Abstract
The inner cell mass (ICM) of the implanting mammalian blastocyst comprises two lineages: the pluripotent epiblast (EPI) and primitive endoderm (PrE). We have identified platelet-derived growth factor receptor alpha (PDGFRα) as an early marker of the PrE lineage and its derivatives in both mouse embryos and ex vivo paradigms of extra-embryonic endoderm (ExEn). By combining live imaging of embryos and embryo-derived stem cells expressing a histone H2B-GFP fusion reporter under the control of Pdgfra regulatory elements with the analysis of lineage-specific markers, we found that Pdgfra expression coincides with that of GATA6, the earliest expressed transcriptional regulator of the PrE lineage. We show that GATA6 is required for the activation of Pdgfra expression. Using pharmacological inhibition and genetic inactivation we addressed the role of the PDGF pathway in the PrE lineage. Our results demonstrate that PDGF signaling is essential for the establishment, and plays a role in the proliferation, of XEN cells, which are isolated from mouse blastocyst stage embryos and represent the PrE lineage. Implanting Pdgfra mutant blastocysts exhibited a reduced number of PrE cells, an effect that was exacerbated by delaying implantation. Surprisingly, we also noted an increase in the number of EPI cells in implantation-delayed Pdgfra-null mutants. Taken together, our data suggest a role for PDGF signaling in the expansion of the ExEn lineage. Our observations also uncover a possible role for the PrE in regulating the size of the pluripotent EPI compartment.
Keywords: Blastocyst, Mouse embryo, Epiblast, Primitive endoderm, Extra-embryonic endoderm, Embryonic stem (ES) cell, XEN cell, PDGF signaling, GATA
INTRODUCTION
During early mammalian development, two extra-embryonic lineages, trophectoderm (TE) and primitive endoderm (PrE), are specified and segregated away from the pluripotent epiblast (EPI). Extra-embryonic tissues serve to pattern the early embryo, as well as providing the essential maternal-fetal connection required to sustain the embryo after implantation.
Stem cell lines that retain properties of each of the three lineages, TE, PrE and EPI, of the implanting (late) blastocyst can be derived and propagated in culture. Embryonic stem (ES) cells represent the EPI (Evans and Kaufman, 1981; Martin, 1981), whereas extra-embryonic endoderm (XEN) cells represent the PrE (Kunath et al., 2005), and trophoblast stem (TS) cells represent the TE (Tanaka et al., 1998). These embryo-derived cell types serve as models for investigating the mechanisms that regulate lineage specification, commitment and maintenance. Lineage choice is governed by the expression of key lineage-specific transcription factors that regulate downstream signaling pathways (reviewed by Arnold and Robertson, 2009; Niwa, 2007; Rossant and Tam, 2009), such that ES cells can be converted into TS-like (Niwa et al., 2005) or XEN-like (Shimosato et al., 2007) cells simply by misexpression of appropriate transcription factors.
In the mouse embryo, commitment to the PrE lineage occurs prior to overt differentiation of the PrE. This model is supported by the salt-and-pepper distribution of PrE markers in cells within the inner cell mass (ICM) of early blastocyst stage embryos (Chazaud et al., 2006; Plusa et al., 2008). A number of transcription factors, including members of the GATA, SOX and HNF families, are expressed by the PrE and its derivatives, as well as by ES cells that are directed to differentiate into extra-embryonic endoderm (ExEn). Loss- and gain-of-function studies have demonstrated that these transcription factors play key roles in ExEn lineage specification, maintenance and differentiation. FGF/MAPK signaling has been shown to regulate Gata6 expression, and so may function upstream of these transcription factors (Arman et al., 1998; Chazaud et al., 2006; Feldman et al., 1995; Goldin and Papaioannou, 2003; Nichols et al., 2009; Yamanaka et al., 2010).
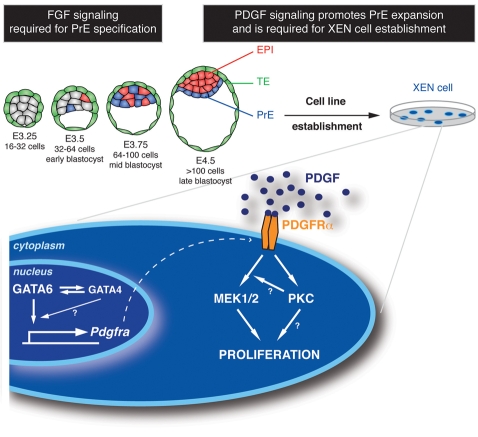
Although the mechanisms of PrE specification have been relatively well studied, little is known about the signaling pathways regulating ExEn lineage expansion and differentiation towards PrE derivatives: visceral endoderm (VE) and parietal endoderm (PE). Higher vertebrates have two platelet-derived growth factor receptors, PDGFRα and PDGFRβ, which form homo- and heterodimers, and at least four PDGF ligands (reviewed by Andrae et al., 2008; Hoch and Soriano, 2003). Owing to this complexity, the effects of PDGF signaling on early development, especially early lineage specification and expansion, have not been comprehensively investigated. We have identified PDGFRα as a marker of the PrE lineage of the blastocyst and its ExEn derivatives, in both embryos and in ex vivo paradigms of the ExEn lineage. From investigating the relationship between PDGFRα and the PrE lineage-determining transcription factors, we propose a model whereby initiation of Pdgfra expression requires GATA6 and its maintenance requires GATA4 and GATA6.
Using pharmacological inhibition and genetic inactivation we addressed the role of PDGF receptor signaling in the ExEn. Our results suggest that the PDGF pathway exerts a mitogenic effect on XEN cells, and that this activity is mediated intracellularly by MEK and PKC signaling. We also noted that implanting Pdgfra mutant blastocysts exhibit a reduction in the number of PrE cells, an effect exacerbated by delaying implantation, suggesting a role for PDGF signaling in expansion/maintenance of the PrE. In addition, implantation-delayed mutant blastocysts exhibited an increase in EPI cell number, uncovering a previously unrecognized role for the PrE in regulating the size of the EPI compartment.
MATERIALS AND METHODS
Embryo collection and in vitro culture
Mice were maintained on a mixed genetic background (129/B6/ICR). Embryos were obtained from ICR or PdgfraH2B-GFP/+ females mated with PdgfraH2B-GFP/+ males (Hamilton et al., 2003). Blastocysts were recovered in M2 media (Chemicon) and cultured for 1-3 days in ES cell media on 0.1% gelatin-coated chambered coverglass slides (Lab-Tek) at 37°C in 5% CO2. Decidua were dissected from uteri in D-MEM/F-12 (Gibco) containing 5% newborn calf serum. Postimplantation embryos were processed for sectioning within decidua. Diapause was induced following intraperitoneal injection of 10 μg tamoxifen (Sigma) and 2-3 mg progesterone (Abraxis) at E2.5. Embryos were recovered 2-3 days later.
ES/XEN cell culture
ES cells were maintained on mitomycin C-treated primary murine embryonic fibroblasts (MEFs) in recombinant leukemia inhibitory factor (LIF) (Mereau et al., 1993) under standard conditions (Nagy et al., 2003). PdgfraH2B-GFP/+ XEN cells were routinely cultured on gelatin coated-dishes in ES cell media in the absence of LIF and feeders and passaged every 2 days at a 1 in 5 dilution.
Inactivation of Pdgfra in XEN cells
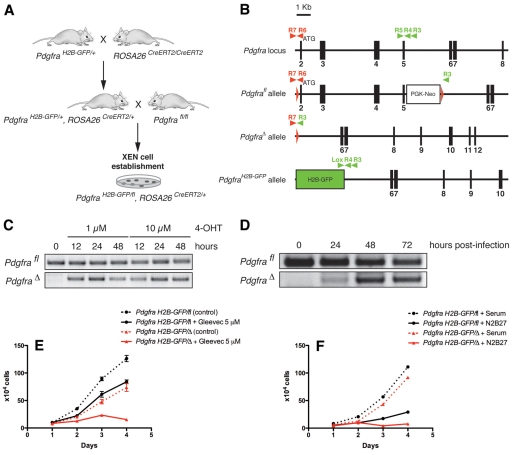
Deletion of the floxed allele in PdgfraH2B-GFP/fl, ROSA26CreERT2/+ XEN cell line was induced either by addition of 4-hydroxytamoxifen (4-OHT, Sigma) or infection with a self-excising Cre-expressing retrovirus (Silver and Livingston, 2001) (following the protocol of L. Le Cam, Institut de Rechercher en Cancérologie de Montpellier, France).
ES/XEN cell line isolation
Blastocysts were collected from matings using KitW-lacZ/+ (Bernex et al., 1996), PdgfraH2B-GFP/+ (Hamilton et al., 2003), Pdgfrafl/fl (Tallquist and Soriano, 2003) and ROSA26CreERT2/CreERT2 (Cheng et al., 2010) alleles. Blastocysts were cultured individually for 5 days in 4-well plates on MEFs in ES cell media containing LIF. ICMs were dissociated in 0.25% trypsin-EDTA, and passaged into 24-well dishes containing MEFs. Media were changed every 2 days. Cells were passaged 7-10 days later into fresh 24-well plates until confluency, then cultured in the absence of MEFs and genotyped by PCR.
