Abstract
Primary cilia function as a sensory signaling compartment in processes ranging from mammalian Hedgehog signaling to neuronal control of obesity. Intraflagellar transport (IFT) is an ancient, conserved mechanism required to assemble cilia and for trafficking within cilia. The link between IFT, sensory signaling, and obesity is not clearly defined, but some novel monogenic obesity disorders may be linked to ciliary defects. The tubby mouse, which presents with adult-onset obesity, arises from mutation in the Tub gene. The tubby-like proteins comprise a related family of poorly understood proteins with roles in neural development and function. We find that specific Tubby family proteins, notably Tubby-like protein 3 (TULP3), bind to the IFT-A complex. IFT-A is linked to retrograde ciliary transport, but, surprisingly, we find that the IFT-A complex has a second role directing ciliary entry of TULP3. TULP3 and IFT-A, in turn, promote trafficking of a subset of G protein-coupled receptors (GPCRs), but not Smoothened, to cilia. Both IFT-A and membrane phosphoinositide-binding properties of TULP3 are required for ciliary GPCR localization. TULP3 and IFT-A proteins both negatively regulate Hedgehog signaling in the mouse embryo, and the TULP3–IFT-A interaction suggests how these proteins cooperate during neural tube patterning.
Keywords: G protein-coupled receptor, TULP3, hedgehog, intraflagellar transport, primary cilia
Primary cilia are dynamic structures, and intraflagellar transport (IFT), itself a motile process, is essential for directing the assembly and function of cilia. IFT consists of trains of multipolypeptide particles that move continuously along axonemal microtubules. Sedimentation analysis of IFT particles identified at least 21 polypeptides organized in two complexes, called complex A (IFT-A) and complex B (IFT-B) (Cole et al. 1998; Rosenbaum and Witman 2002; Cole and Snell 2009). Anterograde transport of these particles is powered by kinesin-II, whereas retrograde transport is mediated by the dynein 2 motor (for review, see Rosenbaum and Witman 2002). Mutations in most known IFT-B subunits present with very short or no cilia and lack IFT, similar to mutations in the anterograde kinesin-II motor. Thus, IFT-B is implicated in anterograde transport. In contrast, mutations in the IFT-A complex subunits and the dynein 2 retrograde motor result in shortened or swollen cilia with accumulation of particles, including IFT-B proteins. For example, in Chlamydomonas, temperature-sensitive mutations in the IFT-A particle subunits IFT144/WDR19 and IFT139/THM1 show decreased retrograde transport velocities, an increased ratio of anterograde to retrograde particles, and an accumulation of complex B proteins in the flagella (Piperno et al. 1998; Iomini et al. 2001, 2009). In Caenorhabditis elegans, mutations in the IFT-A subunit WDR35/IFTA-1 result in truncated cilia with accumulation of IFT-B and kinesin-II and lack of retrograde IFT (Blacque et al. 2006). Accordingly, the IFT-A complex has been implicated in retrograde transport.
Our knowledge of how IFT effectively builds and functions in cilia is fragmentary. IFT transports flagellar precursors to the ciliary tip, and blocking anterograde IFT inhibits tubulin turnover at the tips, causing resorption (Marshall and Rosenbaum 2001; Qin et al. 2004). Retrograde IFT returns the IFT particles, the kinesin-II motor, and turnover products to the ciliary base (Signor et al. 1999; Iomini et al. 2001). Individual IFT-B components, such as IFT46, may have a specific role in outer dynein arm transport in Chlamydomonas (Hou et al. 2007), while IFT172 binds to the microtubule plus-end protein EB1 and remodels the IFT particles at the flagellar tip (Pedersen et al. 2005). A complex of proteins involved in the human ciliopathy Bardet-Biedl syndrome (BBS), called the BBSome, is postulated to function as an IFT cargo, transporting specific ciliary proteins (Ou et al. 2005; Nachury et al. 2007; Berbari et al. 2008; Lechtreck et al. 2009; Jin et al. 2010). The binding of IFT particles to IFT motors and axonemal precursors suggests that the IFT particles link IFT motors and cargo as described for dynein and the dynactin complex (Kardon and Vale 2009). Models notwithstanding, the effectors of IFT-A particles are hitherto unknown.
Primary cilia function as sensory compartments, sensing environmental inputs and transducing intercellular signals (Singla and Reiter 2006). For example, neuronal cilia possess a complement of G protein-coupled receptors (GPCRs), including somatostatin receptor subtype 3 (Sstr3) (Handel et al. 1999), Melanin-concentrating hormone receptor (Mchr1) (Berbari et al. 2008), and downstream effectors including the adenylyl cyclase type 3 (ACIII) (Bishop et al. 2007). Mchr1, the receptor for MCH, is involved in the regulation of feeding and energy balance (Shimada et al. 1998; Chen et al. 2002), and ACIII-deficient mice become obese with age, suggesting that ACIII-mediated cAMP signals are critical in the hypothalamus (Wang et al. 2009). Cilia in mature neurons can also act as extrasynaptic compartments in order to modulate neuronal function. Disruption of IFT in adult mice, possibly acting through the proopiomelanocortin (POMC)-expressing hypothalamic axis, result in hyperphagia-induced obesity (Davenport et al. 2007), while Sstr3 signaling in the hippocampus is important in synaptic plasticity and novelty detection (Einstein et al. 2010). However, our knowledge of the mechanisms by which IFT might modulate sensory signaling in primary cilia is incomplete. IFT particles participate directly in cilium-generated signaling during fertilization in Chlamydomonas (Wang et al. 2006), and are involved in vectorial movement of TRPV channel proteins along Caenorhabditis elegans sensory cilia (Qin et al. 2005). Thus, elucidating the role of IFT in the localization and function of ciliary signaling molecules would add considerably to understanding the link between cilia and neuronal function.
Primary cilia are also important in the mammalian Hedgehog (Hh) signaling machinery, and mutations in IFT components cause two major classes of defects in patterning of the neural tube. Mutations affecting IFT-B subunits and subunits of the IFT kinesin and dynein motors show disruption of Hh pathway activation (for review, see Goetz and Anderson 2010), while mutations of the IFT-A subunit Thm1 and Ift122 show overactivation of the Hh pathway (Tran et al. 2008; Cortellino et al. 2009). It is surprising that mutations in IFT-A subunits differ in phenotype from those of the IFT motor dynein 2, when both are implicated in retrograde IFT. These differences suggest that the IFT-A complex may have functions in addition to its postulated role in retrograde IFT.
Monogenic obesity disorders may be related to ciliary defects. The tubby mouse, arising from a mutation in the Tub gene, has a syndrome characterized by obesity and neurosensory deficits (Kleyn et al. 1996; Noben-Trauth et al. 1996). Tub shares homology with four other tubby-like proteins, Tulp1–Tulp4. The tubby family of proteins plays important roles in nervous system function and development. However, the molecular function of these genes is poorly understood. Tulp3 has been described recently as a negative regulator of Hh signaling in the mouse embryo (Cameron et al. 2009; Norman et al. 2009; Patterson et al. 2009). Genetic epistasis experiments suggest that, similar to the IFT-A subunit Thm1, Tulp3 restricts Gli2 activity in an IFT-dependent manner downstream from Sonic hedgehog (Norman et al. 2009; Patterson et al. 2009). Although Tulp3 and Thm1 act as negative regulators of the Hh pathway, their roles remain unclear.
Here we show that, in addition to its known role in retrograde IFT, IFT-A plays a primary role in providing ciliary access to TULP3 by binding directly to it, thereby coordinately regulating the Hh signaling pathway. TULP3, in turn, regulates trafficking of a subset of ciliary-localized GPCRs. Both IFT-A- and membrane phosphoinositide-binding properties of TULP3 mediate ciliary GPCR localization. TULP3 thus serves as an anchor that links the IFT-A complex to the membrane compartment.
