Abstract
In bilaterians, establishing the correct spatial positioning of structures along the dorsoventral (DV) axis is essential for proper embryonic development. Insects such as Drosophila rely on the Dorsal activity gradient and Bone morphogenetic protein (BMP) signaling to establish cell fates along the DV axis, leading to the distinction between tissues such as mesoderm, neurogenic ectoderm and dorsal ectoderm in the developing embryo. Subsequently, the ventral midline plays a more restricted role in DV patterning by establishing differential cell fates in adjacent regions of the neurogenic ectoderm. In this study, we examine the function of the ventral midline and the midline-associated gene single-minded (Ph-sim) in the amphipod crustacean Parhyale hawaiensis. Remarkably, we found that Ph-sim and the ventral midline play a central role in establishing proper fates along the entire DV axis in this animal; laser ablation of midline cells causes a failure to form neurogenic ectoderm and Ph-sim RNAi results in severely dorsalized embryos lacking both neurogenic ectoderm and the appendage-bearing lateral ectoderm. Furthermore, we hypothesize that this role of midline cells was present in the last common ancestor of crustaceans and insects. We predict that the transition to a Dorsal-dependent DV patterning system in the phylogenetically derived insect lineage leading to Drosophila has led to a more restricted role of the ventral midline in patterning the DV axis of these insects.
Keywords: Parhyale hawaiensis, RNAi, Dorsoventral patterning, Laser ablation, Single-minded, Ventral midline
INTRODUCTION
The organization of tissues along the dorsoventral (DV) axis is well conserved in arthropods from the germ band stage through to adulthood: the central nervous system (CNS) occupies the ventral-most region, appendages develop from ventrolateral ectoderm, and the heart is positioned dorsally (Brusca and Brusca, 2003). Prior to the germ band stage, however, basic morphogenetic processes such as gastrulation and germ rudiment assembly vary considerably among arthropods. Therein lies a fundamental question of evolutionary biology: how are various animals able to construct a similar end product when they begin with such vastly different starting materials?
In the fruit fly Drosophila melanogaster, DV patterning relies on a broad activity gradient of the maternal transcription factor Dorsal that regulates the expression of various tissue-specific target genes in a threshold-dependent fashion (Roth et al., 1989; Jiang et al., 1992; Stathopoulos et al., 2002). Ventrally, high levels of Dorsal activate snail and twist in the presumptive mesoderm, whereas intermediate levels activate the Bone morphogenetic protein (BMP) antagonist short-gastrulation (sog) in the presumptive neurogenic ectoderm (Jiang et al., 1992). Subsequently, secondary DV cell fates, such as the presumptive ventral midline, are established in the embryo. The ventral midline is demarcated by cells expressing the basic helix-loop-helix–Per-ARNT-Sim (bHLH-PAS) transcription factor gene single-minded (sim), which is directly regulated by Dorsal, Snail and Twist, as well as by Notch signaling at the mesoderm-mesectoderm boundary (Thomas et al., 1988; Nambu et al., 1991; Kosman et al., 1991; Kasai et al., 1992; Leptin, 1991; Morel and Schweisguth, 2000). Although sim expression is first visible at the blastoderm stage in two columns of cells flanking the presumptive mesoderm, these cell columns converge during gastrulation to form a single, ventrally located column of cells (Crews et al., 1988). The ventral midline goes on to play a relatively restricted role in subsequent refinement of DV patterning by secreting the EGF ligand Spitz, which helps to ensure proper fate specification within the adjacent neurogenic ectoderm (Golembo et al., 1996; Mayer and Nüsslein-Volhard, 1988; Chang et al., 2000).
By contrast, distinctions between germ layer lineages in the amphipod crustacean Parhyale hawaiensis are made by the eight-cell stage (Price et al., 2010). In this system, gastrulation occurs as visceral and head mesoderm and germline precursors form an aggregation called the rosette, which is internalized under the developing germ disc. Later, somatic mesodermal precursors ingress along the posterior edge of the germ disc (Price and Patel, 2008). After gastrulation, the germ band for body segments posterior to the mandible consists of an ectodermal grid that is assembled progressively from anterior to posterior (Browne et al., 2005).
Furthermore, whereas specification of Drosophila midline cells requires input from Dorsal and mesodermally expressed transcription factors, the ventral midline in amphipods appears to be the first structure to become morphologically and molecularly distinct in the developing germ band (Gerberding and Scholtz, 1999; Browne et al., 2006). Although the ventral midline of Parhyale is assembled in the same manner as the rest of the ectodermal grid, it is assembled from a distinct population of precursor cells, termed midline precursor cells, that can be distinguished from the surrounding ectodermal grid precursor cells by their unique morphology (they are arranged in a wedge shape in the posterior of the embryo) and their expression of the midline marker orthodenticle (Ph-otd-1) (Browne et al., 2006). Non-midline ectoderm is derived from three ectodermal blastomeres (EL, ER and EP), whereas midline ectoderm is derived predominantly from EP (Gerberding et al., 2002). Finally, despite the divergent developmental origins of the ventral midline in Parhyale and Drosophila, the presence of common molecular markers has been used to argue that they are homologous structures (Simanton et al., 2009; Duman-Scheel and Patel, 1999).
Although the Dorsal-dependent DV patterning system seen in Drosophila has been well characterized, it is thought to be evolutionarily derived (novel) with respect to other arthropod groups; even within insects, it has been suggested that some groups rely on Dorsal to lesser degrees to pattern the DV axis (Nunes da Fonseca et al., 2008). Meanwhile, other aspects of DV patterning, such as the role of BMP antagonists in specifying neurogenic ectoderm, appear to be well conserved throughout Bilateria (Holley et al., 1995). When considering the evolution of DV patterning, however, one significant omission has been the lack of functional characterization of sim and the ventral midline in non-insect arthropods. This is possibly owing to the relatively restricted role that they play in Drosophila. As the presence of ventral midline cells in Parhyale represents the first visible manifestation of DV differentiation in the ectodermal grid, we sought to characterize the function of sim and the ventral midline in the overall organization of the DV axis in this animal. We describe here the basic DV fates in the Parhyale embryo, and, through laser ablation of midline cells and knockdown of sim expression, we define the role of the midline in patterning the DV axis of the embryo.
MATERIALS AND METHODS
Fluorescent live imaging
DsRed-NLS
A transgenic line of Parhyale hawaiensis containing the transgene PhHsp70-DsRed-NLS was generated by Melinda Modrell in the Patel laboratory as described previously (Pavlopoulos and Averof, 2005). Embryos were raised at 25°C in filtered artificial seawater. To induce DsRed-NLS expression, stage 8-9 embryos were subject to heat shock at 37°C for 1 hour. After several hours, nuclear DsRed fluorescence was visible within the embryos, allowing visualization of cells and facilitating the targeting of midline cells with the laser.
