Abstract
Cellular uptake of cobalamin (Cbl) is mediated by the transcobalamin receptor (TCblR) that binds and internalizes transcobalamin (TC) saturated with Cbl. These receptors are expressed in actively proliferating cells and are down regulated in quiescent cells. The 5′ region of TCblR gene was analyzed for promoter activity to determine transcriptional regulation of TCblR expression. The region −668 to −455 appears to regulate TCblR expression. We have identified transcription factors MZF-1 (myeloid zinc finger 1) / RREB-1 (Ras-responsive element binding protein 1), C/EBP (CCAAT/enhancer binding protein) / HNF-3ß (hepatocyte nuclear factor 3) and AP-1(activator protein 1) as proteins likely to be involved in this regulation with the former region primarily involved in up regulation and the latter two regions involved in suppression of TCblR expression. These transcription factors are involved in cell proliferation and differentiation. Thus the cell cycle associated expression of TCblR appears to be tightly regulated in synchrony with the proliferative phase of the cell cycle.
Keywords: Transcobalamin, Receptor, TCblR/CD320, vitamin B12, Cobalamin, Promoter, Transcription
1. Introduction
Vitamin B12 (Cobalamin, Cbl) is required for two essential enzymatic reactions: as methylcobalamin for the methylation of homocysteine to methionine by methionine synthase (Weissbach and Taylor, 1966; Taylor and Hanna, 1975) and as 5′-deoxyadenosylcobalamin for the isomerization of methylmalonyl CoA to succinyl CoA by methylmalonyl CoA mutase (Gurnani et al., 1960; Cannata et al., 1965). The first reaction provides active methyl groups for methylation of DNA, RNA, protein, phospholipids and metabolites; it is important for recycling folate, which is required for DNA synthesis (Sakami et al., 1950; Bailey and Gregory, 1999). The second reaction facilitates the catabolism of branched and odd chain fatty acids. Vitamin B12 deficiency causes megaloblastic anemia and neuropathological changes because of its role in the aforementioned metabolic pathways (Weir and Scott, 1995). The highly polar nature of the vitamin prevents its passage across the cell plasma membrane (Hodgkin et al., 1956), and therefore in mammals a complex mechanism has developed for gastrointestinal absorption, blood transport and cellular uptake of dietary Cbl.
The vitamin is transported to the distal ileum bound to gastric intrinsic factor and is absorbed via Cubilin-Amnionless in the ileal enterocyte (Cooper and Castle, 1960; Fyfe et al., 2004). The absorbed Cbl binds to transcobalamin, secreted by the vascular endothelial cells (Quadros et al., 1989; Quadros et al., 1999) and is transported to all tissue cells where it is internalized via receptor-mediated endocytosis (Cooper and Paranchych, 1961; Finkler and Hall, 1967). Even though it was known for more than three decades that TC-Cbl uptake into cells occurs via Ca++ dependent binding of TC to a plasma membrane receptor (Cooper and Paranchych, 1961; Paranchych and Cooper, 1962; DiGirolamo and Huennekens, 1975; Youngdahl-Turner et al., 1978; Youngdahl-Turner et al., 1979), the protein (TCblR) and the gene encoding this receptor was only recently identified (Quadros et al., 2009). Earlier studies had indicated that receptor expression is highest during the log phase of cell growth, suggesting cell-cycle association of this receptor expression (Hall, 1984; Hall et al., 1987; Lindemans et al., 1989; Amagasaki et al., 1990). In this report, we have identified the cis-elements and transcription factors involved in regulation of TCblR expression.
2. Materials and methods
2.1. Constructions of luciferase-based promoter reporter plasmids
Based on the 5′ flanking sequence of the TCblR gene, two fragments, −2009 / −33 and −1062 / −33 upstream of the ATG start codon were amplified by PCR using RP11-259 BAC plasmid (Children’s Hospital Oakland Research Institute) containing human genomic DNA as a template and cloned into luciferase reporter vector pGL2-B (Promega). Plasmid containing fragment −1062 / −33 was digested with enzymes Bsa I, Pml I, Nhe I, Bgl I, and Mlu I (NEB) respectively to obtain 5′ progressive deletion plasmids. Plasmid containing fragment −658 / +4 was digested with enzyme Mlu I to obtain plasmid containing fragment −123 / +4. Promoter region −658 / −455 was amplified and inserted upstream of fragment −123 / +4 to produce chimeric constructs with the −658 / −455 region in the correct orientation and in the inverted orientation. Primer sets used are listed in Table1.
Table 1.
