and MDM produce similar levels of β-chemokines, suggesting that chemokine levels do not contribute to decreased HIV-1 replication [19]. We hypothesized that PM produce different levels of other soluble proteins than do MDM, some of which could contribute to their decreased susceptibility to HIV-1 infection. To test this hypothesis, we utilized surface-enhanced laser desorption ionization time of flight (SELDI-TOF) protein chip assays and LC–MS/MS sequencing to compare protein profiles from PM and MDM supernatants. We found differences in the expression of several important soluble proteins that could provide insights into the mechanisms of inhibition occurring in the placenta.
2. Materials and methods
2.1. Isolation and culture of PM
Full-term placentas from HIV-1, HIV-2 and Hepatitis B seronegative healthy mothers were obtained from the San Juan City Hospital. Placentas with chorioamnionitis were not collected for the study. This study was approved by the University of Puerto Rico Medical Sciences Campus IRB and considered exempt, as there were no identifiers. PM were isolated following the method described before [20,16]. This method yields >90% macrophages [17–19]. In short, several lobules were perfused for 45 min via the fetal side using a solution containing 90% PBS, 5% antimycotic (Sigma, St. Louis, MO), and 5% heparin (1000 U/mL Sigma) in order to eliminate maternal blood. PM were isolated by various enzymatic digestions followed by Ficoll density gradient centrifugation. Cells were then seeded in a 75 cm2 flask at a concentration of 1.5 × 106 cells/mL and supplemented with monocyte medium (70% RPMI, 10% human serum, 20% FBS (Sigma)). Culture medium was exchanged at days 6 and 12 of culture. Cells were washed four times with PBS and cultured in serum-free Dulbecco’s modified Eagle’s medium (DMEM) (Invitrogen) for 24 h prior to supernatant collection for proteomic analysis. PM cultures were monitored for cytopathic effects.
2.2. Isolation and culture of MDM
Peripheral blood mononuclear cells (PBMC) were obtained from the blood of HIV-1 seronegative donors by Ficoll density gradient centrifugation. Cells were plated at a concentration of 1.5 × 106 cells/mL per flask and cultivated in monocyte medium (70% RPMI, 20% FBS, 10% human serum). Monocytes were purified by preferential adherence. Culture medium was exchanged at days 6 and 12 of culture. Cells were, washed four times with PBS, and cultured in serum-free DMEM for 24 h prior to supernatant collection for proteomic analysis. Visual inspection showed adherent cells with no cytopathic effects and the typical elongated morphology at the time of supernatant collection.
2.3. Sample processing for SELDI-TOF profiling
Collected supernatants were centrifuged at 300 × g for 10 min to remove cells from the fluid. Samples were dried and concentrated by centrifugation using a Speed Vac. After concentration, pellets containing proteins were resuspended in lysis buffer (5 mM Tris–HCl buffer, pH 8.0, 0.1% Triton X-100) and dialyzed overnight against water at 4 °C using QuixSep® micro dialyzers (Membrane Filtration Products Inc., Seguin, TX). Protein concentration was determined by using the Bio-Rad BCA assay (Bio-Rad, Hercules, CA).
2.4. SELDI-TOF profiling
Protein Chip® analysis was performed using CM10 (weak cationic exchange) and IMAC30 (metal affinity) chips (Ciphergen Biosystems, Palo Alto, CA). For all experiments, chips were assembled in a 96-well bio-processor; spots were equilibrated with binding buffer (100 mM ammonium acetate, pH 4.0, with Triton X-100 for CM10 chips, and 0.1 M sodium phosphate, 0.5 M NaCl, pH 7.0, for IMAC30 chips). Thereafter, chips were incubated for 5 min at room temperature with shaking. Equal amounts of protein (0.05 μg) were applied to each spot, and the bio-processor was incubated for 30 min at room temperature with shaking. Cationic proteins bind to the CM10 chip while metal (copper)-binding proteins bind to the IMAC30 chip. Unbound proteins were removed by washing with binding buffer and HPLC-grade water. Sinapinic acid (SPA) (Ciphergen Biosystems) was used as the energy-absorbing molecule (EAM). A 50% solution of SPA was prepared using a solvent that contains 100% acetonitrile (ACN) (Sigma), 100% isopropanol (Fisher, Pittsburgh, PA), 1% trifluoroacetic acid (TFA, Sigma), and 10% Triton X-100. Spots were covered with 1 μL of the 50% SPA solution and allowed to air dry. The Protein Chip® Reader was calibrated using a chip with six protein standards of known molecular weights. Mass spectra were collected with the Protein Chip system in the range of 5000–20,000 m/z and peaks were analyzed using the Biomarker Wizard provided within the Protein Chip® software 3.2. Each sample was run in quadruplicate.