Hoechst
Embryos were treated with 10 μg/ml Hoechst 33342 dye (Sigma, St Louis, MO, USA) in artificial sea water for 3 minutes and then washed several times with artificial sea water. Live imaging was carried out by time-lapse videography using Volocity v.5 (Improvision, Waltham, MA, USA) software.
Laser ablation
Laser ablations were carried out using a MicroPoint nitrogen pulsed pumping dye laser (Photonic Instruments, St Charles, IL, USA) using a Coumarin 440 dye cell. Embryos were live-mounted in artificial seawater under a glass coverslip. The laser was attenuated using a graded neutral density filter slider to an intensity that could kill cells without generating vapor bubbles in the embryo. Using a 100× oil-immersion objective, individual nuclei were targeted (identification was made possible by DsRed-NLS fluorescence) and the laser was fired for 500 pulses (∼4 nseconds/pulse) at a frequency of 20 Hz. DsRed fluorescence rapidly fades in cells after they are hit with the laser, allowing us to verify that each cell was targeted correctly.
Cloning
cDNA was generated from mixed stage Parhyale hawaiensis embryos as described previously (Price and Patel, 2008). We cloned Parhyale orthologs of Pax3/7-1 (Ph-Pax3/7-1), prospero (Ph-pros), single-minded (Ph-sim), hedgehog (Ph-hh), and short gastrulation (Ph-sog) from cDNA using the following degenerate primers (5′ to 3′): Ph-Pax3/7-1 forward GGNGGNGTNTTYATHAAYGG, reverse RTTNSWRAACCANACYTG, nested forward MARATHGTNGARATGGC, nested reverse RTANACRTCNGGRTAYTG; Ph-pros forward AARGCNAARYTNATGTTYTT, reverse TCDATRTGRTTRTTNCKRTTRTARTG; Ph-sim forward GGCCCGGACGGNAARATHATG, reverse GGCGCGACDATRCARTGNGG, nested forward GGCGCGAAGCGNAAYGCNGG, nested reverse GGCTGCGGCTCRTANCCNGT; Ph-hh forward GTNATGAAYCARTGGCCNGG, reverse TCRTARTANACCCARTC; and Ph-sog forward GAYYTNGGNCCNCCNTTYG, reverse CNCKNCKCCANACNCCRCA, nested forward MRNAAYATHAARAAYGASTGTCC, nested reverse CCNGGRCANGTYTTGCAGCA. Additional sequence was obtained using 5′ and 3′ RACE (Ambion FirstChoice RLM-RACE Kit). New sequences were deposited in GenBank with accession numbers HM347085 (Ph-Pax3/7-1), HM191476 (Ph-pros), HM191473 (Ph-sim), HM347084 (Ph-hh) and HM191474 (Ph-sog).
Stealth siRNA design
Double-stranded Stealth siRNAs were designed for Ph-sim, Ph-sog and DsRed (negative control) using the BLOCK-IT RNAi program (Invitrogen, Carlsbad, CA, USA). siRNA sequences were as follows (5′ to 3′): Ph-sim 29, CACUGCUCGGGAUACCUCAAGAUCA; Ph-sim 149, GAGAUCAAGAUGCACUCCAACAUGU; Ph-sim 661, CAACAUGGACACAUCACCUUCUUUG; Ph-sog 292, CAGUGCGUCUGUGUCUCGGUACAAA; Ph-sog 1049, CACACAACGUCUCCAU GGUUCUACA; Ph-sog 2024, UGCCUGACGCUUGCUU GCUCGAUAA; DsRed 56, AGGGCUCCGUGAACGGCCACGAGUU; DsRed 197, AGUACGGCUCCAAGGUGUACGUGAA; and DsRed 269, GCUUCAAGUGGGAGCGCGUGAUGAA.
Embryo fixation, histochemical staining and microinjection of siRNAs into single-cell embryos were performed as described previously (Rehm et al., 2009). Some embryos were immersed in boiling water for 1-2 seconds prior to fixation and membrane removal as this allows for faster embryo processing. As boiled embryos retain a spherical shape, we photographed them in multiple focal planes and used Helicon Focus software (Helicon Soft, Kharkov, Ukraine) to generate a single, focused image.
RESULTS
DV regionalization in the Parhyale trunk
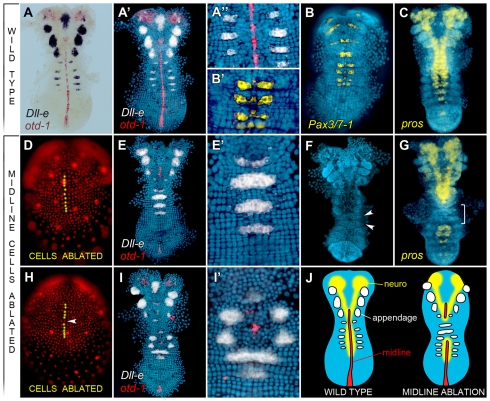
As a first step towards understanding DV patterning in Parhyale, we established molecular and morphological markers for the different DV fates in the ectoderm and mesoderm. The ectodermal grid in Parhyale contains distinct rows and columns, with the latter giving rise to stereotyped tissues along the DV axis of the embryo. The four basic DV fates observed in the ectoderm, from ventral to dorsal, are (1) the ventral midline (ventral-most cell column), (2) the neurogenic ectoderm, (3) a lateral ectodermal region containing appendage primordia, and (4) the dorsal ectoderm that will contribute to the most dorsal epidermis. A fifth (most dorsal) fate is characterized by loosely arranged extraembryonic epithelium.
The ventral midline (column 0) bisects the embryo and expresses Ph-otd-1 (Browne et al., 2006) (Fig. 1A). Neurogenic ectoderm, i.e. that which gives rise to neural precursor cells of the CNS, is derived from the midline along with cells from columns 1-3, as shown by studies following the lineage of Parhyale neurons expressing Engrailed (Browne et al., 2005). For various arthropods, particular Pax3/7 orthologs provide a conserved and earlier expressed marker of the neuroectodermal fate, as their expression is seen in segmental stripes confined to the presumptive neuroectoderm (Davis et al., 2005). We cloned a Parhyale Pax3/7 ortholog (Ph-Pax3/7-1) that showed strongest expression in segmental stripes within cell columns 0-2 (low level expression was also detected in columns 3-4) (Fig. 1B). To visualize the developing neurogenic ectoderm in older embryos, we cloned the Parhyale prospero homolog (Ph-pros) as this gene is expressed in neuroblasts and ganglion mother cells in an evolutionarily conserved fashion (Doe et al., 1991; Oliver et al., 1993). As each body segment matured, Ph-pros expression was seen in neural cells in the ventral regions of the embryo (Fig. 1C). Flanking the neurogenic ectoderm are appendage primordia, revealed molecularly by the conserved appendage-patterning gene Distal-less (Ph-Dll-e) (Panganiban et al., 1995; Liubicich et al., 2009). Ph-Dll-e expression was first seen in segmental stripes within cell columns 3-5 (Fig. 1A). Dorsal ectoderm consists of the most lateral cells within the grid and did not express any of the above-mentioned genes.