Results
TULP3 interacts with the IFT-A complex
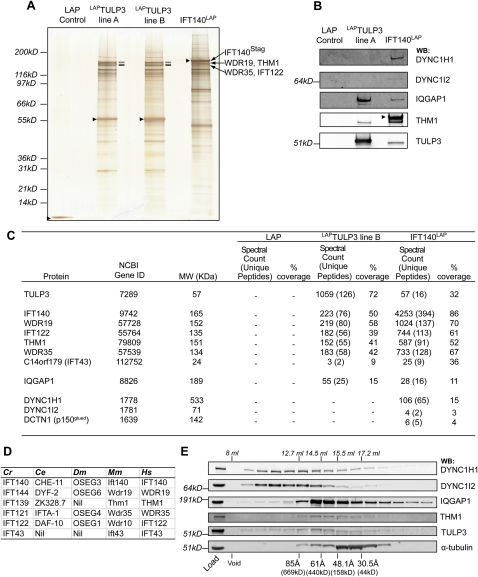
To gain insight into the molecular function of TULP3, we generated localization and tandem affinity purification (LAP)-tagged stable clonal cell lines in telomerase immortalized hTERT RPE-1 (RPE) cells (Nachury et al. 2007). We identified the TULP3-associated proteins by tandem affinity purification followed by mass spectrometry. In both lines (A and B), we found all known components of the IFT-A complex to be associated with TULP3, but not in the control LAP line (Fig. 1A–C). The IFT-A complex proteins recovered were WDR19, IFT140, IFT122, THM1, and WDR35 (Fig. 1A,C, see D and Supplemental Fig. S1B for homologs in other organisms and a comment on the nomenclature). Peptide coverage for all of the IFT-A complex proteins was at least 39%, supporting the high significance of the identifications. IFT43 (C14orf179), reported recently to be a part of the IFT-A complex (Cole and Snell 2009), was also recovered (peptide coverage 9%). The IFT-A complex was similarly associated with LAPTULP3 in murine NIH 3T3 fibroblasts and kidney inner medullary-collecting duct IMCD-3 cells (Supplemental Figs. S1A,B, S2A). Tandem affinity purification of the IFT-A complex using a clonal line expressing IFT140LAP in RPE cells likewise copurified with endogenous TULP3 (Fig. 1A–C). Other copurifying proteins detected included the cytoskeletal regulator IQGAP1 (with both LAPTULP3 and IFT140LAP) (Fig. 1B,C), and components of the cytoplasmic dynein 1 and the Dynactin complex (with IFT140LAP only) (Fig. 1B,C; see the Discussion). Upon gel filtration analyses of RPE cell lysates, TULP3 cofractionated with THM1 (Stokes radius ∼63 Å), suggesting that TULP3 associates with the IFT-A complex, although we also detected TULP3 throughout the gradient (Fig. 1E; see the Discussion).
Figure 1.
TULP3 interacts with the IFT-A complex. (A) Tandem affinity purification of the LAP (GFP-TEV/PreScission-Stag) cassette from control LAP line (without any insert), LAPTULP3 (from two stable clonal lines with varying levels of LAPTULP3), and IFT140LAP in RPE cells. Eluates were resolved on a 4%–12% Bis-Tris NuPAGE gel, and silver-stained. Stagfusion proteins in each lane are marked by arrowheads. The IFT-A complex proteins are marked based on LC MS/MS analysis of gel slices. (B) Western blot of LAP only, LAPTULP3, and IFT140LAP tandem purification eluates using indicated antibodies. The anti-THM1 antibody cross-reacts with Stagfusion proteins, and IFT140Stag protein is shown with an arrowhead. (C) Table showing the peptide spectral count and percent coverage of selected proteins in LAP only, LAPTULP3, and IFT140LAP purification eluates identified by mass spectrometry from RPE cells. (D) Table showing the homologs of IFT-A complex subunits in other organisms. (E) TULP3 partially cofractionates with the IFT-A complex protein THM1. RPE cell lysate was fractionated by size exclusion chromatography, and fractions were immunoblotted with indicated antibodies. See also Supplemental Figure S1.
TULP3 binds to the IFT ‘core’ complex
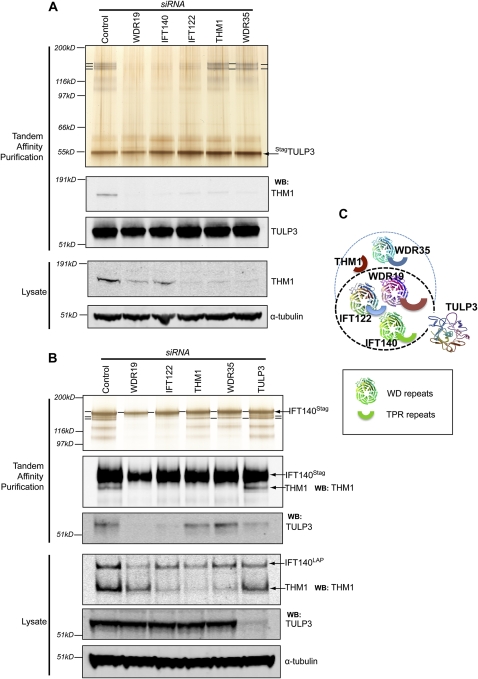
To systematically determine the components of IFT-A binding to TULP3, we depleted each subunit by RNAi in LAPTULP3 and IFT140LAP clonal cell lines (Supplemental Fig. S2B,C), isolated the TULP3-associated proteins by tandem affinity purification, and visualized them by silver staining and immunoblotting for THM1. Depleting WDR19, IFT140, and, to a lesser extent, IFT122 diminished association of LAPTULP3 with the IFT-A complex proteins, including THM1 (Fig. 2A; Supplemental Fig. S2A). Similarly, association of IFT140LAP with TULP3 and all IFT-A complex proteins was remarkably reduced in WDR19- and IFT122-depleted cells (Fig. 2B). However, depletion of THM1 or WDR35 resulted in persistence of a partial IFT-A subcomplex (excluding THM1) that remained associated with LAPTULP3 (Fig. 2A; Supplemental Fig. S2A). Upon depletion of THM1 or WDR35, IFT140LAP remained associated with TULP3 and a subcomplex of IFT-A proteins, but lost binding to THM1 (Fig. 2B). This suggests that WDR19, IFT140, and IFT122 are important in forming a “core” IFT-A subcomplex that binds TULP3, whereas THM1 and WDR35 participate in some additional role distinct from “core” IFT-A complex formation or TULP3 binding (Fig. 2C). The variable decrease in total THM1 protein levels in the cell lysates, upon depletion of other IFT-A complex subunits, reflects its stability contingent on its ability to be part of the IFT-A complex.
Figure 2.
Organization of the IFT-A complex into “core” and accessory subunits. (A) TULP3-associated proteins were purified from LAPTULP3 RPE cells as in Figure 1A, except following 72 h depletion of the indicated proteins by siRNA. Silver-stained gel (top panel) and Western blots (bottom panels) of tandem affinity purifications and total lysates are shown. The IFT-A complex proteins are labeled as in Figure 1A. (B) As in A except RPE IFT140LAP instead of LAPTULP3 cells were used. (C) A summary cartoon of the components of the IFT-A complex and their association with TULP3. The black dotted line encircles the “core” complex subunits, while the blue dashed line contains the accessory subunits. See also Supplemental Figure S2.
TULP3 localizes to the primary cilia, but does not affect ciliogenesis
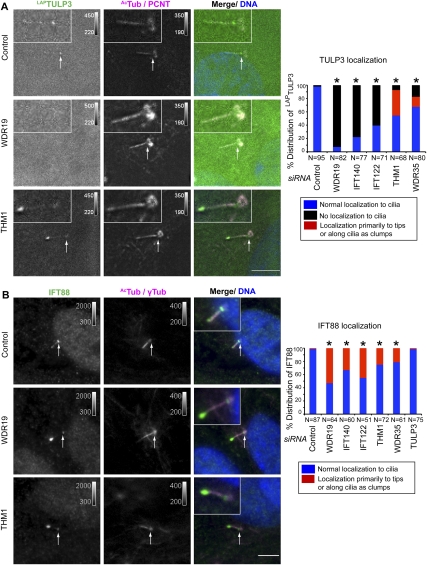
To better understand where TULP3 and IFT-A interact, we next examined localization of stably expressed LAPTULP3 in clonal lines. LAPTULP3 localized to the ciliary base and in punctate spots throughout the cilia in RPE, IMCD-3, and NIH 3T3 cells, consistent with previous reports for the endogenous protein (Fig. 3A; Supplemental Fig. S2D; Norman et al. 2009). Tubby family proteins have been observed to relocalize to the nucleus when displaced from the membrane (Santagata et al. 2001). Upon inhibition of nuclear export using leptomycin-B or mutation of conserved residues important for phosphoinositide binding (K268A and R270A; designated as TULP3KR) (Santagata et al. 2001), LAPTULP3 was detected in the nucleus as well, suggesting that TULP3 is a nucleocytoplasmic shuttling protein (Supplemental Fig. S3A,B). We then checked if TULP3 and IFT-A play a role in ciliogenesis in RPE cells, a well-established system for studying serum starvation-induced ciliogenesis (Pugacheva et al. 2007; Tsang et al. 2008). We found the expected decrease in ciliogenesis upon KIF3A knockdown, but did not see a ciliation defect upon TULP3 or IFT-A component knockdown (Supplemental Fig. S1C). Thus, TULP3 and IFT-A are not required for ciliation in RPE cells.