Primers used for constructing promoter reporter plasmids, amplifying footprinting and gel mobility shift assay probes, and oligonucleotides used for gel mobility shift assay
| Name | Sequence | Position |
|---|---|---|
| Primers for construction of promoter reporter plasmids | ||
| −2009 −33 | forward:AGATCTCTGCCACAAGCCCAGCATAT reverse:AAGCTTTCCAGACCGCTCTCTTATCC |
−2009/ −33 |
| −1062 −33 | forward:AGATCTACAGGAGCATGTTACCACACC reverse:AAGCTTTCCAGACCGCTCTCTTATCC |
−1062/ −33 |
| −1062 +4 | forward:AGATCTACAGGAGCATGTTACCACACC reverse: ACATAAGCTTTCATGCTGTCCCCACA |
−1062/+4 |
| −658 +4 | forward:AGTTAGATCTGCGCCCGGCCTTTATTA reverse:ACATAAGCTTTCATGCTGTCCCCACA |
−658/+4 |
| −658 −455 insert | forward:GCGCCCGGCCTTTATTAT reverse:CTACTGGAGGGCAGGGATCT |
−658/−455 |
| Primers for amplification of footprinting and EMSA probes | ||
| fragment −658 −455 | forward:GCGCCCGGCCTTTATTAT reverse:CTACTGGAGGGCAGGGATCT |
−658/−455 |
| fragment 1 | forward:GCGCCCGGCCTTTATTAT reverse:AAACGTCCATTCTCGGAGTC |
−658/−569 |
| fragment 2 | forward:CAACCCGACTCCGAGAATG reverse:GGATCTGACACATAGTGCTTGA |
−594/−500 |
| fragment 3 | forward:GAGCCTACTATGAGTCAAGCACTATG reverse:CTACTGGAGGGCAGGGATCT |
−535/−455 |
| Oligonucleotides for EMSA | ||
| oligo RREB | forward:TTTCCTTTTTTCCCCCCAAGCAACCCGACTCCG reverse:CGGAGTCGGGTTGCTTGGGGGGAAAAAAGGAAA |
−614/−582 |
| oligo C/EBP | forward:TTTACTCACCAAATGTTTACTAAGAGCC reverse:GGCTCTTAGTAAACATTTGGTGAGTAAA |
−558/−531 |
| oligo AP | forward:TACTATGAGTCAAGCACTATGTGTCAGATCC reverse:AGGATCTGACACATAGTGCTTGACTCATAGTA |
−530/−500 |
| oligo AP2 | forward:GATCGAACTGACCGCCCGCGGCCCGT reverse:ACGGGCCGCGGGCGGTCAGTTCGATC |
nonspecific |
2.2. Site-directed mutagenesis of potential transcription factor binding sites
Introducing mutations into the two AP-1 sites in the promoter region −658 / −455 was performed using the QuikChange II XL Site-Directed Mutagenesis Kit (Stratagene) using primer sets listed in Table 2. An identical strategy was used to delete MZF-1/RREB-1(overlapping), C/EBP/HNF-3β (overlapping), and the two AP-1 binding sites.
Table 2.
Primers used for mutagenesis
| Name | Sequence |
|---|---|
| g135t, c139t (mut1) | |
| sense: | CACCAAATGTTTACTAAGAGCCTACTATTAGTTAAGCACTATGTGTCAGA |
| antisense: | TCTGACACATAGTGCTTAACTAATAGTAGGCTCTTAGTAAACATTTGGTG |
| g149a, c153t (mut2) | |
| sense: | CTAAGAGCCTACTATGAGTCAAGCACTATATGTTAGATCCTGAGAATAA |
| antisense: | TTATTCTCAGGATCTAACATATAGTGCTTGACTCATAGTAGGCTCTTAG |
| del RREB | |
| sense: | ATTGATTTCCTTTCCTTTTTTCCCCCGACTCCGAGAATG |
| antisense: | CATTCTCGGAGTCGGGGGAAAAAAGGAAAGGAAATCAAT |
| del C/EBP | |
| sense: | GGACGTTTCATTTATTCATTTACTCACGTTTACTAAGAGCCTACTATGAC |
| antisense: | CTCATAGTAGGCTCTTAGTAAACGTGAGTAAATGAATAAATGAAACGTCC |
| del AP | |
| sense: | TCACCAAATGTTTACTAAGAGCCTACTATGATCAGATCCTGAGAATA |
| antisense: | TATTCTCAGGATCTGATGATAGTAGGCTCTTAGTAAACATTTGGTGA |
2.3. Luciferase activity assay
HEK 293 cells (6×105) were seeded in complete DMEM with 10% FBS, 100u/ml penicillin, 100ug/ml streptomycin, and 2mM glutamine in six-well plates. After 24h, cells were cotransfected with promoter-luciferase reporter plasmids and pRL-CMV plasmid (Promega) using PolyFect Transfection Reagent (Qiagen) according to the manufacturer’s recommendations. After 24h, cells were lysed and luciferase activity was determined using the Dual-Luciferase Reporter Assay System (Promega). Protein concentration of the lysates was measured using the Bio-Rad Protein Assay reagent. The luciferase activity was corrected for transfection efficiency and protein concentration.