Fig. 1.
Laser ablation of midline cells results in DV mispatterning. All images are ventral views with the anterior at the top. Nuclei were counterstained with DAPI (blue) and a false-color overlay of gene expression patterns was generated. All embryos shown (as well as in Figs 3,4) were processed in a similar manner. (A-A′) Wild-type expression of Ph-Dll-e (black or white, as labeled) in appendage primordia and Ph-otd-1 (red) in midline cells at stage 17. (B,B′) Wild-type expression of Ph-Pax3/7-1 (yellow) in a subset of column 0-2-derived cells. (C) Wild-type expression of Ph-pros (yellow) in neuroblasts and ganglion mother cells at stage 18. (D) Living stage 13 embryo visualized by DsRed-NLS fluorescence. Selected midline cells were ablated using a focused laser (yellow dots). (E,E′) At stage 17, the embryo shown in D shows ventrally fused Ph-Dll-e (white) domains in segments lacking midline. Ph-otd-1 is shown in red. (F,G) Two embryos in which midline cells in three consecutive parasegments were ablated at stage 13. By stage 19 (F), ventrally fused limb buds (arrowheads) are visible in segments lacking midline. At stage 18 (G), segments lacking midline show a lack of Ph-pros staining (yellow) in affected segments (bracket, T3-T4). (H) Trunk midline cells were ablated (yellow dots) at stage 13 except for one midline cell in parasegment 4 (arrowhead). (I,I′) The embryo from H is shown at stage 17. Ph-Dll-e (white) is expressed at a reproducible distance from midline cell clone (large red cell cluster). Note that some scattered red signal is visible; this is associated with debris and does not represent Ph-otd-1 hybridization signal. (J) Illustration of DV fates in wild-type and midline-ablated embryos including midline (red), presumptive neurogenic ectoderm (yellow) and presumptive appendages (white).
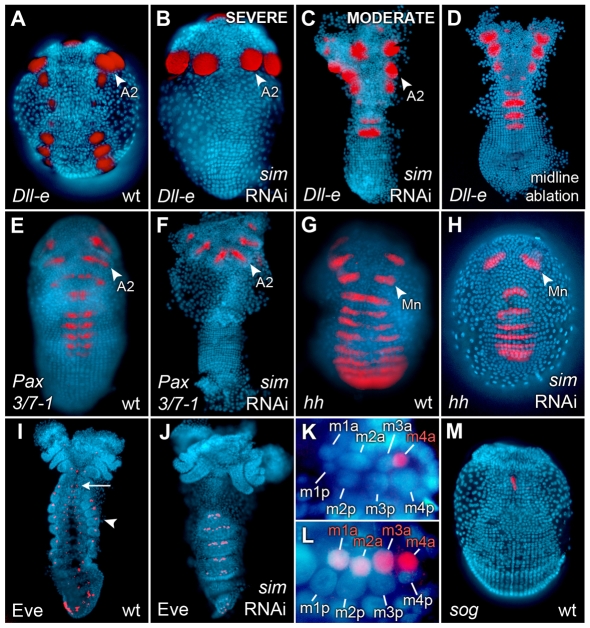
DV regionalization is also observed in the mesoderm of early Parhyale embryos. In the first thoracic segment and posteriorly, mesoderm initially consists of a transverse row of eight mesoblast cells per segment (with four cells on either side of the midline) (Price and Patel, 2008). These mesoblast cells divide along the anteroposterior (AP) axis to form two rows of eight daughter cells per segment, at which point twist (Ph-twi) is expressed bilaterally in the anterior daughters of the column 2 mesodermal cells (m2a) (Price and Patel, 2008). Additionally, using an antibody raised against the Parhyale Even skipped protein (Ph-Eve), we observed that Ph-Eve is expressed bilaterally in the anterior daughters of the dorsal-most mesoblast cells (m4a) (Fig. 4K). This expression pattern is reminiscent of the segmentally repeated clusters of Eve-positive cells observed in the dorsal-most mesoderm in Drosophila that contribute to the larval heart and dorsal somatic muscles (Frasch et al., 1987). In the following sections, we will use these ectodermal and mesodermal DV marker genes to illustrate the effects of laser ablation and Ph-sim knockdown on DV patterning in the Parhyale embryo.
Fig. 4.
Ph-sim RNAi causes dorsalization of embryonic trunk. Nuclei were counterstained with DAPI (blue) and a false-color overlay of gene expression patterns was generated. Where appropriate, the second antennal (A2) or mandibular (Mn) segments are labeled. (A) Ph-Dll-e expression in wild-type embryo. (B) Most Ph-sim RNAi embryos (44 out of 49) lack Ph-Dll-e expression posterior to A2. (C) Some Ph-sim RNAi embryos (4 out of 49) show a more moderate phenotype in which one or more trunk segments contain ventrally fused Ph-Dll-e spots. (D) Midline ablated embryo (from Fig. 1E) showing ventrally fused Ph-Dll-e spots, similar to the phenotype shown in C. (E) Wild-type Ph-Pax3/7-1 expression in a subset of presumptive neuroectodermal cells. (F) Ph-sim RNAi embryos lack Ph-Pax3/7-1 expression posterior to A2 (18 out of 18). (G) Wild-type Ph-hh expression in segmentally reiterated stripes. (H) Ph-sim RNAi embryos show relatively normal Ph-hh expression (16 out of 16). Decreased width of Ph-hh stripes is attributed to the dorsalized germ band of these embryos. (I) Wild-type expression of Ph-Eve in a stage 19 embryo. Protein is detected in dorsal mesoderm (arrowhead), developing neurogenic ectoderm (arrow) and posterior ectoderm. (J) In Ph-sim RNAi embryos, mesodermal Ph-Eve is expressed ectopically in ventral mesoderm suggesting embryonic dorsalization. Neurogenic staining is no longer visible as a result of deletion of this tissue. (K) Magnification of the eight mesodermal cells comprising one hemisegment in stage 18 wild-type embryo. Ventral midline is oriented to the left. Ph-Eve is expressed in the m4a cell. (L) Magnification of one hemisegment in Ph-sim RNAi embryo. Ventral midline is oriented to the left. Ph-Eve is now detected in all four anterior mesodermal cells (m1a-m4a). (M) Wild-type Ph-sog expression in a stage 15 embryo. The highest levels of expression are seen in anterior midline cells in the Mn segment. No Ph-sog expression is detected in the majority of trunk midline cells.
