Abstract
The Arabidopsis basic helix–loop–helix (bHLH) proteins INDEHISCENT (IND) and ALCATRAZ (ALC) specify tissues required for fruit opening that have major roles in seed dispersal and plant domestication. Here, we show that synthesis of the phytohormone gibberellin is a direct and necessary target of IND, and that ALC interacts directly with DELLA repressors, which antagonize ALC function but are destabilized by gibberellin. Thus, the gibberellin/DELLA pathway has a key role in patterning the Arabidopsis fruit, and the interaction between DELLA and bHLH proteins, previously shown to connect gibberellin and light responses, is a versatile regulatory module also used in tissue patterning.
Keywords: Seed dispersal, gibberellin, fruit patterning
Development of the Arabidopsis fruit is a model system for tissue patterning in plants and for understanding the genetic control of seed dispersal, which has a key role in crop domestication and improvement (Purugganan and Fuller 2009). The fruit of Arabidopsis, oilseed rape, and other Brassicaceae is a silique constituted of two valves, protecting the seeds, fused to a central replum by a specific tissue called valve margin (Fig. 1A; Østergaard 2009). Valve margins differentiate into narrow stripes of cells consisting of a lignification layer (LL) and a separation layer (SL). This specialized structure facilitates fruit opening and the efficient release of the seeds: The SL secretes polygalacturonase enzymes to degrade cell walls and allow cell separation, while the LL is believed to provide tension to facilitate the opening mechanism (Petersen et al. 1996; Spence et al. 1996; Mitsuda and Ohme-Takagi 2008). The valve margin identity factors SHATTERPROOF (SHP1/2) and INDEHISCENT (IND) are involved in the differentiation of both the LL and SL (Liljegren et al. 2000, 2004), whereas ALCATRAZ (ALC) is required for SL specification (Rajani and Sundaresan 2001). FRUITFULL (FUL) and REPLUMLESS (RPL) genes, expressed in the valves and replum, respectively, restrict the expression of valve margin identity genes to the valve margins (Ferrandiz et al. 2000; Roeder et al. 2003). Despite the identification of these molecular actors, the precise role of valve margin identity genes—and therefore the set of genes that they control—remains to be elucidated.
Figure 1.
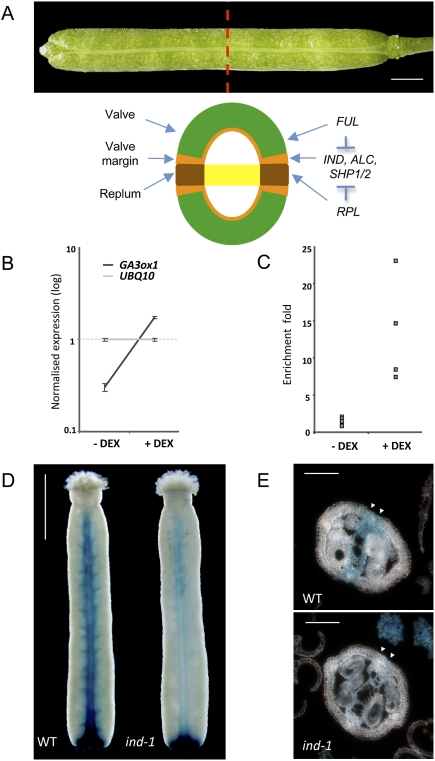
The gibberellin biosynthetic gene GA3ox1 is a direct target of IND. (A) Mature wild-type Arabidopsis fruit and schematic cross-section showing the different tissues. The regulatory network controlling the development of fruit tissues is also indicated. Expression of the valve margin identity genes (IND, ALC, and SHP1/2) is restricted to the valve margins by the FUL and RPL activities in the valve and the replum, respectively. Bar, 1 mm. (B) Transcript profiling assay using 7-d-old 35S∷IND:GR seedlings before (−DEX) and after (+DEX) 6 h of DEX treatment, showing an increase in GA3ox1 mRNA accumulation in response to DEX treatment compared with UBQ10 mRNA as internal control (n = 3). Error bars represent standard deviation (SD). (C) ChIP showing the direct interaction of IND-GR with the GA3ox1 gene. DNA obtained from pull-down with the GR antibody has been analyzed by qPCR using specific primers for the GA3ox1 gene. Values correspond to the ratios between pull-down and input DNA, both initially normalized to the Mu-like transposon. Values for four biological repeats are represented. P-value, <0.01. (D) β-Glucuronidase expression of AtGA3ox1 TC-GUS in young fruit (stage 15 according to Smyth et al. 1990) in the wild type (WT) and ind-1 mutant (ind-1). Bar, 500 μm. (E) β-Glucuronidase expression of AtGA3ox1 TC-GUS in young fruit (stage 15) cross-sections in th wild type and ind-1 mutant (ind-1). Bar, 100 μm.