2.5. 1D SDS-PAGE and trypsin digestion
PM and MDM supernatant fractionation by SDS-PAGE was performed using NuPAGE 12% gradient gel (Invitrogen, Carlsbad, CA). Twenty (20) μg of protein was loaded per lane. After running, the gel was fixed overnight, stained with Coomassie BB (Bio-Rad) for 3–4 h, destained, and digitized using a scanner. Bands were excised from the gel, stored in micro-tubes, and dried by washing two times with 10 mLof ACN. Samples were dried on a vacuum concentrator and distained with 50%, 50 mM NH4HCO3/50% ACN, and 10 mM NH4HCO3/50% ACN, dried and incubated with trypsin (Promega, Madison, WI) overnight at 37 °C. Peptides were extracted by washing gel pieces with 60% ACN and 0.1% trifluoroacetic acid (TFA), transferred to a glass tube and dried. Samples were purified using C18 Zip Tip (Millipore, Billerica, MA) before LC–MS/MS analysis.
2.6. Protein identification
Samples were resuspended in 0.1% formic acid (EMD Biosciences) in HPLC-grade water and peptides were sequenced using an LTQ XL system with a Proteome X configuration (Thermo Electron Corporation, San Jose, CA) as previously described [21]. Data obtained from the LC–MS/MS analysis were matched against the NCBI human fasta protein database using BioWorks 3.2SR (ThermoElectron, Inc.). Protein identifications were accepted as true positives using the following criteria: BioWorks® unified score ≥ 3000, Xcorr for singly charged precursor ion ≥ 2, Xcorr for doubly charged precursor ion ≥ 2.5, Xcorr for triply charged precursor ion ≥ 3, ΔCn ≥ 0.3, and containing more than 60% of fragment ions per sequenced peptide.
2.7. Western blot and ELISA analyses
Samples were separated on 10–20% Tris–HCl Ready Gel and later transferred to a nitrocellulose membrane (Bio-Rad, Hercules, CA) using the semi-dry transfer system. After transfer, the membrane was incubated in blocking buffer (3% BSA in Tris-Buffered Saline + Tween 20 (Fisher)) overnight at 4 °C. Membranes were incubated at 4 °C overnight with the following primary antibodies: monoclonal mouse anti-human peroxiredoxin 5 (AbCam Cambridge, MA, 1:500), polyclonal rabbit anti-human D4-GDI (BD Bioscience, San Jose, CA, 1:1000), and polyclonal rabbit anti-human enolase-1 (Santa Cruz Biotechnologies, 1:2000). Membranes were incubated for 1 h at room temperature with horseradish peroxidase enzyme (HRP)-conjugated secondary antibody at a 1:5000 dilution. Secondary antibodies used were goat anti-mouse (Santa Cruz Biotechnologies, Santa Cruz, CA) and goat anti-rabbit (Pierce, Rockford, IL). Proteins were detected using the Supersignal West Femto system (Pierce). Signal and density of the bands were analyzed using the VersaDoc Imaging System (Bio-Rad Laboratories). Uniformity of the bands was confirmed by Ponceau S staining of the membranes.
MIP-4 levels in PM and MDM supernatants were measured by ELISA using a commercially available CytoSet (Biosource). Assay conditions were as described by the manufacturer.
2.8. Statistical analyses
For analysis of SELDI-TOF spectra, data from the Biomarker wizard were exported to Microsoft Excel and then analyzed using SAS® software 9.0 (SAS Institute, Cary, NC). Analysis of variance (ANOVA) was used to identify peaks with significant differences in the distribution of intensity scores among the groups. The raw intensity values were transformed using the following function: , where ‘X’ is the observed intensity. This was done to stabilize the intensity variance and make the data more normally distributed. It has the advantage (over a log-transformation) that allows negative intensities [22,23]. Each comparison was made using generalized estimating equations (GEE) to account for the correlation among sample replicates by applying the exchangeable correlation structure. The significance level was specified to be 0.05.
For Western blot analyses, an unpaired two-tailed Student’s t test was used to compare the densitometry values of the blots obtained from Quantity One Software (Bio-Rad), and the results were displayed as mean plus one standard error of the mean (SEM). The significance level was specified to be 0.05.