2.4. TCblR mRNA and functional receptor assay
Total RNA from cells was prepared using Trizol reagent (Invitrogen) and TCblR mRNA was quantified by SYBR GreenER (ABI PRISM)based real time PCR (qPCR). Functional TCblR expression in cells was determined by TC[57Co]Cbl binding as previously described (Quadros et al. 1989).
2.5. Preparation of nuclear extracts
HEK293 and SW48 cells were plated in T75 flasks at 1×105 cells per ml. To obtain nuclear proteins from proliferating cells and non-dividing cells, cells were collected after 24h and 120h respectively, washed twice with cold PBS and pelleted at 800g for 10min at 4°C. Cell pellets were suspended in buffer A (10mM Hepes-KOH, pH7.9, 10mM KCl, 1.5mM MgCl2, 1mM DTT, 1mM PMSF) and placed on ice for 10min. NP-40 detergent was added to the cell suspension to a final concentration of 0.6% and vortexed for 15sec. The nuclei were pelleted at 1000g for 10min at 4°C and resuspended in buffer A without NP-40 and pelleted again. Nuclei were suspended in buffer C (20mM Hepes-KOH, pH7.9, 420mM NaCl, 1.5mM MgCl2, 0.2mM EDTA, 25% glycerol, 1mM DTT, 1mM PMSF) and placed on ice for at least 30min. Nuclei were centrifuged at 21,000g for 20min at 4°C and supernatant was collected and dialyzed against buffer D (20mM Hepes-KOH, pH7.9, 100mM KCl, 0.2mM EDTA, 20% glycerol, 1mM DTT, 1mM PMSF) overnight at 4°C. The nuclear extract was centrifuged at 15,000rpm for 15min at 4°C in a Sorvall RC-5B SA-600 rotor, aliquoted and frozen at −80°C.
2.6. Electrophoretic mobility shift assay
The promoter fragment −658/ −455 and three overlapping fragments (f1: −658 / −569, f2: −594 / −500, and f3: −535 / −455) of less than 100bp spanning the region −658 / −455 were generated by PCR amplification using four pair of primers shown in table1. Each fragment was radiolabeled using [32P] – dATP (MP Biomedicals) in the PCR reactions and purified by native PAGE (29:1, 12% gel). The nuclear extract (2.5ug protein) was incubated for 15min at RT in 1x binding buffer (10mM Hepes-KOH, pH 7.9, 75mM KCl, 1mM MgCl2, 1mM EDTA, 10% glycerol, 1mM DTT, 50ug/ml BSA) and 1.2ug Poly (dI-dC).The [32P]-radiolabeled probe (10,000cpm) was added and incubated for 20min at RT. After incubation, reactions were subjected to electrophoresis in a 6% polyacylamide gel in 0.5xTBE buffer, the gel was dried and the protein-DNA complexes formed were identified by autoradiography. Specificity of binding was established by competing out the binding with molar excess of unlabeled fragments or motif specific oligonucleotide.
2.7. DNase I footprinting assay
The promoter fragment −658 / −455 was generated by PCR; the forward primer was end-labeled using T4-kinase and 32P-ATP to generate a 5′ end labeled probe. DNase I footprinting assay was performed by incubating 32P-labeled probe (30,000cpm) with 40 ug HEK293 nuclear extract, in the same reagent component and same procedure as EMSA. After the incubation and addition of Ca++/Mg++ solution (10mM CaCl2, 50mM MgCl2), the samples were digested with 0.05u DNase I (Amresco) for free probe and 2u DNase I for probe with nuclear extract for 1min at RT. The reaction was terminated by adding DNase I stop solution (0.5M EDTA, 1M Tris-HCl, 10mg/ml tRNA). All samples were phenol-extracted, ethanol-precipitated and separated in a 8% polyacrylamide/7M urea sequencing gel. DNA ladders were generated by Sequenase Version 2.0 DNA Sequencing Kit (USB) with forward primer: GCGCCCGGCCTTTATTAT and plasmid containing fragment −668 / −33.
3. Results
3.1. The promoter region − 668 to − 455 contains regulatory elements
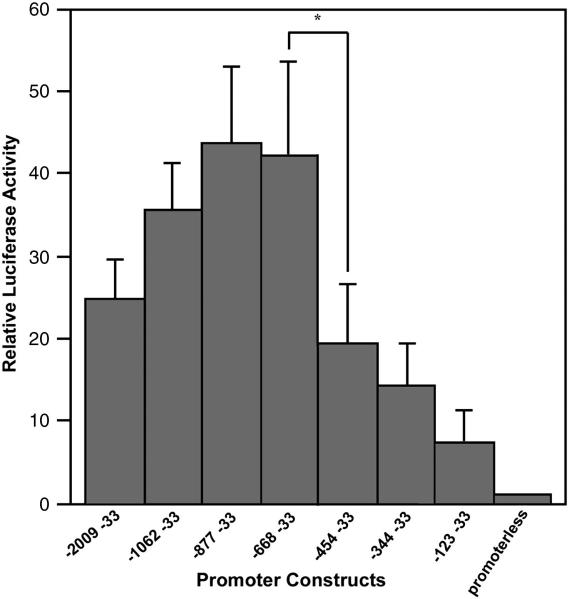
To identify the regulatory elements involved in controlling the TCblR expression, various deletion fragments from the 5′ flanking ~2000bp region of TCblR gene were investigated by luciferase reporter assay in HEK 293 cells. The promoter region − 877 to −33 showed highest promoter activity (Fig.1); deleting the region − 877 to − 668 didn’t affect activity. Deletion of the region − 668 to − 455 resulted in a 58% decrease in luciferase activity. The − 123 to − 33 region had some basal promoter activity and the region −2009 to −1062 had some regulation that suppressed promoter activity.