Results and Discussion
IND directly activates a key gibberellin biosynthesis gene
To investigate the role of IND in the patterning of valve margins, we used a dexamethasone (DEX)-inducible version of the IND gene translationally fused to the glucocorticoid receptor (GR) under the control of the CaMV35S promoter (35S∷IND:GR) (Sorefan et al. 2009). Global transcriptomic profiling comparing DEX-induced and noninduced plants was used to reveal putative targets of IND. We observed that the gene GA3ox1 was induced after 6 h of DEX treatment compared with the internal control UBQ10 (Fig. 1B). This result was confirmed using quantitative PCR (qPCR) analysis (Supplemental Fig. S1A). We then used cycloheximide to prevent de novo protein synthesis and therefore reveal immediate IND targets (Sablowski and Meyerowitz 1998). The DEX treatment was still able to trigger the accumulation of GA3ox1 mRNA without active translational machinery, suggesting that GA3ox1 is an immediate target of IND (Supplemental Fig. S1B). To investigate whether this regulation is direct, we performed a chromatin immunoprecipitation (ChIP) assay using the GR antibody followed by qPCR analysis. GA3ox1 was significantly enriched in DEX-induced 35S∷IND:GR plants compared with no DEX control (Fig. 1C), showing that GA3ox1 was bound by IND:GR in vivo.
GA3ox1 encodes a Gibberellin 3-oxidase, which catalyses the last step in the biosynthesis of bioactive gibberellins (GA1 and GA4) (Talon et al. 1990). GAs are important regulators of plant growth through both cell division and cell elongation (Hedden and Phillips 2000; Sun and Gubler 2004; Yamaguchi 2008). It has been shown that the expression pattern of GA3ox1 (and other members of the GA3ox family) corresponds to the sites where active GAs are produced (Itoh et al. 1999; Mitchum et al. 2006). To confirm that endogenous IND participates in establishing the GA3ox1 expression pattern, we used the AtGA3ox1 TC-GUS reporter, which has been validated previously in a detailed analysis of GA3ox1 expression in seedlings (Mitchum et al. 2006; Hu et al. 2008). Examination of stage 15 gynoecia showed expression of AtGA3ox1 TC-GUS in valve margins and the septum (Fig. 1D,E; stages defined in Smyth et al. 1990), overlapping with the expression pattern of IND (Liljegren et al. 2004). Importantly, this specific aspect of AtGA3ox1 TC-GUS expression depended on IND: In the ind-1 mutant, expression was significantly reduced in valve margins but remained comparable in the gynophore (at the base of the developing fruit) (Fig. 1D). In conclusion, the IND-dependent AtGA3ox1 TC-GUS expression, the microarray, and the ChIP results together indicate that IND directly activates GA3ox1 in medial tissues of the fruit, which include valve margins.
Gibberellin is required for valve margin development
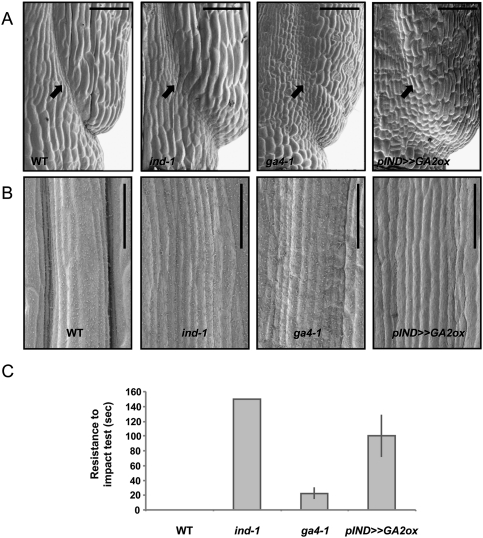
Fruit development involves extensive GA-activated cell elongation, which is partially dependent on GA3ox1 (Koornneef and Van der Veen 1980; Chiang et al. 1995). IND, however, is not required for fruit elongation (Liljegren et al. 2004), raising the question of whether GAs might have additional roles during fruit development. To test whether GAs have specific roles downstream from IND during valve margin development, we first analyzed valve margin morphology in the ga4-1 mutant. This mutant contains a point mutation in the coding sequence of GA3ox1 (Koornneef and Van der Veen 1980; Chiang et al. 1995), and has reduced levels of bioactive GAs in the shoot (Talon et al. 1990). Scanning electron microscopy (SEM) showed that valve margins were not properly defined in the ga4-1 mutant compared with the wild type (Fig. 2A,B). To avoid the pleiotropic effects of gibberellin-deficient mutants and reveal the consequences of lowering gibberellin levels specifically in the valve margins, we also generated pIND >> GA2ox plants, in which the IND promoter directed expression of the gibberellin-inactivating GA2ox2 gene (Rieu et al. 2008). As observed with ga4-1, these plants also showed valve margin defects (Fig. 2A,B), confirming that localized gibberellin levels control differentiation of valve margins.
Figure 2.