Figure 3.
TULP3 localizes to the primary cilium dependent on the IFT-A complex. (A) RPE LAPTULP3 line A cells were transfected with siRNAs (indicated to the left of the micrograph panels) for 72 h and serum-starved for the last 24 h before fixing and staining for pericentrosomal pericentrin (PCNT, magenta), the axonemal marker Ac-tubulin (AcTub, magenta), and DNA (blue). White arrows indicate the ciliary base. Bars, 5 μm. Quantification of localization of LAPTULP3 to the cilia in similar assays is shown to the right. (*) P < 0.0001 with respect to control, using a χ2 test. (B) RPE cells were transfected with the indicated siRNAs for 72 h and serum-starved for the last 24 h before fixing and staining for IFT88 (green), pericentrosomal γ-tubulin (γTub, magenta), the axonemal marker Ac-tubulin (AcTub, magenta), and DNA (blue). White arrows indicate the ciliary base. Bars, 5 μm. Quantification and statistical significance are as in A.
The IFT-A ‘core’ complex provides ciliary access to TULP3
The IFT-A complex has been suggested to function in retrograde IFT of certain ciliary proteins, including the IFT-B complex proteins. Consistent with this model, we find that IFT88 (an IFT-B subunit) accumulates at cilia tips upon knockdown of any IFT-A subunit (Fig. 3B). In contrast, depletion of the “core” IFT-A complex proteins WDR19, IFT140, and IFT122 resulted in a loss of detectable localization of LAPTULP3 to the cilia (Fig. 3A). In contrast, association between TULP3 and the IFT-A “core” complex persists upon depletion of the peripheral components THM1 or WDR35 (Fig. 2C), and resulted in accumulation of LAPTULP3 primarily at the tips of the cilia (Fig. 3A). The above experiments suggest two separate roles for the IFT-A complex: The “core” IFT-A is important to localize TULP3 to the cilia, whereas an intact complex regulates TULP3's retrograde transport. Conversely, TULP3 depletion did not affect organization of the IFT-A complex (Fig. 2B) or ciliary localization of IFT140LAP (Supplemental Fig. S2E), suggesting that IFT140 localizes independently of TULP3. Besides, TULP3 depletion did not affect the ciliary distribution of IFT88, in sharp contrast to depletion of IFT-A complex subunits (Fig. 3B).
Fine mapping of the IFT-A interaction domain of TULP3
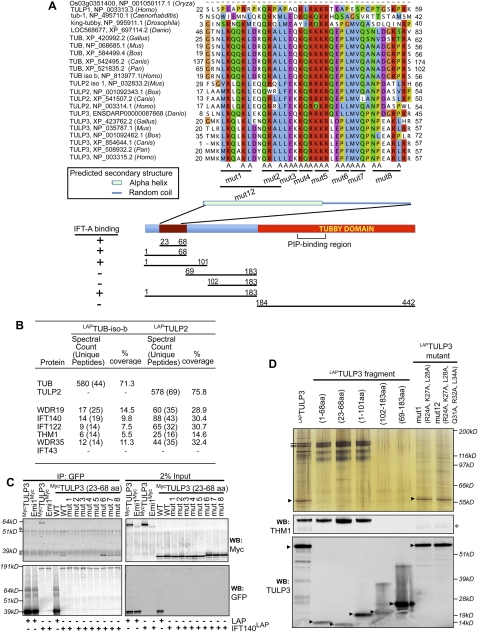
TULP3 belongs to the extended family of tubby-like proteins (Supplemental Fig. S4). These proteins share a conserved C-terminal “tubby domain” (Santagata et al. 2001); however, the N-terminal domains do not, upon first comparison, look closely related. We generated clonal LAP-tagged lines for all of the Tubby family proteins in RPE cells, including both isoforms of the human Tubby homolog (which are named a and b [NCBI] and bear slight differences in their N-terminal regions, with the b isoform being conserved in mice). Whereas LAPTULP1 and LAPTUB (isoform a) were localized to both the nucleus and the cytoplasm, LAPTUB (isoform b), LAPTULP2, and LAPTULP4 were exclusively cytoplasmic (data not shown), with only LAPTULP3 showing ciliary localization. Upon tandem affinity purification and identification by mass spectrometry, IFT-A complex proteins were found associated with both LAPTUB (isoform b) and LAPTULP2 (Fig. 4B), albeit at much lower levels than with LAPTULP3 (the binding is undetectable by silver staining and immunoblotting). IFT-A was not detected with either LAPTUB (isoform a), LAPTULP1, or LAPTULP4 (data not shown), suggesting that only TULP3, and, to a lesser extent, TUB (isoform b) and TULP2 bind the IFT-A complex.
Figure 4.
Fine mapping of the IFT-A interaction domain of TULP3. (A) A cartoon depicting different domains of TULP3 is shown below the alignments, along with the different sets of mutations evaluated and their binding properties. (B) Table showing the peptide spectral count and percent coverage of selected proteins in LAPTUB isoform b, and LAPTULP2 tandem affinity purification eluates identified by mass spectrometry. (C) Coimmunoprecipitation of IFT140 with TULP3 in HEK293T cells. Cells were cotransfected with IFT140LAP or LAP empty cassette and the indicated Myc-tagged constructs for 72 h. GFP immunoprecipitates (left panels), and total lysates (right panels) were immunoblotted for GFP and Myc tag. Cross-reacting bands in the anti-Myc blots are marked with asterisks. (D) TULP3-associated proteins were purified from RPE cells expressing different LAPTULP3 fragments, as in Figure 1A, and tandem affinity purification eluates were silver-stained (top panel) or immunoblotted (bottom panels). The IFT-A complex proteins are labeled as in Figure 1A. Arrowheads and asterisk indicate StagTULP3 full-length or fragment forms and a cross-reacting band (in the anti-THM1 blot), respectively. See also Supplemental Figure S3.
To map the IFT-A-binding domain of TULP3, we first compared which binding proteins associated with the LAP-tagged versions of the TULP3 N-terminal (amino acids 1–183) and C-terminal domains (amino acids 184–442) by mass spectrometry and silver staining. We found that only the N-terminal fragment of TULP3 bound efficiently to the IFT-A complex (Fig. 4A). Deletion analysis further narrowed the IFT-A-binding domain to a 46-amino-acid stretch (23–68 amino acids) (Fig. 4C) predicted to have an α-helical structure (Fig. 4A; Rost 1996). Consistent with our tandem affinity purifications, re-examination of the Tubby family N-terminal domains showed that this region is conserved between TULP3, TULP2, and TUB, but not the nonbinding TULP1 and TULP4 (Fig. 4A; data not shown for TULP4). Coimmunoprecipitation of TULP3 variants with mutated amino acid triplets in this region (denoted mut1 to mut8) with IFT140LAP in HEK293T cells permitted fine mapping of the IFT-A-binding domain to amino acids 23–43 (mut1 to mut5) (Fig. 4C). Mutating these amino acids (for example, mut1 or mut1 and mut2 combined [mut12]) in the context of full-length TULP3 and generating stable lines confirmed the IFT-A-binding site, each of them completely abrogating binding to IFT-A complex proteins (Fig. 4D) and ciliary localization (Supplemental Fig. S3C). Thus, a conserved helical region in the N terminus of TULP3 mediates interaction with the IFT-A complex, and fine mapping of this domain allowed us to develop a tool to perturb and test the importance of the TULP3–IFT-A interaction (see the last section).
TULP3 regulates localization of a subset of ciliary GPCRs via IFT-A-dependent trafficking
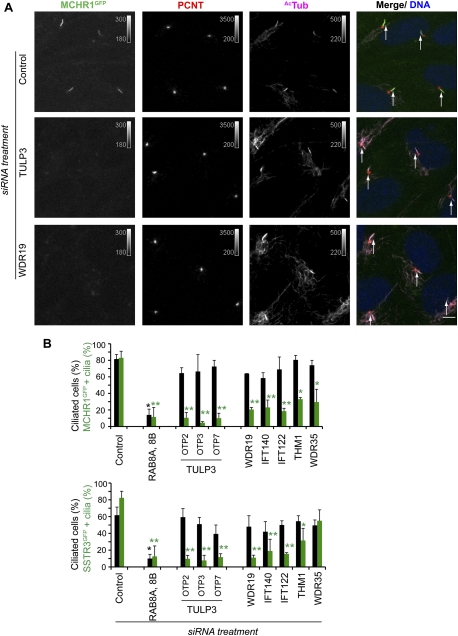
The phenotypes of Tub and Tulp1 knockout mice suggest a role for these proteins in intracellular vesicular trafficking of the rhodopsin GPCR in photoreceptors (Hagstrom et al. 1999; Ikeda et al. 2000). Since GPCRs similar to Rhodopsin, such as Sstr3 and Mchr1, localize to primary cilia, we tested whether TULP3 plays a role in their localization. While ∼80% of the cilia in serum-depleted RPE cells displayed stably expressing C-terminal GFP-tagged versions of these receptors (Fig. 5B), siRNA-mediated depletion of TULP3 strongly inhibited their ciliary localization, without affecting ciliogenesis (Fig. 5A,B). Because IFT-A regulates localization of TULP3 to the cilia, we tested whether IFT-A complex proteins modulate ciliary localization of these GPCRs. Indeed, depleting the “core” complex proteins WDR19, IFT140, and IFT122 strongly affected ciliary localization of both SSTR3GFP and MCHR1GFP, whereas depletion of the accessory subunits THM1 and WDR35 had variable effects (Fig. 5A,B). Notably, other ciliary proteins, such as RAB8a, were unaffected by depletion of either TULP3 or the IFT-A complex (Supplemental Fig. S5A), whereas depletion of RAB8 decreased ciliogenesis as well as localization of these receptors to the remaining cilia (Fig. 5B).