The ventral midline is required to specify ventral fates in Parhyale trunk
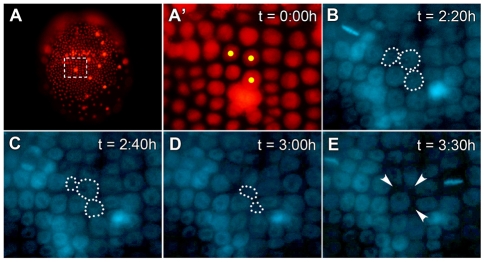
To investigate the potential patterning role of ventral midline cells in Parhyale, we performed laser ablations of these cells in developing embryos. This technique can be used to kill specific cells in the living embryo without causing peripheral damage to neighboring cells. Within 2-3 hours of ablation, targeted cells are absorbed into the yolk and are no longer visible by Hoechst fluorescence, whereas untargeted cells appear to be unaffected (Fig. 2).
Fig. 2.
Visualization of cell death following laser ablation. (A) Living stage 13 embryo visualized by DsRed-NLS fluorescence. (A′) Magnification of the region indicated in A. Cells marked with yellow dots were targeted for laser ablation. (B-E) Two hours after ablation, the embryo was treated with Hoechst dye and recorded by fluorescent time-lapse videography. Times shown represent total time elapsed (t) since ablation. Ablated cells have dotted outlines. (E) By 3:30 hours after ablation, all three targeted cells were no longer visible by Hoechst fluorescence. The space previously occupied by these cells (arrowheads) was minimized as surrounding cells moved in to fill the gaps.
When ventral midline cells were ablated at developmental stage 13, a localized defect in both Ph-Dll-e and Ph-pros expression was observed by stage 17. In segments lacking ventral midline cells, Ph-Dll-e-expressing domains were shifted ventrally to produce a single domain of expression (Fig. 1D,E). Accordingly, by stage 19, segments lacking midline formed a single ventrally fused limb bud, whereas non-manipulated segments formed normally positioned, bilateral limb buds (Fig. 1F). We also observed a complete loss of Ph-pros expression in segments in which the midline had been ablated, although Ph-pros was expressed normally in non-manipulated segments (Fig. 1G). To verify that these results represented a mis-specification of DV cell fates (as opposed to the death of presumptive neuroectodermal cells, for example), we followed midline-ablated embryos by time-lapse videography. Only cells targeted by the laser were observed to die, whereas adjacent cells continued to proliferate (see Movie 1 in the supplementary material). These results suggest that midline cells are required to establish the neurogenic ectoderm (a flanking ventral fate) in surrounding cells. In their absence, cells that would normally be fated as neurogenic ectoderm are transformed to a lateral ectodermal fate. Cells adjacent to the midline (column 1 cells) were ablated as a control and did not produce a similar DV phenotype (data not shown).
To further test the effect of midline cell loss, we ablated the majority of the midline in stage 13 embryos, leaving a single midline cell unablated (Fig. 1H). By stage 17, the progeny of the non-ablated cell had formed a small cluster of Ph-otd-1-positive cells. Remarkably, such small clusters of midline cells rescued DV patterning, as seen by Ph-Dll-e expression, not only in the segment containing these midline cells, but also in an adjacent segment. Ph-Dll-e was still expressed in the appropriate segmental rows, but expression always occurred at a fixed distance from the remaining midline cells (Fig. 1I). This result illustrates that, within the ectoderm, the distance of a cell from the ventral midline appears to be the primary determinant in its decision to adopt a given DV fate.
Ph-sim is expressed in the ventral midline and is required for midline differentiation
As a first step towards determining the molecular properties of the ventral midline, we cloned the Parhyale ortholog of single-minded (Ph-sim). In Drosophila, sim mutants fail to form a functional midline and the mesectodermal cells instead take on the fate of neighboring neuroectodermal cells, whereas ectopic sim is sufficient to induce expression of midline genes in a cell-autonomous fashion (Xiao et al., 1996; Nambu et al., 1991).
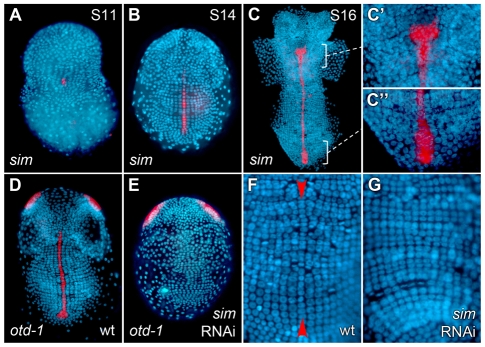
Ph-sim expression was first seen at stage 11 in ventral midline cells, coincident with the initiation of germ band condensation (Fig. 3A). During germ band formation, Ph-sim transcript was observed throughout the ventral midline, as well as in the midline precursor cells located in the posterior of the developing germ band (Fig. 3B,C). The anterior boundary of Ph-sim is initially coincident with the morphologically visible anterior boundary of midline cells (mandibular segment). By stage 16, we observed an expanded expression domain in the second antennal segment that was not confined to a single column of cells (Fig. 3C′).
Fig. 3.
Ph-sim is expressed in midline cells and is required for midline differentiation. Nuclei were counterstained with DAPI (blue) and a false-color overlay of gene expression patterns was generated. (A-C) Wild-type expression of Ph-sim. (A) Stage 11. Ph-sim is first visible in midline cells during initial condensation of germ band. (B) Stage 14. Ph-sim is expressed throughout the ventral midline. (C) Ph-sim expression at stage 16. (C′,C′) Magnification of regions indicated by brackets in C. (C′) Ph-sim expression is now visible in the second antennal segment in the expanded medial domain. (C′) Magnification showing Ph-sim expression in posterior midline precursor cells (not confined to a single column of cells). (D) Wild-type expression of Ph-otd-1 in midline cells as well as in two domains in the head. (E) Ph-otd-1 expression in Ph-sim RNAi embryo. Midline staining is absent (in 23 out of 24 embryos), whereas staining in the head is unaffected (24 out of 24). (F) Ectodermal grid in wild-type embryo. The ventral midline (red arrowheads) bisects the embryo and is discernible based on morphology. (G) Ph-sim RNAi embryo. Midline cells and midline precursor cells are no longer visible.