Low GA levels disrupt valve margin development and inhibit fruit opening. (A) SEM images showing the base of Arabidopsis fruit (stage 17b) of the wild type (WT), ind-1, ga4-1, and pIND >> GA2ox. Bar, 50 μm. (B) SEM images showing a close-up of valve margin and replum tissues. Pictures were taken in the middle of an Arabidopsis fruit (stage 17b) of the wild type, ind-1, ga4-1, and pIND >> GA2ox. Bar, 50 μm. (C) Shattering quantification of wild-type, ind-1, ga4-1, and pIND >> GA2ox fruits. Values represent the mean of three biological repeats (n = 3) (20 siliques each). Error bars represent standard deviation (SD).
To verify that the valve margin defects caused by gibberellin depletion affected fruit opening, we developed a random impact test (RIT) for quantifying shatter resistance in Arabidopsis fruits (see the Materials and Methods for details). Shattering measurements revealed that the fruits of both the ga4-1 mutant and the pIND >> GA2ox transgenic line were more resistant to opening than the Landsberg erecta wild type, which is too sensitive for the assay to obtain a value greater than zero (Fig. 2C). This shatter-resistant phenotype was confirmed using another mutant allele for GA3ox1, the ga4-3 mutant in a Columbia background (Supplemental Fig. S2). The pIND >> GA2ox transgenic line showed more pronounced shatter resistance than the ga4-1 mutant, suggesting that other members of the GA3ox family (Mitchum et al. 2006) might contribute to gibberellin production during valve margin formation. In agreement with this observation, expression of a gene encoding a GA3ox2 enzyme (At1g80340) was also found to be significantly up-regulated in response to DEX induction in the transcriptomic profiling described above (data not shown). In contrast to the partial shatter resistance of ga4 mutants and the pIND >> GA2ox line, fruits of the ind-1 mutant never opened during the analysis (Fig. 2C), confirming that fruit opening is promoted by additional IND functions apart from activating gibberellin synthesis (Sorefan et al. 2009). Nevertheless, the results above showed that local production of gibberellins promoted by IND is required for the differentiation of fully functional valve margins.
Valve margin defects caused by low gibberellin resemble the alc phenotype
Next, we used transmission electron microscopy (TEM) to reveal in cellular detail how gibberellin depletion affected valve margin development. In wild-type fruits, the LL and SL were clearly recognizable: LL cells exhibited thick cell walls, whereas SL cells were small and nonlignified (Fig. 3A,B) as described previously (Rajani and Sundaresan 2001; Wu et al. 2006). Small SL cells were found directly adjacent to the LL. In the ga4-1 mutant, cells forming the SL could not be identified. Instead, larger cells resembling replum cells were found adjacent to the LL. The valve margin defect observed in the ga4-1 mutant background is thus due to the lack of a proper SL, whereas the LL appears to be unaffected compared with the wild type (Fig. 3C). Since IND is involved in the formation of both the SL and LL (Liljegren et al. 2004), this result may explain why the fruit-opening defects were less severe in ga4-1 compared with ind-1.
Figure 3.
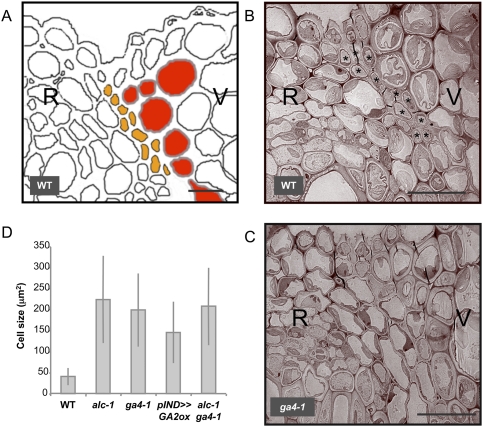
Low GA levels cause SL defects that resemble those of the alc mutant. (A) Diagram (based on a mPS-PI/confocal image) of the different cell layers in the wild-type valve margins: SL (orange) and LL (red). (B,C) TEM images showing valve margin tissues (cross-section) of the wild type (WT) (B) and ga4-1 mutant (C). SL cells are marked with asterisks (*). (R) Replum; (V) valve. Bar, 10 μm. (D) Quantification of cell sizes in the cell layer corresponding to the SL in the wild type (n = 38), alc-1 (n = 46), ga4-1 (n = 41), pIND >> GA2ox (n = 34), and alc-1; ga4-1 double mutant (n = 49). Error bars represent standard deviation (SD).