3. Results
3.1. Protein profiling
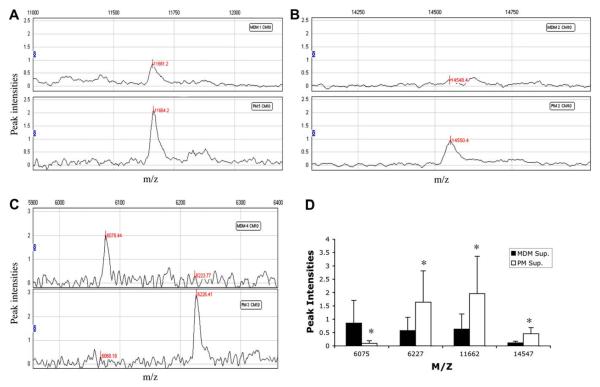
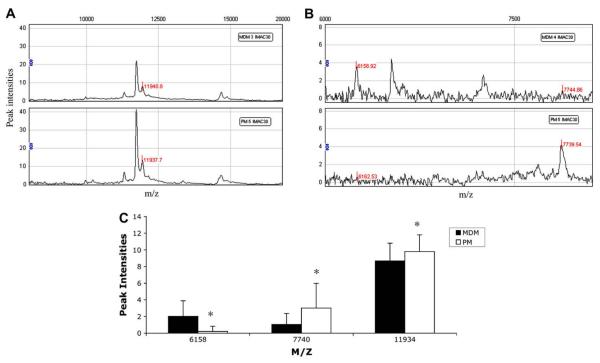
In order to investigate the natural resistance of PM to HIV-1 infection, culture supernatants from PM and MDM collected at day 12 of culture were analyzed using SELDI-TOF to determine protein profiles. The CM10 (weak cationic exchange) and IMAC30 (metal affinity) protein chip arrays were used since these yielded spectra with increased resolution and intensity. Samples from 4 PM and 4 MDM cultures were analyzed using the CM10 chip array. Analysis of the spectra shows significant differences between the two cell types in the expression of four protein peaks with an m/z of 6065, 6227, 11,661, and 14,547. Representative spectra from PM and MDM samples for each differentially expressed peak are shown (Fig. 1A–C). The protein peak of 6065 was underexpressed in PM, and the protein peaks of 6227, 11,661, and 14,547 were overexpressed in PM as compared with MDM (Fig. 1D). Using the IMAC30 chip array samples from 5 PM and 5 MDM cultures were analyzed and significant differences in the expression of three protein peaks were found. Representative spectra from PM and MDM samples for each differentially expressed peak are shown (Fig. 2A and B). The protein peaks with an m/z of 6158 were underexpressed in PM and the 7740 and 11,934 peaks overexpressed in PM as compared with MDM (Fig. 2C).
Fig. 1.
Differentially expressed protein peaks between PM and MDM. SELDI-TOF representative spectra of PM and MDM supernatants on CM10 chip showing protein peaks with m/z of 11,662 (A), 14,547 (B), and 6075, 6227 (C). Spectra analysis of mean intensities of PM and MDM supernatants on CM10 chip (D). *p value <0.05, generalized estimating equations. Results are representative of 4 PM and 4 MDM samples.
Fig. 2.
Differentially expressed peaks between PM and MDM. SELDI-TOF representative spectra of PM and MDM supernatants on IMAC30 chip showing peaks with m/z of 11,934 (A), and 6158, 7740 (B). Spectra analysis of mean intensities of PM and MDM supernatants on IMAC30 chip (C). Results are representative of 5 PM and 5 MDM samples. *p value <0.05, generalized estimating equations.
3.2. Identification of differentially expressed proteins
SELDI-TOF profiling showed differentially expressed peaks between PM and MDM samples. On the basis of these findings, we proceeded to separate supernatants from both cell types by 1D SDS-PAGE in order to identify differentially secreted proteins. After separation, bands corresponding to the molecular weights of interest found by SELDI-TOF were excised. These bands were then digested with trypsin and peptides were sequenced by LC–MS/MS. Proteins were identified with high confidence following the parameters described in Section ***2. In total, 148 proteins were identified. Table 1A shows proteins that were common to both groups and that were identified with at least two peptides. Table 1B shows proteins that were found to be different between PM and MDM and were identified in at least two of the samples. Worth noting are peroxiredoxin 5 (Prx 5), MIP-4, α-enolase, Rho-GDI 2, fatty acid-binding protein (FAB)-3, immunophilin FKBP 12, and thioredoxin. Peroxiredoxin 5 and MIP-4 were identified mostly in PM samples while α-enolase and Rho-GDI 2 were identified only in MDM. Three identified proteins had molecular masses that corresponded to protein peaks that were overexpressed in PM over MDM by SELDI-TOF analyses (Table 2); these were fatty acid-binding protein 3 (FAB; 14,858 Da) corresponding to the 14,547 m/z SELDI-TOF protein peak, FKBP 12 (11,951 Da) corresponding to the 11,934 m/z protein peak, and thioredoxin (11,737 Da), another antioxidant protein in PM that closely correlates with the 11,662 m/z protein peak.
Table 1A.
Proteins identified as common in PM and MDM supernatants.