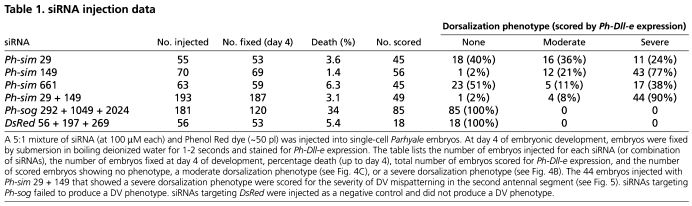
To test the developmental role of Ph-sim, embryonic RNAi was performed. Three individual siRNAs targeting different regions of the Ph-sim transcript were used, giving similar results with varying penetrance (Table 1). A combination of two siRNAs (Ph-sim 29 and Ph-sim 149) produced maximum penetrance and was used to generate the results shown here.
Table 1.
siRNA injection data
Ph-sim RNAi embryos lacked expression of the midline marker Ph-otd-1 in the ectodermal grid, whereas expression in the head was unaffected (Fig. 3E). When visualized by DAPI fluorescence, the midline in wild-type embryos can be identified by its distinctive morphology; midline cells are initially smaller and slightly recessed from the plane of the ectodermal grid (Fig. 3F). In Ph-sim RNAi embryos, however, no midline cells could be identified (Fig. 3G). These results suggest a failure to differentiate ventral midline cells in Ph-sim RNAi embryos, as well as implicating a conserved role for this gene in specifying the midline fate in Parhyale.
Ph-sim RNAi causes severe dorsalization of the embryonic trunk
A small number of Ph-sim RNAi embryos (4 out of 49) showed a moderate dorsalization phenotype, similar to that seen in our midline ablation experiments; the ventral neuroectodermal fate was lost in these embryos and the more dorsal appendage fate was shifted to occupy the ventral-most cells in the germ band (Fig. 4C,D). In these embryos, one or more segments showed a single ventral domain of Ph-Dll-e expression. However, the majority of Ph-sim RNAi embryos (44 out of 49) displayed a more severe dorsalization phenotype. In these embryos, no Ph-Dll-e expression was observed posterior to the second antennal segment (Fig. 4B). In some embryos, the second antennal appendage buds were fused ventrally (Fig. 5C) or were missing altogether (Fig. 5B). Additionally, Ph-sim RNAi embryos failed to express the early presumptive neuroectodermal marker Ph-Pax3/7-1 posterior to the second antennal segment, confirming the failure to specify this tissue (Fig. 4F).
Fig. 5.
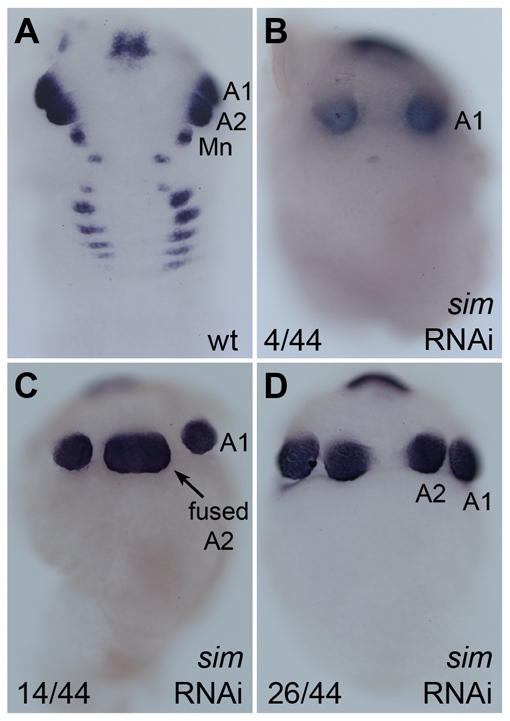
Head morphology variations in sim RNAi embryos. Where appropriate, the first and second antennal (A1, A2) and mandibular (Mn) segments are labeled (A) Ph-Dll-e expression in wild-type embryo. (B) In 4 out of 44 embryos showing a severe DV phenotype, the A2 appendages are completely missing. (C) In 14 out of 44 embryos, DV patterning in A2 is moderately affected leading to a single, ventrally fused appendage bud (arrow). (D) In 26 out of 44 embryos, DV patterning in the A2 segment is minimally affected and appendage buds appear distinctly separate (same embryo as shown in Fig. 4B).
We interpret the lack of Ph-Dll-e and Ph-Pax3/7-1 expression in the developing germ band as a complete transformation of the ectodermal grid to the dorsal ectoderm fate (dorsal ectoderm is the only tissue that fails to express both genes in the wild-type embryo). Although we have not identified any positive markers for dorsal ectoderm, Ph-Eve is a robust marker of the dorsal mesoderm. In wild-type embryos, Ph-Eve was detected in the m4a mesodermal cell and its progeny, and was visible at stage 19 in small clusters of cells at the periphery (dorsal-most regions) of the embryo (Fig. 4I). In Ph-sim RNAi embryos, however, Ph-Eve expression was seen in a larger number of cells traversing the ventral-most regions of the embryo (Fig. 4J). To show that this was a result of ectopic Ph-Eve expression rather than increased proliferation of m4a, we looked in stage 17 embryos at segments in which the mesoderm still consisted of 16 cells (immediately following the first division of mesoblast cells). At this stage, Ph-sim RNAi embryos showed ectopic expression of Ph-Eve in more ventral mesoblast daughter cells (21 out of 23), and, occasionally, in all four anterior daughter cells (m1a-m4a) on a given side of the embryo (2 out of 23) (Fig. 3L). This result further supports the hypothesis that ventral and lateral tissues transform to a dorsal fate in Ph-sim RNAi embryos. Additionally, the ventral midline appears to be either directly or indirectly involved in generating DV pattern in both the ectodermal and mesodermal germ layers. We also observed that the ectodermal grid in Ph-sim RNAi embryos frequently appeared narrower than that seen in wild-type embryos. We interpret this as an expansion of dorsal extra-embryonic tissues, which is likely to be a consequence of overall dorsalization of the DV axis.
Finally, we looked at the effect of Ph-sim RNAi on segmental (AP) patterning using the segment polarity gene hedgehog. We cloned the Parhyale ortholog (Ph-hh) and observed that it was expressed in segmental stripes in wild-type embryos (Fig. 4G). Ph-hh showed similar segmental expression in the developing germ band of Ph-sim RNAi embryos, with minor aberrations attributed to the affected morphology of these embryos, suggesting that AP patterning was largely unaffected by the loss of a functional midline (Fig. 4H). siRNAs designed against the fluorescent protein DsRed were used as a negative control; DsRed RNAi embryos showed no difference in expression of Ph-otd-1 (8 out of 8), Ph-Dll-e (18 out of 18), or Ph-Pax3/7-1 (8 out of 8) from wild-type embryos (data not shown).