To our knowledge, ALC is the only gene shown to be specifically involved in the differentiation of the SL. Interestingly, the valve margin phenotype of ga4-1 resembles the alc-1 phenotype described by Rajani and Sundaresan (2001). To compare quantitatively the cellular defects seen in alc-1 and in plants with reduced gibberellin levels, we analyzed optical cross-sections of Arabidopsis fruits stained with a modified pseudo-Schiff propidium iodide (mPS-PI) method (Truernit et al. 2008). Using this technique, the SL is visible in the wild type as a layer of small, thin-walled cells adjacent to the LL (Supplemental Fig. S3). In contrast, the SL in the alc-1 mutant was not differentiated, and large cells were located close to the lignified tissue as described previously (Rajani and Sundaresan 2001). Similar defects were seen in the ga4-1 mutant and in the pIND >> GA2ox transgenic line (Supplemental Fig. S3). Measurement of the size of the cells directly adjacent to the LL in the wild-type, alc-1, ga4-1, and pIND >> GA2ox transgenic lines confirmed that all three genotypes had significantly larger cells at the site of separation than the wild type (Fig. 3D). The similar phenotypes suggested that ALC and GA3ox1 could act in the same pathway to control SL differentiation. This was confirmed by generating the alc-1; ga4-1 double mutant: The size of SL cells in the alc-1; ga4-1 double mutant was not significantly different from SL cell size of alc-1 and ga4-1 single mutants (Fig. 3D).
Gibberellin releases ALC from DELLA repression at the valve margin
ALC is a basic helix–loop–helix (bHLH) transcription factor belonging to group VII, according to the bHLH classification described by Heim et al. (2003). This group also contains PHYTOCHROME-INTERACTING FACTOR 3 (PIF3) and PIF4. In the absence of GA, PIF transcription factors are prevented from activating their targets through interaction via their DNA interaction domain with the growth-repressing DELLA proteins, whereas the presence of GA relieves this inhibition (de Lucas et al. 2008; Feng et al. 2008). Notably, ALC, PIF3, and PIF4 share the same H-E-R motif in their DNA interaction domains (Heim et al. 2003). To test if ALC could be regulated in a similar manner, we analyzed the potential interaction of ALC with DELLA proteins. Yeast two-hybrid experiments revealed that ALC indeed interacts with the Arabidopsis DELLA proteins GAI, RGA, and RGL2 (Fig. 4A; Supplemental Fig. S4A). In order to confirm this interaction in planta, we performed bimolecular fluorescence complementation (BiFC) assays. Interaction between ALC and RGA was again detected in nuclei of transiently transformed Nicotiana benthamiana leaf cells (Supplemental Fig. S4B).
Figure 4.
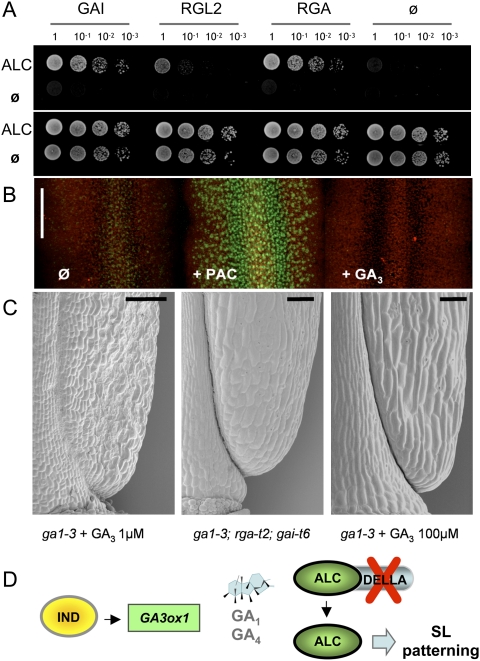
DELLA proteins interact with ALC and inhibit valve margin development in the absence of GA. (A) Yeast two-hybrid interaction using ALC as a bait and GAI, RGA, and RGL2 as prey. (Top and bottom panels) Dilution series plated on selective medium (−Leu Trp His Ade) and control medium (−Leu Trp), respectively. (B) Projections of confocal sections showing expression of pRGA∷GFP-RGA in medial tissues of stage 15 gynoecium: untreated (Ø), treated with 1 μM PAC (+PAC), or treated with 10 μM gibberellin (+GA3) for 24 h. (C) SEM images showing the base of a stage 17b fruit of the ga1-3 mutant treated with 1 μM GA3 (left panel) or 100 μM GA3 (right panel), and of the ga-3; rga-t2; gai-t6 triple mutant (middle panel). Bar = 100 μm. (D) Model of DELLA-mediated control of ALC by IND. IND induces GA biosynthesis via the direct activation of GA3ox1. Without GAs, ALC interacts with DELLA proteins. During valve margin development, IND-induced local GA production leads to DELLA degradation and release of ALC, which then controls SL genes.
Interaction with ALC in vivo would also be consistent with the reported expression of GAI, RGA, and RGL2 in flowers and siliques (Lee et al. 2002). To confirm that DELLA proteins are expressed in the tissues where ALC functions during valve margin formation, we analyzed in detail the expression pattern of the previously characterized pRGA∷GFP:RGA line (Silverstone et al. 2001). Because GFP-RGA expressed in the valve margins was predicted to be degraded by locally produced GA, we compared pRGA∷GFP:RGA expression in untreated controls and in developing fruits treated with the GA biosynthesis inhibitor Paclobutrazol (PAC) or exogenous GA. PAC treatment revealed strong expression of GFP-RGA throughout the medial tissues of young fruits, including valve margins (Fig. 4B). Consistent with the idea that locally produced GA leads to degradation of DELLAs in developing valve margins, untreated controls showed much lower GFP-RGA expression, which was abolished after treatment with GA3 (Fig. 4B).