| Proteina | Peptides |
MWb | SwissProtc | |
|---|---|---|---|---|
| PM | MDM | |||
| Redox | ||||
| Glutathione transferase | 2 | 3 | 23,355 | P09211 |
| Superoxide dismutase [Cu–Zn] | 1 | 1 | 15,918 | P00441 |
| Ferritin, light polypeptide | 3 | 3 | 20,019 | P02792 |
| Peroxiredoxin 1 | 2 | 3 | 22,110 | Q06830 |
| Peroxiredoxin 3 | 1 | 2 | 27,693 | P30048 |
| Thioredoxin | 2 | 2 | 11,721 | P10599 |
| Enzyme | ||||
| Cyclophilin B | 3 | 3 | 22,742 | P23284 |
| Cyclophilin A | 3 | 2 | 17,881 | P62937 |
| Metabolic | ||||
| Glyceraldehyde-3-phosphate dehydrogenase |
2 | 2 | 36,053 | P04406 |
| Pyruvate kinase 3 isoform 1 | 2 | 6 | 57,937 | P14618 |
| Apolipoprotein E precursor | 2 | 2 | 36,154 | P02649 |
| Phosphoglycerate kinase 1 | 1 | 2 | 44,614 | P00558 |
| Fatty acid-binding protein 3 | 1 | 2 | 14,858 | P05413 |
| Apolipoprotein A-I preproprotein | 2 | 3 | 30,777 | P02647 |
| Structural | ||||
| Cofilin 1 | 3 | 3 | 18,502 | P23528 |
| Vimentin | 4 | 6 | 53,652 | P08670 |
| Profilin-1 | 4 | 4 | 14,852 | P07737 |
| Myosin regulatory light chain Mrcl2 | 2 | 3 | 19,779 | P19105 |
| Filamin-A | 1 | 2 | 280,757 | P21333 |
| Actin-related protein 2/3 complex subunit 4 |
2 | 3 | 19,667 | P59998 |
| Actin, cytoplasmic 1 | 1 | 2 | 41,777 | P60709 |
| Regulatory | ||||
| Ribosomal protein P2 | 2 | 2 | 11,665 | P05387 |
| S100 calcium-binding protein A9 | 1 | 2 | 13,242 | P06702 |
| Cystatin A | 1 | 2 | 11,006 | P01040 |
| Cystatin B | 2 | 3 | 11,140 | P04080 |
| Dual specificity phosphatase 3 | 2 | 1 | 20,478 | P51452 |
| Interleukin 1 receptor antagonist isoform 1 precursor |
1 | 2 | 20,055 | P18510 |
| Myosin, light polypeptide 6 | 2 | 2 | 16,961 | P60660 |
| MIP-4 | 1 | 1 | 9849 | P55774 |
Proteins included in this table were identified with 2 or more peptides.
Molecular weight.
SwissProt accession number.
Table 1B.
Proteins identified as unique to PM or MDM supernatant.
| Proteina | PM | MDM | MWb | SwissProtc |
|---|---|---|---|---|
| Regulatory | ||||
| Rho GDP dissociation inhibitor beta | − | + | 22,987 | P52566 |
| Transmembrane protein 4 | + | − | 20,652 | Q9Y2B0 |
| Destrin isoform a | + | − | 18,506 | P60981 |
| Cold inducible RNA-binding protein | + | − | 18,648 | Q14011 |
| Thrombospondin-1 precursor | + | − | 129,412 | P07996 |
| Structural | ||||
| Histone H2B | − | + | 13,936 | Q99877 |
| H2A histone member Z | − | + | 13,552 | P0C0S5 |
| Tubulin beta | − | + | 48,879 | P07437 |
| Tubulin alpha 6 | − | + | 49,895 | Q9BQE3 |
| Tubulin beta-2C chain | − | + | 49,831 | P68371 |
| Glial fibrillary acidic protein | − | + | 49,880 | P14136 |
| Metabolic | ||||
| Enolase-1 | − | + | 47,168 | P06733 |
| Fatty acid-binding protein 4 | − | + | 14718.8 | P15090 |
| Redox | ||||
| Peroxiredoxin 5 | + | − | 22,026 | P30044 |
Proteins included in this table were identified in at least 2 samples.
Molecular weight.
SwissProt accession number.
Table 2.
Identified proteins correlating with differentially expressed SELDI-TOF peaks.
| m/z SELDI-TOF | MW | Protein | Function |
|---|---|---|---|
| 11,662 | 11,721 | Thioredoxin | Participates in various redox reactions |
| 11,934 | 11,803 | Immunophilin FKBP 12 |
PPIases accelerate the folding of proteins |
| 14,547 | 14,857 | Fatty acid-binding protein 3 |
FABP plays a role in the intracellular transport of long-chain fatty acids and their acyl-CoA esters |
3.3. Validation of differentially expressed proteins
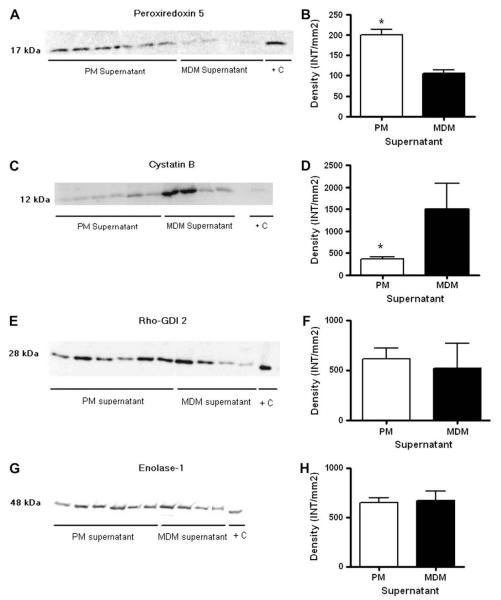
After protein identification we chose four proteins for validation (peroxiredoxin 5, MIP-4, Rho-GDI 2, and α-enolase) that were differentially expressed by tandem mass spectrometry and one (Cystatin B) for which the relative abundance of sequenced peptides was higher in MDM supernatants (Fig. 3). Proteins were selected if they were identified with high confidence in samples from at least two donors, and based on their relationship to the immune response. Cystatin B was chosen based on previous studies that found it to be more abundant in MDM than PM cell lysates [24]. Western blot analysis showed peroxiredoxin 5 to be significantly more abundant in supernatants from PM than from MDM (Fig. 3A and B), whereas cystatin B was found to be significantly more abundant in MDM supernatants (Fig. 3C and D). However, Rho-GDI 2 and α-enolase were found to be in equal abundance in supernatants of both cell types (Fig. 3E–H).
