Abstract
Glioblastoma multiforme (GBM) is a lethal brain tumor characterized by intense apoptosis resistance and extensive necrosis. Bcl2L12 (for Bcl2-like 12) is a cytoplasmic and nuclear protein that is overexpressed in primary GBM and functions to inhibit post-mitochondrial apoptosis signaling. Here, we show that nuclear Bcl2L12 physically and functionally interacts with the p53 tumor suppressor, as evidenced by the capacity of Bcl2L12 to (1) enable bypass of replicative senescence without concomitant loss of p53 or p19Arf, (2) inhibit p53-dependent DNA damage-induced apoptosis, (3) impede the capacity of p53 to bind some of its target gene promoters, and (4) attenuate endogenous p53-directed transcriptomic changes following genotoxic stress. Correspondingly, The Cancer Genome Atlas profile and tissue protein analyses of human GBM specimens show significantly lower Bcl2L12 expression in the setting of genetic p53 pathway inactivation. Thus, Bcl2L12 is a multifunctional protein that contributes to intense therapeutic resistance of GBM through its ability to operate on two key nodes of cytoplasmic and nuclear signaling cascades.
Keywords: Glioblastoma multiforme, Bcl2L12, p53
Glioblastoma multiforme (GBM) is the most aggressive form of malignant glioma and is characterized by rapid tumor cell proliferation, intense apoptosis resistance, florid necrosis, and robust angiogenesis. These tumor biological properties underlie the poor clinical outcome by conferring strong resistance to chemotherapy and radiotherapy, and by promoting a neurologically debilitating course leading to death within 12–18 mo post-diagnosis (for review, see Zhu and Parada 2002; Furnari et al. 2007).
Recent comprehensive resequencing analyses of primary GBM samples established TP53 deletion/mutation as a pivotal and common genetic event in the development of human primary GBM, with 29%–38% of samples harboring prototypical TP53 mutations (The Cancer Genome Atlas Research Network 2008; Zheng et al. 2008). Accordingly, CNS-specific deletion of p53 and Pten provokes an acute-onset high-grade malignant phenotype in the mouse, with clinical and pathobiological resemblance to the human disease (Kwon et al. 2008; Zheng et al. 2008; Alcantara Llaguno et al. 2009). The importance of p53 in gliomagenesis is reinforced by the occurrence of astrocytomas in Li-Fraumeni cancer syndrome, which stems from germline p53 mutations (Malkin et al. 1990; Srivastava et al. 1990). Across many cancer types, a large body of evidence has established that the tumor suppressor activity of p53 derives primarily from its transcriptional control of many (>2500) genes governing cell proliferation, cell survival, metabolism, and angiogenesis, among other cancer-relevant processes (Hoh et al. 2002; Levine et al. 2006). Several notable p53 target genes include the cell cycle regulators p21/CDKN1 (el-Deiry et al. 1993, 1994) and GADD45 (Hollander et al. 1993; Carrier et al. 1999), and the proapoptosis factors Bax (Miyashita et al. 1994; Zhan et al. 1994; Miyashita and Reed 1995), Puma (Nakano and Vousden 2001), and Noxa (Oda et al. 2000).
The importance of impaired p53 signaling in malignant glioma is highlighted further by genetic alterations targeting key components of the p53 pathway. Specifically, 11% of primary GBMs have Mdm2 amplification, which opposes the function of p53 directly by promoting its degradation, and indirectly through interaction with the E2F1 and Rb family proteins p107 and Rb1 (Dubs-Poterszman et al. 1995; Martin et al. 1995; Xiao et al. 1995; Haupt et al. 1997; Kubbutat et al. 1997). In addition, the Mdm2-related gene Mdm4, which inhibits p53 transcription and enhances the ubiquitin ligase activity of Mdm2, is amplified in 4% of GBM in the absence of TP53 mutation and Mdm2 amplification (Shvarts et al. 1996; Riemenschneider et al. 1999; Gu et al. 2002; Linares et al. 2003; The Cancer Genome Atlas Research Network 2008). Other mutations affect p14Arf (p19Arf in mouse) (Kamijo et al. 1997), which blocks the degradation of p53 through direct binding to Mdm2, thereby stabilizing p53 (Kamijo et al. 1997; Pomerantz et al. 1998; Stott et al. 1998; Zhang et al. 1998).
We recently identified Bcl2L12 (Bcl2-like 12) as a critical cell death regulator in glial cells that shows near universal overexpression in primary human GBM tumor specimens and contributes to important phenotypic hallmarks of the disease, such as resistance toward chemotherapy-induced apoptosis and enhanced necrogenesis (Stegh et al. 2007). Bcl2L12's potent anti-apoptotic and pronecrotic activities stem from its ability to block post-mitochondrial apoptosis signaling at the level of effector caspase-3 and caspase-7 activation. Direct physical interaction with the proenzyme matches well with Bcl2L12's ability to inhibit caspase-7 (Stegh et al. 2007), while up-regulation of αB-crystallin, a small heat-shock protein and known caspase-3-specific inhibitor, is instrumental for its caspase-3 inhibitory function (Stegh et al. 2008b).
Several lines of evidence prompted us to explore a possible physical and functional link between Bcl2L12 and p53, including their prominent roles in glioma pathogenesis, Bcl2L12's nuclear localization, and the capacity of other Bcl-2 family proteins (e.g., Bax, Bak, Bcl-xL, and Bcl-2) to interact with and functionally impact p53 (Mihara et al. 2003; Chipuk et al. 2004, 2005; Leu et al. 2004; Petros et al. 2004). Here, we show that Bcl2L12 colocalizes with and binds p53 within the nucleus to neutralize p53-directed transcriptional and growth arrest activities by blocking p53 binding to a large subset of its gene target promoter elements. As with p19Arf loss or Mdm2/4 amplification, Bcl2L12 overexpression compromises p53 tumor suppressor function, thus revealing another dimension of Bcl2L12's versatile progliomagenic activity profile.
Results
Bcl2L12 impedes the p53-dependent processes of replicative senescence and DNA damage-induced apoptosis
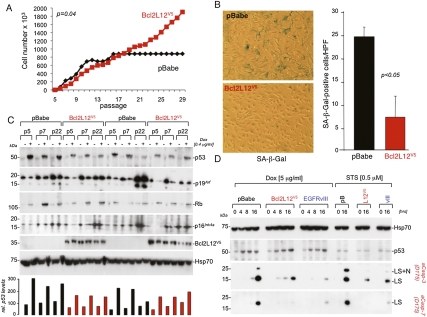
To evaluate the biological relevance of a potential Bcl2L12-p53 signaling axis, we assessed whether Bcl2L12 expression could neutralize the classical p53-dependent process of passage-induced cellular senescence. Bypass of the senescence checkpoint in primary murine cells has been shown to require loss of p53 or p19Arf (Serrano et al. 1996; Kamijo et al. 1997). Passage 2 (p2) wild-type mouse embryonic fibroblasts (MEFs) were retrovirally transduced with Bcl2L12 or vector (pBabe) control, and their proliferative capacity was monitored upon serial passage. Bcl2L12-expressing MEFs exhibited unabated growth for >35 passages, whereas control cultures showed significantly reduced growth rates or underwent growth arrest (Fig. 1A, P = 0.04; Supplemental Fig. S1A for three representative, independent transfectants) with coincidental increase in senescence-associated β-galactosidase (SA-β-Gal) activity (Fig. 1B, P < 0.05). On the molecular level, these long-term passaged Bcl2L12-expressing MEFs retained p53 on genomic DNA (Supplemental Fig. S1B) and protein (Fig. 1C; Supplemental Fig. S1C) levels, and remained intact for p19Arf, Rb, and p16Ink4a (Fig. 1C; Supplemental Fig. S1C). Of note, early-passaged, DNA-damaged Bcl2L12-expressing cultures expressed p53 at slightly lower levels in comparison with pBabe control (1.5-fold to twofold) (see histogram in Fig. 1C for densitometric quantification of corresponding p53 Western blots, shown in the top panel), implying that Bcl2L12 may modestly impact p53 protein stability (see below). These findings suggest that Bcl2L12 enables bypass of cellular senescence, possibly via functional neutralization of p53, and prompted an in-depth assessment of the impact of Bcl2L12 expression on additional p53-dependent activities.
Figure 1.