To test whether DELLA expression during valve margin formation is functionally relevant, we compared valve margin morphology in the ga1-3 mutant and the ga1-3; rga-t2; gai-t6 triple mutant. The GA1 gene encodes a key enzyme in the GA biosynthetic pathway, and the ga1-3 mutant presents severe developmental defects due to very low levels of GA (Sun et al. 1992). Applications of GA3 were necessary to restore plant growth and obtain fruits. Consistent with our previous results, fruits of ga1-3 mutant treated with low GA3 concentration (1 μM) presented the same valve margin defects as ga4-1 and pIND >> GA2ox plants (Fig. 4C). This valve margin defect was corrected in both the ga1-3 treated with high GA3 (100 μm) and the ga1-3; rga-t2; gai-t6 triple mutant (Fig. 4C). We conclude that the DELA proteins RGA and GAI act as repressors of valve margin formation in the absence of GA.
Together, all of the results described above support the following model for GA-mediated specification of SL in Arabidopsis (Fig. 4D). We propose that IND directly activates the expression of GA3ox1 and, consequently, the production of bioactive GAs in developing valve margins. Prior to SL specification, we hypothesize that ALC is bound to DELLA proteins, preventing activation of its targets. Local gibberellin synthesis leads to degradation of DELLA proteins, releasing ALC to modulate the expression of its target genes and direct the differentiation of the SL. In this model, activation of ALC by GA biosynthesis occurs post-transcriptionally and not through regulation of ALC gene expression. To test if this is the case, we analyzed the ALC expression pattern in the ga4-1 mutant (Supplemental Fig. S5). No alterations in ALC expression were observed in the ga4-1 mutant background, consistent with our hypothesis that GAs control ALC at a post-transcriptional level.
Conclusions
Our results show that the GA/DELLA pathway is an important component of the regulatory network controlling fruit opening and seed dispersal. de Lucas et al. (2008) and Feng et al. (2008) demonstrated that the interaction between DELLA and PIF3/PIF4 transcription factors coordinate light and gibberellin responses, raising the possibility that interactions with DELLA proteins could control the activity of additional plant bHLH transcription factors. Accordingly, two other bHLH transcription factors, PIL5 and SPT, have also been involved in light and GA signaling (Oh et al. 2007; Penfield et al. 2005), making them potential targets for DELLA repressors. Here we show that this DELLA/bHLH regulatory module is not only involved in the integration of light and gibberellin signals, but is implicated in fruit patterning as well, supporting the idea that it is a versatile regulatory device used in multiple environmental and developmental responses.
An emerging feature of plant development is that major regulatory genes often function as orchestrators of localized hormone synthesis or response. The results reported here reveal IND as a central coordinator of different hormonal pathways. Our demonstration that IND-promoted biosynthesis of GA is necessary for fruit opening and seed dispersal echoes the previous implication of another plant hormone, auxin, in this mechanism (Sorefan et al. 2009). This raises the question of how GA and auxin pathways interact to form the valve margin. In fact, these two pathways appear to function independently: The auxin minima flanking the replum (Sorefan et al. 2009) were still present in the ga4-1 mutant (Supplemental Fig. S6), suggesting that establishment of the auxin minima is independent of GA biosynthesis. IND therefore controls multiple hormonal pathways to ensure the proper development of the valve margins and possibly to coordinate valve margin differentiation with fruit growth. This may reflect the location of valve margins at a developmental boundary, between the valves and the replum. Coordinated regulation of multiple intercellular signals is characteristic of boundary regions, seen, for example, during anteroposterior patterning of the Drosophila wing imaginal disc (Bangi and Wharton 2006).
Materials and methods
Plant material and growth conditions
Plants were grown on soil in long days (16 h light/8 h dark). The mutant lines ind-1 (Liljegren et al. 2004), ga4-1 (Koornneef and Van der Veen 1980), and alc-1 (Rajani and Sundaresan 2001) were in Landsberg erecta background, and the ga4-3 mutant was in Columbia background (Mitchum et al. 2006). AtGA3ox1 TC-GUS plants were in Columbia background (Mitchum et al. 2006). The 35S∷IND:GR line has been described (Sorefan et al. 2009). Constructs were generated according to standard techniques. Details for constructs are given in the Supplemental Material.
For expression assays, 35S∷IND:GR seeds (∼20) were germinated in 0.5% Murashige and Skoog medium with constant shaking. After 7 d of growth under constant light, seedlings were treated with DEX for 6 h and then snap-frozen in liquid nitrogen. When cycloheximide was used, DEX treatment was reduced to 2 h. DEX (Sigma, D1756) was dissolved in DMSO and used at a final concentration of 10 μM. Cycloheximide (Sigma, C4859) was dissolved in ethanol and used at a final concentration of 100 μM.