Loss of the midline phenotype suggests involvement of BMP antagonists
Based on the results of our laser ablation experiments, we predicted that the midline produces one or more secreted proteins capable of inducing ventral fates (e.g. neurogenic ectoderm) in the surrounding tissue. Furthermore, comparisons with Drosophila and other insects suggest that these secreted proteins are likely to function by antagonizing BMPs. The link between extracellular BMP antagonism and neural specification appears to be widely conserved and has been demonstrated in Drosophila, Tribolium, the chelicerate Achaearanea, and the annelid Platynereis, as well as in vertebrate systems (Francois et al., 1994; van der Zee et al., 2006; Akiyama-Oda and Oda, 2006; Denes et al., 2007; Harland, 2000).
Drosophila DV patterning involves various secreted proteins that contribute to a BMP activity gradient by the gastrula stage (Little and Mullins, 2006). Among these, the BMP antagonist short gastrulation (sog) is activated in the presumptive neuroectoderm via a threshold response to the Dorsal gradient (Jiang et al., 1992) and, interestingly, is also activated in midline cells by sim (Zinzen et al., 2006). Furthermore, a sog ortholog is expressed in midline cells in the branchiopod crustacean Artemia franciscana (Akiyama-Oda and Oda, 2006). We therefore considered sog to be a strong candidate for mediating midline function in Parhyale, and cloned the Parhyale ortholog of this gene (Ph-sog). We were able to detect Ph-sog transcript in midline cells in Parhyale, but, surprisingly, it was restricted to a group of midline cells in the anterior, primarily in the presumptive mandibular segment. Posterior to this, no localized expression was observed in the midline (Fig. 4M). Furthermore, Ph-sog knockdown was attempted via embryonic siRNA injection, but this failed to generate the dorsalization phenotype seen in Ph-sim RNAi embryos (Table 1).
DISCUSSION
Parhyale midline cells pattern surrounding tissue
This work provides the first functional study of sim and the ventral midline in a non-insect arthropod system. We show that within the ectodermal grid of Parhyale, the relative distance of a cell from the ventral midline is the primary determinant of its eventual DV fate. This long-range patterning function of the midline is analogous to vertebrate structures such as the Spemann organizer and the floor plate of the CNS (Harland and Gerhart, 1997; Placzek and Briscoe, 2005). Interestingly, there appears to be no single structure in the insect embryo that is as prominently required for patterning the DV axis. Furthermore, we consider this to be strong evidence that the ventral midline of Parhyale secretes one or more morphogenic proteins that are capable of patterning the surrounding tissue. In Drosophila, the ventral midline produces two distinct secreted signals: the BMP antagonist Sog and the EGF ligand Spitz. Although it has been shown that Drosophila Egfr (DER) mutants show aberrant expression of Dll (Kubota et al., 2000), EGF signaling alone is unlikely to explain the complete loss of ventral and lateral ectoderm in Ph-sim RNAi embryos. For example, various Drosophila mutations that interfere with Spitz signaling cause defects in proper neuroblast delamination and fate specification, but still contain neurogenic ectoderm (Chang et al., 2000). Instead, the dorsalization phenotype appears to be more consistent with a loss of BMP antagonism in ventral tissues.
Our data argue against the idea that Ph-sog alone is responsible for mediating the function of the Parhyale midline. However, it is likely that additional BMP antagonists are present in Parhyale and that these might be key factors in mediating the developmental role of midline cells. DV patterning in vertebrates, for example, involves multiple secreted BMP antagonists and Tribolium contains orthologs to many of these vertebrate BMP antagonists that are not present in the Drosophila genome (Little and Mullins, 2006; Van der Zee et al., 2008). It would, therefore, be interesting to characterize additional BMP antagonists in Parhyale in order to determine whether they contribute to the function of midline cells.
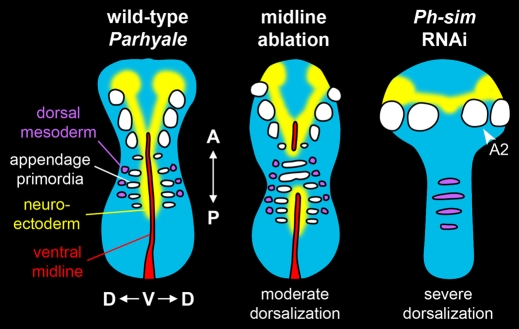
Although we were able to generate dorsalization phenotypes in embryos through either laser ablation of midline cells or Ph-sim RNAi, the DV phenotype observed in Ph-sim RNAi embryos was more severe (Fig. 6). We believe that this was a result of the timing of the loss of midline function; laser ablation of midline cells is performed after the cells have been present (and presumably functional) in the germ band for several hours. By contrast, Ph-sim RNAi prevents the formation of midline altogether. We therefore predict that the latter represents the true loss of midline phenotype in Parhyale, and that the difference in phenotypic severity suggests that the timing and/or duration of signaling from the midline contribute to differential fates along the DV axis.
Fig. 6.
Dorsalization phenotypes. Wild-type, midline-ablated, and Ph-sim RNAi embryos are depicted. In midline-ablated embryos, segments lacking midline failed to generate neuroectoderm and ventral cells were mis-specified as lateral ectoderm (bearing appendage primordia). The location of dorsal mesoderm in midline-ablated embryos was not examined, but is inferred from ectodermal markers. In Ph-sim RNAi embryos, the ventral midline, neuroectoderm and appendage primordia were absent posterior to the second antennal segment (A2). Dorsalization was confirmed by the presence of dorsal mesoderm in ventral regions of embryos.
Finally, Ph-sim RNAi generated a dramatic DV phenotype in the Parhyale trunk, but the first antennal segment was typically unaffected, and the second antennal segment was dorsalized with lower penetrance and varying degrees of severity. This is consistent with the observation that the ventral midline does not extend into the first antennal segment and that Ph-sim is only expressed in the second antennal segment later in development (stage 16). Because these anterior segments are the first to form in the embryo, it is possible that midline cells function by propagating existing DV patterning information to newly formed body segments, using anterior segments as a template. However, the mechanism for generating DV pattern in the anterior-most segments of Parhyale remains unclear.
Evolution of midline function and DV patterning in arthropods
It has previously been argued that the ventral midline in Parhyale and Drosophila are homologous structures (Simanton et al., 2009). The experimental results showing that Ph-sim specifies the midline fate in Parhyale supports this idea and further suggests that this developmental role of sim dates back to at least the last common ancestor of insects and malacostracan crustaceans. Furthermore, expression of At-sim in the midline of Achaearanea and in ventrally located cell clusters in Platynereis indicates that this might be a plesiomorphy of all protostomes (Akiyama-Oda and Oda, 2006; Denes et al., 2007).