Bcl2L12 inhibits passage-induced senescence and p53-dependent apoptosis. (A) MEF immortalization assays. pBabe and Bcl2L12-infected (wild-type) MEF cultures were serially passaged and cell numbers were determined. Shown is one representative growth curve documenting unabated growth of Bcl2L12-infected MEF cultures at >30 passages. (B) Staining for and quantification of senescence-associated β-galactosidase (SA-β-Gal) in representative pBabe and Bcl2L12 MEF lines. Histogram shows quantification of β-Gal-positive cells in pBabe versus Bcl2L12 transfectants; three high-power fields (HPFs) were counted. Data are represented as mean ± SD. (C) Molecular analysis of p53–Rb signaling components. pBabe and Bcl2L12 MEFs of indicated passages were treated with Dox (0.4 μg/mL) for 16 h to assess p53, p19Arf, and Rb protein levels. p53 proteins levels were quantified densitometrically. (D) Bcl2L12 potently blocked Dox-induced effector caspase activation. Cultures were treated with Dox (5 μg/mL) and STS (0.5 μM) for the indicated periods of time, and post-mitochondrial caspase-3 and caspase-7 activation was followed by Western blot analysis. Of note, p53 protein was induced in pBabe (pB), Bcl2L12 (L12V5), and EGFRvIII (vIII) astrocytic cultures upon Dox, but not STS, treatment. The migration positions of cleaved caspase-3 and caspase-7 species ([LS] large subunits; [LS+N] large subunits with N-peptide) are indicated. Hsp70 is shown as a loading control.
To this end, we analyzed DNA damage-induced programmed cell death in primary murine cortical astrocytes null for the glioma tumor suppressor Ink4a/Arf. In the context of these experiments, it is worth noting that p53 and p19Arf have nonoverlapping functions in tumor surveillance, as p19Arf can act independently of the Mdm2–p53 axis, likely through its ability to inhibit gene expression by impacting the activity of other transcription factors besides p53 (for review, see Sherr 2006). Consequently, the utilization of Ink4a/Arf-deficient astrocytes relates not only to the importance of this tumor suppressor for GBM pathogenesis (Furnari et al. 2007), but also to the validity of this cell culture model for studying p53-dependent processes. Upon treatment with the DNA-damaging agent doxorubicin (Dox, 5 μg/mL), pBabe (pB) control, Bcl2L12 (L12V5)-transduced, and EGFRvIII (vIII)-transduced Ink4a/Arf−/− astrocytes showed robust induction of p53 (Fig. 1D). In contrast, the pan-kinase inhibitor staurosporine (STS), which causes cell death largely via p53-independent and effector caspase-dependent processes (Yamasaki et al. 2003), expectedly failed to induce p53 protein levels (Fig. 1D). The strict dependence of Dox-instigated apoptosis on functional p53 was further confirmed in DNA fragmentation assays contrasting apoptosis rates in p53−/− and Ink4a/Arf−/− astrocytes: Dox-treated p53−/− astrocytic cultures showed significantly reduced subG1 DNA subpopulations compared with Ink4a/Arf−/− cells (DNA fragmentation: p53−/− = 23.45% ± 2.1% vs. Ink4a/Arf−/− = 63.4% ± 2%, P = 0.004). In accordance with its p53-independent mechanism of action, STS was equally effective in provoking apoptosis in both genotypes (DNA fragmentation: p53−/− = 42.33% ± 1.49% vs. Ink4a/Arf−/− = 34.03% ± 3.1%, P = 0.01). Comparing effector caspase activation in DNA-damaged astrocytes with enforced Bcl2L12 and EGFRvIII expression revealed profound and comparable inhibition of post-mitochondrial effector caspase activation in Dox- and STS-treated cells relative to pBabe controls (Fig. 1D). Similar results were obtained using actinomycin D (ActD), another DNA damage-inducing agent, which triggers p53-dependent apoptosis (data not shown). Together, these findings further substantiate and expand previous studies documenting broad anti-apoptotic and caspase inhibitory activities of Bcl2L12 at levels comparable with the signature anti-apoptotic oncoprotein EGFRvIII (Nagane et al. 1998; Yamasaki et al. 2003; Stegh et al. 2007).
Bcl2L12 is less robustly amplified/overexpressed in GBM tumors with compromised p53 signaling
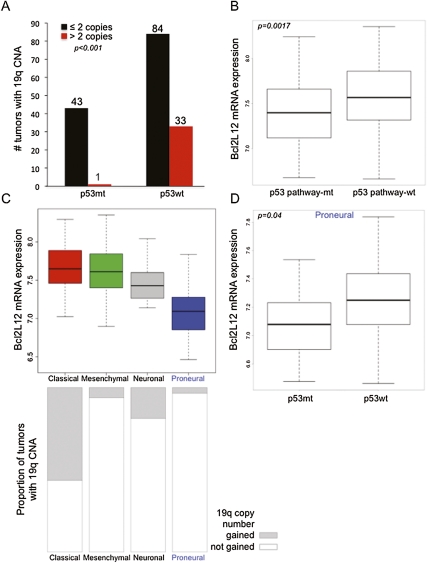
To provide genetic evidence of Bcl2L12 and p53 interaction in human GBM specimens, we explored whether Bcl2L12 overexpression represents an alternative event enabling neutralization of p53 tumor suppression. In silico analysis of 272 primary GBM samples of the multidimensional data set from The Cancer Genome Atlas (TCGA) (http://cancergenome.nih.gov/dataportal) revealed a highly significant correlation of Bcl2L12 genomic gain/amplification (nonfocal 19q event) and elevated Bcl2L12 mRNA expression with an intact p53 signaling pathway (P = 0.00014 and 0.0017, respectively) (Fig. 2A,B). A recent integrated analysis of TCGA data has identified distinct molecular subclasses of GBM defined by both transcriptomal and genomic alterations (Verhaak et al. 2010). The “proneural” GBM subtype is enriched for p53 mutations, and consequently also shows both reduced Bcl2L12 copy number gain and expression (Fig. 2C). To exclude the trivial possibility that the Bcl2L12–p53 correlation is a direct consequence of, or is driven by, association with one GBM tumor subtype, we examined Bcl2L12 expression in p53 wild-type versus mutant tumors of the proneural subtype. Bcl2L12 mRNA levels were significantly higher even in proneural GBMs with intact p53 signaling (P = 0.04) (Fig. 2D), suggesting that the correlation of Bcl2L12 expression with the p53 mutational status is not wholly due to mutual association with transcriptome-defined GBM subclasses.
Figure 2.
Reduced Bcl2L12 amplification and expression in GBM tumors with compromised p53 pathways. (A) Gain of 19q including the Bcl2L12 locus is seen more commonly in tumors with wild-type p53 (p53wt), and is notably rare in the presence of TP53 mutation. Shown are tumors with more than two Bcl2L12 copies (red bars) and two or fewer Bcl2L12 copies (black bars). (B) mRNA expression of Bcl2L12 is significantly elevated in tumors with intact p53 pathways, as defined by absence of TP53 mutation and Mdm2 or Mdm4 amplifications (log2 levels shown). Of note, Bcl2L12 mRNA levels are significantly elevated across all TCGA tumor specimens. Specimens with p53 pathway inactivation, however, showed significantly less Bcl2L12 abundance relative to tumors with functional p53 signaling, suggesting that the degree of Bcl2L12 overexpression correlates with the p53 status. (C) Bcl2L12 expression varies among transcriptomal subclasses of GBM recently identified from TCGA (Verhaak et al. 2010). Expression is lowest in the proneural subclass, which is enriched for p53 mutations (50.0% p53 mutant [p53mt] vs. 18.8% in nonproneural tumors), and in which 19q gains encompassing the Bcl2L12 locus are rare. (D) Analysis of Bcl2L12 expression in proneural tumors documenting its significant association with TP53 mutation. The expression analysis in all TCGA samples (B) considered Mdm2/4 amplification/overexpression, whereas the copy number alteration (CNA) (A) and expression analysis in proneural tumors (C) included only p53 mutations without taking into account the Mdm2/4 status.