Figure 5.
TULP3 and IFT-A coregulate localization of ciliary GPCRs. (A) RPE MCHR1GFP stable cells were transfected with the indicated siRNAs for 78 h and serum-starved for the last 30 h before fixing and staining for pericentrin (PCNT, red), Ac-tubulin (AcTub, magenta), and DNA (blue). White arrows indicate cilia. Bars, 5 μm. (B) Percentages of total cilia and GFP-positive cilia in assays similar to A were counted in two to three independent experiments with RPE MCHR1GFP and RPE SSTR3GFP stable cells. Error bars represent SEM. (*) P < 0.05; (**) P < 0.001 with respect to control in each group. See also Supplemental Figure S5.
Tulp3 and IFT-A mutants cause increased Hh signaling in the mammalian neural tube. We therefore examined if localization of known Hh pathway components were perturbed by TULP3 and IFT-A. Interestingly, although knockdown of the IFT motor dynein 2 heavy chain (Dync2h1) caused accumulation of Smoothened (Smo) in cilia of Hh untreated cells as shown before (Ocbina and Anderson 2008), there was no appreciable defect in Smo trafficking upon single or double knockdowns of Tulp3 and any other IFT-A complex subunit (Supplemental Fig. S5B). In the absence of Hh, Gli3 is proteolytically processed into Gli3R (Wang et al. 2000), which is inhibited by Hh treatment, causing rapid accumulation of full-length Gli3 at the tips of primary cilia (Supplemental Fig. S5C; Wen et al. 2010). Knockdown of Dync2h1 decreased Gli3R levels and increased the full-length Gli3 in untreated cells, and Gli3 processing remained unchanged upon Hh treatment (Supplemental Fig. S5D–F). However, single or double knockdowns of Tulp3 and/or IFT-A subunits, or stable expression of the TULP3 N-terminal dominant-negative fragment (which disrupts the TULP3–IFT-A interaction; see the next section), showed normal Gli3 processing and ciliary translocation upon Hh treatment (Supplemental Fig. S5C–F; data not shown). Double knockdowns of IFT retrograde motor subunit Dync2h1 with Tulp3 or IFT-A subunits showed effects similar to Dync2h1 knockdown in both Smo translocation and Gli3 processing assays (Supplemental Fig. S5B,D–F). Thus, TULP3 or IFT-A functions distinctly from dynein 2 in the Hh pathway (see the Discussion). Besides, TULP3, through its association with IFT-A, mediates ciliary localization of only a subset of ciliary GPCRs, not including Smo.
Both IFT-A- and phosphoinositide-binding properties of TULP3 regulate ciliary GPCR trafficking
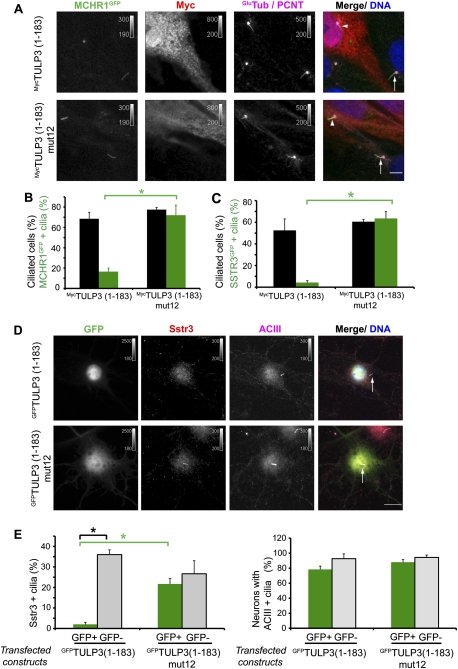
To confirm whether the TULP3 N-terminal domain (required for interaction with the IFT-A complex) plays a role in GPCR localization, we expressed a Myc-tagged variant of the IFT-A-binding N-terminal fragment (1–183 amino acids) of TULP3 by stable integration or transient transfection in RPE cells stably expressing SSTR3GFP and MCHR1GFP (Figs. 6A–C, 7B). In each case, we detected a strong decrease in ciliary localization of these receptors (Fig. 6A–C). Importantly, ciliary localization of SSTR3GFP and MCHR1GFP was unaffected upon expression of the non-IFT-A-binding mut12 N-terminal mutant (Fig. 6A–C), suggesting that the N-terminal fragment of TULP3 could be dominantly affecting the regulation of GPCRs through its binding with IFT-A. To assess the role of TULP3 in the localization of endogenous GPCRs, we cultured primary murine hippocampal neurons that bear cilia containing ACIII and Sstr3 (Berbari et al. 2008). Transient transfection of the GFP-tagged N-terminal half of TULP3, but not of the corresponding IFT-A-binding mutant, resulted in decreased localization of endogenous Sstr3, but not ACIII, to the neuronal cilia (Fig. 6D,E). Thus, the N-terminal, IFT-A-binding fragment of TULP3 perturbs trafficking of specific endogenous GPCRs, but does not affect the localization of other ciliary proteins, such as the membrane-associated ACIII, that presumably traffic to cilia by distinct pathways.
Figure 6.
TULP3 N-terminal fragment regulates localization of ciliary GPCRs. (A) RPE MCHR1GFP cells stably expressing indicated myc-tagged N-terminal TULP3 fragments were serum-starved for 30 h before fixing and immunostaining for myc (red), pericentrin (PCNT, magenta), axonemal marker Glu-tubulin (GluTub, magenta), and DNA (blue). White arrowheads and arrows indicate cilia in myc-positive cells and nonexpressing cells, respectively. Bar, 5 μm. (B) Percentages of total cilia and GFP-positive cilia in myc-positive cells in assays similar to A were counted in two independent experiments. Error bars represent SD. (*) P < 0.05. (C) RPE SSTR3GFP stable cells were transfected with the indicated myc-tagged constructs for 78 h and serum-starved for the last 30 h before fixing and staining as in A. Percentages of total cilia and GFP-positive cilia in myc-positive cells were counted in two independent experiments. Error bars represent SD. (*) P < 0.05. (D) Effect of transfecting indicated GFP-tagged constructs on endogenous Sstr3 localization in mouse primary hippocampal neurons. DIV5 primary hippocampal neurons cultured from E16 mice were transfected. Three days later, neurons were fixed and immunostained for endogenous Sstr3 (red), ACIII (magenta), and DNA (blue). White arrows indicate cilia. Bar, 10 μm. (E) Percentages of ACIII-positive cilia coexpressing Sstr3 (left) or neurons with ACIII-positive cilia (right) in transfected (GFP+) or untransfected (GFP−) primary hippocampal neurons in assays similar to D were counted in three independent experiments. Error bars represent SEM. (*) P < 0.01.
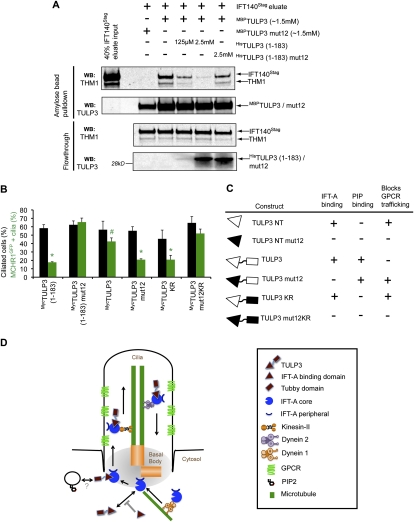
Figure 7.