Fig. 3.
Western blot and densitometry analysis of peroxiredoxin 5 (A, B), cystatin B (C, D) Rho-GDI 2 (E, F), and enolase-α (G, H). 20 μg of protein from PM and MDM supernatant was loaded per lane. Densitometry analysis is the average of two replicate gels with 6 PM and 4 MDM samples. *p value <0.05, t-test.
The results from MIP-4 ELISA showed that MDM supernatants have significantly higher levels of this protein than do PM supernatants (Fig. 4).
Fig. 4.
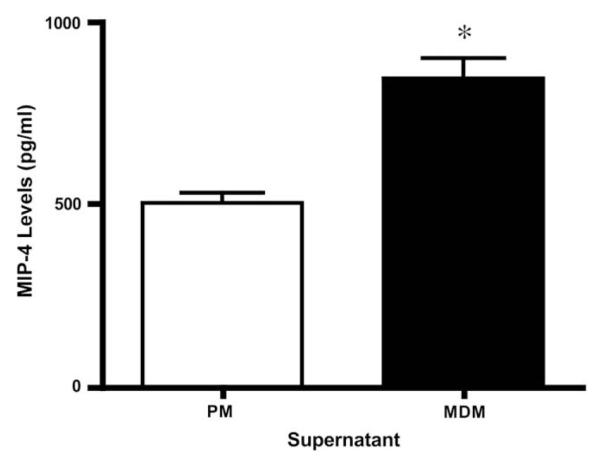
MIP-4 protein levels. Protein levels in PM and MDM supernatants were measured by ELISA. Values are the average of 6 PM and 4 MDM samples. *p value <0.05, t-test.
4. Discussion
This study is unique because it explores the secretome of PM, an important cell type that protects the fetus against infection during pregnancy. By applying “state of the art” proteomics approaches to compare the secretomes of PM and MDM, we could identify novel soluble factors that would help to elucidate the role of PM in the inhibition of HIV-1 infection occurring in PM and gain a better understanding of the mechanisms of HIV-1 restriction in macrophages. Supernatant from PM in their basal state was used in this study because we wanted to characterize the PM secretome as it would be in a normal placenta. Proteomics has been used successfully to study the proteome and secretome of HIV-1-infected macrophages [21,25,26]. In the latter work the authors found differences between control and HIV-1-infected MDM in secreted proteins having a wide range of functions including structural, redox, enzymatic and regulatory functions. In the current study, we explored differences between PM and MDM using SELDI-TOF technology as an initial screening tool since it allows us to rapidly screen the secretome of the cell at the 5–20 kDa m/z range. Using this technology, we identified seven differentially expressed protein peaks, a finding that confirms our hypothesis that PM and MDM secrete different proteins. The majority of these peaks (71%) were seen overexpressed in PM. Differences were seen in metal-binding and positive-charged proteins, which may have a diverse range of biological functions.
After confirming our hypothesis by using SELDI-TOF, we proceeded to separate PM and MDM samples by 1D SDS-PAGE and sequenced the trypsinized samples corresponding to the SELDI-TOF area of interest by LC–MS/MS in order to identify differentially expressed proteins between the two cell types. Five proteins were chosen for validation on the basis of their absence in one cell type relative to the other and their importance in immune processes. Western blots of two of the proteins, α-enolase, which has been found to be increased in MDM after HIV-1 infection, and Rho-GDI 2, important in signal transduction in T-cells and macrophages, showed similar abundance in supernatant from PM and MDM. However, peroxiredoxin 5 was found to be significantly more abundant in supernatant from PM. Peroxiredoxins (Prxs) are a family of antioxidant molecules that eliminate H2O2 from the cell by transferring electrons from thioredoxin. Prx 5 has been found to be widely distributed in mammalian cell lines and to be localized to the cytosol, mitochondria, and peroxisomes [27]. It has also been shown to be secreted by THP-1 cells, a monocytic cell line [28]. Peroxiredoxin 3 was found to be up-regulated in mouse placenta upon stimulation with LPS, suggesting a role for Prxs in inflammatory processes in the placenta. Prxs are important in a wide variety of diseases including HIV-1. Prxs 1, 2, and 4 have antiviral activity against HIV-1 [29,30]. Prxs 1 and 6 were found to be associated to HIV-1 virion, and T-cells from HIV-1-exposed uninfected individuals express higher levels of Prx 2 [31,32]. Prxs suppress TNF-α-induced NF-κB transcriptional activity [33], and this has been suggested as the mechanism for their antiviral activity since NF-κB is the main activator of viral transcription [29,30].Prx 5 has been found to negatively regulate TNF-α signaling, and so it could also suppress NF-κB activity. Thus higher expression of Prx 5 in PM could be one of the mechanisms by which HIV-1 replicates inefficiently in these cells.