While Bcl2L12 is expressed robustly in nearly all primary GBM specimens, its expression does vary over a wide range, prompting examination of Bcl2L12 protein expression levels in relation to the p53 status in primary GBM surgical specimens—seven with wild-type TP53, and four with either TP53 mutation or Mdm4 amplification (Supplemental Fig. S2). Using highly quantitative laser scanning cytometry (LSC), we documented lower Bcl2L12 protein expression in TP53 mutant versus TP53 wild-type tumors (P = 0.026) (Supplemental Fig. S2). Together with the molecular analysis of Bcl2L12-driven immortalization and cell death processes in primary murine cell cultures, these genomic and immunohistological data suggest a potential link between Bcl2L12 and p53, prompting further studies on the biochemical and functional levels.
Bcl2L12 colocalizes and interacts with p53
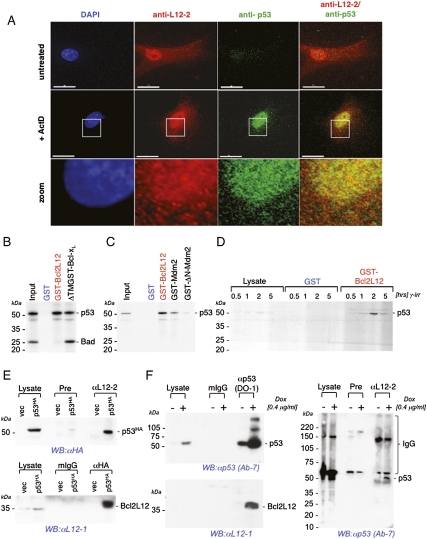
As a first step in assessing whether Bcl2L12:p53 complex formation is involved in repression of p53 activity, we assessed their cellular distribution and physical interaction via deconvolution immunofluorescence (IF) microscopy, subcellular fractionation, and coimmunoprecipitation studies of transduced and endogenous proteins. In IF studies, reconstitution of p53-null LNZ308 glioma cells with HA-tagged p53, followed by anti-Bcl2L12 and anti-HA immunostainings, revealed a pattern consistent with colocalization of p53HA and endogenous Bcl2L12, predominantly in the cell nucleus (Supplemental Fig. S3), further confirming previous observations of significant nuclear localization of Bcl2L12 (Stegh et al. 2007). Similarly, IF studies of untreated and ActD-treated (p53 wild-type) HCT116 cells (Supplemental Fig. S3) and U87MG cells (Fig. 3A) showed predominant colocalization of endogenous Bcl2L12 and p53 in the cell nucleus of ActD-treated cells. In accord with the IF findings, subcellular fractionation assays confirmed the coexistence of Bcl2L12 and p53 in the nuclear fractions of DNA-damaged U87MG cells (data not shown).
Figure 3.
Bcl2L12 colocalizes with and binds to p53 in vitro and in vivo. (A) Deconvolution IF studies of untreated and ActD-treated U87MG (p53 wild-type) cells showed predominant colocalization of endogenous Bcl2L12 and p53 in the cell nucleus of ActD-treated cells. Cells were stained with polyclonal anti-L12-2 (red) and monoclonal anti-p53 (green) antibodies. DAPI was used as a DNA counterstain. Bar, 15 μm. (B) In vitro GST interaction assays compared the binding affinity of Bcl2L12 with Bcl-xL, a well-characterized p53-binding Bcl-2 family member. GST-Bcl2L12 and GST-ΔTM-Bcl-xL were incubated with a mix of [35S]-labeled in vitro translated p53 and Bad, followed by SDS-PAGE and autoradiography. Migration positions of p53 and Bad are indicated. (C) The robustness of this in vitro Bcl2L12:p53 interaction was assessed by comparing the affinity of p53 with Bcl2L12 and Mdm2. GST-Bcl2L12, GST-Mdm2, and GST-ΔN-Mdm2 lacking the p53-binding interface were incubated with [35S]-labeled in vitro translated p53. Pull-down of p53 was quantified by SDS-PAGE and subsequent autoradiography. (D) Bcl2L12:p53 complex formation in vivo. Cellular lysates derived from γ-irradiated Ink4a/Arf-deficient astrocytes were incubated with GST or GST-Bcl2L12 recombinant proteins. Pull-down of p53 was assessed by anti-p53 Western blot analysis. (E) Reciprocal anti-HA and anti-Bcl2L12 coimmunoprecipitations from p53HA-transfected LNZ308 cells documented efficient complex formation between endogenous Bcl2L12 and ectopic p53. Migration position of Bcl2L12 and HA-tagged p53 are indicated. (F) Precipitation of endogenous p53 and Bcl2L12 levels in Dox-treated U87MG cells using a monoclonal anti-p53 antibody (Ab-6/DO-1) and a polyclonal Bcl2L12 antibody (L12-2), followed by SDS-PAGE and Western blot analysis for p53 and Bcl2L12, revealed robust Bcl2L12:p53 complex formation on endogenous protein levels. The migration positions of Bcl2L12, p53, and IgG are indicated. (Pre) Anti-L12-2 preimmune serum. Inputs in E and F represent one out of 100 lysates (corresponding to 10 μg) used for immunoprecipitations.
Next, in vitro interaction assays compared the binding affinity of Bcl2L12 with Bcl-xL, a well-characterized p53-binding Bcl-2 family member. GST-Bcl2L12 and GST-Bcl-xL lacking the transmembrane (TM) region (GST-ΔTM-Bcl-xL) were incubated with a mix of [35S]-labeled in vitro translated p53 and Bad. GST-ΔTM-Bcl-xL bound proapoptotic Bad and p53 as reported previously (Yang et al. 1995; Kelekar et al. 1997; Mihara et al. 2003), whereas GST-Bcl2L12 bound p53, but not Bad (Fig. 3B). The selectivity and specificity of Bcl2L12:p53 complex formation was further supported by reduced binding of thermodynamically and kinetically unstable, transcriptionally inactive V143 p53 point mutants to GST-Bcl2L12 compared with p53 wild type (Supplemental Fig. S6A). Notably, this in vitro Bcl2L12:p53 interaction appears to be as robust as the p53:Mdm2 association in that GST-Bcl2L12 and GST-Mdm2 bound p53 comparably, whereas the mutant protein ΔN-Mdm2 deficient in p53 binding expectedly failed to coprecipitate p53 (Fig. 3C). In addition, coprecipitation of p53 in cellular lysates of γ-irradiated mouse astrocytes revealed the presence of cellular p53 in GST-Bcl2L12, but not in GST control precipitates (Fig. 3D).
Subsequently, we examined cellular protein interactions. We first documented efficient complex formation between endogenous Bcl2L12 and ectopic p53 in reciprocal anti-HA and anti-Bcl2L12 coimmunoprecipitations from p53HA-reconstituted LNZ308 cells (Fig. 3E). Using a monoclonal anti-p53 (Ab-6/DO-1) and a polyclonal anti-Bcl2L12 antibody (αL12-2) (see Stegh et al. 2007 for antibody validation), reciprocal coprecipitation of endogenous p53 and Bcl2L12 in Dox-treated U87MG cells revealed robust interaction (Fig. 3F), demonstrating the presence of an endogenous Bcl2L12:p53 complex formation under conditions of p53 activation.
Bcl2L12 selectively represses p53-dependent transcription
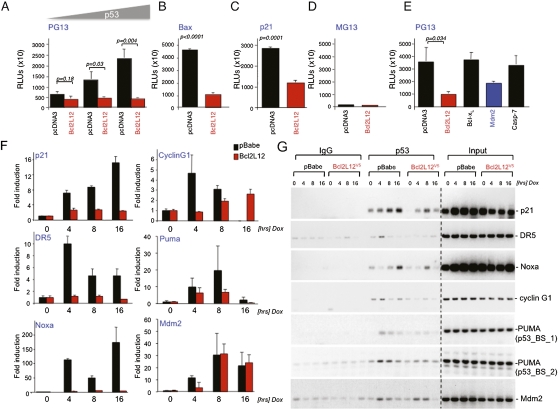
The capacity of p53 to activate the expression of genes governing cellular growth and survival is fundamental to its tumor suppressor activity (for review, see Harris and Levine 2005). To provide molecular validation of the Bcl2L12–p53 interaction, we examined the ability of Bcl2L12 to influence p53-directed reporter activity. Primary Ink4a/Arf−/− astrocytes were cotransfected with p53-responsive reporters, Bcl2L12, and p53 expression constructs. Bcl2L12 suppressed the activity of several different reporters, including PG13 and others driven by p53-binding elements of the Bax and p21 gene promoters (Fig. 4A–C; see Supplemental Fig. S7A for corresponding p53 and Bcl2L12 expression levels). These reporter systems are strictly dependent on p53 activity, as reflected by undetectable reporter activity in the absence of a consensus p53-binding site (MG13) (Fig. 4D). It is worth noting that Bcl2L12 suppressed p53 transactivation potential to a level comparable with or greater than Mdm2; in contrast, Bcl-xL or caspase-7 (a cytoplasmic binding partner of Bcl2L12) did not influence reporter activity, indicating that repression of p53 transactivation is not a general activity of the Bcl-2 family (Fig. 4E).