The ga1-3 mutant and ga1-3; rga-t2; gai-t6 triple mutant were grown under continuous light. GA (Sigma, G7645) was applied every 2 d.
Array hybridization and analysis of expression data
For microarray analysis, total RNA were first isolated using RNAeasy Kit (Qiagen), then hybridized to Affymetrix ATH1 array according to the manufacturer's instructions. Three biological repeats were analyzed. The microarray results were visualized and normalized using Genespring GX 7.3 software (Agilent). Normalization was performed by RMA (robust multichip average).
ChIP
ChIP experiments were performed using a GR antibody as described previously (Sorefan et al. 2009). qPCR was performed using SYBR Green JumpStart Taq ReadyMix in a Bio-Rad Chromo4 Q-PCR machine and using the primers GA3OX1F, GA3OX1R, Mu-likeF, and Mu-likeR. The values correspond to the ratios between pull-down DNA and input with the GR antibody, both initially normalized by Mu-like transposon.
Expression analysis
GUS assays were performed according to the protocol described by Rodrigues-Pousada et al. (1993). Plants were fixed in 90% acetone on ice for 20 min, then rinsed with a rinse buffer containing 0.5 mM K-ferrocyanide (Sigma, P-8131) and from 0 to 0.2 mM K-ferricyanide (Sigma, P-9387) in 50 mM sodium phosphate buffer (pH 7.2). Samples were then incubated for 24–48 h at 37°C in this rinse buffer containing 2 mM 5-bromo-4-chloro-3-indolyl p-D-glucuronide (Melford, MB1121).
SEM
Plants were fixed in FAA (3.7% of formaldehyde, 5% acetic acid, 50% ethanol) overnight at 4°C, then dehydrated in ethanol series. Tissues were critical-point-dried in liquid CO2, sputter-coated with gold, and analyzed with a Philips XL 30 FEG SEM.
TEM
Stage 17b fruits were fixed in 2.5% (v/v) glutaraldehyde and 0.05 M Na cacodylate (pH 7.2), vacuum-infiltrated, and left overnight at room temperature. Samples were post-fixed in 1% osmium tetroxide and 0.05 M Na cacodylate for 1h, washed briefly with water, and dehydrated in ethanol series. Samples were then infiltrated in London Resin White resin (London Resin Co., Ltd.) and sectioned for TEM imaging with an FEI Technai G2 20 Twin TEM.
mPS-PI staining, confocal microscopy, and cell size quantification
Stage 17b fruits were cut into small pieces (≈0.5 mm) and stained as described previously (Truernit et al. 2008), then incubated for 5 h in 50 μg/mL PI. Confocal microscopy was performed using a Zeiss Axo Imager M1 upright microscope. PI and GFP were excited using the 488-nm argon ion laser and collected between 600 and 656 nm and 505 nm and 530 nm, respectively. Images were analyzed using the Zeiss LSM 510 software. SL cell size quantification was achieved by measuring the size (in square microns) of cells adjacent to the LL. For each genotype, a total of six different images were used, and SL cells were measured from both sides of the central replum using ImageJ. For RGA-GFP expression, the same settings were used to generate maximum intensity projections from image stacks for each treatment.
Assessment of dehiscence using an Arabidopsis RIT (ARIT)
An assay enabling measurement of the strength required to initiate dehiscence in oilseed rape pods has been described previously (Morgan et al. 1998; Bruce et al. 2002). This RIT was modified to enable quantification of shatter resistance in Arabidopsis. Silique samples at stage 18 or older (Smyth et al. 1990) were selected randomly from wild-type and mutant plants. Fruits were placed in an equilibration chamber at 25°C and 50% relative humidity for a minimum of 3 d. Three replicate samples of 20 siliques were then subjected to the ARIT. Siliques were placed together with five 2-mm steel balls (weighing ∼275 mg) in a 60-mm diameter glass petri dish, attached to an eppendorf shaker. The petri dish was agitated for 5-sec intervals from 0 to 60 sec and 10-sec intervals from 60 to 150 sec, until all fruit had dehisced or 150 sec of shaking had elapsed. After each interval, the frequency of intact/dehisced siliques was recorded. Fruits were considered to have dehisced only when both valves had detached. The time point at which 50% of siliques had dehisced, estimated using a fitting curve, was used as a comparative measure of shattering between different lines.
Yeast two-hybrid
The GAI, RGA, RGL2, and ALC coding regions were PCR-amplified with the primers described in Supplemental Table 1, and were cloned in pGAD424 and pGBT9 vectors (Clontech Laboratories, Inc.). The yeast two-hybrid experiment was performed according to the manufacturer's instructions. Further details are given in the Supplemental Material.