Both IFT-A- and phosphoinositide-binding properties of TULP3 regulate GPCR-trafficking. (A) The TULP3 N terminus prevents recruitment of full-length TULP3 to an intact preformed IFT-A complex. PreScission eluates from TULP3 siRNA-treated IFT140LAP RPE cells were added to MBPTULP3 or MBPTULP3mut12 beads in the presence of HisTULP3 (1–183) or HisTULP3 (1–183) mut12. MBPTULP3 or MBPTULP3mut12-bound proteins and corresponding flowthroughs were immunoblotted for THM1 and TULP3. The anti-THM1 antibody cross-reacts with Stagfusion proteins, and allowed us to detect the IFT140Stag protein. For details, see the Supplemental Material. (B) RPE MCHR1GFP stable cells were transfected with the indicated myc-tagged TULP3 constructs and serum-starved for 30 h before fixing and immunostaining for myc, pericentrin, axonemal marker Glu-tubulin, and DNA. Percentages of total cilia and GFP-positive cilia in myc-positive cells were counted in at least three independent experiments. Error bars represent SEM. (*) P < 0.05 with respect to MCHR1GFP-positive cilia in MycTULP3 (1–183) mut12, MycTULP3, or MycTULP3mut12KR-expressing cells; (#) P < 0.05 with respect to MCHR1GFP-positive cilia in MycTULP3 (1–183) mut12-expressing cells. All other values are not significant with respect to each other. (C) Summary table of different TULP3 constructs and their effects on IFT-A binding, phosphoinositide binding, and GPCR trafficking. The mutant domains (for the image key, see D) are shown in black. (D) Model depicting the role of IFT-A and TULP3 in GPCR trafficking. IFT-A “core” complex associates with and provides ciliary access to TULP3. TULP3, in turn, is required for trafficking of certain ciliary-localized GPCRs. GPCR trafficking may be facilitated by loading of IFT-A onto preciliary vesicles (see also Sedmak and Wolfrum 2010) via the association of TULP3 with membrane phosphoinositides or novel interacting proteins. Dynein-1 also binds to the IFT-A complex, and may have roles in preciliary transport. Knocking down the IFT-A “core” complex proteins prevents TULP3 from localizing to the cilia, thereby inhibiting GPCR trafficking. Knocking down TULP3 affects ciliary localization of GPCRs, but not of IFT-A or IFT-B. Expression of the TULP3 N-terminal fragment prevents endogenous TULP3 from being recruited to the IFT-A complex, and possibly prevents loading of the IFT-A complex to the preciliary vesicles. See also Supplemental Figure S6.
In order to delineate the mechanism by which the TULP3 N-terminal fragment inhibits GPCR trafficking and IFT-A function, we first reconstituted the binding of TULP3 to the IFT-A complex in vitro. We purified the IFT-A complex from TULP3 siRNA-treated IFT140LAP RPE cells, and added this to recombinant MBPTULP3 or MBPTULP3mut12 beads. We detected binding of the IFT-A complex to MBPTULP3, but not to MBPTULP3mut12, suggesting that TULP3 can be recruited to an intact IFT-A complex (Supplemental Fig. S6A). Interestingly, although IFT140LAP was somewhat overexpressed compared with the other IFT-A subunits, the IFT-A complex subunits bound to MBPTULP3 were stoichiometric in silver-stained gels (Supplemental Fig. S6A). Addition of the recombinant HisTULP3 N terminus, but not of the HisTULP3mut12 N terminus, prevented IFT-A recruitment to MBPTULP3 in these reactions, suggesting that the N terminus prevents the full-length TULP3 from being associated with the IFT-A complex (Fig. 7A). Furthermore, stable expression of the Myc-tagged N-terminal TULP3 construct, but not of the corresponding IFT-A-binding mutant, decreased ciliary localization of LAPTULP3 (Supplemental Fig. S6B), suggesting that the N-terminal domain displaces full-length TULP3 from localizing to the cilia in vivo. These experiments argue that the TULP3 N terminus dominantly affects endogenous TULP3 localization, probably by preventing its recruitment to an intact IFT-A complex, thereby affecting GPCR trafficking.
How does TULP3 determine the specificity of trafficking of a subset of ciliary components? The Tubby domain binds to phosphoinositides (Santagata et al. 2001); however, the specifics of binding of full-length TULP3 to phosphoinositides have not been determined. We directly assayed phosphoinositide specificity of the recombinant GSTTULP3 on PIP and membrane lipid blots, and found that it binds most strongly to PtdIns(4,5)P2, followed by PtdIns(3,4)P2 and PtdIns(3,4,5)P3 (Supplemental Fig. S6C). A phosphoinositide-binding mutant of TULP3 (TULP3KR), however, retained binding to IFT-A (data not shown). We next transfected the Myc-tagged full-length TULP3 constructs with mutations in either the IFT-A (TULP3mut12) or phosphoinositide-binding regions (TULP3KR) in MCHR1GFP cells. Expression of MycTULP3mut12 (which retains phosphoinositide binding) or MycTULP3KR (which retains IFT-A binding) resulted in a strong decrease in ciliary localization of MCHR1GFP with respect to expression of MycTULP3 or MycTULP3mut12KR mutant (which does not bind to either IFT-A or phosphoinositides) (Fig. 7B,C). This suggests that both the IFT-A- and phosphoinositide-binding properties of TULP3 regulate trafficking of MCHR1GFP to the cilia (Fig. 7C,D).
Discussion
TULP3 promotes the IFT-A complex-mediated trafficking of a subset of ciliary GPCRs
In this study, we show that TULP3 interacts directly with the IFT-A complex—specifically with the “core” proteins IFT122, IFT140, and WDR19—and is provided ciliary access through this interaction. A conserved helical region in the N terminus of TULP3 mediates this interaction. TULP3, in turn, promotes localization of a subset of GPCRs to the cilia in an IFT-A-dependent process. IFT-A thus has a second role in providing ciliary access to TULP3 and subsequently regulating GPCR trafficking in addition to its well-known function in retrograde IFT.
We consider TULP3 as an effector rather than a bona fide component of the IFT-A complex for the reasons stated below. First, TULP3 can be recruited to an intact preformed IFT-A complex in vitro, and does not disrupt the IFT-A complex upon being depleted. Second, TULP3 localization to the cilia depends on the IFT-A “core,” but not vice versa. Third, in contrast to knockdown of any of the IFT-A subunits, TULP3 knockdown does not disrupt retrograde transport of the IFT-B complex protein IFT88, suggesting that TULP3 does not function in retrograde IFT. Fourth, expression of the TULP3 N terminus dominantly affects endogenous TULP3 localization, probably by preventing its recruitment to an intact IFT-A complex, thereby affecting GPCR trafficking. Fifth, gel filtration experiments suggest that only a fraction of cellular TULP3 cofractionates with the IFT-A complex. Finally, although TULP3 is a nucleocytoplasmic shuttling protein, IFT140LAP does not localize to the nucleus in Leptomycin B-treated cells (data not shown). The dynamics of IFT-A complex assembly in vivo are not known; however, based on the aforementioned data, it would be reasonable to predict that a fraction of cellular TULP3 is recruited to the IFT-A complex later during assembly. IFT140 (but not TULP3) also strongly associates with dynein 1 and dynactin components, most likely enabling the dynein 1–dynactin complex to transport IFT-A and other cytoplasmic minus-end microtubule-based traffic close to the ciliary basal body (Fig. 7D; Sedmak and Wolfrum 2010).
Studies on the specialized mechanosensory chordotonal cilia in Drosophila also suggest a role of IFT-A in providing ciliary access to proteins (Lee et al. 2008). In these cilia, IFT140 normally localizes to the ciliary dilation (a highly structured axonemal inclusion of unknown function), which is lacking in the IFT140 mutant. While the IFT-B subunit IFT88 accumulates in these mutant cilia, they lack the TRPV ion channel, which normally localizes proximal to the dilation. In contrast, the IFT dynein mutant expresses the TRPV ion channel, although mislocalizing it beyond the disorganized ciliary dilation. So, it seems that, in these specialized cilia, in addition to its role in retrograde transport, IFT-A also functions in localizing the TRPV channel, and organizing the ciliary dilation.
Similarly, if the IFT-A complex functioned only in retrograde IFT, we would predict that ciliary signaling in the mammalian Hh pathway would be similarly affected in IFT-A and dynein 2 mutants. It is surprising, however, that mammalian neural tube phenotypes of mutants of IFT-A subunits differ from those of the IFT motor dynein 2 (Tran et al. 2008; Cortellino et al. 2009; Goetz and Anderson 2010), while Tulp3 alleles also show overactivation of the Hh pathway similar to the IFT-A mutants (Norman et al. 2009; Patterson et al. 2009). IFT-A associates with TULP3, and has both an early role in delivering TULP3 to cilia and another role in retrograde traffic of TULP3, compromising TULP3 function in either case. The regulation of TULP3's localization to the cilia by IFT-A provides a simplified model for their role in the Hh pathway.