How could the seemingly disparate roles of Parhyale and Drosophila midline cells in DV axis patterning have evolved? We hypothesize that the common ancestor used ventral midline cells as a localized source of secreted BMP antagonists, including sog, and that these cells played an ancestral role in patterning the DV axis. The fact that midline function in Parhyale does not appear to be mediated by Ph-sog could be explained if there were multiple midline-associated BMP antagonists present in the common ancestor. If this were true, the lack of Ph-sog in the majority of Parhyale midline cells could simply be a derived feature (apomorphy) of this lineage, as it appears to be retained in both Artemia and Drosophila.
Furthermore, it has been theorized that the evolution of holometabolous insects has been accompanied by a functional shift in which Toll signaling and the Dorsal activity gradient have gradually replaced BMP signaling as the primary determinants of DV axis formation (Nunes da Fonseca et al., 2008). We propose that once Dorsal assumed regulatory control of sog in the insect lineage, it would have bypassed the ancestral requirement of the ventral midline as a discrete source of BMP antagonists. Consequently, the ventral midline in insects could have evolved to play a more restricted role in DV patterning. This would explain the fact that no functional role for sog in the Drosophila midline has been described. Implicitly, we would also expect that Dorsal plays only a small role, if any, in the DV patterning of crustaceans such as Parhyale. This work provides an important step towards understanding the evolution of DV patterning in arthropods. However, further analysis of the function of ventral midline cells and BMP antagonists in additional arthropod systems will be required to test our predictions and to gain further insight into ancestral DV patterning mechanisms.
Supplementary Material
Acknowledgments
We thank Melinda Modrell for supplying the Hsp70-DsRed-NLS animal line, Elaine Kwan for generating the Ph-Eve antibody, and Henrique Marques-Souza, Mike Perry and Crystal Chaw for helpful comments on the manuscript. N.H.P. is an Investigator of the Howard Hughes Medical Institute. Deposited in PMC for release after 6 months.
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material for this article is available at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.055160/-/DC1
References
- Akiyama-Oda Y., Oda H. (2006). Axis specification in the spider embryo: dpp is required for radial-to-axial symmetry transformation and sog for ventral patterning. Development 133, 2347-2357 [DOI] [PubMed] [Google Scholar]
- Browne W. E., Price A. L., Gerberding M., Patel N. H. (2005). Stages of embryonic development in the amphipod crustacean, Parhyale hawaiensis. Genesis 42, 124-149 [DOI] [PubMed] [Google Scholar]
- Browne W. E., Schmid B. G., Wimmer E. A., Martindale M. Q. (2006). Expression of otd orthologs in the amphipod crustacean, Parhyale hawaiensis. Dev. Genes Evol. 216, 581-595 [DOI] [PubMed] [Google Scholar]
- Brusca R. C., Brusca G. J. (2003). Invertebrates. Sunderland, MA: Sinauer Associates; [Google Scholar]
- Chang J., Kim I. O., Ahn J. S., Kwon J. S., Jeon S. H., Kim S. H. (2000). The CNS midline cells coordinate proper cell cycle progression and identity determination of the Drosophila ventral neuroectoderm. Dev. Biol. 227, 307-323 [DOI] [PubMed] [Google Scholar]
- Crews S. T., Thomas J. B., Goodman C. S. (1988). The Drosophila single-minded gene encodes a nuclear protein with sequence similarity to the per gene product. Cell 52, 143-151 [DOI] [PubMed] [Google Scholar]
- Davis G. K., D'Alessio J. A., Patel N. H. (2005). Pax3/7 genes reveal conservation and divergence in the arthropod segmentation hierarchy. Dev. Biol. 285, 169-184 [DOI] [PubMed] [Google Scholar]
- Denes A. S., Jékely G., Steinmetz P. R., Raible F., Snyman H., Prud'homme B., Ferrier D. E., Balavoine G., Arendt D. (2007). Molecular architecture of annelid nerve cord supports common origin of nervous system centralization in bilateria. Cell 129, 277-288 [DOI] [PubMed] [Google Scholar]
- Doe C. Q., Chu-LaGraff Q., Wright D. M., Scott M. P. (1991). The prospero gene specifies cell fates in the Drosophila central nervous system. Cell 65, 451-464 [DOI] [PubMed] [Google Scholar]
- Duman-Scheel M., Patel N. H. (1999). Analysis of molecular marker expression reveals neuronal homology in distantly related arthropods. Development 126, 2327-2334 [DOI] [PubMed] [Google Scholar]
- Francois V., Solloway M., O'Neill J. W., Emery J., Bier E. (1994). Dorsal-ventral patterning of the Drosophila embryo depends on a putative negative growth factor encoded by the short gastrulation gene. Genes Dev. 8, 2602-2616 [DOI] [PubMed] [Google Scholar]
- Frasch M., Hoey T., Rushlow C., Doyle H., Levine M. (1987). Characterization and localization of the even-skipped protein of Drosophila. EMBO J. 6, 749-759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerberding M., Scholtz G. (1999). Cell lineage of the midline cells in the amphipod crustacean Orchestia cavimana (Crustacea, Malacostraca) during formation and separation of the germ band. Dev. Genes Evol. 209, 91-102 [DOI] [PubMed] [Google Scholar]
- Gerberding M., Browne W. E., Patel N. H. (2002). Cell lineage analysis of the amphipod crustacean Parhyale hawaiensis reveals an early restriction of cell fates. Development 129, 5789-5801 [DOI] [PubMed] [Google Scholar]
- Golembo M., Raz E., Shilo B. Z. (1996). The Drosophila embryonic midline is the site of Spitz processing, and induces activation of the EGF receptor in the ventral ectoderm. Development 122, 3363-3370 [DOI] [PubMed] [Google Scholar]
- Harland R. (2000). Neural induction. Curr. Opin. Genet. Dev. 10, 357-362 [DOI] [PubMed] [Google Scholar]
- Harland R., Gerhart J. (1997). Formation and function of Spemann's organizer. Annu. Rev. Cell Dev. Biol. 13, 611-667 [DOI] [PubMed] [Google Scholar]