Figure 1. Deletion analysis of TCblR promoter in HEK293 cells.
Two ug of various promoter reporter constructs and 1 ng of Renilla luciferase reporter construct, as internal control, were cotransfected into HEK293 cells using PolyFect reagent. Firefly luciferase activity was normalized to Renilla luciferase activity. Results are expressed as the means of relative luciferase activity (relative to the activity of pGL-2 basic plasmid, arbitrarily set as 1) from five separate experiments, each performed in triplicate. The S.D. is indicated by the error bar. *, p<0.01 for construct −668 to −33 versus construct −454 to −33.
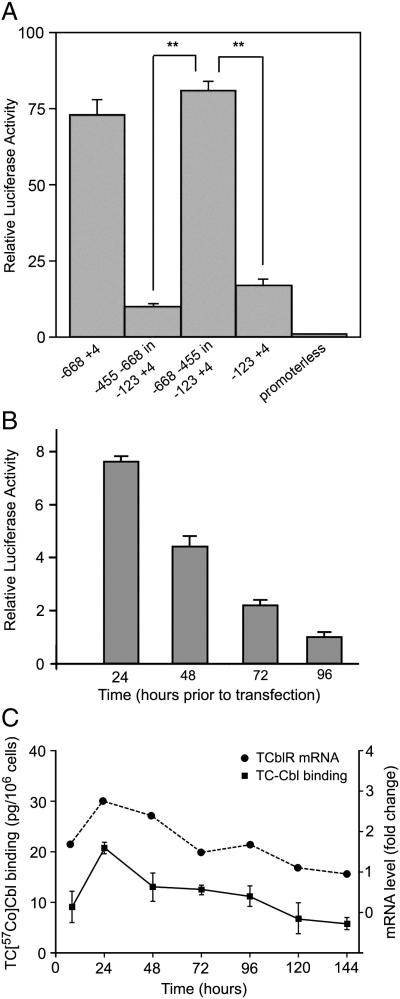
Further confirmation of the promoter activity in the −668 to −455 region was obtained when this region was inserted upstream of the sequence −123 to +4 in pGL2-basic vector which showed full promoter activity in the correct orientation and no activity in the reverse orientation (Fig. 2A). This promoter activity was cell cycle associated as evident from highest promoter activity at 24h after seeding when the cells were in the log phase of growth (Fig. 2B). The peak promoter activity also coincided with peak TCblR mRNA level and TC-Cbl binding in cells (Fig. 2C).
Figure 2. A. Confirmation of promoter activity in the TCblR gene −668 to −455 region.
Two ug of promoter reporter plasmids and 1 ng Renilla luciferase plasmid as internal control were cotransfected into HEK293 cells. Results are presented as the means of relative luciferase activity (relative to the activity of pGL-2 basic plasmid, arbitrarily set as 1) from three separate experiments, each performed in triplicate. The S.D. is indicated by the error bar. **, p<0.001 for the construct in the correct orientation versus the construct in the reverse orientation.
B. Luciferase activity in cells seeded for various time periods shows maximum activity in cells 24h after seeding and lowest activity in confluent cultures.
C. TCblR mRNA and TC-Cbl binding at various time periods after seeding shows highest uptake at 24h in actively proliferating cells.
3.2. The promoter region from −668 to −455 shows complex transcription factor binding
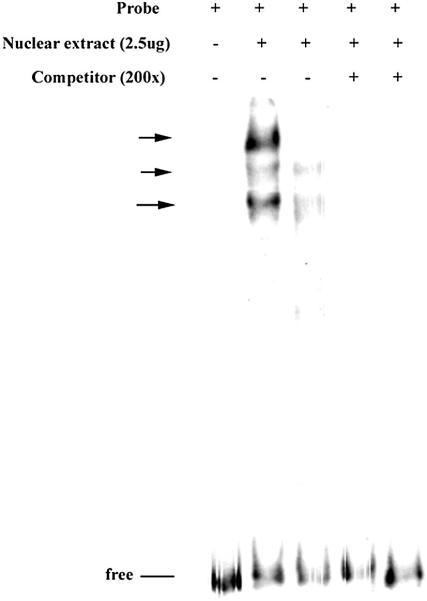
Binding of transcription factors to the region from −668 to −455 was evaluated using nuclear extracts from both actively proliferating and confluent HEK293 cells. As expected, binding of nuclear proteins was evident from the multiple DNA – protein complexes formed. From the intensity of the shifted bands, it was clear that more of the binding factors were present in nuclear extracts from actively dividing cells (Fig. 3). Further analysis of this binding was done using smaller overlapping fragments of DNA, which showed multiple discrete bands as a result of nuclear factors binding to each of the DNA probes. Competition with unlabeled fragments confirmed the specificity of binding (Fig. 4).