ES cell differentiation assays
ES cells were maintained on gelatin in the absence of MEFs prior to differentiation. PdgfraH2B-GFP/+ cells (Hamilton et al., 2003) were plated at 2×105 cells per 35-mm dish containing gelatin-coated glass coverslips. Next day, ES cell media were replaced with LIF-free media containing 10% FCS and 1 μM trans-retinoic acid (Sigma). Media were changed daily. Transfections of plasmids pCMV5-Flag (gift of M. E. Donohoe, Cornell University, NY, USA), pCMV-Tag2 Gata4 and pCMV-Tag2 Gata6 (gift of Y. Hayashi, Nagoya University, Japan) were carried out using Lipofectamine 2000 (Invitrogen). Embryoid bodies were generated by plating 1×106 cells onto a 10-cm Petri dish (VWR) with culture in LIF-free medium containing 10% FBS. Media were changed every 2 days.
Proliferation assay
XEN cells were plated at 1×104-105 cells per well of a 24-well plate (Falcon). Media were changed daily and counts were performed in triplicate. Inhibitor compounds used were: Gleevec (gift of P. Besmer, Sloan-Kettering Institute, NY, USA) and bisindolylmaleimide (BIS), LY294002 and U0126 (all from Cell Signaling). Recombinant human PDGF-AA (R&D Systems) and mouse SCF proteins (R&D Systems) were added at 10 ng/ml and 20 ng/ml, respectively, to N2B27 medium (Ying and Smith, 2003).
Immunostaining
Blastocysts and cells cultured on coverslips were immunostained as previously described (Artus et al., 2005). Primary antibodies used were: CDX2 (Biogenex), FOXA2 (1:400, Abcam), GATA4 (1:300, Santa Cruz), GATA6 (1:100, R&D Systems), Ki67 (1:400, Vector Laboratories), NANOG (1:700, Cosmo Bio), OCT4 (1:200, Santa Cruz), PDGFRα (1:100, eBioscience), SOX7 (1:100, R&D Systems) and SOX17 (1:400, R&D Systems). Alexa Fluor-conjugated secondary antibodies (Invitrogen) were used at 1:200. DNA was counterstained with Hoechst 33342 (1:200, Invitrogen). Blastocysts were genotyped after analysis by PCR (Artus et al., 2005) using the primers detailed in Fig. 5B and Table S1 in the supplementary material. Coverslips were mounted in Vectashield (Vector Laboratories). Decidua-containing embryos were fixed overnight in 4% paraformaldehyde (PFA) at 4°C, embedded in PBS containing 4% agarose and 5% sucrose and sectioned at 200 μm using a vibrating microtome (VT1000S, Leica). F-actin was visualized with Alexa Fluor-phalloidin (1:500, Invitrogen). Embryoid bodies were fixed overnight in 4% PFA at 4°C, cryoprotected in PBS containing 30% sucrose, embedded in O.C.T. compound (Tissue-Tek) and sectioned at 12 μm using a cryostat (CM3050S, Leica).
Fig. 5.
Conditional inactivation of Pdgfra in XEN cells impairs their proliferation. (A) Strategy for isolating PdgfraH2B-GFP/fl; ROSA26CreERT2/+ XEN cells. (B) Schematic representation of Pdgfra alleles and genotyping primers used (arrowheads). (C) PCR detection of floxed and deleted alleles upon addition of 4-OHT for the indicated periods of time. (D) PCR detection of floxed and deleted alleles after infection with Cre-expressing retrovirus. (E,F) Proliferation curves of PdgfraH2B-GFP/fl (black) and PdgfraH2B-GFP/+ (red) XEN cell lines depict (E) the effect of 5 μM Gleevec and (F) the failure of PdgfraH2B-GFP/+ XEN cells to proliferate in N2B27 medium. Error bars indicate s.e.m.
Image acquisition and processing
Laser-scanning confocal data were acquired on a Zeiss LSM510 META. Fluorescence was excited with a 405 nm diode laser (Hoechst), a 488 nm argon laser (GFP) or a 543 nm HeNe laser (Alexa Fluor 546, 568). Images were acquired using Plan Apochromat 20×/NA0.75, Plan Neofluar 40×/NA1.3 or Plan Apochromat 63×/NA1.4 objectives. Optical sections ranged from 0.38-4 μm. Raw data were processed using Zeiss AIM (Carl Zeiss Microsystems), IMARIS 6.3.1 (Bitplane), Photoshop CS2 and Illustrator CS2 (Adobe).
RT-PCR and qPCR
Total RNA was extracted using TRIzol (Invitrogen), reverse transcribed using the Superscript III First-Strand Synthesis Kit (Invitrogen) and 50 ng of the RNA used as template for PCR amplification. Quantitative (q) PCR was performed using SYBR Green (Roche) on a LightCycler 480 real-time PCR instrument and analyzed with LightCycler 480 software (version 1.5.0.39). For sequences of primers and cycling conditions, see Table S1 in the supplementary material.
RESULTS
Pdgfra is expressed in the PrE and its derivatives
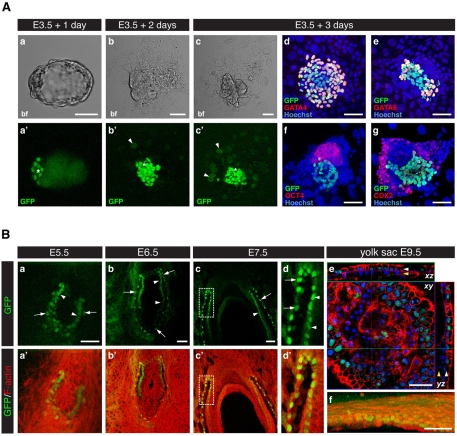
We have identified Pdgfra as one of the earliest markers of the PrE lineage in the mouse blastocyst (Plusa et al., 2008). To determine whether Pdgfra is expressed in later PrE derivatives, we investigated the expression of the PdgfraH2B-GFP reporter in explanted blastocysts. Under the conditions used, embryos hatched from their zona pellucida after 1 day of culture, then attached to the dish and formed outgrowths. The TE spread onto the bottom of the dish and eventually differentiated into trophoblast giant cells (TGCs). Concomitantly, the ICM expanded to produce an outgrowth (Fig. 1Aa-c). Nuclear-localized GFP was detected within the ICM after 1 to 3 days in culture (Fig. 1Aa′-c′). After 2 days in culture, weak GFP signal was also detected in cells located on the bottom of the dish outside of the outgrowth. These weakly GFP-expressing cells had large nuclei or were bi-nucleated, and were likely to be TGCs (Fig. 1Ab′,c′).
Fig. 1.
Pdgfra is expressed in the primitive endoderm (PrE) and its extra-embryonic endoderm (ExEn) derivatives. (A) Pdgfra expression during mouse blastocyst outgrowth. (a-c) Single bright-field (bf) optical sections of PdgfraH2B-GFP/+ blastocyst cultured in vitro for 3 days. (a′-c′) GFP (green) is expressed in the inner cell mass (ICM; asterisk). (b′,c′) GFP is weakly detected in trophoblast giant cells (TGCs; arrowheads). (d,e) After 3 days in culture, GFP colocalizes with PrE markers GATA4 (d, red) and GATA6 (e, red). (f,g) Mutually exclusive expression of GFP, OCT4 (f, red) and CDX2 (g, red) after 3 days culture. (a'-c',d-g) Three-dimensional projections of z-stacks. (B) (a-d′) Pdgfra expression in the PrE derivatives parietal endoderm (PE, arrows) and visceral endoderm (VE, arrowheads) at E5.5 (a), E6.5 (b) and E7.5 (c,d). (c,c′) Extra-embryonic region of an E7.5 embryo. (a-c′) Three-dimensional projections of z-stacks. (d,d′) Magnified view of the boxed region from c and c′. (e,f) GFP is detected in the endoderm of the visceral yolk sac at E9.5. (e) Orthogonal views of z-stack of an E9.5 yolk sac. (f) Three-dimensional projection of e. White arrowhead, VE; yellow arrowhead, mesoderm derivatives of the yolk sac. Blue, Hoechst; green, GFP; red, F-actin. Scale bars: 50 μm.
We next confirmed that, within ICM outgrowths, GFP colocalized with PrE-specific markers, including the transcription factors GATA4 (Fig. 1Ad) and GATA6 (Fig. 1Ae). Of the GFP-positive cells, 91% were GATA4 positive (out of 291 GFP-positive cells, 264 were GATA4 positive; n=2 embryos) and 56% were GATA6 positive (out of 383 GFP-positive cells, 215 were GATA6 positive; n=4 embryos). Although the majority of GFP-expressing cells were also GATA4 positive, we noted the presence of a minor population of GFP-positive cells that did not express GATA4. These cells could represent later PrE derivatives or, alternatively, mesoderm derivatives, which have been reported to express PDGFRα (see Fig. S1A2 in the supplementary material) (Orr-Urtreger and Lonai, 1992). Notably, GFP reporter expression did not colocalize with either EPI- or TE-specific transcription factors (Fig. 1Af,g).