The finding that cystatin B was more abundant in MDM supernatant confirms previous results using MDM and PM cell lysates found in our laboratory. In that study, downregulation of cystatin B in MDM, using siRNA, significantly reduced HIV-1 replication in these cells, thus suggesting a role for this protein in promoting HIV-1 replication [24]. Cystatin B has also been found to be secreted in higher quantities by HIV-1-infected macrophages than by uninfected controls [21]. This protein increases nitric oxide production in macrophages, and it has been suggested that it does so by upregulation of TNF-α [34,35]. In addition, previous studies have shown that nitric oxide synthesis enhances HIV-1 replication in macrophages and HIV-1-infected T-cells [36,37]. All of these results suggest that lower expression of cystatin B in PM could impair their capacity to replicate HIV-1.
MIP-4 (also known as CCL18) was found in higher levels in MDM supernatant. This protein is chemotactic for CD4+ and CD8+ T-cells [38] and has been found to activate macrophages [39]. MIP-4 was identified in more PM samples but was found by ELISA to be increased more in MDM than in PM. This confirms that LC–MS/MS is very good as an identification method but that protein abundance should be validated by other methods such as Western blot or ELISA.
Taken together, these results are important since for the first time we have elucidated the secretome of PM as compared with MDM and uncovered proteins that could contribute to HIV-1 restriction in the placenta. Future investigations in our laboratory will study the secretome of HIV-1-infected PM as compared to HIV-1-infected MDM in order to determine the effects of the virus on these cells. This will also give us insight into the mechanisms occurring in PM that lead to its lower susceptibility to HIV-1 replication. Proteomics offers novel ways to study the cells in different tissues and allows us to further understand the role of the placenta in fighting infection and protecting the fetus.
Acknowledgments
This work was made possible by Dr. Meléndez’s laboratory staff, and was supported by the following grants from the National Institutes of Health: NIGMS-MBRS-SCORE S06GM0822, NINDS-SNRP-U54NS430, and NCRR-RCMI-G12RR-03051 that funds the UPR-MSC Clinical Proteomics Discovery Core Facility. Ms. Katia Garcia received MBRS-RISE NIH-NIGMS-GM61838 during her graduate studies.
References
- [1].Nahmias A, Abramowsky C, Dobronyi I, Ibegbu C, Henderson S. Infection and immunity at the maternal–placental–fetal interface: focus on HIV-1. Trophoblast Res. 1998;12:103–24. [Google Scholar]
- [2].Anderson VM. The placental barrier to maternal HIV infection. Obstet Gynecol Clin North Am. 1997;24:797–820. doi: 10.1016/s0889-8545(05)70345-4. [DOI] [PubMed] [Google Scholar]
- [3].Mofenson LM. Mother–child HIV-1 transmission: timing and determinants. Obstet Gynecol Clin North Am. 1997;24:759–84. doi: 10.1016/s0889-8545(05)70343-0. [DOI] [PubMed] [Google Scholar]
- [4].Gustafsson C, Mjosberg J, Matussek A, Geffers R, Matthiesen L, Berg G, et al. Gene expression profiling of human decidual macrophages: evidence for immunosuppressive phenotype. PLoS ONE. 2008;3:e2078. doi: 10.1371/journal.pone.0002078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Szekeres-Bartho J. Regulation of NK cell cytotoxicity during pregnancy. Reprod Biomed Online. 2008;16:211–7. doi: 10.1016/s1472-6483(10)60576-7. [DOI] [PubMed] [Google Scholar]
- [6].Kammerer U, Eggert AO, Kapp M, McLellan AD, Geijtenbeek TB, Dietl J, et al. Unique appearance of proliferating antigen-presenting cells expressing DC-SIGN (CD209) in the decidua of early human pregnancy. Am J Pathol. 2003;162:887–96. doi: 10.1016/S0002-9440(10)63884-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Patterson BK, Tjernlund A, Andersson J. Endogenous inhibitors of HIV: potent anti-HIV activity of leukemia inhibitory factor. Curr Mol Med. 2002;2:713–22. doi: 10.2174/1566524023361817. [DOI] [PubMed] [Google Scholar]
- [8].Kondapi AK, Hafiz MA, Sivaram T. Anti-HIV activity of a glycoprotein from first trimester placental tissue. Antiviral Res. 2002;54:47–57. doi: 10.1016/s0166-3542(01)00212-1. [DOI] [PubMed] [Google Scholar]