Figure 3. Binding of transcription factors to the TCblR gene −668 to −455 region.
The 32P-labeled probe was incubated with nuclear extracts from proliferating (24h culture) and resting (120h culture) HEK293 cells. From left to right, lane1, free probe; lane2, proliferating nuclear extract; lane3, resting nuclear extract; lane4 and 5, competition with unlabeled fragment. The protein-DNA complexes formed are identified by arrow. The samples were analyzed in a 6% native polyacrylamide gel and visualized by autoradiography.
Figure 4. Binding of transcription factors to specific regions within the −668 to −455 fragment.
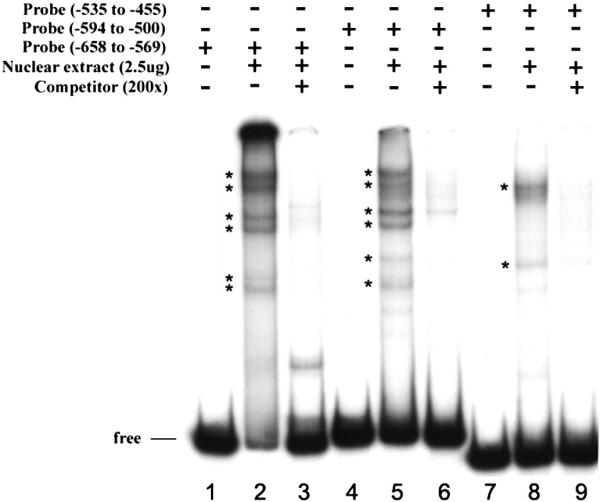
Three overlapping fragments labeled with 32P were incubated with nuclear extract from actively proliferating HEK293 cells and were subjected to mobility shift assay. The lanes for each probe are arranged in the same order from left to right: lane1, free probe; lane2, proliferative nuclear extract; and lane3, addition of unlabeled fragment. The protein-DNA complexes formed are identified by asterisk. The binding reactions were separated in a 6% native polyacrylamide gel, dried, and exposed to X-ray film.
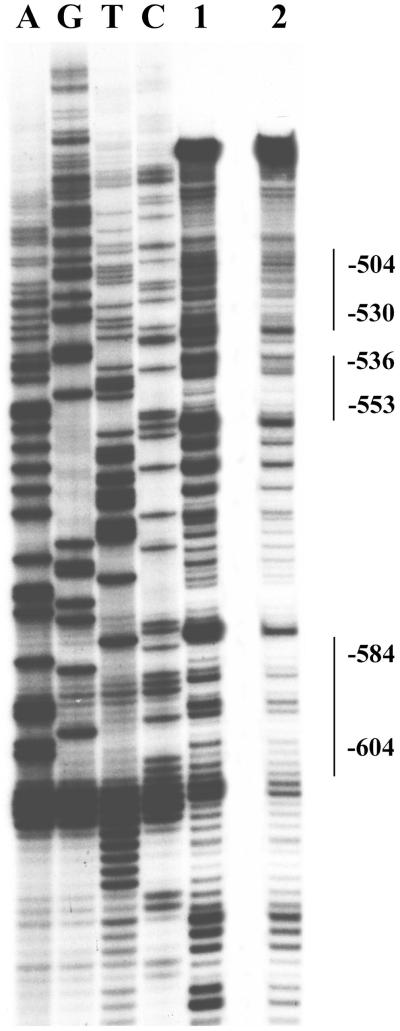
3.3. DNase I footprinting analysis
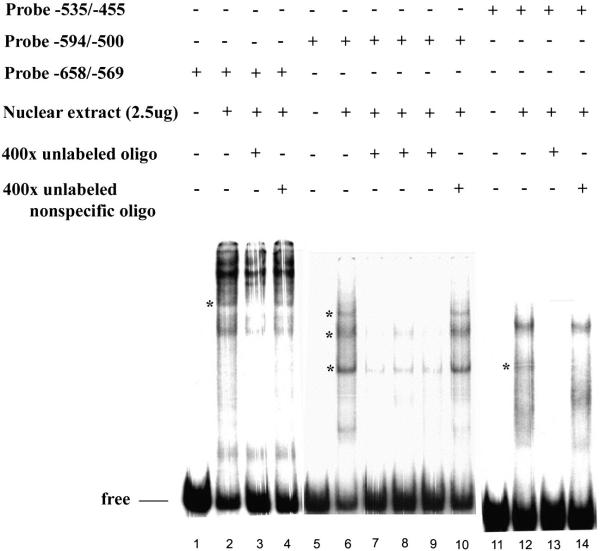
The specific sequences to which transcription factors bind were identified by resistance to digestion with DNase I. Three separate regions (−604 to −584), (−553 to −536), and (−530 to −504) corresponding respectively to transcription factors MZF-1/RREB-1, C/EBP/HNF-3β, and AP-1 binding sites were observed (Fig. 5). The specificity of binding and sequence recognition was determined by competing with synthetic oligonucleotides designed to cover all the protected and part of the non-protected regions within the −668 to −455 region. Oligo RREB −614/ −582 corresponds to MZF-1/RREB-1 sites, oligo C/EBP −558/ −531 corresponds to C/EBP/HNF-3β sites, and oligo AP −530/ −500 corresponds to two AP-1 sites. Oligo RREB competed out the specific band from fragment −658/ −569; oligo C/EBP and oligo AP competed out three bands from fragment −594/ −500; oligo AP competed out the specific band from fragment −535/ −455 (Fig. 6).