Figure 4.
Bcl2L12 bocks p53 transactivation potential by preventing p53 binding to target promoters. (A–E) To assay the capacity of Bcl2L12 to modulate p53-mediated transactivation, primary Ink4a/Arf-deficient astrocytes were cotransfected with Bcl2L12 and p53 expression constructs together with the luciferase reporters PG13 (A), Bax (B), p21 (C), and MG13 (D). Reporter activity was measured luminometrically. Data are represented as mean ± SD. (E) To comparatively quantify Bcl2L12's capacity to repress p53 transactivation, Bcl2L12, Bcl-xL, Mdm2, and caspase-7 cDNAs were transfected into astrocytes together with a PG13 luciferase reporter and a p53 cDNA. Reporter activity was measured luminometrically as in A–D and expressed as relative luminescence units (RLUs) normalized to concomitantly assessed β-Gal activity. Data are represented as mean ± SD. (F) Bcl2L12 blocks mRNA transcription of selected p53 target genes. pBabe and Bcl2L12-expressing astrocytes were treated with Dox (0.4 μg/mL) for the indicated periods of time, and p53 target gene expression was determined by qRT–PCR. Data are represented as mean ± SD. (G) ChIP assay for p21, DR5, Noxa, cyclin G1, Puma, and Mdm2 in Dox-treated astrocytic cultures. pBabe control and Bcl2L12-expresing Ink4a/Arf-deficient astrocytes were treated for the indicated periods of time with Dox (0.4 μg/mL). Chromatin was prepared, followed by p53 immunoprecipitation and PCRs for selected p53 target gene promoter fragments.
To obtain further evidence of Bcl2L12 regulation of p53 transactivation potential, we examined the impact of Bcl2L12 overexpression on well-established p53 target genes—including p21, DR5, Noxa, cyclin G1, and Puma—in Dox-treated astrocytes. In line with the above reporter assays, these endogenous p53-responsive genes were significantly repressed in the presence of Bcl2L12 on the mRNA (Fig. 4F; see also Supplemental Fig. S4 for additional p53 targets) and protein levels (data not shown). Similar results were obtained using camptothecin (CPT) as another well-known p53 inducer (Supplemental Fig. S4). Notably, while Bcl2L12 repressed many p53-induced apoptosis and cell cycle regulatory genes, this target gene repression was selective. In particular, Mdm2 expression was not affected, as evidenced by quantitative RT–PCR (qRT–PCR) analyses of Mdm2 mRNA levels in pBabe versus Bcl2L12-expressing astrocytes showing similar induction upon Dox treatment (Fig. 4F). These findings indicate a selective impact of Bcl2L12 on p53-directed transactivation across its gene targets.
Bcl2L12 selectively impedes p53 binding to its target promoter sequences
To determine the mechanism of Bcl2L12-mediated repression of p53 target gene induction, we assessed whether Bcl2L12 impairs p53 binding to its target gene promoters in chromatin immunoprecipitation (ChIP) assays using a highly specific polyclonal rabbit anti-p53 antibody (clone FL-393). Upon confirmation of comparable chromatin pull-down in pBabe control and Bcl2L12 lysates by anti-histone H3 ChIP (Supplemental Fig. S5A,B) and p53 protein precipitation (∼1.5-fold reduction in p53 pull-down) (Supplemental Fig. S5C), we found impaired p53 promoter occupancy in the presence of Bcl2L12 (Fig. 4G; Supplemental Fig. S5A,B), as documented by reduced anti-p53 pull-down of p21, DR5, Noxa, cyclin G1, and Puma promoter elements. These studies suggest that repression of p53 transactivation by Bcl2L12 is a direct consequence of reduced/abrogated binding of p53 to target gene promoter elements. Mirroring similar levels of Mdm2 mRNA induction in DNA-damaged control and Bcl2L12-expressing astrocytes (Fig. 4F), p53 occupancy of the Mdm2 promoter remained largely unaffected by Bcl2L12 (Fig. 4G, bottom panel), further confirming the selective impact of Bcl2L12 on p53 promoter occupancy and transcriptomes.
The capacity of Bcl2L12 to interfere with p53 binding to gene promoter elements of its target genes is consistent with GST pull-down studies establishing that Bcl2L12 can interact with p53 domains critical for its transactivational activity. Specifically, mutants lacking the tetradimerization domain (TET), the extreme C terminus (CT), and the transactivation domain (TAD) showed reduced binding to GST-Bcl2L12, suggesting a complex, nonlinear interaction interface between Bcl2L12 and p53 (Supplemental Fig. S6B). Conversely, reciprocal pull-down experiments using recombinant GST-p53 and various [35]S-labeled in vitro-translated Bcl2L12 mutants identified the Bcl-2 homology domain 2 (BH2) and a C-terminal region (amino acids 98–115) of the Bcl2L12 polypeptide as p53-interacting regions (Supplemental Fig. S6C). In line with previous studies documenting multiple binding interfaces between Bcl-2 family proteins (e.g., Bak, Bax, Bcl-2, and Bcl-xL) and p53 (Leu et al. 2004; Petros et al. 2004), Bcl2L12:p53 complex formation is mediated via nonlinear binding epitopes.
Bcl2L12 reduces p53 protein stability
Prompted by the molecular p53 pathway analyses in DNA-damaged MEFs showing reduced p53 accumulation in the presence of Bcl2L12 (Fig. 1C; Supplemental Fig. S1C), we further probed the impact of Bcl2L12 on p53 protein stability in Bcl2L12-driven (cDNA complementation) gain-of-function and (RNAi) loss-of-function systems. Enforced transient or stable Bcl2L12 expression in Ink4a/Arf-deficient astrocytes revealed modest (average, ∼1.5-fold) reductions in p53 protein (Supplemental Fig. S7A,B, top panel). Consistent with acetylation and ATM/ATR-directed phosphorylation as important p53-stabilizing signals in response to DNA damage (for review, see Ashcroft and Vousden 1999), we found reduced p53 phosphorylation (Ser18 and Ser389) and acetylation (acK379) in the presence of Bcl2L12 (Supplemental Fig. S7B). Attenuated p53 accumulation in ectopic expression studies is mirrored by enhanced p53 protein stability and phosphorylation/acetylation in RNAi loss-of-function experiments (Supplemental Fig. S7D). Specifically, stable transduction of U87G cells with shRNAs targeting Bcl2L12 (shL12-1 and shL-2) resulted in reduced endogenous Bcl2L12 protein levels (Supplemental Fig. S7C) that translated into ∼1.7-fold enhanced p53 accumulation (Supplemental Fig. S7D) and target gene induction (Supplemental Fig. S7E) in DNA-damaged U87MG cultures. We hypothesize that reduced p53 phosphorylation in Bcl2L12-expressing cells in the setting of intact Mdm2 (Fig. 4F,G) may contribute to p53 destabilization (see the Discussion below) as an additional Bcl2L12-driven mechanism to attenuate p53. In summary, multiple lines of evidence establish that Bcl2L12 is a potent regulator of p53 in primary and transformed cells and tumor specimens. Bcl2L12 binds p53 in vitro and in vivo, and potently inhibits p53-dependent processes. We propose that Bcl2L12's impact on p53 protein stability contributes to reduced p53 binding to target promoter sequences such as p21, DR5, Noxa, PUMA, and cyclin G1. Despite modestly reduced p53 levels, however, p53 binding to the Mdm2 promoter, and, consequently, induction of Mdm2 mRNA levels, remained similar in Bcl2L12-expressing Ink4a/Arf-deficient astrocytes compared with control cultures, suggesting selective Bcl2L12-directed inhibition of p53 promoter occupancy as a pivotal means to attenuate p53 function.