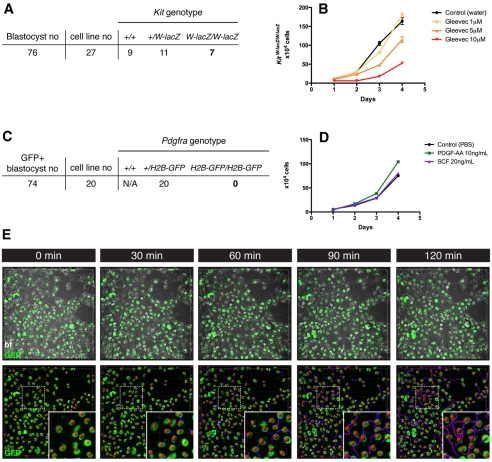
Next, to determine whether Pdgfra is expressed in XEN cells, which represent the PrE lineage, we established XEN cell lines from the PdgfraH2B-GFP strain. Live imaging has revealed that even though they are clonal, XEN cells exhibit heterogeneity in their morphology (Kunath et al., 2005). The PdgfraH2B-GFP reporter was expressed in all cells at equivalent levels (see Fig. 4E), suggesting that Pdgfra is homogeneously expressed in XEN cells irrespective of morphological transitions.
Fig. 4.
PDGFRα, but not KIT, signaling is required for XEN cell establishment. (A) Isolation of Kit-deficient XEN cell lines. (B) Dose-dependent effect of Gleevec in the absence of Kit. Kit-deficient XEN cells were plated at 5×104 cells/well (24-well plate) and treated with Gleevec at final concentrations of 0 (black), 1 (yellow), 5 (orange) and 10 (red) μM. (C) Failure to isolate Pdgfra-deficient XEN cell lines. (D) Effect of the addition of recombinant PDGF-AA (green) or SCF (purple) on XEN cells, compared with control (black). Wild-type XEN cells plated at 5×104 cells/well (24-well plate); 10 ng/ml PDGF-AA or 20 ng/ml SCF was added to the serum-free N2B27 cell culture medium. Error bars indicate s.e.m. (E) Four-dimensional time-lapse imaging of PdgfraH2B-GFP/+ XEN cells showing homogeneous levels of fluorescence during the acquisition period. Nuclear GFP staining allows cell tracking. Insets show the boxed region (dashed line) at higher magnification. z-stacks were acquired every 15 minutes for a total of 15 hours. Green, GFP; red, tracked nucleus; purple, dragon tail.
We next determined that Pdgfra expression was maintained within the PE and VE, the two PrE derivative tissues, encompassing the period preceding and during gastrulation, from embryonic day (E) 5.5 to 7.5 (Fig. 1B and see Fig. S1 in the supplementary material). PdgfraH2B-GFP reporter expression was downregulated at ∼E6.5 in VE cells overlying the epiblast, coincident with the onset of gastrulation (Fig. 1Bb), such that a gradient of GFP signal was detected along the proximal-distal axis (see Fig. S1B,B2 in the supplementary material), as confirmed by PDGFRα localization (see Fig. S1A1 in the supplementary material). At E7.5, GFP was visualized in the endoderm layer of the visceral yolk sac, confirming that Pdgfra expression is maintained in this ExEn derivative (Fig. 1Be,f). These observations indicate that Pdgfra is dynamically expressed in vivo in the PrE and its derivatives.
Pdgfra expression in ex vivo models of ExEn
We next determined whether Pdgfra expression serves as an ExEn marker in various ES cell models of ExEn formation. These paradigms have proved invaluable for studying diverse aspects of ExEn biology (Coucouvanis and Martin, 1999; Fujikura et al., 2002; Rula et al., 2007; Shimosato et al., 2007; Yang et al., 2007). We used the PdgfraH2B-GFP reporter (Hamilton et al., 2003; Plusa et al., 2008) to visualize Pdgfra expression in these assays.
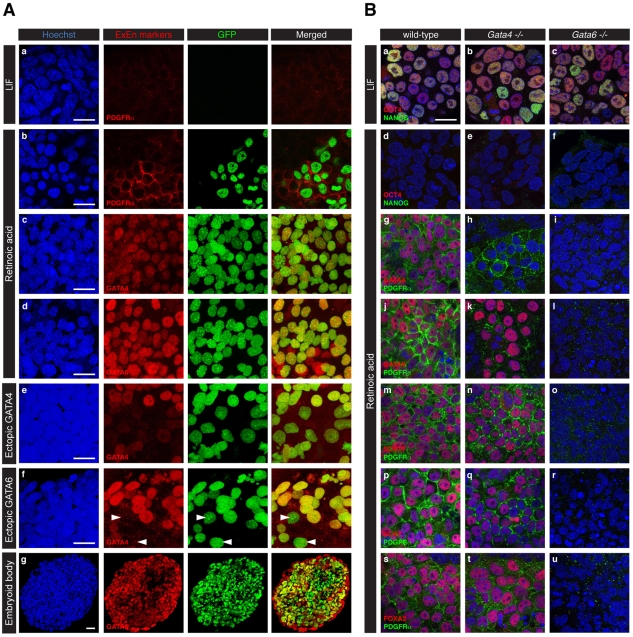
Retinoic acid (RA) promotes the differentiation of ES cells into various cell types, including ExEn derivatives (Soprano et al., 2007). PdgfraH2B-GFP/+ ES cells propagated in the presence of LIF, and thus grown in an undifferentiated state, expressed neither PDGFRα nor the GFP reporter (Fig. 2Aa). However, a 4-day treatment with 1 μM RA induced the expression of both endogenous PDGFRα protein and the GFP reporter (Fig. 2Ab). GFP was detected at low levels 2 days after RA addition (see Fig. S2Ac,h in the supplementary material) and had increased by 3 days (see Fig. S2Ad,i in the supplementary material). GFP-positive cells expressed GATA4 (Fig. 2Ac) and GATA6 (Fig. 2Ad), suggesting that they had acquired an ExEn identity. Their identity was confirmed using additional markers, including SOX17, SOX7 and FOXA2 (Fig. 2B). Immunodetection of AFP as a VE marker (Dziadek and Adamson, 1978) and SPARC as a PE marker (Holland et al., 1987; Mason et al., 1986) revealed that RA treatment induced a heterogeneous population containing both VE- and PE-like cells (see Fig. S3 in the supplementary material).
Fig. 2.
Pdgfra expression in ex vivo models of PrE formation is regulated by GATA6. (A) (a) PdgfraH2B-GFP/+ mouse ES cells propagated in the presence of LIF do not express PDGFRα protein or GFP reporter. (b-d) Upon retinoic acid (RA) treatment, both nuclear-localized GFP and endogenous PDGFRα protein are detected (b). GFP-positive cells express GATA4 (c) and GATA6 (d). (e,f) Expression of GFP and GATA4 is detected 48 hours after ectopic expression of GATA4 (e) or GATA6 (f). Few GFP-positive cells do not express GATA4 (arrowheads) upon GATA6 misexpression. (g) Section through an embryoid body at 5 days of differentiation. Cells in the outer layer express GATA6 but not the GFP reporter. (a,b) Single optical section. (c-g) Three-dimensional projections of the z-stacks. Blue, Hoechst; green, GFP; red, PDGFRα, GATA4 or GATA6. (B) Requirement for GATA6, but not GATA4, for Pdgfra expression upon RA treatment. (a-c) Wild-type (a), Gata4–/– (b) and Gata6–/– (c) ES cells cultured in the presence of LIF express the pluripotency markers OCT4 and NANOG. (d-f) Upon RA treatment, ES cell differentiation is visualized by loss of Oct4 and Nanog expression. (g-u) ExEn formation induced by RA treatment is visualized by co-expression of PDGFRα, GATA4, GATA6, SOX17, SOX7 and FOXA2. Like wild-type ES cells (left column), Gata4–/– ES cells differentiate into ExEn (middle column). In the absence of Gata6, ES cells fail to differentiate into ExEn derivatives (right column). (a-u) Single optical sections. Blue, Hoechst; green, NANOG (a-f) or PDGFRα (g-u); red, OCT4 (a-f), GATA4 (g-i), GATA6 (j-l), SOX17 (m-o), SOX7 (p-r) or FOXA2 (s-u). Scale bars: 20 μm.
Ectopic transient expression of GATA4 or GATA6 in ES cells is sufficient to induce ExEn differentiation (Fujikura et al., 2002; Shimosato et al., 2007). Ectopic expression of either GATA4 or GATA6 was sufficient to induce the GFP reporter (Fig. 2Ae,f), which was detected from 15 hours post-transfection (see Fig. S2Ba,f in the supplementary material). These data demonstrate that activation of the PdgfraH2B-GFP/+ reporter occurred concomitant with ExEn specification.