Acknowledgments
We thank Sue Bunnewell (John Innes Centre, Norwich) for TEM, Vicente Balanzá and Alejandro Ferrando for providing BiFC vectors, Valérie Nicaise and Myriam Charpentier for BiFC advice, and Tai-ping Sun, Nick Harberd, Martin Yanofsky, and The Nottingham Arabidopsis Stock Centre for seeds. Work in the R.S. laboratory and the L.Ø. laboratory is funded by the Biotechnology and Biological Sciences Research Council (CISCODE project) and the European Union.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.593410.
Supplemental material is available at http://www.genesdev.org.
References
- Bangi E, Wharton K 2006. Dpp and Gbb exhibit different effective ranges in the establishment of the BMP activity gradient critical for Drosophila wing patterning. Dev Biol 295: 178–193 [DOI] [PubMed] [Google Scholar]
- Bruce DM, Farrent JW, Morgan CL, Child RD 2002. Determining the oilseed rape pod strength needed to reduce seed loss due to pod shatter. Biosys Eng 81: 179–184 [Google Scholar]
- Chiang HH, Hwang I, Goodman HM 1995. Isolation of the Arabidopsis GA4 locus. Plant Cell 7: 195–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lucas M, Daviere JM, Rodriguez-Falcon M, Pontin M, Iglesias-Pedraz JM, Lorrain S, Fankhauser C, Blazquez MA, Titarenko E, Prat S 2008. A molecular framework for light and gibberellin control of cell elongation. Nature 451: 480–484 [DOI] [PubMed] [Google Scholar]
- Feng S, Martinez C, Gusmaroli G, Wang Y, Zhou J, Wang F, Chen L, Yu L, Iglesias-Pedraz JM, Kircher S, et al. 2008. Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451: 475–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrandiz C, Liljegren SJ, Yanofsky MF 2000. Negative regulation of the SHATTERPROOF genes by FRUITFULL during Arabidopsis fruit development. Science 289: 436–438 [DOI] [PubMed] [Google Scholar]
- Hedden P, Phillips AL 2000. Gibberellin metabolism: New insights revealed by the genes. Trends Plant Sci 5: 523–530 [DOI] [PubMed] [Google Scholar]
- Heim MA, Jakoby M, Werber M, Martin C, Weisshaar B, Bailey PC 2003. The basic helix–loop–helix transcription factor family in plants: A genome-wide study of protein structure and functional diversity. Mol Biol Evol 20: 735–747 [DOI] [PubMed] [Google Scholar]
- Hu J, Mitchum MG, Barnaby N, Ayele BT, Ogawa M, Nam E, Lai WC, Hanada A, Alonso JM, Ecker JR, et al. 2008. Potential sites of bioactive gibberellin production during reproductive growth in Arabidopsis. Plant Cell 20: 320–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh H, Tanaka-Ueguchi M, Kawaide H, Chen X, Kamiya Y, Matsuoka M 1999. The gene encoding tobacco gibberellin 3β-hydroxylase is expressed at the site of GA action during stem elongation and flower organ development. Plant J 20: 15–24 [DOI] [PubMed] [Google Scholar]
- Koornneef M, Van der Veen JH 1980. Induction and analysis of gibberellin sensitive mutants in Arabidopsis thaliana (L.) heynh. Theor Appl Genet 58: 257–263 [DOI] [PubMed] [Google Scholar]
- Lee S, Cheng H, King KE, Wang W, He Y, Hussain A, Lo J, Harberd NP, Peng J 2002. Gibberellin regulates Arabidopsis seed germination via RGL2, a GAI/RGA-like gene whose expression is up-regulated following imbibition. Genes Dev 16: 646–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liljegren SJ, Ditta GS, Eshed Y, Savidge B, Bowman JL, Yanofsky MF 2000. SHATTERPROOF MADS-box genes control seed dispersal in Arabidopsis. Nature 404: 766–770 [DOI] [PubMed] [Google Scholar]
- Liljegren SJ, Roeder AH, Kempin SA, Gremski K, Østergaard L, Guimil S, Reyes DK, Yanofsky MF 2004. Control of fruit patterning in Arabidopsis by INDEHISCENT. Cell 116: 843–853 [DOI] [PubMed] [Google Scholar]
- Mitchum MG, Yamaguchi S, Hanada A, Kuwahara A, Yoshioka Y, Kato T, Tabata S, Kamiya Y, Sun TP 2006. Distinct and overlapping roles of two gibberellin 3-oxidases in Arabidopsis development. Plant J 45: 804–818 [DOI] [PubMed] [Google Scholar]
- Mitsuda N, Ohme-Takagi M 2008. NAC transcription factors NST1 and NST3 regulate pod shattering in a partially redundant manner by promoting secondary wall formation after the establishment of tissue identity. Plant J 56: 768–778 [DOI] [PubMed] [Google Scholar]