Discussion
Previous work has established that p53 plays pivotal roles in cell death execution following genotoxic stress (for review, see Aylon and Oren 2007), and, consequently, is intricately intertwined with canonical apoptosis signaling pathways, particularly those executed by Bcl-2 family proteins. Correspondingly, p53-dependent transcriptional up-regulation of the proapoptotic Bcl-2 family members Puma and Noxa is essential for DNA damage-mediated apoptosis via induction of mitochondrial membrane permeabilization (Jeffers et al. 2003; Villunger et al. 2003; Shibue et al. 2006). Independently of its transactivational activity, cytoplasmic p53 can interact physically with Bcl-2 proteins such as Bcl-2, Bcl-xL, and Bax to trigger cytochrome c release (for review, see Vaseva and Moll 2009). Notably, nuclear/transactivational and cytoplasmic activities may be linked, as p53 transcriptionally induces Puma that can bind Bcl-xL to displace p53 from a p53:Bcl-xL complex. Subsequently, released p53 directly interacts with and activates Bax to induce mitochondrial membrane permeabilization (Chipuk et al. 2005).
Bcl2L12 is an atypical Bcl-2 family protein that represses p53 transactivation
Reflecting its atypical domain structure with only focally restricted homology with known Bcl-2 family proteins, Bcl2L12's attributes are quite distinct from other members of the Bcl-2 family, including its prominent nucleoplasmic localization (Stegh et al. 2007, 2008b). Unlike canonical Bcl-2 family proteins with mitochondrial/cytoplasmic distribution, Bcl2L12 functions uniquely to modulate p53's nuclear transactivational activity rather than its cytoplasmic functions of impacting mitochondrial membrane integrity. Mechanistically, Bcl2L12's colocalization with and binding to nuclear p53 matches well with its ability to prevent p53 from binding promoter elements of its target genes, such as the proapoptotic genes DR5, Puma, and Noxa as key regulators of extrinsic and intrinsic apoptosis signaling, respectively, and the cell cycle regulators p21 and cyclin G1. Notably, some p53 target genes, such as Mdm2, were not repressed by Bcl2L12, pointing to a selective impact of Bcl2L12 on p53 gene target induction.
Precisely how Bcl2L12 prevents p53 binding to selective promoters and distinctively represses a subset of p53-induced genes remains to be determined. Interestingly, p53 can differentially transactivate promoters based on (1) the affinity of their p53-binding sites; (2) its post-translational modification status, such as acetylation (for review, see Lee and Gu 2010); and (3) its complex formation with cofactors capable of enhancing the interaction with certain promoter elements, such as p63/p73 (Flores et al. 2002), Bbp (Iwabuchi et al. 1998), and ASPP proteins (Samuels-Lev et al. 2001). The detailed analysis of Bcl2L12-orchestrated p53 post-translational modifications, together with proteomics to determine the composition of the Bcl2L12:p53 complex and the requirement of distinct transcriptional cofactors in the context of specific promoters and chromatin modification, will be needed to understand the full complexity of the cellular transcriptional response to p53 pathway activation in the context of Bcl2L12 (over)expression and various types of cellular stress.
Bcl2L12 impacts p53 protein stability
Besides its selective impact on p53 promoter occupancy as a major means to regulate p53 activity, Bcl2L12 modestly reduces p53 protein stability in response to DNA damage, as evidenced by attenuated (enhanced) p53 accumulation in cDNA complementation (RNAi loss-of-function) experiments. Modulating the activity of p53-degrading ubiquitin ligases—foremost Mdm2—represents a pivotal mechanism to regulate its tumor suppressor activity. Therefore, selectively preventing interaction between p53 and Mdm2 through phosphorlyation of p53's N terminus is a well-characterized signaling mechanism that leads to an increase in p53 protein abundance. Several reports suggest potential roles for Ser15, Ser20, and Ser33 phosphorylation in stabilizing p53. Consistent with studies documenting ATM/ATR-mediated phosphorylation of Ser15 (Ser18 in murine p53) as a pivotal p53 stabilization pathway in response to DNA damage (Kastan et al. 1992; Lu and Lane 1993; Siliciano et al. 1997; Khanna et al. 1998; Nakagawa et al. 1999; Tibbetts et al. 1999), we found reduced Ser18 phosphorylation in murine Bcl2L12-transduced astrocytic cultures and enhanced Ser15 phosphorylation in human U87MG cells stably transduced with Bcl2L12 targeting shRNAs. Together with the modulation of other factors regulating p53 stability (such as JNK, pRb, and cAbl signaling) (for review, see Ashcroft and Vousden 1999), we propose that reduced p53 phosphorylation in Bcl2L12-expressing cells in the setting of intact Mdm2 may contribute to the reduction in p53 levels and effects on overall p53 transactivation/promoter binding of gene targets such as DR5, Puma, Noxa, p21, and cyclin G1.
Bcl2L12 impacts p53-dependent replicative senescence and apoptosis induction upon DNA damage
We functionally validated Bcl2L12-mediated inhibition of p53 by detailed analyses of replicative senescence and DNA damage-induced apoptosis. Loss of growth-suppressive genes and the ensuing immortalization processes are caused by multiple mechanisms, including chromosomal amplification/deletion/recombination, point mutations, and epigenetic silencing targeting diverse cellular pathways, including IFN, IGF, MAP kinase, and, most importantly, the p53–Mdm2-p19Arf signaling axes (for review, see Lowe et al. 2004). Consequently, p53 or p19Arf-deficient MEFs do not senesce and can be propagated indefinitely (Serrano et al. 1996; Kamijo et al. 1997). Ectopic expression of Bcl2L12 resulted in enhanced proliferative capacity of MEF cultures in the absence p53, p19Arf, p16Ink4a, or Rb loss, further supporting the direct inactivation of p53 by Bcl2L12.
Oncogenomic and proteomic analyses document reduced Bcl2L12 amplification/expression in p53 mutant GBM tumors
Quantification of Bcl2L12 gene amplification, mRNA, and protein expression in primary GBM specimens with annotated TP53 mutational status revealed lower Bcl2L12 levels in tumors with impaired p53 signaling. These studies parallel oncogenomic efforts assessing Mdm2 and TP53 status. Mdm2, a p53-specific and E3 ubiquitin ligase, is the principal cellular antagonist of p53, acting to limit the p53 growth-suppressive function through complex formation and subsequent ubiquitinylation and proteosomal degradation (for review, see Toledo and Wahl 2006). Genetically verifying the Mdm2:p53 complex formation, studies in GBM (e.g., Reifenberger et al. 1993) and other cancer types, such as sarcoma (Oliner et al. 1992), demonstrated that Mdm2 amplification and TP53 mutation/deletion are mutually exclusive events.
Recent resequencing efforts, together with population-based and TCGA studies, established significant occurrences of TP53 mutations in not only secondary, but also primary, GBM (Ohgaki et al. 2004; Fukushima et al. 2006; The Cancer Genome Atlas Research Network 2008; Zheng et al. 2008). In addition, mutational analyses documenting genetic aberrations of other p53 pathway components in GBM (such as Mdm2, Mdm4, and p19Arf), together with functional and mouse modeling studies (e.g., Kwon et al. 2008; Zheng et al. 2008; Alcantara Llaguno et al. 2009), have established the p53 signaling network as an integral tumor suppressor pathway in GBM pathogenesis that impacts diverse cellular processes such as cell cycle control, cell death execution, and differentiation (for review, see Louis 2006). This study establishes a novel potent link between nuclear p53 and Bcl2L12, a Bcl-2-like effector caspase inhibitor that recapitulates the glioma-relevant phenotypic hallmarks of apoptosis (chemotherapy) resistance and necrogenesis (Stegh et al. 2007, 2008a,b). We propose that overexpression of Bcl2L12 in GBM represents an additional mechanism besides TP53 deletion/mutation, Mdm2/4 amplification, and p19Arf deletion to inactivate the tumor-suppressive function of the p53 pathway.