When cultured without LIF under non-adherent conditions, ES cells form aggregates called embryoid bodies (EBs), which recapitulate several developmental events (Coucouvanis and Martin, 1995; Martin et al., 1977). These include the formation of an outer layer of cells that resembles VE (Coucouvanis and Martin, 1995; Coucouvanis and Martin, 1999). Upon EB formation at 5 days of differentiation, we noted that the outer layer of cells expressed GATA6, but did not express the PdgfraH2B-GFP reporter (Fig. 2Ag). Our observation that Pdgfra is downregulated in the distal VE at E6.5 might suggest that the outer layer of cells in these EBs resembles the distal portion of the VE that overlies the epiblast. To further explore the identity of the surface endoderm layer of EBs, we investigated the presence of other ExEn markers at 3, 5 and 7 days of EB formation (see Fig. S4 in the supplementary material). No ExEn markers were detected after 3 days of EB formation. However, by 5 days, robust localization of GATA6, FOXA2 and SOX17 and weak localization of GATA4 and HNF4α were observed in the outer layer of cells. After 7 days, all ExEn markers analyzed were detected in the outer layer of cells and their distribution appeared relatively homogeneous, except for HNF4α, which was distributed in patches. Interestingly, SOX7, which is detected in the proximal VE overlying the extra-embryonic ectoderm at E6.5-7.5 (Kanai-Azuma et al., 2002), was not detected in cells in the outer layer of EBs, supporting the notion that the outer layer of EBs resembles the distal VE overlying the epiblast. Collectively, these observations reveal that Pdgfra is expressed in ExEn cell types formed upon ES cell differentiation. By contrast, Pdgfra is not expressed in the ExEn cells that constitute the outer layer of EBs, suggesting that these cells are likely to represent a subdomain of the VE.
GATA6 regulates Pdgfra expression
GATA4 and GATA6 are considered to be the key regulators of ExEn identity based on the finding that their misexpression is sufficient to direct ES cells towards an ExEn fate (Shimosato et al., 2007), as confirmed using the PdgfraH2B-GFP reporter (Fig. 2Ae,f). However, the respective roles of these two transcription factors in the regulation of Pdgfra expression has remained unclear. Previous studies have shown that GATA binding sites within cis-regulatory sequences upstream of the Pdgfra gene are crucial for expression and can be bound by GATA4 (Wang and Song, 1996). Having previously demonstrated that in the mouse embryo, PDGFRα and GATA6 are the earliest known markers of the PrE lineage and that their expression precedes that of GATA4 (Plusa et al., 2008), we suggest a model whereby GATA6 is involved in the activation of Pdgfra expression, and GATA4 and GATA6 are involved in its maintenance. In support of such a model, we noted that 48 hours after GATA6 misexpression in ES cells, a small percentage of GFP-positive cells did not express GATA4 (Fig. 2Af, arrowheads).
We analyzed the kinetics of expression of the endogenous Gata4, Gata6 and Pdgfra genes 24 hours after forced expression of the genes encoding to GATA transcription factors. Upon Gata4 misexpression, a small percentage of GFP-positive cells did not express detectable levels of GATA4 (see Fig. S5A in the supplementary material). However, all GFP-positive cells expressed GATA6 (see Fig. S5B in the supplementary material). By contrast, upon Gata6 misexpression, all GFP-positive cells expressed GATA6 (see Fig. S5D in the supplementary material), but most did not express GATA4 (see Fig. S5C in the supplementary material, white arrowheads). These observations suggest that activation of Pdgfra expression is preceded by, and might require, GATA6.
Next, we reasoned that if GATA6 is an important regulator of Pdgfra expression then, in the absence of GATA6, Pdgfra should not be expressed. We investigated Pdgfra expression upon RA treatment of Gata4 (Soudais et al., 1995) or Gata6 (Morrisey et al., 1998) mutant ES cells. It has been shown that differentiation of ES cells into ExEn induced by RA treatment requires Gata6 but not Gata4 (Capo-Chichi et al., 2005). As expected, RA treatment of wild-type, Gata4- and Gata6-deficient ES cells induced their differentiation, as indicated by the absence of detectable levels of OCT4 (POU5F1 – Mouse Genome Informatics) and NANOG proteins (Fig. 2Bd-f). Wild-type RA-treated ES cells acquired an ExEn identity (Fig. 2B). In Gata4-deficient differentiated cells (Fig. 2Bh), PDGFRα colocalized with GATA6 (Fig. 2Bk), SOX17 (Fig. 2Bn), SOX7 (Fig. 2Bq) and FOXA2 (Fig. 2Bt). By contrast, Gata6-deficient ES cells (Fig. 2Bl) failed to differentiate into ExEn and expressed neither Pdgfra nor PrE markers (Fig. 2B). These data led us to propose that Gata6 expression is required for Pdgfra expression, both for its initial activation and for its maintenance in the early mouse embryo and in cell-based assays of PrE formation (see Fig. 7).
Fig. 7.
Model for the role of PDGF signaling in the PrE lineage of the mouse blastocyst. Early mouse development is characterized by progressive lineage restriction ensuring the segregation of the ICM cells (gray) into epiblast (EPI, red) and primitive endoderm (PrE, blue). XEN cells are derived from the PrE of the blastocyst. GATA6, which is considered a key regulator of PrE identity, may control the expression of genes, including Gata4 and Pdgfra. The PDGF pathway exerts a mitogenic effect through MEK/PKC signaling and promotes in vivo and ex vivo PrE lineage expansion and XEN cell establishment.
Inhibition of RTK, MEK1/2 or PKC signaling affects ExEn cell proliferation
Several observations suggest that receptor tyrosine kinase (RTK) signaling mediated by MAPK is necessary for the formation of the PrE. The most compelling data come from the analysis of GRB2 adaptor molecule-deficient embryos and EBs in which PrE fails to form (Chazaud et al., 2006; Cheng et al., 1998). To investigate a potential role for PDGF signaling in the PrE, we adopted a pharmacological approach using small-molecule compounds to inhibit signal transduction. Gleevec (also know as Imatinib) occupies the active site of tyrosine kinases, leading to a decrease in their activity (Carroll et al., 1997). Gleevec inhibits several kinases, including PDGFR, KIT and ABL proteins. Addition of Gleevec to embryos affected blastocyst outgrowth, especially the TE and its derivatives, and so could not be used to investigate PrE formation (data not shown). Several explanations could account for the deleterious effects of Gleevec in early embryos. Pdgfra is expressed in TGCs (Fig. 1Ab′-c′). Additionally, the KIT receptor is also expressed in TGCs and a role for KIT signaling in the TE lineage is supported by KIT ligand promotion of TE outgrowth (Mitsunari et al., 1999).
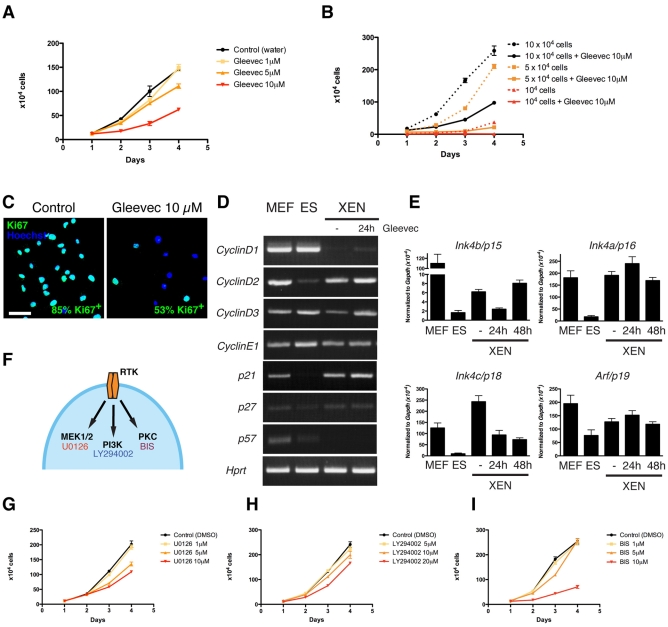
To address the role of RTK signaling in the PrE, we analyzed the effect of Gleevec on XEN cells. We first assayed the effect of an increasing dose of Gleevec on the proliferation of wild-type XEN cells (Fig. 3A). We estimated the doubling time to be 21 (control), 20 (1 μM), 25 (5 μM) and 31 (10 μM) hours. We then determined whether the effect of Gleevec on XEN cell proliferation is also cell density-dependent (Fig. 3B). At 10×104 cells per well, 10 μM Gleevec increased cell doubling time by 1.4-fold, whereas at 5×104 and 1×104 cells per well the doubling time was increased by 2.7- and 4.2-fold, respectively. Thus, Gleevec treatment affects XEN cell proliferation in a dose- and cell density-dependent manner.
Fig. 3.