- [9].Chakraborty PD, Bhattacharyya D. In vitro growth inhibition of microbes by human placental extract. Curr Sci. 2005;88:782–6. [Google Scholar]
- [10].Kim HS, Cho JH, Park HW, Yoon H, Kim MS, Kim SC. Endotoxin-neutralizing antimicrobial proteins of the human placenta. J Immunol. 2002;168:2356–64. doi: 10.4049/jimmunol.168.5.2356. [DOI] [PubMed] [Google Scholar]
- [11].Nahmias AJ, Kourtis AP. The great balancing acts. The pregnant woman, placenta, fetus, and infectious agents. Clin Perinatol. 1997;24:497–521. [PubMed] [Google Scholar]
- [12].Kesson AM, Fear WR, Kazazi F, Mathijs JM, Chang J, King NJ, et al. Human immunodeficiency virus type 1 infection of human placental macrophages in vitro. J Infect Dis. 1993;168:571–9. doi: 10.1093/infdis/168.3.571. [DOI] [PubMed] [Google Scholar]
- [13].Kesson AM, Fear WR, Williams L, Chang J, King NJ, Cunningham AL. HIV infection of placental macrophages: their potential role in vertical transmission. J Leukoc Biol. 1994;56:241–6. doi: 10.1002/jlb.56.3.241. [DOI] [PubMed] [Google Scholar]
- [14].McGann KA, Collman R, Kolson DL, Gonzalez-Scarano F, Coukos G, Coutifaris C, et al. Human immunodeficiency virus type 1 causes productive infection of macrophages in primary placental cell cultures. J Infect Dis. 1994;169:746–53. doi: 10.1093/infdis/169.4.746. [DOI] [PubMed] [Google Scholar]
- [15].Fear WR, Kesson AM, Naif H, Lynch GW, Cunningham AL. Differential tropism and chemokine receptor expression of human immunodeficiency virus type 1 in neonatal monocytes, monocyte-derived macrophages, and placental macrophages. J Virol. 1998;72:1334–44. doi: 10.1128/jvi.72.2.1334-1344.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Melendez-Guerrero LM, Holmes R, Backé E, Polliotti B, Ibegbu C, Lee F, et al. In vitro infection of Hofbauer cells with a monocyte-tropic strain of HIV-1. Trophoblast Res. 1994;8:33–45. [Google Scholar]
- [17].Plaud-Valentin M, Delgado R, Garcia V, Zorrilla C, Gandia J, Melendez-Guerrero LM. HIV infection of placental macrophages: effect on the secretion of HIV stimulatory cytokines. Cell Mol Biol (Noisy-le-grand) 1999;45:423–31. [PubMed] [Google Scholar]
- [18].Torres G, Garcia V, Sanchez E, Segarra A, Patterson BK, Melendez-Guerrero LM. Expression of the HIV-1 co-receptors CCR5 and CXCR4 on placental macrophages and the effect of IL-10 on their expression. Placenta. 2001;22(Suppl. A):S29–33. doi: 10.1053/plac.2001.0652. [DOI] [PubMed] [Google Scholar]
- [19].Melendez J, Garcia V, Sanchez E, Delgado R, Torres G, Melendez-Guerrero LM. Is decreased HIV-1 infectivity of placental macrophages caused by high levels of beta-chemokines? Cell Mol Biol (Noisy-le-grand) 2001;47:OL51–9. [Online Pub] [PubMed] [Google Scholar]
- [20].Uren S, Boyle W. Isolation of macrophages from human placenta. J Immunol Methods. 1985;78:25–34. doi: 10.1016/0022-1759(85)90326-6. [DOI] [PubMed] [Google Scholar]
- [21].Ciborowski P, Kadiu I, Rozek W, Smith L, Bernhardt K, Fladseth M, et al. Investigating the human immunodeficiency virus type 1-infected monocyte-derived macrophage secretome. Virology. 2007;363:198–209. doi: 10.1016/j.virol.2007.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Huber W, von Heydebreck A, Sultmann H, Poustka A, Vingron M. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics. 2002;18(Suppl. 1):S96–104. doi: 10.1093/bioinformatics/18.suppl_1.s96. [DOI] [PubMed] [Google Scholar]
- [23].Beyer S, Walter Y, Hellmann J, Kramer PJ, Kopp-Schneider A, Kroeger M, et al. Comparison of software tools to improve the detection of carcinogen induced changes in the rat liver proteome by analyzing SELDI-TOF-MS spectra. J Proteome Res. 2006;5:254–61. doi: 10.1021/pr050279o. [DOI] [PubMed] [Google Scholar]
- [24].Luciano-Montalvo C, Ciborowski P, Duan F, Gendelman H, Meléndez L. Proteomic analyses associate cystatin B with restricted HIV-1 replication in placental macrophages. Placenta. 29:1016–23. doi: 10.1016/j.placenta.2008.09.005. [Epub 2008 Oct 31] [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Luo X, Carlson KA, Wojna V, Mayo R, Biskup TM, Stoner J, et al. Macrophage proteomic fingerprinting predicts HIV-1-associated cognitive impairment. Neurology. 2003;60:1931–7. doi: 10.1212/01.wnl.0000064396.54554.26. [DOI] [PubMed] [Google Scholar]