Bcl2L12 and p53 are important, functionally intertwined gliomagenic proteins
We identified Bcl2L12 as a potent direct inhibitor of post-mitochondrial effector caspase activation with anti-apoptotic and pronecrogenic activities (Stegh et al. 2007, 2008a,b). By blocking p53-dependent apoptosis through repression of p53-transactivated proapoptosis genes—foremost DR5, Noxa, and Puma—this study unravels yet another dimension of Bcl2L12's multifaceted cell death regulatory activities. Interestingly, and similar to other p53-reponsive apoptosis regulators, Bcl2L12 contains several evolutionarily conserved p53-binding sites, as revealed by TRANSFAC motif library searches (p53 binding sites at positions −3587, −3647, −2682, −2366, and +68 relative to the transcriptional start site) (http://www.motif.genome.jp), suggesting that, similar to Bcl-2 family proteins Puma, Noxa, and Bax, Bcl2L12 might be under p53-dependent transcriptional control. Expanding its role in cell death signaling beyond apoptosis regulation, p53 has been identified recently as a modulator of programmed necrosis (Tu et al. 2009), as it transcriptionally up-regulates cathepsin Q, which cooperates with reactive oxygen species (ROS) to provoke the necrotic signature of cellular and nuclear swelling, membrane rupture, and organelle disintegration. Future studies aim to understand how Bcl2L12, as an important player in programmed necrosis signaling, impacts necrogenesis under conditions of p53 inactivation.
Based on Bcl2L12's versatile oncogenic profile, we propose that targeting the Bcl2L12:p53 axis alone or in combination with stress-inducing therapies to reactivate p53-dependent growth arrest and cell death programs in GBM may provide an additional point for therapeutic intervention for a significant fraction of GBM cases that retain intact p53.
Materials and methods
Cell culture
Wild-type MEFs were generated from 13.5-d post-coitum embryos. Cortical Ink4a/Arf-deficient murine astrocytes were isolated as described (Bachoo et al. 2002). pBabe/Bcl2L12V5-expressing astrocytes were generated and characterized previously (Stegh et al. 2007). All cell lines and primary cultures were propagated in Dulbecco's Modification of Eagle's Medium (Invitrogen) supplemented with 10% FBS (Invitrogen). For 3T3 analyses, 3 × 105 MEFs (five independent transfectants) retrovirally transduced with pBabe or pBabe-Bcl2L12V5 according to established protocols using the pBabe/pCL system (Naviaux et al. 1996) were passaged into six-well plates every 3 d. Senescence was determined using the SA-β-Gal staining kit (Abcam) according to the manufacturer's instructions and quantified by counting SA-β-Gal-positive cells per high-power field (HPF). Generation of shCo, shL12-1, and shL12--2-expressing U87MG cells was done as reported previously (Stegh et al. 2007).
Integrated analysis of GBM data from TCGA
A description of TCGA data types, platforms, and analyses are as described previously (The Cancer Genome Atlas Research Network 2008). Processed genomic data sets were downloaded from TCGA public data portal (http://www.cancergenome.nih.gov/dataportal) as available on July 20, 2009. Specific data sources were as follows: Expression data: “Level 3” normalized gene expression was derived from the Cancer Genome Characterization Center (CGCC) at Lawrence Berkeley National Laboratory (Affymetrix Exon 1.0). Expression data were inverse log2-transformed. Transcriptomal cluster assignments for TCGA tumors were as described recently (Verhaak et al. 2010). Array CGH (aCGH) data: “Level 3” normalized and segmented copy number data were obtained from the CGCC at Memorial Sloan-Kettering Cancer Center (Agilent 244K CGH Array). For aCGH data, one unique profile was selected for each tumor based on the highest signal-to-noise estimate. Sequencing data: All available sequencing data summaries were in “multiple alignment format” (MAF) files current as of July 20, 2009 (http://www.broadinstitute.org, GBM.ABI.1.32maf; http://genome.wustl.edu, GBM.ABI.53.maf; and http://hgsc.bcm.tmc.edu, GBM.ABI.1.24.maf, and http://hgsc.bcm.tmc.edu, GBM.ABI.2.7.maf). Mutations were further filtered by excluding events, which were classified as “germline,” “synonymous” or “silent,” or “unvalidated.” Level 3 aCGH data were used for local copy number estimation. Amplification was defined by regional log2 ratio >2.0 (more than eight copies). Copy number at the Bcl2L12 locus was defined as two or fewer copies (log2 ratio ≤ 0.25), or gained more than two copies (log2 ratio > 0.25). TP53 mutation was defined as somatic nonsynonymous validated SNP or indel. TP53 pathway alteration was defined as TP53 mutation and/or amplification of Mdm1, Mdm2, or Mdm4. We did not include Arf loss, as this is nearly always inclusive of Ink4a loss and is not found to be mutually exclusive of either TP53 mutation or Mdm amplification.
Acknowledgments
We thank Dr. William Kaelin (Dana-Farber Cancer Institute [DFCI], Boston) for providing us with cDNAs encoding various p53 deletion mutants, and Dr. Stanley Korsmeyer (DFCI, Boston) for the GST-Bcl-xL and Bad expression constructs. A.H.S. was supported by NIH grant 5R00CA129172-04 and a Zell Scholar award. C.B. was supported by the NIOH grants 5P01CA95616 and U24 CA126543-01. Grant support derives from the Ben and Catherine Ivy Foundation (to R.A.D. and L.C.), the Goldhirsh Foundation (to R.A.D.), and NIH grants RO1CA99041 (to L.C) and 5P01CA95616 (to L.C. and R.A.D.). R.A.D. is an American Cancer Society Research Professor, and is supported by the Robert A. and Renee E. Belfer Foundation Institute for Innovative Cancer Science. The results published here are based in part upon data generated by The Cancer Genome Atlas pilot project established by the NCI and National Human Genome Research Institute. Information about TCGA and the investigators and institutions who constitute the The Cancer Genome Atlas Research Network can be found at http://www.cancergenome.nih.gov. A.H.S. dedicates this study to the memory of his father, Theodor Stegh, who passed away in May 2009 after a brave battle with ALS.
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1924710.
Supplemental material is available at http://www.genesdev.org.