Inhibition of RTK activity, MEK1/2 or PKC signaling affects XEN cell proliferation. (A) Dose-dependent effect of Gleevec. Gleevec concentrations: 0 (black), 1 (yellow), 5 (orange) and 10 (red) μM. (B) Cell density-dependent effect of Gleevec. Cells were plated at 1×104 (red), 5×104 (orange) and 10×104 (black) cells/well (24-well plate) in the presence of 10 μM Gleevec (solid lines; dotted lines indicate controls). (C) Reduction in the number of Ki67-positive cells after 48 hours of Gleevec treatment. Blue, Hoechst; green, Ki67. Scale bar: 50 μm. (D,E) RT-PCR (D) and qPCR (E) analyses of cell cycle regulator expression in mouse embryonic fibroblast (MEF), ES and extra-embryonic endoderm (XEN) cells in the presence and absence of Gleevec. (F) Schematic representation of signal transduction pathways activated upon ligand binding to RTK; the inhibitors used to block their activities are indicated. (G-I) Proliferation curves depicting the effect of 1-10 μM U0126 (G), 5-20 μM LY294002 (H) and 1-10 μM BIS (I). Cells were split at 5×104 cells/well (24-well plate). Error bars indicate s.e.m.
We next investigated whether the reduction in cell proliferation was due to cell death, cell cycle exit, or both. Immunodetection of the active, cleaved form of caspase 3 in cells treated with Gleevec did not reveal an increase in apoptosis (data not shown). However, we observed a decrease in the percentage of Ki67-positive cells: 53% of cells treated for 48 hours with 10 μM Gleevec as compared with 85% of untreated cells (Fig. 3C). These data suggest that, in response to Gleevec, XEN cells exit the cell cycle. We determined the expression of genes encoding cell cycle regulators of the G1 to S transition, including cyclins and the CDK inhibitors of the CIP/KIP and INK4 families. We noted that, in contrast to ES cells, which lack a G1 phase and do not express any CDK inhibitors, XEN cells expressed p21Cip1 (Cdkn1a) and low levels of p15INK4b (Cdkn2b), p16INK4a (Cdkn2a), p18INK4c (Cdkn2c) and p19ARF (Cdkn2a) (Fig. 3D,E). Upon 24 and 48 hours of Gleevec treatment, we did not observe a significant upregulation in the levels of CDK inhibitors that could explain, at a transcriptional level, the inhibition of XEN cell proliferation in response to Gleevec.
Upon ligand-mediated receptor activation, RTKs signal through various second messengers (Andrae et al., 2008). To determine the second messenger(s) through which RTKs exert their mitogenic effect, we employed a pharmacological approach using U0126 [MEK1/2 (MAP2K1/2) inhibitor], LY294002 (PI3K inhibitor) and BIS (PKC inhibitor) (Fig. 3F). We confirmed the specificity and lack of cytotoxicity of the compounds (see Fig. S6 in the supplementary material). Inhibition of MEK1/2 using U0126 at 10 μM led to a decrease in cell proliferation (Fig. 3G). Inhibition of PI3K activity by LY294002 produced a moderate effect on cell proliferation (Fig. 3H), whereas PKC inhibition using 10 μM BIS led to a dramatic decrease in proliferation (Fig. 3I). Based on these observations, we propose that in the PrE, RTK activity promotes cell cycle progression through MAPK and PKC signaling (see Fig. 7). Since Gleevec treatment inhibits several kinases, including PDGFRα, KIT and ABL, these data did not allow us to determine the respective role of each RTK in XEN cells.
The PDGF pathway is essential for XEN cell establishment and is involved in proliferation
Expression analyses suggest that both Pdgfra and Kit, but not Abl1, genes are expressed in PrE and in XEN cells (Brown et al., 2010; Kunath et al., 2005). To investigate the functional requirement for PDGFRα and KIT in XEN cells, we determined whether we could isolate Kit- or Pdgfra-deficient cells. Seven KitW-lacZ/W-lacZ XEN cell lines were isolated from blastocysts recovered from KitW-lacZ/+ heterozygous intercrosses (Bernex et al., 1996), indicating that Kit is not required for the establishment or maintenance of XEN cells (Fig. 4A). Furthermore, KitW-lacZ/W-lacZ XEN cells proliferated at a similar rate to wild-type XEN cells in serum and serum-free culture conditions (data not shown). To determine whether we could recapitulate the mitogenic effect of Gleevec in the absence of KIT signaling, Kit-deficient XEN cells were cultured in the presence of Gleevec (Fig. 4B). Their proliferation was comparable to that of wild-type XEN cells.
By contrast, no homozygous PdgfraH2B-GFP/H2B-GFP XEN cells were recovered (χ2=9.9 with 1 degree of freedom, P=0.0016) (Fig. 4C). To further validate a role for the PDGF pathway in XEN cell proliferation we tested the effect of PDGF-AA on wild-type XEN cells cultured in serum-free conditions and noted that it enhanced proliferation (Fig. 4D). By contrast, addition of recombinant kit ligand (KITL; SCF) had no effect.
The failure to isolate Pdgfra-deficient XEN cell lines suggested that PDGF signaling is required for XEN cell establishment, but prevented us from determining whether it is required for the maintenance of XEN cells in culture. To investigate the requirement for PDGF signaling in established XEN cell lines, we derived XEN cells in which Pdgfra could be conditionally inactivated. This cell line carried two Pdgfra alleles, the PdgfraH2B-GFP null allele and a conditional mutant Pdgfrafl allele that is hypomorphic in the unexcised configuration (Tallquist and Soriano, 2003), as well as a ubiquitous inducible Cre-ERT2 transgene (Cheng et al., 2010) (Fig. 5A). Induction of Cre activity by addition of 4-OHT resulted in mosaic deletion of the Pdgfrafl allele (Fig. 5C). In addition, infection with a self-excising Cre-expressing retrovirus (Silver and Livingston, 2001) also induced mosaic deletion of the Pdgfrafl allele in cultured XEN cells (Fig. 5D). To overcome the mosaic genotype (PdgfraH2B-GFP/fl versus PdgfraH2B-GFP/+), cells were subcloned after 4-OHT treatment. PdgfraH2B-GFP/+ XEN cell lines proliferated in the absence of MEFs, and the rate of cell division progressively increased when cells were maintained for extended passages (Fig. 5E). Treatment with 5 μM Gleevec affected proliferation even in the absence of Pdgfra, suggesting that other RTKs inhibited by Gleevec might adapt to the loss of PDGF signaling. However, in serum-free conditions, Pdgfra-deficient XEN cells failed to proliferate (Fig. 5F). This phenotype could not be rescued by addition of FGF4 or SCF (data not shown). These data suggest that PDGF signaling is crucial for the propagation of ExEn cells in vitro, and that this effect is masked in serum-containing culture conditions.
The PDGF pathway plays a role in PrE lineage expansion around the time of implantation
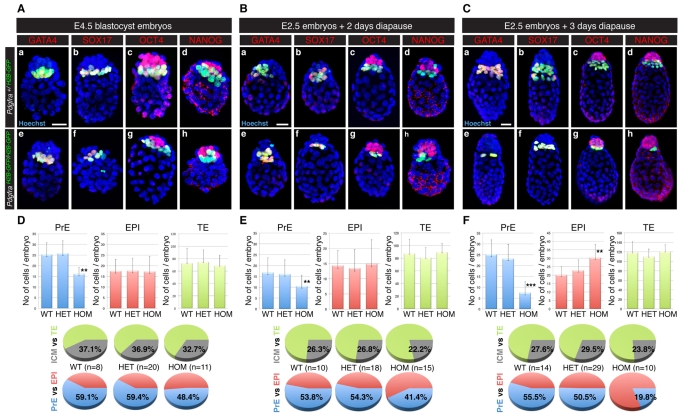
Our observations suggesting a role for the PDGF pathway in XEN cells cultured in vitro contrast with the apparent absence of a detectable phenotype in preimplantation Pdgfra-null mutant embryos (Hamilton et al., 2003; Soriano, 1997). We therefore determined the respective composition of PrE, EPI and TE lineages in implanting wild-type and mutant blastocysts (E4.5) (Fig. 6A,D; see Movies 1 and 2 in the supplementary material). Although mutant embryos had the same mean TE cell number (68.7±17.3 TE cells versus 74.4±14.1 for heterozygotes and 72.6±9.7 for wild type) or EPI cell number (17.3±6.6 EPI cells versus 17.7±6.1 for heterozygotes and 17.5±5.0 for wild type), they exhibited a significant reduction in the number of PrE cells (16.2±6.8 PrE cells versus 25.8±5.6 for heterozygotes and 25.3±5.3 for wild type) (Fig. 6D). These findings suggest that the PDGF pathway might be important in the maintenance of the PrE.
Fig. 6.
Implantation-delayed blastocysts lacking PDGFRα exhibit defects in the PrE and pluripotent epiblast (EPI) lineages. (Aa-Ch) PdgfraH2B-GFP/+ (top row) and PdgfraH2B-GFP/H2B-GFP (bottom row) mouse embryos at E4.5 (A), 2 days (B) and 3 days (C) after tamoxifen injection. Blue, Hoechst; green, GFP; red, GATA4 (a,e), SOX17 (b,f), OCT4 (c,g) and NANOG (d,h). Scale bar: 20 μm. (D-F) Distribution of PrE (blue), EPI (red) and trophectoderm (TE; green) cells in Pdgfra+/+ (WT), PdgfraH2B-GFP/+ (HET) and PdgfraH2B-GFP/H2B-GFP (HOM) embryos at E4.5 (D), 2 days (E) and 3 days (F) after tamoxifen injection. Gray, ICM. **, P<0.007; ***, P<0.0001. Error bars indicate s.d.