How do Tulp3 and the IFT-A complex act as negative regulators of the Hh pathway to pattern the mammalian neural tube? In the absence of Hh, inactive Smo is transported laterally from the adjacent plasma membrane to the ciliary membrane at a baseline level, and knockdown of Dync2h1 causes Smo to accumulate in the untreated cilia (Ocbina and Anderson 2008; Milenkovic et al. 2009). However, Tulp3 and the IFT-A complex knockdowns do not cause accumulation of Smo in untreated cells, and genetic epistasis experiments suggest that Tulp3 function is independent of Smo (Norman et al. 2009; Patterson et al. 2009). In addition, unlike Dync2h1 knockdowns, Tulp3 and IFT-A complex knockdowns do not affect Gli3 processing or ciliary translocation (see also Norman et al. 2009; Patterson et al. 2009). These experiments suggest that Tulp3 and the IFT-A complex have functions distinct from Dync2h1 in the Hh signaling pathway. Tubby family proteins have been shown to bind to dsDNA, and the tubby domain of TULP1 apparently can act as a transcriptional activator (Boggon et al. 1999; Hu et al. 2009). Although TULP3 shuttles to the nucleus, TULP3 does not bind directly to an extensive set of DNA motifs in a protein microarray-based strategy (Hu et al. 2009). A more intriguing possibility is that Tulp3 and IFT-A would regulate a novel endogenous GPCR to be trafficked to the cilia of neural tube cells, thereby regulating patterning. We are currently looking into candidate GPCRs that could be important in neural tube patterning. In addition, although we do not detect any direct association between TULP3 and known players in Hh signaling [such as Gli1/2/3, Kif7, or Su(Fu)], IFT-A and TULP3 could function via their association with novel factors regulating Hh signaling. We generated mass spectrometry-based proteomic networks of some of these interesting candidates, and are in the process of generating mouse knockouts to further define their role in Hh signaling.
TULP3 regulates GPCR trafficking to primary cilia through its IFT-A- and phosphoinositide-binding properties
Tubby associates with G protein αq (Gαq) family proteins. GPCR-mediated activation of Gαq and phospholipase C-β triggers depletion of membrane PI(4,5)P2, detachment of tubby from the plasma membrane and subsequent nuclear translocation (Santagata et al. 2001). Similarly, mutating conserved residues important for phosphoinositide binding in TULP3 results in its redistribution to the nucleus. However, we do not detect any direct interactions between LAPTULP3 and G proteins or GPCRs, and its localization is not affected by overexpressing constitutively active Gα proteins in RPE cells (S Mukhopadhyay and PK Jackson, unpubl.). Instead, TULP3 appears to regulate trafficking of Rhodopsin family ciliary GPCRs, including SSTR3 and MCHR1, to the cilia in heterologous cell cultures and primary hippocampal cultures. Thus, TULP3 or “core” components of the IFT-A complex, important in providing ciliary access to TULP3, probably determine a critical step in GPCR trafficking. Both Tub and Tulp1 knockout mice exhibit early-onset progressive retinal degeneration (Hagstrom et al. 1999; Ikeda et al. 2000), and the role of TULP3 in GPCR trafficking is thus strongly reminiscent of the role of Tulp1 in intracellular vesicular trafficking of rhodopsin in vertebrate photoreceptors (Hagstrom et al. 1999). Other ciliary-localized proteins—such as the membrane-associated ACIII, RAB8a, and the Hh-induced Frizzled/Smoothened family membrane receptor Smo—remain unaffected, suggesting that numerous pathways gate constituents of the ciliary membrane. Out of this multitude of pathways, TULP3 effectively constitutes a sorting tag for a subset of GPCRs.
IFT complex proteins share a common architecture with components of coat protein I (COPI) of the coated vesicle transport pathways (Jekely and Arendt 2006), and are localized in defined preciliary target domains associated with transport vesicles (Sedmak and Wolfrum 2010). In addition, electron tomographic analysis of IFT particles show them to be closely apposed to the inner surface of the ciliary membrane (Pigino et al. 2009). Present evidence for preciliary transport of ciliary membrane proteins includes the AP1-dependent transport of ODR-10 in C. elegans (Dwyer et al. 2001), the presence of IFT20 at the Golgi apparatus and its role in polycystin-2 trafficking in mammalian cells (Follit et al. 2006), and the Rab8-dependent transport of rhodopsin in vertebrate photoreceptors (Moritz et al. 2001). In addition, depletion of ARL6/BBS3 and components of BBSome in CNS neurons show defects in trafficking of ciliary GPCRs (Berbari et al. 2008; Jin et al. 2010), and the flagella in a Chlamydomonas bbs4 mutant show abnormal accumulation of several signaling proteins, disrupting phototaxis (Lechtreck et al. 2009). It is possible that the C-terminal tubby domain of TULP3 facilitates the association of the IFT-A complex with preciliary vesicles and the ciliary membrane through its binding to phosphoinositides (see also Sedmak and Wolfrum 2010). In fact, mutations in either the IFT-A-binding domain or the phosphoinositide-binding domain of TULP3 dominantly affect GPCR localization in heterologous cell systems, suggesting that both of these functions of TULP3 simultaneously regulate trafficking. TULP3 selectively binds to PI(4,5)P2 in lipid overlay assays. Although there may be no selective enrichment of phosphoinositides in the ciliary membrane (Lobasso et al. 2010), depletion of PI(4,5)P2 affects axonemal outgrowth during Drosophila spermatogenesis (Wei et al. 2008). Biochemical reconstitution and liposome-binding experiments are currently under way to test whether TULP3 tethers IFT-A to membrane vesicles. We are also studying additional PH domain-containing proteins that interact with TULP3 for their potential roles in enhancing interactions with the ciliary membrane and/or preciliary vesicles.
GPCR-mediated signaling in the primary cilia in CNS neurons enables them to act as an extrasynaptic compartment in regulating neuronal function (Shimada et al. 1998; Chen et al. 2002; Einstein et al. 2010). The role of TULP3 in regulating neuronal GPCR trafficking thus provides an intriguing avenue for future investigation into the role of cilia in neurons. Current studies are under way in determining the role of TULP3 in neuronal activity.
Materials and methods
Antibodies and reagents
Antibodies were raised in rabbits against recombinant MBPTULP3, and were affinity-purified on a GSTTULP3 column using standard protocols. Antibodies against Thm1 (Tran et al. 2008), Smo, and Gli3 (Wen et al. 2010) were published previously. Details of commercial antibodies and plasmids are provided in the Supplemental Material.
Cell culture and transfections
Stable RPE, NIH 3T3, or IMCD-3 cell lines were generated by retroviral infection or Flp-in technology. Details are in the Supplemental Material. All siRNAs were predesigned On-target Plus (OTP) siRNA duplexes (except custom-designed OTPs 5–8 for TULP3), shown to yield a reduced frequency of off-target effects (Dharmacon) (Jackson et al. 2006). The most potent siRNA duplex was selected after assessment of mRNA knockdown by immunoblotting or qRT–PCR, and sequences are available on request. RPE cells were plated on coverslips and transfected with 50 nM siRNA duplexes using Lipofectamine RNAiMAX (Invitrogen). Cells were shifted from 10% serum to 0.2% serum 48 h after transfection to induce ciliation, and were fixed 24–30 h post-transfection. Plasmids were transfected into RPE cells using Fugene 6 (Roche).
Hippocampal neuronal cultures
Hippocampi were dissected from embyronic day 16 (E16) C57BL6 mice (Charles River Laboratories) and incubated for 5 min at 37°C in Hank's Balanced Salt Solution with 20 U/mL papain (Worthington Biochemical Corp.). Next, the cells were dissociated by trituration and plated on poly-D-lysine and laminin-coated coverslips (BD Biosciences) in Neurobasal medium with B27 supplement (Invitrogen). Cells were transfected with DNA constructs expressing GFPTULP3 fragments under the chicken β-actin promoter after 5 d in culture, using Lipofectamine 2000 (Invitrogen). Cells were fixed in 4% paraformaldehyde with 3% sucrose after a further 3 d in culture and processed for IF. Staining with anti-β-Tubulin III antibodies (which is a marker of neurons), and differences in nuclear morphology were used to distinguish glia from neurons in culture.