Figure 5. DNase I footprinting analysis of the TCblR gene −668 to −455 region.
The reactions were performed as described in “Experimental Procedures”. From left to right, laneA,G,T,C sequencing reactions for the top strand; lane 1, free probe; lane 2, addition of 40ug nuclear extracts from proliferating HEK293 cells; Protected regions are indicated by a vertical lines.
Figure 6. The specificity of transcription factors binding as determined by competition with unlabeled oligonucleotides.
The three overlapping 32P-labeled probes (f1: −658/ −569; f2: −594/ −500; f3: −535/ −455) within −668 to −455 region of the TCblR gene containing binding sites for transcription factors were incubated with SW48 nuclear extract. lane1, free probe −658/ −569; lane 2, with nuclear extract; lane 3, with 400x unlabeled oligo −614/−582; lane 4, with 400x unlabeled nonspecific oligo AP 2; lane 5, free probe −594/ −500; lane 6,with nuclear extract; lane 7, with 400x unlabeled oligo −558/−531; lane 8, with 400x unlabeled oligo −614/−582; lane 9, with 400x unlabeled oligo −530/−500; lane10, with 400x unlabeled nonspecific oligo AP 2 (ns); lane11, free probe −535/ −455; lane12, with nuclear extract; lane13, with 400x unlabeled oligo −530/−500; lane14, with 400x unlabeled nonspecific oligo AP2. The specific bands are identified by asterisk. The DNA-protein complexes were separated in a 6% native PAGE and visualized by exposure to X-ray film.
3.4. Mutation analysis of AP-1 binding sites
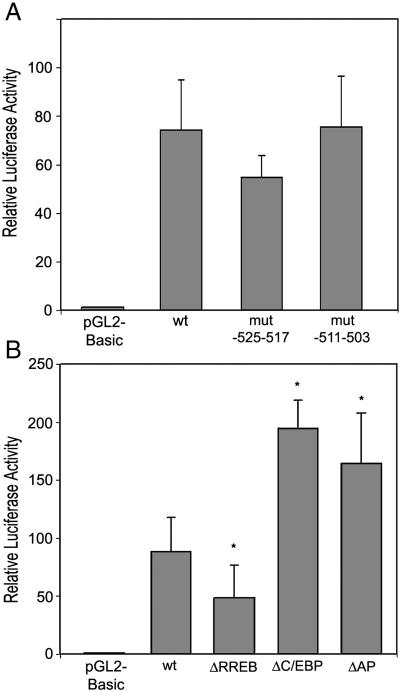
To study if the AP-1 sites are functional binding sites, we generated two mutants in which only one AP-1 site was mutated. HEK293 cells were transfected with these plasmids and luciferase activity was measured. The results were normalized for transfection efficiency and lysate concentration. Mutating either of two AP-1 sites does not significantly affect promoter activity compared with the wild-type promoter (Fig. 7A).
Figure 7. Mutagenesis analysis of potential transcription factor binding sites in the promoter region −668 to −455.
A. Two plasmids, each with one of the two AP1 binding sites mutated. B. Three plasmids, each with MZF-1/RREB, C/EBP/HNF-3β or two AP1 binding sites deleted. Two ug promoter reporter plasmids and 1 ng Renilla luciferase plasmid as internal control were cotransfected into HEK293 cells. Firefly luciferase activity was normalized to Renilla luciferase activity.Results are expressed as the means of relative luciferase activity (relative to the activity of pGL-2 basic plasmid, arbitrarily set as 1) from three separate experiments, each performed in triplicate. The S.D. is indicated by the error bar. *, p<0.05 for comparison of wild type and mutant promoter constructs.