Artificially delaying implantation through the induction of a period of diapause has been used to demonstrate the role of LIF receptor signaling in the maintenance of the EPI lineage despite the absence of a phenotype at the blastocyst stage (Li et al., 1995; Nichols et al., 2001; Ware et al., 1995). Diapause preserves the general topology of the three lineages of the blastocyst, but promotes an expansion in cell number (Fig. 6B-F; see Movie 3 in the supplementary material). In PdgfraH2B-GFP/+ 3-day diapause blastocysts, the PrE layer was maintained at the interface between the blastocoel and the EPI, and expressed the PdgfraH2B-GFP reporter as well as GATA4 (Fig. 6Ca) and SOX17 (Fig. 6Cb). The EPI lineage of these implantation-delayed blastocysts was encapsulated by TE and PrE, and expressed OCT4 (Fig. 6Cc) and low levels of NANOG (Fig. 6Cd) (Batlle-Morera et al., 2008). Implantation-delayed blastocyst stage PdgfraH2B-GFP/H2B-GFP mutant embryos also contained GATA4 and SOX17-positive PrE cells with robust nuclear-localized GFP signal that were in contact with the blastocoel (Fig. 6Ce-h). However, the number of PrE cells was reduced (7.5±5.1 PrE cells versus 23.2±6.6 for heterozygotes and 25.1±6.7 for wild type) (Fig. 6F), such that in the most affected embryos the PrE layer was present as a small patch of cells that did not line the entire interface between the EPI and blastocoel (Fig. 6Cg,h and see Movie 4 in the supplementary material). These data suggest that PDGF signaling is required for maintenance of the PrE lineage in implantation-delayed blastocysts (Fig. 7).
PrE limits the size of the EPI compartment
Interestingly, although the absence of Pdgfra resulted in a reduction of the number of PrE cells, the number of EPI cells was increased in PdgfraH2B-GFP/H2B-GFP mutants (30.3±7.9 EPI cells versus 22.8±6.2 for heterozygotes and 20.1±4.8 for wild type) (Fig. 6F). This imbalance could result from an alteration in cell fate specification of the ICM, whereby the wild-type balance is skewed towards EPI. Alternatively, if cell fate specification is unaffected, reciprocal tissue interactions might be essential earlier than previously believed, with the PrE layer functioning to regulate the size of the EPI. To distinguish between these possibilities, we analyzed the distribution of EPI and PrE cells in 2-day diapause embryos (Fig. 6E). Although we observed a reduction in the number of PrE cells in mutant embryos (10.5±4.7 PrE cells versus 16.1±3.5 for heterozygotes and 16.8±2.3 for wild type), the number of EPI cells did not deviate between mutant (14.9±4.4 cells), heterozygous (13.5±4.1 cells) and wild-type (14.4±4.4 cells) embryos. We therefore conclude that between 2 and 3 days of diapause, the EPI compartment exhibits inappropriate expansion in the absence of Pdgfra or a PrE layer. Based on this observation, we suggest that either PDGF signaling or PrE tissue might be required for regulating the size of the EPI.
DISCUSSION
In this study, we have characterized the expression, regulation and function of Pdgfra in the PrE and its derivatives. We previously reported that in early preimplantation stage mouse embryos, Pdgfra is initially expressed in a subset of blastomeres at the 16-cell stage and is then progressively restricted to the nascent PrE layer in the blastocyst stage embryo prior to implantation (Plusa et al., 2008). Here we show that Pdgfra expression is maintained after implantation in the two PrE derivatives, the PE and VE, eventually being downregulated in the distal VE, which overlies the epiblast. This localization correlates with our recent findings for other genes and transgenes that are initially expressed throughout the VE at early postimplantation stages, but later downregulated in the distal VE [which is also referred to as embryonic visceral endoderm (emVE) (Mesnard et al., 2006)] overlying the epiblast by E6.5, including Afp (Dziadek and Adamson, 1978; Kwon et al., 2006), Hnf4a (Duncan et al., 1994; Kwon et al., 2008) and Ttr (Kwon and Hadjantonakis, 2009; Makover et al., 1989).
We investigated the expression of Pdgfra in various cellular models of ExEn formation including ES cell differentiation and XEN cells. Collectively, our data suggest that GATA6 is a key regulator of Pdgfra (Fig. 7). This model is supported by our observation that Pdgfra is not expressed upon RA treatment of Gata6-deficient ES cells, whereas it is expressed in RA-treated Gata4-deficient ES cells. This model validates our in vivo studies in which we reported the onset of Gata6 expression at the ∼8-cell stage, Pdgfra at the ∼8- to 16-cell stage and Gata4 at the ∼64-cell stage (Plusa et al., 2008).
Our studies have uncovered a role for PDGF signaling in the proliferation of ExEn cells, acting via MEK and PKC (Fig. 7). Our failure to establish Pdgfra-deficient XEN cell lines, as well as the decreased proliferative capacity of conditionally Pdgfra-deficient XEN cells, support such a function.
Intriguingly, Pdgfra-deficient ES cells were able to differentiate into PrE, suggesting that the PDGF pathway is not required in the determination of PrE identity (see Fig. S7 in the supplementary material). An unresolved question concerns the upstream signal(s) that regulate MAPK activity within the PrE lineage. Several studies have reported an essential role for FGF signaling in PrE formation (Arman et al., 1998; Feldman et al., 1995; Goldin and Papaioannou, 2003; Nichols et al., 2009; Yamanaka et al., 2010). However, XEN cells do not appear to require FGF signaling, as they can be maintained in serum-free conditions. Furthermore, exogenous addition of FGF4 protein does not elicit any detectable change in XEN cell proliferation or morphology (our unpublished data). By contrast, FGF4 is required to derive and maintain TS cells in an undifferentiated state (Tanaka et al., 1998). In conclusion, our data support a model whereby PDGF pathway signaling via MEK and PKC plays a key role in XEN cell propagation. However, the exact and respective roles of MEK and PKC remain unclear, and we cannot exclude cross-talk, as has been reported in various cell systems (Lee et al., 2006; Okazaki et al., 2000; Robin et al., 2004). Future studies will be required to address the specific and combinatorial roles of these signal transducers.
Genetic inactivation of members of the PDGF pathway does not affect the PrE in embryos (reviewed by Andrae et al., 2008; Hoch and Soriano, 2003), arguing in favor of a model whereby the PDGF pathway is not required for the acquisition of PrE identity. Several explanations could account for this apparent disparity. First, the presence of maternal stores of protein or mRNA accumulated in the oocyte could compensate for the absence of zygotic expression. However, Pdgfra transcripts are only detected from the 8-cell stage onwards by RT-PCR (Plusa et al., 2008), and we failed to detect GFP reporter expression in PdgfraH2B-GFP/+ oocytes (see Fig. S8 in the supplementary material) or in embryos prior to the 16-cell stage (Plusa et al., 2008). Alternatively, PDGFRβ, the second member of the PDGF receptor family, could be expressed during this period. Pdgfrb expression has not been reported during the pre/peri-implantation period in wild-type embryos (Hamatani et al., 2004; Wang et al., 2004). However, Pdgfra and Pdgfrb double-mutant embryos can be recovered at postimplantation stages (P. Soriano, personal communication). Lastly, signaling through other RTKs could compensate for the absence of PDGF signaling. Candidates include KIT, owing to its expression in XEN cells (Brown et al., 2010; Kunath et al., 2005). However, our data suggest that KIT signaling is not essential in XEN cells, as demonstrated by our isolation of Kit-deficient XEN cell lines, as well as the absence of a mitogenic effect when wild-type XEN cells were cultured in the presence of SCF. Nevertheless, we cannot exclude the possibility that, in vivo in Pdgfra mutant embryos, KIT signaling or signaling through another RTK might compensate for the absence of PDGFRα.
The role that we report for PDGF signaling in the establishment of the XEN cells and in the regulation of their proliferation might reveal a hitherto unsuspected requirement for this pathway in the maintenance of an exclusively in vitro cell type that has no identical in vivo counterpart. XEN cells were only isolated recently, and to date they are not well characterized. XEN cells might indeed represent the PrE lineage in aspects of their general morphological characteristics, gene expression and developmental potential (Kunath et al., 2005), but they might not be identical to any cell type in the embryo. It is therefore possible, as with the LIF signaling pathway in mouse ES cells (Li et al., 1995; Smith et al., 1988; Ware et al., 1995; Williams et al., 1988), that XEN cell signaling requirements are distinct from those of the PrE lineage in vivo.
However, our studies have revealed that E4.5 Pdgfra-deficient embryos exhibit a reduction in the number of PrE cells, a situation that is exacerbated when embryos are implantation delayed. We suggest that PDGF signaling functions in PrE lineage expansion and maintenance. In addition, because we observed an expansion of the EPI compartment in implantation-delayed embryos lacking Pdgfra, we propose that the PrE regulates the size of the EPI compartment. Thus, a reduction in the number of PrE cells results in a reciprocal expansion of the EPI. This hypothesis is supported by a previous report demonstrating that the efficiency of ES cell isolation from the ICM is increased upon PrE removal (Brook and Gardner, 1997). Indeed, these observations warrant further investigation as they suggest that juxtaposition of, and therefore reciprocal signaling between, PrE and EPI tissues might serve to regulate compartment size within the peri-implantation mammalian embryo.