References
- Alcantara Llaguno S, Chen J, Kwon CH, Jackson EL, Li Y, Burns DK, Alvarez-Buylla A, Parada LF 2009. Malignant astrocytomas originate from neural stem/progenitor cells in a somatic tumor suppressor mouse model. Cancer Cell 15: 45–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashcroft M, Vousden KH 1999. Regulation of p53 stability. Oncogene 18: 7637–7643 [DOI] [PubMed] [Google Scholar]
- Aylon Y, Oren M 2007. Living with p53, dying of p53. Cell 130: 597–600 [DOI] [PubMed] [Google Scholar]
- Bachoo RM, Maher EA, Ligon KL, Sharpless NE, Chan SS, You MJ, Tang Y, DeFrances J, Stover E, Weissleder R, et al. 2002. Epidermal growth factor receptor and Ink4a/Arf: Convergent mechanisms governing terminal differentiation and transformation along the neural stem cell to astrocyte axis. Cancer Cell 1: 269–277 [DOI] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Research Network 2008. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455: 1061–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrier F, Georgel PT, Pourquier P, Blake M, Kontny HU, Antinore MJ, Gariboldi M, Myers TG, Weinstein JN, Pommier Y, et al. 1999. Gadd45, a p53-responsive stress protein, modifies DNA accessibility on damaged chromatin. Mol Cell Biol 19: 1673–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chipuk JE, Kuwana T, Bouchier-Hayes L, Droin NM, Newmeyer DD, Schuler M, Green DR 2004. Direct activation of Bax by p53 mediates mitochondrial membrane permeabilization and apoptosis. Science 303: 1010–1014 [DOI] [PubMed] [Google Scholar]
- Chipuk JE, Bouchier-Hayes L, Kuwana T, Newmeyer DD, Green DR 2005. PUMA couples the nuclear and cytoplasmic proapoptotic function of p53. Science 309: 1732–1735 [DOI] [PubMed] [Google Scholar]
- Dubs-Poterszman MC, Tocque B, Wasylyk B 1995. MDM2 transformation in the absence of p53 and abrogation of the p107 G1 cell-cycle arrest. Oncogene 11: 2445–2449 [PubMed] [Google Scholar]
- el-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, Lin D, Mercer WE, Kinzler KW, Vogelstein B 1993. WAF1, a potential mediator of p53 tumor suppression. Cell 75: 817–825 [DOI] [PubMed] [Google Scholar]
- el-Deiry WS, Harper JW, O'Connor PM, Velculescu VE, Canman CE, Jackman J, Pietenpol JA, Burrell M, Hill DE, Wang Y, et al. 1994. WAF1/CIP1 is induced in p53-mediated G1 arrest and apoptosis. Cancer Res 54: 1169–1174 [PubMed] [Google Scholar]
- Flores ER, Tsai KY, Crowley D, Sengupta S, Yang A, McKeon F, Jacks T 2002. p63 and p73 are required for p53-dependent apoptosis in response to DNA damage. Nature 416: 560–564 [DOI] [PubMed] [Google Scholar]
- Fukushima T, Favereaux A, Huang H, Shimizu T, Yonekawa Y, Nakazato Y, Ohagki H 2006. Genetic alterations in primary glioblastomas in Japan. J Neuropathol Exp Neurol 65: 12–18 [DOI] [PubMed] [Google Scholar]
- Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, Stegh A, Hahn WC, Ligon KL, Louis DN, Brennan C, et al. 2007. Malignant astrocytic glioma: Genetics, biology, and paths to treatment. Genes Dev 21: 2683–2710 [DOI] [PubMed] [Google Scholar]
- Gu J, Kawai H, Nie L, Kitao H, Wiederschain D, Jochemsen AG, Parant J, Lozano G, Yuan ZM 2002. Mutual dependence of MDM2 and MDMX in their functional inactivation of p53. J Biol Chem 277: 19251–19254 [DOI] [PubMed] [Google Scholar]
- Harris SL, Levine AJ 2005. The p53 pathway: Positive and negative feedback loops. Oncogene 24: 2899–2908 [DOI] [PubMed] [Google Scholar]
- Haupt Y, Maya R, Kazaz A, Oren M 1997. Mdm2 promotes the rapid degradation of p53. Nature 387: 296–299 [DOI] [PubMed] [Google Scholar]
- Hoh J, Jin S, Parrado T, Edington J, Levine AJ, Ott J 2002. The p53MH algorithm and its application in detecting p53-responsive genes. Proc Natl Acad Sci 99: 8467–8472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollander MC, Alamo I, Jackman J, Wang MG, McBride OW, Fornace AJ Jr 1993. Analysis of the mammalian gadd45 gene and its response to DNA damage. J Biol Chem 268: 24385–24393 [PubMed] [Google Scholar]
- Iwabuchi K, Li B, Massa HF, Trask BJ, Date T, Fields S 1998. Stimulation of p53-mediated transcriptional activation by the p53-binding proteins, 53BP1 and 53BP2. J Biol Chem 273: 26061–26068 [DOI] [PubMed] [Google Scholar]
- Jeffers JR, Parganas E, Lee Y, Yang C, Wang J, Brennan J, MacLean KH, Han J, Chittenden T, Ihle JN, et al. 2003. Puma is an essential mediator of p53-dependent and -independent apoptotic pathways. Cancer Cell 4: 321–328 [DOI] [PubMed] [Google Scholar]
- Kamijo T, Zindy F, Roussel MF, Quelle DE, Downing JR, Ashmun RA, Grosveld G, Sherr CJ 1997. Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell 91: 649–659 [DOI] [PubMed] [Google Scholar]
- Kastan MB, Zhan Q, el-Deiry WS, Carrier F, Jacks T, Walsh WV, Plunkett BS, Vogelstein B, Fornace AJ Jr 1992. A mammalian cell cycle checkpoint pathway utilizing p53 and GADD45 is defective in ataxia-telangiectasia. Cell 71: 587–597 [DOI] [PubMed] [Google Scholar]
- Kelekar A, Chang BS, Harlan JE, Fesik SW, Thompson CB 1997. Bad is a BH3 domain-containing protein that forms an inactivating dimer with Bcl-XL. Mol Cell Biol 17: 7040–7046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna KK, Keating KE, Kozlov S, Scott S, Gatei M, Hobson K, Taya Y, Gabrielli B, Chan D, Lees-Miller SP, et al. 1998. ATM associates with and phosphorylates p53: Mapping the region of interaction. Nat Genet 20: 398–400 [DOI] [PubMed] [Google Scholar]
- Kubbutat MH, Jones SN, Vousden KH 1997. Regulation of p53 stability by Mdm2. Nature 387: 299–303 [DOI] [PubMed] [Google Scholar]
- Kwon CH, Zhao D, Chen J, Alcantara S, Li Y, Burns DK, Mason RP, Lee EY, Wu H, Parada LF 2008. Pten haploinsufficiency accelerates formation of high-grade astrocytomas. Cancer Res 68: 3286–3294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JT, Gu W 2010. The multiple levels of regulation by p53 ubiquitination. Cell Death Differ 17: 86–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leu JI, Dumont P, Hafey M, Murphy ME, George DL 2004. Mitochondrial p53 activates Bak and causes disruption of a Bak–Mcl1 complex. Nat Cell Biol 6: 443–450 [DOI] [PubMed] [Google Scholar]
- Levine AJ, Hu W, Feng Z 2006. The P53 pathway: What questions remain to be explored? Cell Death Differ 13: 1027–1036 [DOI] [PubMed] [Google Scholar]
- Linares LK, Hengstermann A, Ciechanover A, Muller S, Scheffner M 2003. HdmX stimulates Hdm2-mediated ubiquitination and degradation of p53. Proc Natl Acad Sci 100: 12009–12014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis DN 2006. Molecular pathology of malignant gliomas. Annu Rev Pathol 1: 97–117 [DOI] [PubMed] [Google Scholar]
- Lowe SW, Cepero E, Evan G 2004. Intrinsic tumour suppression. Nature 432: 307–315 [DOI] [PubMed] [Google Scholar]
- Lu X, Lane DP 1993. Differential induction of transcriptionally active p53 following UV or ionizing radiation: Defects in chromosome instability syndromes? Cell 75: 765–778 [DOI] [PubMed] [Google Scholar]
- Malkin D, Li FP, Strong LC, Fraumeni JF Jr, Nelson CE, Kim DH, Kassel J, Gryka MA, Bischoff FZ, Tainsky MA, et al. 1990. Germ line p53 mutations in a familial syndrome of breast cancer, sarcomas, and other neoplasms. Science 250: 1233–1238 [DOI] [PubMed] [Google Scholar]
- Martin K, Trouche D, Hagemeier C, Sorensen TS, La Thangue NB, Kouzarides T 1995. Stimulation of E2F1/DP1 transcriptional activity by MDM2 oncoprotein. Nature 375: 691–694 [DOI] [PubMed] [Google Scholar]
- Mihara M, Erster S, Zaika A, Petrenko O, Chittenden T, Pancoska P, Moll UM 2003. p53 has a direct apoptogenic role at the mitochondria. Mol Cell 11: 577–590 [DOI] [PubMed] [Google Scholar]
- Miyashita T, Reed JC 1995. Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell 80: 293–299 [DOI] [PubMed] [Google Scholar]
- Miyashita T, Krajewski S, Krajewska M, Wang HG, Lin HK, Liebermann DA, Hoffman B, Reed JC 1994. Tumor suppressor p53 is a regulator of bcl-2 and bax gene expression in vitro and in vivo. Oncogene 9: 1799–1805 [PubMed] [Google Scholar]