3.5. Effect of deleting MZF-1/RREB-1, C/EBP/HNF-3β, and AP-1 binding sites on promoter activity
To further investigate if MZF-1/RREB-1, C/EBP/HNF-3β and the two AP-1 are functional binding sites, three mutants, each with a single binding site deleted, were generated by site-directed mutagenesis. HEK293 cells were transfected with these plasmids and luciferase activity was measured. Deleting MZF-1/RREB-1 site decreased promoter activity by 45%, deleting C/EBP/HNF-3β site increased promoter activity by 122% and deleting two AP-1 sites increased promoter activity 86% (Fig. 7B).
4. Discussion
The role of Cbl as a cofactor for MS enzyme in the conversion of methyl-THF to THF serves the essential function of making available various forms of reduced folates for key single carbon exchange reactions, purine /pyrimidine synthesis and for the de novo synthesis of thymidine from deoxyuridine. The latter pathways provide the basic building blocks for DNA synthesis and therefore, both Cbl and folate deficiency lead to megaloblastic anemia (Weir and Scott, 1995; Bailey and Gregory, 1999) Earlier observation of cell cycle association of TCblR expression (Hall, 1984; Hall et al., 1987; Lindemans et al., 1989; Amagasaki et al., 1990) is biologically and metabolically relevant since this ensures adequate intracellular Cbl during DNA synthesis. Therefore, the transcriptional regulation of TCblR expression and its association with the proliferative phase of the cell cycle is of particular interest both for normal as well as neoplastic cell proliferation. This study is the first effort to identify the proximal promoter of the TCblR gene. The ~2000bp 5′ flanking sequence of TCblR contains numerous potential cis elements to which transcription factors could bind to drive the transcription of TCblR (Fig. 8). Transient transfection of deletion constructs located all of the promoter activity within the immediate 5′ 1000bp sequence. Loss of >50% of the activity upon deleting the region −668 to −455, suggested that cis elements within this region play a major role in transcriptional regulation of TCblR. This conclusion was supported by mobility shift and DNase I footprinting assays, which identified specific regions within this sequence to which nuclear proteins bound. Based on known consensus sequences, transcription factors MZF-1/RREB-1, C/EBP/HNF-3ß and AP-1 were identified as factors likely to bind within this regulatory region. Competition with specific oligos, mutagenesis and deletion of these sequences confirmed the role of these motifs in the regulation of TCblR expression. The MZF-1/RREB-1 appears to be a positive regulator since deletion of this sequence decreased promoter activity by 45%. The C/EBP/HNF-3β and AP-1 sites appear to be involved in suppression or down-regulation of TCblR expression since deletion of these regions increased promoter activity by 122% and 86% respectively.
Figure 8. Partial DNA sequence of the human TCblR promoter with the start codon.
The start codon is set as +1. The region containing tested transcription factor binding sites are underlined.
Both MZF-1 and RREB-1 are Krüppel type zinc finger proteins. MZF-1 is primarily expressed in myeloid progenitor cells and is involved in the growth, proliferation, differentiation, and apoptosis of myeloid progenitors (Hromas et al., 1991; Morris et al., 1994). RREB-1 binds to Ras responsive elements in promoters and may exert its control via the Ras/Raf pathway (Thiagalingam et al., 1996; Zhang et al., 1999). The C/EBP transcription factors are a family of basic region leucine zipper (bZIP) transcription factors that are composed of six members C/EBPα to C/EBPζ that bind to consensus motifs within promoters and enhancers. These can form homodimers or heterodimers with members of their family or with other transcription factors that may or may not contain the leucine zipper domain. C/EBP family members interact and cooperate with many other transcription factors, including p50, Myb, CREB/ATF, and AP-1. C/EBP proteins play roles in the control of cellular proliferation, growth, differentiation, metabolism, and immune modulation (Ramji and Foka, 2002; Cai et al., 2008). HNF-3β is a member of the forkhead/winged-helix Foxa family of proteins and can be involved in regulation of specific hepatic and/or pancreatic genes. HNF-3β plays an important role in regulating cellular proliferation, differentiation, longevity, and transformation (Wu et al., 1997; Wang et al., 2002). The negative regulation by AP-1 is likely to be complex since this can be both a positive as well a negative regulator orchestrated by a group of bZIP proteins that consist of Jun, Fos, ATF and JDP protein family members. Typically AP-1 regulates genes containing TPA response elements and is activated by the Ras cascades. This activation involves the formation of homodimers or heterodimers and interaction with other transcription factors, the composition of which varies in different genes (Glover and Harrison, 1995; Shaulian and Karin, 2001; Hess et al., 2004). Since AP-1 is involved in regulating a multitude of cellular processes including proliferation, differentiation, and apoptosis, the suppression of TCblR transcription by AP-1 must involve recruitment of other proteins at various stages of the cell cycle to up regulate expression in proliferating cells and down regulate expression in quiescent cells. Due to MZF-1 and HNF-3β specificity for myeloid progenitor and liver genes, their role in TCblR expression is not clear. We speculate that RREB-1, C/EBP, and AP-1 may coordinately regulate TCblR expression.