Supplementary Material
Acknowledgments
We thank Alexandra Joyner and Philippe Soriano for the ROSA26CreERT2 and Pdgfra mouse strains, respectively; Michael Parmacek, Philippe Soriano and David Wilson for ES cell lines; Mary Donohoe, Yoshitaka Hayashi and Daniel Silver for plasmids; Peter Besmer for Gleevec; Florence Bernex and Geneviève Aubin-Houzelstein for KitW-lacZ husbandry; Laurent Le Cam for the retroviral infection protocol; Kemar Brown and Ann Foley for advice on qPCR; Ferdinando Rossi and Yasemin Yozgat for advice on immunoblotting; Ann Foley, Tilo Kunath and Jennifer Nichols for valuable discussions; and Florence Bernex, Mary Donohoe and Ann Foley for critical reading and comments on the manuscript. Work in A.-K.H.'s laboratory is supported by the National Institutes of Health (RO1-HD052115 and RO1-DK084391) and NYSTEM. Deposited in PMC for release after 12 months.
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material for this article is available at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.050864/-/DC1
References
- Andrae J., Gallini R., Betsholtz C. (2008). Role of platelet-derived growth factors in physiology and medicine. Genes Dev. 22, 1276-1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arman E., Haffner-Krausz R., Chen Y., Heath J. K., Lonai P. (1998). Targeted disruption of fibroblast growth factor (FGF) receptor 2 suggests a role for FGF signaling in pregastrulation mammalian development. Proc. Natl. Acad. Sci. USA 95, 5082-5087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold S. J., Robertson E. J. (2009). Making a commitment: cell lineage allocation and axis patterning in the early mouse embryo. Nat. Rev. Mol. Cell Biol. 10, 91-103 [DOI] [PubMed] [Google Scholar]
- Artus J., Vandormael-Pournin S., Frodin M., Nacerddine K., Babinet C., Cohen-Tannoudji M. (2005). Impaired mitotic progression and preimplantation lethality in mice lacking OMCG1, a new evolutionarily conserved nuclear protein. Mol. Cell. Biol. 25, 6289-6302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batlle-Morera L., Smith A., Nichols J. (2008). Parameters influencing derivation of embryonic stem cells from murine embryos. Genesis 46, 758-767 [DOI] [PubMed] [Google Scholar]
- Bernex F., De Sepulveda P., Kress C., Elbaz C., Delouis C., Panthier J. J. (1996). Spatial and temporal patterns of c-kit-expressing cells in WlacZ/+ and WlacZ/WlacZ mouse embryos. Development 122, 3023-3033 [DOI] [PubMed] [Google Scholar]
- Brook F. A., Gardner R. L. (1997). The origin and efficient derivation of embryonic stem cells in the mouse. Proc. Natl. Acad. Sci. USA 94, 5709-5712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown K., Legros S., Artus J., Doss M. X., Khanin R., Hadjantonakis A.-K., Foley A. (2010). A comparative analysis of extra-embryonic endoderm cell lines. PLoS ONE 5, e12016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capo-Chichi C. D., Rula M. E., Smedberg J. L., Vanderveer L., Parmacek M. S., Morrisey E. E., Godwin A. K., Xu X. X. (2005). Perception of differentiation cues by GATA factors in primitive endoderm lineage determination of mouse embryonic stem cells. Dev. Biol. 286, 574-586 [DOI] [PubMed] [Google Scholar]
- Carroll M., Ohno-Jones S., Tamura S., Buchdunger E., Zimmermann J., Lydon N. B., Gilliland D. G., Druker B. J. (1997). CGP 57148, a tyrosine kinase inhibitor, inhibits the growth of cells expressing BCR-ABL, TEL-ABL, and TEL-PDGFR fusion proteins. Blood 90, 4947-4952 [PubMed] [Google Scholar]
- Chazaud C., Yamanaka Y., Pawson T., Rossant J. (2006). Early lineage segregation between epiblast and primitive endoderm in mouse blastocysts through the Grb2-MAPK pathway. Dev. Cell 10, 615-624 [DOI] [PubMed] [Google Scholar]
- Cheng A. M., Saxton T. M., Sakai R., Kulkarni S., Mbamalu G., Vogel W., Tortorice C. G., Cardiff R. D., Cross J. C., Muller W. J., et al. (1998). Mammalian Grb2 regulates multiple steps in embryonic development and malignant transformation. Cell 95, 793-803 [DOI] [PubMed] [Google Scholar]
- Cheng Y., Sudarov A., Szulc K. U., Sgaier S. K., Stephen D., Turnbull D. H., Joyner A. L. (2010). The Engrailed homeobox genes determine the different foliation patterns in the vermis and hemispheres of the mammalian cerebellum. Development 137, 519-529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coucouvanis E., Martin G. R. (1995). Signals for death and survival: a two-step mechanism for cavitation in the vertebrate embryo. Cell 83, 279-287 [DOI] [PubMed] [Google Scholar]
- Coucouvanis E., Martin G. R. (1999). BMP signaling plays a role in visceral endoderm differentiation and cavitation in the early mouse embryo. Development 126, 535-546 [DOI] [PubMed] [Google Scholar]
- Duncan S. A., Manova K., Chen W. S., Hoodless P., Weinstein D. C., Bachvarova R. F., Darnell J. E., Jr (1994). Expression of transcription factor HNF-4 in the extraembryonic endoderm, gut, and nephrogenic tissue of the developing mouse embryo: HNF-4 is a marker for primary endoderm in the implanting blastocyst. Proc. Natl. Acad. Sci. USA 91, 7598-7602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziadek M., Adamson E. (1978). Localization and synthesis of alphafoetoprotein in post-implantation mouse embryos. J. Embryol. Exp. Morphol. 43, 289-313 [PubMed] [Google Scholar]
- Evans M. J., Kaufman M. H. (1981). Establishment in culture of pluripotential cells from mouse embryos. Nature 292, 154-156 [DOI] [PubMed] [Google Scholar]
- Feldman B., Poueymirou W., Papaioannou V. E., DeChiara T. M., Goldfarb M. (1995). Requirement of FGF-4 for postimplantation mouse development. Science 267, 246-249 [DOI] [PubMed] [Google Scholar]
- Fujikura J., Yamato E., Yonemura S., Hosoda K., Masui S., Nakao K., Miyazaki J.-I., Niwa H. (2002). Differentiation of embryonic stem cells is induced by GATA factors. Genes Dev. 16, 784-789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldin S. N., Papaioannou V. E. (2003). Paracrine action of FGF4 during periimplantation development maintains trophectoderm and primitive endoderm. Genesis 36, 40-47 [DOI] [PubMed] [Google Scholar]
- Hamatani T., Carter M. G., Sharov A. A., Ko M. S. (2004). Dynamics of global gene expression changes during mouse preimplantation development. Dev. Cell 6, 117-131 [DOI] [PubMed] [Google Scholar]
- Hamilton T. G., Klinghoffer R. A., Corrin P. D., Soriano P. (2003). Evolutionary divergence of platelet-derived growth factor alpha receptor signaling mechanisms. Mol. Cell. Biol. 23, 4013-4025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoch R. V., Soriano P. (2003). Roles of PDGF in animal development. Development 130, 4769-4784 [DOI] [PubMed] [Google Scholar]
- Holland P. W., Harper S. J., McVey J. H., Hogan B. L. (1987). In vivo expression of mRNA for the Ca++-binding protein SPARC (osteonectin) revealed by in situ hybridization. J. Cell Biol. 105, 473-482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai-Azuma M., Kanai Y., Gad J. M., Tajima Y., Taya C., Kurohmaru M., Sanai Y., Yonekawa H., Yazaki K., Tam P. P., et al. (2002). Depletion of definitive gut endoderm in Sox17-null mutant mice. Development 129, 2367-2379 [DOI] [PubMed] [Google Scholar]
- Kunath T., Arnaud D., Uy G. D., Okamoto I., Chureau C., Yamanaka Y., Heard E., Gardner R. L., Avner P., Rossant J. (2005). Imprinted X-inactivation in extra-embryonic endoderm cell lines from mouse blastocysts. Development 132, 1649-1661 [DOI] [PubMed] [Google Scholar]
- Kwon G. S., Hadjantonakis A. K. (2009). Transthyretin mouse transgenes direct RFP expression or Cre-mediated recombination throughout the visceral endoderm. Genesis 47, 447-455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon G. S., Fraser S. T., Eakin G. S., Mangano M., Isern J., Sahr K. E., Hadjantonakis A. K., Baron M. H. (2006). Tg(Afp-GFP) expression marks primitive and definitive endoderm lineages during mouse development. Dev. Dyn. 235, 2549-2558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon G. S., Viotti M., Hadjantonakis A. K. (2008). The endoderm of the mouse embryo arises by dynamic widespread intercalation of embryonic and extraembryonic lineages. Dev. Cell 15, 509-520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. Y., Park S. H., Lee Y. J., Heo J. S., Lee J. H., Han H. J. (2006). EGF-induced inhibition of glucose transport is mediated by PKC and MAPK signal pathways in primary cultured chicken hepatocytes. Am. J. Physiol. Gastrointest. Liver Physiol. 291, G744-G750 [DOI] [PubMed] [Google Scholar]