Immunofluorescence and microscopy
Immunofluorescence was performed according to standard protocols after fixation in 4% PFA, and post-fixed with 100% methanol for 10 min at −20°C (only for γ-tubulin staining). Images were acquired on an Everest deconvolution workstation (Intelligent Imaging Innovations) equipped with a Zeiss AxioImager.Z1 microscope and a CoolSnapHQ cooled CCD camera (Roper Scientific) and a 40× PlanApochromat NA 1.3 objective or 63× PlanApochromat NA 1.4 objective. Between eight and 14 Z sections at 0.3- to 0.5-mm intervals were acquired, and Z stacks were deconvolved using Slidebook 5 software (Intelligent Imaging Innovation). For quantitative analysis of ciliated RPE cell lines (Fig. 5B; Supplemental Fig. S1C), stacks of images were acquired from at least five to seven consecutive fields with confluent cells by looking into the DAPI channel, and percentages of ciliated cells were counted. Counting for localization of LAPTULP3 and IFT88 in RPE cell lines (Fig. 3) was performed blinded. For quantitative analysis of MCHR1GFP or SSTR3GFP RPE-positive cilia (Fig. 5), background correction for each experiment was done using the same parameters according to the Slidebook manual. For quantitative analysis of ciliated hippocampal neurons (Fig. 6D,E), stacks of images were acquired from fields with GFP+ neurons, and percentages of Sstr3- or ACIII-positive ciliated neurons were counted blinded for transfected or nontransfected cells.
Tandem affinity purification, mass spectrometry, biochemical fractionations, and in vitro protein-binding assays
LAP purification with anti-GFP antibodies, TEV/PreScission protease cleavage, and subsequent Protein S agarose affinity purification were performed according to Cheeseman and Desai (2005). Details of mass spectrometry, biochemical fractionations, and in vitro protein binding assays are provided in the Supplemental Material.
Statistical analyses
Statistical analyses of receptor localization experiments were performed using one-way ANOVA and Tukey-Kramer's post hoc multiple comparison tests between all possible pairs in each data set (Figs. 5B, 7B), while a Student's t-test was used for comparing two groups of treated cells (Fig. 6) using the JMP 8 statistical analysis package.
Acknowledgments
We thank David Beier for the THM1 antibody, and Christopher J. Westlake for the stable clonal RPE cell line expressing GFPRAB8a. We also thank Alexander Loktev and Mindan Sfakianos for the rabbit polyclonal antibody used for LAP purifications. We thank Piali Sengupta, Rachel Giles, Kevin Wright, and other members of the Jackson laboratory for critical review of the manuscript and for stimulating discussions.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1966210.
Supplemental material is available at http://www.genesdev.org.
References
- Berbari NF, Lewis JS, Bishop GA, Askwith CC, Mykytyn K 2008. Bardet-Biedl syndrome proteins are required for the localization of G protein-coupled receptors to primary cilia. Proc Natl Acad Sci 105: 4242–4246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop GA, Berbari NF, Lewis J, Mykytyn K 2007. Type III adenylyl cyclase localizes to primary cilia throughout the adult mouse brain. J Comp Neurol 505: 562–571 [DOI] [PubMed] [Google Scholar]
- Blacque OE, Li C, Inglis PN, Esmail MA, Ou G, Mah AK, Baillie DL, Scholey JM, Leroux MR 2006. The WD repeat-containing protein IFTA-1 is required for retrograde intraflagellar transport. Mol Biol Cell 17: 5053–5062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boggon TJ, Shan WS, Santagata S, Myers SC, Shapiro L 1999. Implication of tubby proteins as transcription factors by structure-based functional analysis. Science 286: 2119–2125 [DOI] [PubMed] [Google Scholar]
- Cameron DA, Pennimpede T, Petkovich M 2009. Tulp3 is a critical repressor of mouse hedgehog signaling. Dev Dyn 238: 1140–1149 [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, Desai A 2005. A combined approach for the localization and tandem affinity purification of protein complexes from metazoans. Sci STKE 2005: pl1 doi: 10.1126/stke.2662005pl1 [DOI] [PubMed] [Google Scholar]
- Chen Y, Hu C, Hsu CK, Zhang Q, Bi C, Asnicar M, Hsiung HM, Fox N, Slieker LJ, Yang DD, et al. 2002. Targeted disruption of the melanin-concentrating hormone receptor-1 results in hyperphagia and resistance to diet-induced obesity. Endocrinology 143: 2469–2477 [DOI] [PubMed] [Google Scholar]
- Cole DG, Snell WJ 2009. SnapShot: Intraflagellar transport. Cell 137: 784–784.e1 [DOI] [PubMed] [Google Scholar]
- Cole DG, Diener DR, Himelblau AL, Beech PL, Fuster JC, Rosenbaum JL 1998. Chlamydomonas kinesin-II-dependent intraflagellar transport (IFT): IFT particles contain proteins required for ciliary assembly in Caenorhabditis elegans sensory neurons. J Cell Biol 141: 993–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortellino S, Wang C, Wang B, Bassi MR, Caretti E, Champeval D, Calmont A, Jarnik M, Burch J, Zaret KS, et al. 2009. Defective ciliogenesis, embryonic lethality and severe impairment of the Sonic Hedgehog pathway caused by inactivation of the mouse complex A intraflagellar transport gene Ift122/Wdr10, partially overlapping with the DNA repair gene Med1/Mbd4. Dev Biol 325: 225–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davenport JR, Watts AJ, Roper VC, Croyle MJ, van Groen T, Wyss JM, Nagy TR, Kesterson RA, Yoder BK 2007. Disruption of intraflagellar transport in adult mice leads to obesity and slow-onset cystic kidney disease. Curr Biol 17: 1586–1594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwyer ND, Adler CE, Crump JG, L'Etoile ND, Bargmann CI 2001. Polarized dendritic transport and the AP-1 mu1 clathrin adaptor UNC-101 localize odorant receptors to olfactory cilia. Neuron 31: 277–287 [DOI] [PubMed] [Google Scholar]
- Einstein EB, Patterson CA, Hon BJ, Regan KA, Reddi J, Melnikoff DE, Mateer MJ, Schulz S, Johnson BN, Tallent MK 2010. Somatostatin signaling in neuronal cilia is critical for object recognition memory. J Neurosci 30: 4306–4314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follit JA, Tuft RA, Fogarty KE, Pazour GJ 2006. The intraflagellar transport protein IFT20 is associated with the Golgi complex and is required for cilia assembly. Mol Biol Cell 17: 3781–3792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz SC, Anderson KV 2010. The primary cilium: A signalling centre during vertebrate development. Nat Rev Genet 11: 331–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagstrom SA, Duyao M, North MA, Li T 1999. Retinal degeneration in tulp1−/− mice: Vesicular accumulation in the interphotoreceptor matrix. Invest Ophthalmol Vis Sci 40: 2795–2802 [PubMed] [Google Scholar]
- Handel M, Schulz S, Stanarius A, Schreff M, Erdtmann-Vourliotis M, Schmidt H, Wolf G, Hollt V 1999. Selective targeting of somatostatin receptor 3 to neuronal cilia. Neuroscience 89: 909–926 [DOI] [PubMed] [Google Scholar]
- Hou Y, Qin H, Follit JA, Pazour GJ, Rosenbaum JL, Witman GB 2007. Functional analysis of an individual IFT protein: IFT46 is required for transport of outer dynein arms into flagella. J Cell Biol 176: 653–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S, Xie Z, Onishi A, Yu X, Jiang L, Lin J, Rho HS, Woodard C, Wang H, Jeong JS, et al. 2009. Profiling the human protein–DNA interactome reveals ERK2 as a transcriptional repressor of interferon signaling. Cell 139: 610–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda S, Shiva N, Ikeda A, Smith RS, Nusinowitz S, Yan G, Lin TR, Chu S, Heckenlively JR, North MA, et al. 2000. Retinal degeneration but not obesity is observed in null mutants of the tubby-like protein 1 gene. Hum Mol Genet 9: 155–163 [DOI] [PubMed] [Google Scholar]
- Iomini C, Babaev-Khaimov V, Sassaroli M, Piperno G 2001. Protein particles in Chlamydomonas flagella undergo a transport cycle consisting of four phases. J Cell Biol 153: 13–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iomini C, Li L, Esparza JM, Dutcher SK 2009. Retrograde intraflagellar transport mutants identify complex A proteins with multiple genetic interactions in Chlamydomonas reinhardtii. Genetics 183: 885–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson AL, Burchard J, Leake D, Reynolds A, Schelter J, Guo J, Johnson JM, Lim L, Karpilow J, Nichols K, et al. 2006. Position-specific chemical modification of siRNAs reduces “off-target” transcript silencing. RNA 12: 1197–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jekely G, Arendt D 2006. Evolution of intraflagellar transport from coated vesicles and autogenous origin of the eukaryotic cilium. Bioessays 28: 191–198 [DOI] [PubMed] [Google Scholar]