- [26].Wojna V, Carlson KA, Luo X, Mayo R, Melendez LM, Kraiselburd E, et al. Proteomic fingerprinting of human immunodeficiency virus type 1-associated dementia from patient monocyte-derived macrophages: A case study. J Neurovirol. 2004;10(Suppl. 1):74–81. doi: 10.1080/753312756. [DOI] [PubMed] [Google Scholar]
- [27].Seo MS, Kang SW, Kim K, Baines IC, Lee TH, Rhee SG. Identification of a new type of mammalian peroxiredoxin that forms an intramolecular disulfide as a reaction intermediate. J Biol Chem. 2000;275:20346–54. doi: 10.1074/jbc.M001943200. [DOI] [PubMed] [Google Scholar]
- [28].Krutilina RI, Kropotov AV, Leutenegger C, Serikov VB. Migrating leukocytes are the source of peroxiredoxin V during inflammation in the airways. J Inflamm (Lond) 2006;3:13. doi: 10.1186/1476-9255-3-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Geiben-Lynn R, Kursar M, Brown NV, Addo MM, Shau H, Lieberman J, et al. HIV-1 antiviral activity of recombinant natural killer cell enhancing factors, NKEF-A and NKEF-B, members of the peroxiredoxin family. J Biol Chem. 2003;278:1569–74. doi: 10.1074/jbc.M209964200. [DOI] [PubMed] [Google Scholar]
- [30].Jin DY, Chae HZ, Rhee SG, Jeang KT. Regulatory role for a novel human thioredoxin peroxidase in NF-kappaB activation. J Biol Chem. 1997;272:30952–61. doi: 10.1074/jbc.272.49.30952. [DOI] [PubMed] [Google Scholar]
- [31].Chertova E, Chertov O, Coren LV, Roser JD, Trubey CM, Bess JW, Jr, et al. Proteomic and biochemical analysis of purified human immunodeficiency virus type 1 produced from infected monocyte-derived macrophages. J Virol. 2006;80:9039–52. doi: 10.1128/JVI.01013-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Misse D, Yssel H, Trabattoni D, Oblet C, Lo Caputo S, Mazzotta F, et al. IL-22 participates in an innate anti-HIV-1 host-resistance network through acute-phase protein induction. J Immunol. 2007;178:407–15. doi: 10.4049/jimmunol.178.1.407. [DOI] [PubMed] [Google Scholar]
- [33].Kang SW, Rhee SG, Chang TS, Jeong W, Choi MH. 2-Cys peroxiredoxin function in intracellular signal transduction: therapeutic implications. Trends Mol Med. 2005;11:571–8. doi: 10.1016/j.molmed.2005.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Verdot L, Lalmanach G, Vercruysse V, Hartmann S, Lucius R, Hoebeke J, et al. Cystatins up-regulate nitric oxide release from interferon-gamma-activated mouse peritoneal macrophages. J Biol Chem. 1996;271:28077–81. doi: 10.1074/jbc.271.45.28077. [DOI] [PubMed] [Google Scholar]
- [35].Verdot L, Lalmanach G, Vercruysse V, Hoebeke J, Gauthier F, Vray B. Chicken cystatin stimulates nitric oxide release from interferon-gamma-activated mouse peritoneal macrophages via cytokine synthesis. Eur J Biochem. 1999;266:1111–7. doi: 10.1046/j.1432-1327.1999.00964.x. [DOI] [PubMed] [Google Scholar]
- [36].Blond D, Raoul H, Le Grand R, Dormont D. Nitric oxide synthesis enhances human immunodeficiency virus replication in primary human macrophages. J Virol. 2000;74:8904–12. doi: 10.1128/jvi.74.19.8904-8912.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Jimenez JL, Gonzalez-Nicolas J, Alvarez S, Fresno M, Munoz-Fernandez MA. Regulation of human immunodeficiency virus type 1 replication in human T lymphocytes by nitric oxide. J Virol. 2001;75:4655–63. doi: 10.1128/JVI.75.10.4655-4663.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Schutyser E, Richmond A, Van Damme J. Involvement of CC chemokine ligand 18 (CCL18) in normal and pathological processes. J Leukoc Biol. 2005;78:14–26. doi: 10.1189/jlb.1204712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Schraufstatter I, Takamori H, Sikora L, Sriramarao P, DiScipio RG. Eosinophils and monocytes produce pulmonary and activation-regulated chemokine, which activates cultured monocytes/macrophages. Am J Physiol Lung Cell Mol Physiol. 2004;286:L494–501. doi: 10.1152/ajplung.00323.2002. [DOI] [PubMed] [Google Scholar]