- Holley S. A., Jackson P. D., Sasai Y., Lu B., De Robertis E. M., Hoffmann F. M., Ferguson E. L. (1995). A conserved system for dorsal-ventral patterning in insects and vertebrates involving sog and chordin. Nature 376, 249-253 [DOI] [PubMed] [Google Scholar]
- Jiang J., Rushlow C. A., Zhou Q., Small S., Levine M. (1992). Individual dorsal morphogen binding sites mediate activation and repression in the Drosophila embryo. EMBO J. 11, 3147-3154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasai Y., Nambu J. R., Lieberman P. M., Crews S. T. (1992). Dorsal-ventral patterning in Drosophila: DNA binding of snail protein to the single-minded gene. Proc. Natl. Acad. Sci. USA 89, 3414-3418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosman D., Ip Y. T., Levine M., Arora K. (1991). Establishment of the mesoderm-neuroectoderm boundary in the Drosophila embryo. Science 254, 118-122 [DOI] [PubMed] [Google Scholar]
- Kubota K., Goto S., Eto K., Hayashi S. (2000). EGF receptor attenuates Dpp signaling and helps to distinguish the wing and leg cell fates in Drosophila. Development 127, 3769-3776 [DOI] [PubMed] [Google Scholar]
- Leptin M. (1991). twist and snail as positive and negative regulators during Drosophila mesoderm development. Genes Dev. 5, 1568-1576 [DOI] [PubMed] [Google Scholar]
- Little S. C., Mullins M. C. (2006). Extracellular modulation of BMP activity in patterning the dorsoventral axis. Birth Defects Res. C Embryo Today 78, 224-242 [DOI] [PubMed] [Google Scholar]
- Liubicich D. M., Serano J. M., Pavlopoulos A., Kontarakis Z., Protas M. E., Kwan E., Chatterjee S., Tran K. D., Averof M., Patel N. H. (2009). Knockdown of Parhyale Ultrabithorax recapitulates evolutionary changes in crustacean appendage morphology. Proc. Natl. Acad. Sci. USA 106, 13892-13896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer U., Nüsslein-Volhard C. (1988). A group of genes required for pattern formation in the ventral ectoderm of the Drosophila embryo. Genes Dev. 2, 1496-1511 [DOI] [PubMed] [Google Scholar]
- Morel V., Schweisguth F. (2000). Repression by suppressor of hairless and activation by Notch are required to define a single row of single-minded expressing cells in the Drosophila embryo. Genes Dev. 14, 377-388 [PMC free article] [PubMed] [Google Scholar]
- Nambu J. R., Lewis J. O., Wharton K. A., Crews S. T. (1991). The Drosophila single-minded gene encodes a helix-loop-helix protein that acts as a master regulator of CNS midline development. Cell 67, 1157-1167 [DOI] [PubMed] [Google Scholar]
- Nunes da Fonseca R., von Levetzow C., Kalscheuer P., Basal A., van der Zee M., Roth S. (2008). Self-regulatory circuits in dorsoventral axis formation of the short-germ beetle Tribolium castaneum. Dev. Cell 14, 605-615 [DOI] [PubMed] [Google Scholar]
- Oliver G., Sosa-Pineda B., Geisendorf S., Spana E. P., Doe C. Q., Gruss P. (1993). Prox 1, a prospero-related homeobox gene expressed during mouse development. Mech. Dev. 44, 3-16 [DOI] [PubMed] [Google Scholar]
- Panganiban G., Sebring A., Nagy L., Carroll S. (1995). The development of crustacean limbs and the evolution of arthropods. Science 270, 1363-1366 [DOI] [PubMed] [Google Scholar]
- Pavlopoulos A., Averof M. (2005). Establishing genetic transformation for comparative developmental studies in the crustacean Parhyale hawaiensis. Proc. Natl. Acad. Sci. USA 102, 7888-7893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Placzek M., Briscoe J. (2005). The floor plate: multiple cells, multiple signals. Nat. Rev. Neurosci. 6, 230-240 [DOI] [PubMed] [Google Scholar]
- Price A. L., Patel N. H. (2008). Investigating divergent mechanisms of mesoderm development in arthropods: the expression of Ph-twist and Ph-mef2 in Parhyale hawaiensis. J. Exp. Zool. B Mol. Dev. Evol. 310, 24-40 [DOI] [PubMed] [Google Scholar]
- Price A. L., Modrell M. S., Hannibal R. L., Patel N. H. (2010). Mesoderm and ectoderm lineages in the crustacean Parhyale hawaiensis display intra-germ layer compensation. Dev. Biol. 341, 256-266 [DOI] [PubMed] [Google Scholar]
- Rehm E. J., Hannibal R. L., Chaw R. C., Vargas-Vila M. A., Patel N. H. (2009). The Crustacean Parhyale hawaiensis: a new model for arthropod development. Cold Spring Harbor Protoc. 2009, pdb.emo114 [DOI] [PubMed] [Google Scholar]
- Roth S., Stein D., Nüsslein-Volhard C. (1989). A gradient of nuclear localization of the dorsal protein determines dorsoventral pattern in the Drosophila embryo. Cell 59, 1189-1202 [DOI] [PubMed] [Google Scholar]
- Simanton W., Clark S., Clemons A., Jacowski C., Farrell-VanZomeren A., Beach P., Browne W. E., Duman-Scheel M. (2009). Conservation of arthropod midline netrin accumulation revealed with a cross-reactive antibody provides evidence for midline cell homology. Evol. Dev. 11, 260-268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stathopoulos A., Van Drenth M., Erives A., Markstein M., Levine M. (2002). Whole-genome analysis of dorsal-ventral patterning in the Drosophila embryo. Cell 111, 687-701 [DOI] [PubMed] [Google Scholar]
- Thomas J. B., Crews S. T., Goodman C. S. (1988). Molecular genetics of the single-minded locus: a gene involved in the development of the Drosophila nervous system. Cell 52, 133-141 [DOI] [PubMed] [Google Scholar]
- Van der Zee M., Stockhammer O., von Levetzow C., Nunes da Fonseca R., Roth S. (2006). Sog/Chordin is required for ventral-to-dorsal Dpp/BMP transport and head formation in a short germ insect. Proc. Natl. Acad. Sci. USA 103, 16307-16312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Zee M., da Fonseca R. N., Roth S. (2008). TGFbeta signaling in Tribolium: vertebrate-like components in a beetle. Dev. Genes Evol. 218, 203-213 [DOI] [PubMed] [Google Scholar]
- Xiao H., Hrdlicka L. A., Nambu J. R. (1996). Alternate functions of the single-minded and rhomboid genes in development of the Drosophila ventral neuroectoderm. Mech. Dev. 58, 65-74 [DOI] [PubMed] [Google Scholar]
- Zinzen R. P., Cande J., Ronshaugen M., Papatsenko D., Levine M. (2006). Evolution of the ventral midline in insect embryos. Dev. Cell 11, 895-902 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.