- Jin H, White SR, Shida T, Schulz S, Aguiar M, Gygi SP, Bazan JF, Nachury MV 2010. The conserved Bardet-Biedl syndrome proteins assemble a coat that traffics membrane proteins to cilia. Cell 141: 1208–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardon JR, Vale RD 2009. Regulators of the cytoplasmic dynein motor. Nat Rev Mol Cell Biol 10: 854–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleyn PW, Fan W, Kovats SG, Lee JJ, Pulido JC, Wu Y, Berkemeier LR, Misumi DJ, Holmgren L, Charlat O, et al. 1996. Identification and characterization of the mouse obesity gene tubby: A member of a novel gene family. Cell 85: 281–290 [DOI] [PubMed] [Google Scholar]
- Lechtreck KF, Johnson EC, Sakai T, Cochran D, Ballif BA, Rush J, Pazour GJ, Ikebe M, Witman GB 2009. The Chlamydomonas reinhardtii BBSome is an IFT cargo required for export of specific signaling proteins from flagella. J Cell Biol 187: 1117–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E, Sivan-Loukianova E, Eberl DF, Kernan MJ 2008. An IFT-A protein is required to delimit functionally distinct zones in mechanosensory cilia. Curr Biol 18: 1899–1906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobasso S, Lopalco P, Angelini R, Baronio M, Fanizzi FP, Babudri F, Corcelli A 2010. Lipidomic analysis of porcine olfactory epithelial membranes and cilia. Lipids 45: 593–602 [DOI] [PubMed] [Google Scholar]
- Marshall WF, Rosenbaum JL 2001. Intraflagellar transport balances continuous turnover of outer doublet microtubules: Implications for flagellar length control. J Cell Biol 155: 405–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milenkovic L, Scott MP, Rohatgi R 2009. Lateral transport of Smoothened from the plasma membrane to the membrane of the cilium. J Cell Biol 187: 365–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz OL, Tam BM, Hurd LL, Peranen J, Deretic D, Papermaster DS 2001. Mutant rab8 Impairs docking and fusion of rhodopsin-bearing post-Golgi membranes and causes cell death of transgenic Xenopus rods. Mol Biol Cell 12: 2341–2351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachury MV, Loktev AV, Zhang Q, Westlake CJ, Peranen J, Merdes A, Slusarski DC, Scheller RH, Bazan JF, Sheffield VC, et al. 2007. A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell 129: 1201–1213 [DOI] [PubMed] [Google Scholar]
- Noben-Trauth K, Naggert JK, North MA, Nishina PM 1996. A candidate gene for the mouse mutation tubby. Nature 380: 534–538 [DOI] [PubMed] [Google Scholar]
- Norman RX, Ko HW, Huang V, Eun CM, Abler LL, Zhang Z, Sun X, Eggenschwiler JT 2009. Tubby-like protein 3 (TULP3) regulates patterning in the mouse embryo through inhibition of Hedgehog signaling. Hum Mol Genet 18: 1740–1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocbina PJ, Anderson KV 2008. Intraflagellar transport, cilia, and mammalian Hedgehog signaling: Analysis in mouse embryonic fibroblasts. Dev Dyn 237: 2030–2038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou G, Blacque OE, Snow JJ, Leroux MR, Scholey JM 2005. Functional coordination of intraflagellar transport motors. Nature 436: 583–587 [DOI] [PubMed] [Google Scholar]
- Patterson VL, Damrau C, Paudyal A, Reeve B, Grimes DT, Stewart ME, Williams DJ, Siggers P, Greenfield A, Murdoch JN 2009. Mouse hitchhiker mutants have spina bifida, dorso-ventral patterning defects and polydactyly: Identification of Tulp3 as a novel negative regulator of the Sonic hedgehog pathway. Hum Mol Genet 18: 1719–1739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen LB, Miller MS, Geimer S, Leitch JM, Rosenbaum JL, Cole DG 2005. Chlamydomonas IFT172 is encoded by FLA11, interacts with CrEB1, and regulates IFT at the flagellar tip. Curr Biol 15: 262–266 [DOI] [PubMed] [Google Scholar]
- Pigino G, Geimer S, Lanzavecchia S, Paccagnini E, Cantele F, Diener DR, Rosenbaum JL, Lupetti P 2009. Electron-tomographic analysis of intraflagellar transport particle trains in situ. J Cell Biol 187: 135–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno G, Siuda E, Henderson S, Segil M, Vaananen H, Sassaroli M 1998. Distinct mutants of retrograde intraflagellar transport (IFT) share similar morphological and molecular defects. J Cell Biol 143: 1591–1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugacheva EN, Jablonski SA, Hartman TR, Henske EP, Golemis EA 2007. HEF1-dependent Aurora A activation induces disassembly of the primary cilium. Cell 129: 1351–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin H, Diener DR, Geimer S, Cole DG, Rosenbaum JL 2004. Intraflagellar transport (IFT) cargo: IFT transports flagellar precursors to the tip and turnover products to the cell body. J Cell Biol 164: 255–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin H, Burnette DT, Bae YK, Forscher P, Barr MM, Rosenbaum JL 2005. Intraflagellar transport is required for the vectorial movement of TRPV channels in the ciliary membrane. Curr Biol 15: 1695–1699 [DOI] [PubMed] [Google Scholar]
- Rosenbaum JL, Witman GB 2002. Intraflagellar transport. Nat Rev Mol Cell Biol 3: 813–825 [DOI] [PubMed] [Google Scholar]
- Rost B 1996. PHD: Predicting one-dimensional protein structure by profile-based neural networks. Methods Enzymol 266: 525–539 [DOI] [PubMed] [Google Scholar]
- Santagata S, Boggon TJ, Baird CL, Gomez CA, Zhao J, Shan WS, Myszka DG, Shapiro L 2001. G-protein signaling through tubby proteins. Science 292: 2041–2050 [DOI] [PubMed] [Google Scholar]
- Sedmak T, Wolfrum U 2010. Intraflagellar transport molecules in ciliary and nonciliary cells of the retina. J Cell Biol 189: 171–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada M, Tritos NA, Lowell BB, Flier JS, Maratos-Flier E 1998. Mice lacking melanin-concentrating hormone are hypophagic and lean. Nature 396: 670–674 [DOI] [PubMed] [Google Scholar]
- Signor D, Wedaman KP, Orozco JT, Dwyer ND, Bargmann CI, Rose LS, Scholey JM 1999. Role of a class DHC1b dynein in retrograde transport of IFT motors and IFT raft particles along cilia, but not dendrites, in chemosensory neurons of living Caenorhabditis elegans. J Cell Biol 147: 519–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singla V, Reiter JF 2006. The primary cilium as the cell's antenna: Signaling at a sensory organelle. Science 313: 629–633 [DOI] [PubMed] [Google Scholar]
- Tran PV, Haycraft CJ, Besschetnova TY, Turbe-Doan A, Stottmann RW, Herron BJ, Chesebro AL, Qiu H, Scherz PJ, Shah JV, et al. 2008. THM1 negatively modulates mouse sonic hedgehog signal transduction and affects retrograde intraflagellar transport in cilia. Nat Genet 40: 403–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang WY, Bossard C, Khanna H, Peranen J, Swaroop A, Malhotra V, Dynlacht BD 2008. CP110 suppresses primary cilia formation through its interaction with CEP290, a protein deficient in human ciliary disease. Dev Cell 15: 187–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Fallon JF, Beachy PA 2000. Hedgehog-regulated processing of Gli3 produces an anterior/posterior repressor gradient in the developing vertebrate limb. Cell 100: 423–434 [DOI] [PubMed] [Google Scholar]
- Wang Q, Pan J, Snell WJ 2006. Intraflagellar transport particles participate directly in cilium-generated signaling in Chlamydomonas. Cell 125: 549–562 [DOI] [PubMed] [Google Scholar]
- Wang Z, Li V, Chan GC, Phan T, Nudelman AS, Xia Z, Storm DR 2009. Adult type 3 adenylyl cyclase–deficient mice are obese. PLoS ONE 4: e6979 doi: 10.1371/journal.pone.0006979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei HC, Rollins J, Fabian L, Hayes M, Polevoy G, Bazinet C, Brill JA 2008. Depletion of plasma membrane PtdIns(4,5)P2 reveals essential roles for phosphoinositides in flagellar biogenesis. J Cell Sci 121: 1076–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen X, Lai CK, Evangelista M, Hongo JA, de Sauvage FJ, Scales SJ 2010. Kinetics of hedgehog-dependent full-length Gli3 accumulation in primary cilia and subsequent degradation. Mol Cell Biol 30: 1910–1922 [DOI] [PMC free article] [PubMed] [Google Scholar]