- Morgan CL, Bruce DM, Child R, Ladbrooke ZL, Arthur AE 1998. Genetic variation for pod shatter resistance among lines of oilseed rape developed from synthetic B-Napus. Field Crops Res 58: 153–165 [Google Scholar]
- Oh E, Yamaguchi S, Hu J, Yusuke J, Jung B, Paik I, Lee HS, Sun TP, Kamiya Y, Choi G 2007. PIL5, a phytochrome-interacting bHLH protein, regulates gibberellin responsiveness by binding directly to the GAI and RGA promoters in Arabidopsis seeds. Plant Cell 19: 1192–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Østergaard L 2009. Don't ‘leaf’ now. The making of a fruit. Curr Opin Plant Biol 12: 36–41 [DOI] [PubMed] [Google Scholar]
- Penfield S, Josse EM, Kannangara R, Gilday AD, Halliday KJ, Graham IA 2005. Cold and light control seed germination through the bHLH transcription factor SPATULA. Curr Biol 15: 1998–2006 [DOI] [PubMed] [Google Scholar]
- Petersen M, Sander L, Child R, van Onckelen H, Ulvskov P, Borkhardt B 1996. Isolation and characterisation of a pod dehiscence zone-specific polygalacturonase from Brassica napus. Plant Mol Biol 31: 517–527 [DOI] [PubMed] [Google Scholar]
- Purugganan MD, Fuller DQ 2009. The nature of selection during plant domestication. Nature 457: 843–848 [DOI] [PubMed] [Google Scholar]
- Rajani S, Sundaresan V 2001. The Arabidopsis myc/bHLH gene ALCATRAZ enables cell separation in fruit dehiscence. Curr Biol 11: 1914–1922 [DOI] [PubMed] [Google Scholar]
- Rieu I, Eriksson S, Powers SJ, Gong F, Griffiths J, Woolley L, Benlloch R, Nilsson O, Thomas SG, Hedden P, et al. 2008. Genetic analysis reveals that C19-GA 2-oxidation is a major gibberellin inactivation pathway in Arabidopsis. Plant Cell 20: 2420–2436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues-Pousada RA, De Rycke R, Dedonder A, Van Caeneghem W, Engler G, Van Montagu M, Van Der Straeten D 1993. The Arabidopsis 1-aminocyclopropane-1-carboxylate synthase gene 1 is expressed during early development. Plant Cell 5: 897–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder AH, Ferrandiz C, Yanofsky MF 2003. The role of the REPLUMLESS homeodomain protein in patterning the Arabidopsis fruit. Curr Biol 13: 1630–1635 [DOI] [PubMed] [Google Scholar]
- Sablowski RW, Meyerowitz EM 1998. A homolog of NO APICAL MERISTEM is an immediate target of the floral homeotic genes APETALA3/PISTILLATA. Cell 92: 93–103 [DOI] [PubMed] [Google Scholar]
- Silverstone AL, Jung HS, Dill A, Kawaide H, Kamiya Y, Sun TP 2001. Repressing a repressor: Gibberellin-induced rapid reduction of the RGA protein in Arabidopsis. Plant Cell 13: 1555–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth DR, Bowman JL, Meyerowitz EM 1990. Early flower development in Arabidopsis. Plant Cell 2: 755–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorefan K, Girin T, Liljegren SJ, Ljung K, Robles P, Galvan-Ampudia CS, Offringa R, Friml J, Yanofsky MF, Østergaard L 2009. A regulated auxin minimum is required for seed dispersal in Arabidopsis. Nature 459: 583–586 [DOI] [PubMed] [Google Scholar]
- Spence J, Vercher Y, Gates P, Harris N 1996. ‘Pod shatter’ in Arabidopsis thaliana, Brassica napus and B. juncea. J Microsc 181: 195–203 [Google Scholar]
- Sun TP, Gubler F 2004. Molecular mechanism of gibberellin signaling in plants. Annu Rev Plant Biol 55: 197–223 [DOI] [PubMed] [Google Scholar]
- Sun T, Goodman HM, Ausubel FM 1992. Cloning the Arabidopsis GA1 locus by genomic subtraction. Plant Cell 4: 119–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talon M, Koornneef M, Zeevaart JA 1990. Endogenous gibberellins in Arabidopsis thaliana and possible steps blocked in the biosynthetic pathways of the semidwarf ga4 and ga5 mutants. Proc Natl Acad Sci 87: 7983–7987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truernit E, Bauby H, Dubreucq B, Grandjean O, Runions J, Barthelemy J, Palauqui JC 2008. High-resolution whole-mount imaging of three-dimensional tissue organization and gene expression enables the study of Phloem development and structure in Arabidopsis. Plant Cell 20: 1494–1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Mori A, Jiang X, Wang Y, Yang M 2006. The INDEHISCENT protein regulates unequal cell divisions in Arabidopsis fruit. Planta 224: 971–979 [DOI] [PubMed] [Google Scholar]
- Yamaguchi S 2008. Gibberellin metabolism and its regulation. Annu Rev Plant Biol 59: 225–251 [DOI] [PubMed] [Google Scholar]