- Li M., Sendtner M., Smith A. (1995). Essential function of LIF receptor in motor neurons. Nature 378, 724-727 [DOI] [PubMed] [Google Scholar]
- Makover A., Soprano D. R., Wyatt M. L., Goodman D. S. (1989). An in situ-hybridization study of the localization of retinol-binding protein and transthyretin messenger RNAs during fetal development in the rat. Differentiation 40, 17-25 [DOI] [PubMed] [Google Scholar]
- Martin G. R. (1981). Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc. Natl. Acad. Sci. USA 78, 7634-7638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G. R., Wiley L. M., Damjanov I. (1977). The development of cystic embryoid bodies in vitro from clonal teratocarcinoma stem cells. Dev. Biol. 61, 230-244 [DOI] [PubMed] [Google Scholar]
- Mason I. J., Murphy D., Munke M., Francke U., Elliott R. W., Hogan B. L. (1986). Developmental and transformation-sensitive expression of the Sparc gene on mouse chromosome 11. EMBO J. 5, 1831-1837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mereau A., Grey L., Piquet-Pellorce C., Heath J. K. (1993). Characterization of a binding protein for leukemia inhibitory factor localized in extracellular matrix. J. Cell Biol. 122, 713-719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesnard D., Guzman-Ayala M., Constam D. B. (2006). Nodal specifies embryonic visceral endoderm and sustains pluripotent cells in the epiblast before overt axial patterning. Development 133, 2497-2505 [DOI] [PubMed] [Google Scholar]
- Mitsunari M., Harada T., Tanikawa M., Iwabe T., Taniguchi F., Terakawa N. (1999). The potential role of stem cell factor and its receptor c-kit in the mouse blastocyst implantation. Mol. Hum. Reprod. 5, 874-879 [DOI] [PubMed] [Google Scholar]
- Morrisey E. E., Tang Z., Sigrist K., Lu M. M., Jiang F., Ip H. S., Parmacek M. S. (1998). GATA6 regulates HNF4 and is required for differentiation of visceral endoderm in the mouse embryo. Genes Dev. 12, 3579-3590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy A., Gertsenstein M., Vintersten K., Behringer R. (2003). Manipulating the Mouse Embryo. A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; [Google Scholar]
- Nichols J., Chambers I., Taga T., Smith A. (2001). Physiological rationale for responsiveness of mouse embryonic stem cells to gp130 cytokines. Development 128, 2333-2339 [DOI] [PubMed] [Google Scholar]
- Nichols J., Silva J., Roode M., Smith A. (2009). Suppression of Erk signalling promotes ground state pluripotency in the mouse embryo. Development 136, 3215-3222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H. (2007). How is pluripotency determined and maintained? Development 134, 635-646 [DOI] [PubMed] [Google Scholar]
- Niwa H., Toyooka Y., Shimosato D., Strumpf D., Takahashi K., Yagi R., Rossant J. (2005). Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell 123, 917-929 [DOI] [PubMed] [Google Scholar]
- Okazaki J., Mawatari K., Liu B., Kent K. C. (2000). The effect of protein kinase C and its alpha subtype on human vascular smooth muscle cell proliferation, migration and fibronectin production. Surgery 128, 192-197 [DOI] [PubMed] [Google Scholar]
- Orr-Urtreger A., Lonai P. (1992). Platelet-derived growth factor-A and its receptor are expressed in separate, but adjacent cell layers of the mouse embryo. Development 115, 1045-1058 [DOI] [PubMed] [Google Scholar]
- Plusa B., Piliszek A., Frankenberg S., Artus J., Hadjantonakis A. K. (2008). Distinct sequential cell behaviours direct primitive endoderm formation in the mouse blastocyst. Development 135, 3081-3091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robin P., Boulven I., Bole-Feysot C., Tanfin Z., Leiber D. (2004). Contribution of PKC-dependent and -independent processes in temporal ERK regulation by ET-1, PDGF, and EGF in rat myometrial cells. Am. J. Physiol. Cell Physiol. 286, C798-C806 [DOI] [PubMed] [Google Scholar]
- Rossant J., Tam P. P. (2009). Blastocyst lineage formation, early embryonic asymmetries and axis patterning in the mouse. Development 136, 701-713 [DOI] [PubMed] [Google Scholar]
- Rula M. E., Cai K. Q., Moore R., Yang D. H., Staub C. M., Capo-Chichi C. D., Jablonski S. A., Howe P. H., Smith E. R., et al. (2007). Cell autonomous sorting and surface positioning in the formation of primitive endoderm in embryoid bodies. Genesis 45, 327-338 [DOI] [PubMed] [Google Scholar]
- Shimosato D., Shiki M., Niwa H. (2007). Extra-embryonic endoderm cells derived from ES cells induced by GATA factors acquire the character of XEN cells. BMC Dev. Biol. 7, 80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver D. P., Livingston D. M. (2001). Self-excising retroviral vectors encoding the Cre recombinase overcome Cre-mediated cellular toxicity. Mol. Cell 8, 233-243 [DOI] [PubMed] [Google Scholar]
- Smith A. G., Heath J. K., Donaldson D. D., Wong G. G., Moreau J., Stahl M., Rogers D. (1988). Inhibition of pluripotential embryonic stem cell differentiation by purified polypeptides. Nature 336, 688-690 [DOI] [PubMed] [Google Scholar]
- Soprano D. R., Teets B. W., Soprano K. J. (2007). Role of retinoic acid in the differentiation of embryonal carcinoma and embryonic stem cells. Vitam. Horm. 75, 69-95 [DOI] [PubMed] [Google Scholar]
- Soriano P. (1997). The PDGF alpha receptor is required for neural crest cell development and for normal patterning of the somites. Development 124, 2691-2700 [DOI] [PubMed] [Google Scholar]
- Soudais C., Bielinska M., Heikinheimo M., MacArthur C. A., Narita N., Saffitz J. E., Simon M. C., Leiden J. M., Wilson D. B. (1995). Targeted mutagenesis of the transcription factor GATA-4 gene in mouse embryonic stem cells disrupts visceral endoderm differentiation in vitro. Development 121, 3877-3888 [DOI] [PubMed] [Google Scholar]
- Tallquist M. D., Soriano P. (2003). Cell autonomous requirement for PDGFRalpha in populations of cranial and cardiac neural crest cells. Development 130, 507-518 [DOI] [PubMed] [Google Scholar]
- Tanaka S., Kunath T., Hadjantonakis A. K., Nagy A., Rossant J. (1998). Promotion of trophoblast stem cell proliferation by FGF4. Science 282, 2072-2075 [DOI] [PubMed] [Google Scholar]
- Wang C., Song B. (1996). Cell-type-specific expression of the platelet-derived growth factor alpha receptor: a role for GATA-binding protein. Mol. Cell. Biol. 16, 712-723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q. T., Piotrowska K., Ciemerych M. A., Milenkovic L., Scott M. P., Davis R. W., Zernicka-Goetz M. (2004). A genome-wide study of gene activity reveals developmental signaling pathways in the preimplantation mouse embryo. Dev. Cell 6, 133-144 [DOI] [PubMed] [Google Scholar]
- Ware C. B., Horowitz M. C., Renshaw B. R., Hunt J. S., Liggitt D., Koblar S. A., Gliniak B. C., McKenna H. J., Papayannopoulou T., Thoma B., et al. (1995). Targeted disruption of the low-affinity leukemia inhibitory factor receptor gene causes placental, skeletal, neural and metabolic defects and results in perinatal death. Development 121, 1283-1299 [DOI] [PubMed] [Google Scholar]
- Williams R. L., Hilton D. J., Pease S., Willson T. A., Stewart C. L., Gearing D. P., Wagner E. F., Metcalf D., Nicola N. A., Gough N. M. (1988). Myeloid leukaemia inhibitory factor maintains the developmental potential of embryonic stem cells. Nature 336, 684-687 [DOI] [PubMed] [Google Scholar]
- Yamanaka Y., Lanner F., Rossant J. (2010). FGF signal-dependent segregation of primitive endoderm and epiblast in the mouse blastocyst. Development 137, 715-724 [DOI] [PubMed] [Google Scholar]
- Yang D. H., Cai K. Q., Roland I. H., Smith E. R., Xu X. X. (2007). Disabled-2 is an epithelial surface positioning gene. J. Biol. Chem. 282, 13114-13122 [DOI] [PubMed] [Google Scholar]
- Ying Q. L., Smith A. G. (2003). Defined conditions for neural commitment and differentiation. Methods Enzymol. 365, 327-341 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.