- Nagane M, Levitzki A, Gazit A, Cavenee WK, Huang HJ 1998. Drug resistance of human glioblastoma cells conferred by a tumor-specific mutant epidermal growth factor receptor through modulation of Bcl-XL and caspase-3-like proteases. Proc Natl Acad Sci 95: 5724–5729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa K, Taya Y, Tamai K, Yamaizumi M 1999. Requirement of ATM in phosphorylation of the human p53 protein at serine 15 following DNA double-strand breaks. Mol Cell Biol 19: 2828–2834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano K, Vousden KH 2001. PUMA, a novel proapoptotic gene, is induced by p53. Mol Cell 7: 683–694 [DOI] [PubMed] [Google Scholar]
- Naviaux RK, Costanzi E, Haas M, Verma IM 1996. The pCL vector system: Rapid production of helper-free, high-titer, recombinant retroviruses. J Virol 70: 5701–5705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda E, Ohki R, Murasawa H, Nemoto J, Shibue T, Yamashita T, Tokino T, Taniguchi T, Tanaka N 2000. Noxa, a BH3-only member of the Bcl-2 family and candidate mediator of p53-induced apoptosis. Science 288: 1053–1058 [DOI] [PubMed] [Google Scholar]
- Ohgaki H, Dessen P, Jourde B, Horstmann S, Nishikawa T, Di Patre PL, Burkhard C, Schuler D, Probst-Hensch NM, Maiorka PC, et al. 2004. Genetic pathways to glioblastoma: A population-based study. Cancer Res 64: 6892–6899 [DOI] [PubMed] [Google Scholar]
- Oliner JD, Kinzler KW, Meltzer PS, George DL, Vogelstein B 1992. Amplification of a gene encoding a p53-associated protein in human sarcomas. Nature 358: 80–83 [DOI] [PubMed] [Google Scholar]
- Petros AM, Gunasekera A, Xu N, Olejniczak ET, Fesik SW 2004. Defining the p53 DNA-binding domain/Bcl-x(L)-binding interface using NMR. FEBS Lett 559: 171–174 [DOI] [PubMed] [Google Scholar]
- Pomerantz J, Schreiber-Agus N, Liegeois NJ, Silverman A, Alland L, Chin L, Potes J, Chen K, Orlow I, Lee HW, et al. 1998. The Ink4a tumor suppressor gene product, p19Arf, interacts with MDM2 and neutralizes MDM2's inhibition of p53. Cell 92: 713–723 [DOI] [PubMed] [Google Scholar]
- Reifenberger G, Liu L, Ichimura K, Schmidt EE, Collins VP 1993. Amplification and overexpression of the MDM2 gene in a subset of human malignant gliomas without p53 mutations. Cancer Res 53: 2736–2739 [PubMed] [Google Scholar]
- Riemenschneider MJ, Buschges R, Wolter M, Reifenberger J, Bostrom J, Kraus JA, Schlegel U, Reifenberger G 1999. Amplification and overexpression of the MDM4 (MDMX) gene from 1q32 in a subset of malignant gliomas without TP53 mutation or MDM2 amplification. Cancer Res 59: 6091–6096 [PubMed] [Google Scholar]
- Samuels-Lev Y, O'Connor DJ, Bergamaschi D, Trigiante G, Hsieh JK, Zhong S, Campargue I, Naumovski L, Crook T, Lu X 2001. ASPP proteins specifically stimulate the apoptotic function of p53. Mol Cell 8: 781–794 [DOI] [PubMed] [Google Scholar]
- Serrano M, Lee H, Chin L, Cordon-Cardo C, Beach D, DePinho RA 1996. Role of the INK4a locus in tumor suppression and cell mortality. Cell 85: 27–37 [DOI] [PubMed] [Google Scholar]
- Sherr CJ 2006. Divorcing ARF and p53: An unsettled case. Nat Rev Cancer 6: 663–673 [DOI] [PubMed] [Google Scholar]
- Shibue T, Suzuki S, Okamoto H, Yoshida H, Ohba Y, Takaoka A, Taniguchi T 2006. Differential contribution of Puma and Noxa in dual regulation of p53-mediated apoptotic pathways. EMBO J 25: 4952–4962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shvarts A, Steegenga WT, Riteco N, van Laar T, Dekker P, Bazuine M, van Ham RC, van der Houven van Oordt W, Hateboer G, van der Eb AJ, et al. 1996. MDMX: A novel p53-binding protein with some functional properties of MDM2. EMBO J 15: 5349–5357 [PMC free article] [PubMed] [Google Scholar]
- Siliciano JD, Canman CE, Taya Y, Sakaguchi K, Appella E, Kastan MB 1997. DNA damage induces phosphorylation of the amino terminus of p53. Genes Dev 11: 3471–3481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava S, Zou ZQ, Pirollo K, Blattner W, Chang EH 1990. Germ-line transmission of a mutated p53 gene in a cancer-prone family with Li-Fraumeni syndrome. Nature 348: 747–749 [DOI] [PubMed] [Google Scholar]
- Stegh AH, Kim H, Bachoo RM, Forloney KL, Zhang J, Schulze H, Park K, Hannon GJ, Yuan J, Louis DN, et al. 2007. Bcl2L12 inhibits post-mitochondrial apoptosis signaling in glioblastoma. Genes Dev 21: 98–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegh AH, Chin L, Louis DN, DePinho RA 2008a. What drives intense apoptosis resistance and propensity for necrosis in glioblastoma? A role for Bcl2L12 as a multifunctional cell death regulator. Cell Cycle 7: 2833–2839 [DOI] [PubMed] [Google Scholar]
- Stegh AH, Kesari S, Mahoney JE, Jenq HT, Forloney KL, Protopopov A, Louis DN, Chin L, DePinho RA 2008b. Bcl2L12-mediated inhibition of effector caspase-3 and caspase-7 via distinct mechanisms in glioblastoma. Proc Natl Acad Sci 105: 10703–10708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stott FJ, Bates S, James MC, McConnell BB, Starborg M, Brookes S, Palmero I, Ryan K, Hara E, Vousden KH, et al. 1998. The alternative product from the human CDKN2A locus, p14(ARF), participates in a regulatory feedback loop with p53 and MDM2. EMBO J 17: 5001–5014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibbetts RS, Brumbaugh KM, Williams JM, Sarkaria JN, Cliby WA, Shieh SY, Taya Y, Prives C, Abraham RT 1999. A role for ATR in the DNA damage-induced phosphorylation of p53. Genes Dev 13: 152–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo F, Wahl GM 2006. Regulating the p53 pathway: In vitro hypotheses, in vivo veritas. Nat Rev Cancer 6: 909–923 [DOI] [PubMed] [Google Scholar]
- Tu HC, Ren D, Wang GX, Chen DY, Westergard TD, Kim H, Sasagawa S, Hsieh JJ, Cheng EH 2009. The p53–cathepsin axis cooperates with ROS to activate programmed necrotic death upon DNA damage. Proc Natl Acad Sci 106: 1093–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaseva AV, Moll UM 2009. The mitochondrial p53 pathway. Biochim Biophys Acta 1787: 414–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, et al. 2010. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 17: 98–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villunger A, Michalak EM, Coultas L, Mullauer F, Bock G, Ausserlechner MJ, Adams JM, Strasser A 2003. p53- and drug-induced apoptotic responses mediated by BH3-only proteins puma and noxa. Science 302: 1036–1038 [DOI] [PubMed] [Google Scholar]
- Xiao ZX, Chen J, Levine AJ, Modjtahedi N, Xing J, Sellers WR, Livingston DM 1995. Interaction between the retinoblastoma protein and the oncoprotein MDM2. Nature 375: 694–698 [DOI] [PubMed] [Google Scholar]
- Yamasaki F, Hama S, Yoshioka H, Kajiwara Y, Yahara K, Sugiyama K, Heike Y, Arita K, Kurisu K 2003. Staurosporine-induced apoptosis is independent of p16 and p21 and achieved via arrest at G2/M and at G1 in U251MG human glioma cell line. Cancer Chemother Pharmacol 51: 271–283 [DOI] [PubMed] [Google Scholar]
- Yang E, Zha J, Jockel J, Boise LH, Thompson CB, Korsmeyer SJ 1995. Bad, a heterodimeric partner for Bcl-XL and Bcl-2, displaces Bax and promotes cell death. Cell 80: 285–291 [DOI] [PubMed] [Google Scholar]
- Zhan Q, Fan S, Bae I, Guillouf C, Liebermann DA, O'Connor PM, Fornace AJ Jr 1994. Induction of bax by genotoxic stress in human cells correlates with normal p53 status and apoptosis. Oncogene 9: 3743–3751 [PubMed] [Google Scholar]
- Zhang Y, Xiong Y, Yarbrough WG 1998. ARF promotes MDM2 degradation and stabilizes p53: ARF–INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell 92: 725–734 [DOI] [PubMed] [Google Scholar]
- Zheng H, Ying H, Yan H, Kimmelman AC, Hiller DJ, Chen AJ, Perry SR, Tonon G, Chu GC, Ding Z, et al. 2008. p53 and Pten control neural and glioma stem/progenitor cell renewal and differentiation. Nature 455: 1129–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Parada LF 2002. The molecular and genetic basis of neurological tumours. Nat Rev Cancer 2: 616–626 [DOI] [PubMed] [Google Scholar]