The highest promoter activity observed in cells 24 hours after seeding and the decrease observed during the next 96 hours when cell division has slowed, supports the conclusion that TCblR transcription is up regulated in actively dividing cells and likely precedes the proliferative phase of the cell cycle. Even though the precise phase of the cell cycle associated with optimum TCblR expression is not known, high level of functional TCblR and mRNA in cells between 10 - 24 hours after seeding, when the cells are in early log phase of growth would suggest optimum uptake of Cbl during DNA synthesis. Cancer cells and especially those that are highly proliferative, are likely to have high expression of TCblR. The regulation and over expression of TCblR in various cancers is of particular interest in light of Cbl requirement for folate recycling and DNA synthesis. Down regulation of TCblR or blocking Cbl uptake in cancer cells are strategies likely to arrest proliferation of cancer cells (McLean et al., 1997; Walker et al., 1997). Identifying cancers with sustained high levels of TCblR expression, would also permit delivery of drugs and toxins preferentially to these tumors. In conclusion, this report describes the identification of cis elements within the promoter region of TCblR/CD320 gene and that are involved in the transcriptional regulation of TCblR expression.
Acknowledgments
This study was supported by NIH grant DK 064732
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amagasaki T, Green R, Jacobsen DW. Expression of transcobalamin II receptors by human leukemia K562 and HL-60 cells. Blood. 1990;76:1380–6. [PubMed] [Google Scholar]
- Bailey LB, Gregory JF., 3rd Folate metabolism and requirements. J Nutr. 1999;129:779–82. doi: 10.1093/jn/129.4.779. [DOI] [PubMed] [Google Scholar]
- Cai DH, Wang D, Keefer J, Yeamans C, Hensley K, Friedman AD. C/EBP alpha:AP-1 leucine zipper heterodimers bind novel DNA elements, activate the PU.1 promoter and direct monocyte lineage commitment more potently than C/EBP alpha homodimers or AP-1. Oncogene. 2008;27:2772–9. doi: 10.1038/sj.onc.1210940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannata JJ, Focesi A, Jr., Mazumder R, Warner RC, Ochoa S. Metabolism of Propionic Acid in Animal Tissues. Xii. Properties of Mammalian Methylmalonyl Coenzyme a Mutase. J Biol Chem. 1965;240:3249–57. [PubMed] [Google Scholar]
- Cooper BA, Castle WB. Sequential mechanisms in the enhanced absorption of vitamin B12 by intrinsic factor in the rat. J Clin Invest. 1960;39:199–214. doi: 10.1172/JCI104019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper BA, Paranchych W. Selective uptake of specifically bound cobalt-58 vitamin B12 by human and mouse tumour cells. Nature. 1961;191:393–5. doi: 10.1038/191393a0. [DOI] [PubMed] [Google Scholar]
- DiGirolamo PM, Huennekens FM. Transport of vitamin B12 into mouse leukemia cells. Arch Biochem Biophys. 1975;168:386–93. doi: 10.1016/0003-9861(75)90267-2. [DOI] [PubMed] [Google Scholar]
- Finkler AE, Hall CA. Nature of the relationship between vitamin B12 binding and cell uptake. Arch Biochem Biophys. 1967;120:79–85. doi: 10.1016/0003-9861(67)90600-5. [DOI] [PubMed] [Google Scholar]
- Fyfe JC, Madsen M, Hojrup P, Christensen EI, Tanner SM, de la Chapelle A, He Q, Moestrup SK. The functional cobalamin (vitamin B12)-intrinsic factor receptor is a novel complex of cubilin and amnionless. Blood. 2004;103:1573–9. doi: 10.1182/blood-2003-08-2852. [DOI] [PubMed] [Google Scholar]
- Glover JN, Harrison SC. Crystal structure of the heterodimeric bZIP transcription factor c-Fos-c-Jun bound to DNA. Nature. 1995;373:257–61. doi: 10.1038/373257a0. [DOI] [PubMed] [Google Scholar]
- Gurnani S, Mistry SP, Johnson BC. Function of vitamin B12 in methylmalonate metabolism. I. Effect of a cofactor form of B12 on the activity of methylmalonyl-CoA isomerase. Biochim Biophys Acta. 1960;38:187–8. doi: 10.1016/0006-3002(60)91224-5. [DOI] [PubMed] [Google Scholar]
- Hall CA. The uptake of vitamin B12 by human lymphocytes and the relationships to the cell cycle. J Lab Clin Med. 1984;103:70–81. [PubMed] [Google Scholar]
- Hall CA, Colligan PD, Begley JA. Cyclic activity of the receptors of cobalamin bound to transcobalamin II. J Cell Physiol. 1987;133:187–91. doi: 10.1002/jcp.1041330125. [DOI] [PubMed] [Google Scholar]
- Hess J, Angel P, Schorpp-Kistner M. AP-1 subunits: quarrel and harmony among siblings. J Cell Sci. 2004;117:5965–73. doi: 10.1242/jcs.01589. [DOI] [PubMed] [Google Scholar]
- Hodgkin DC, Kamper J, Mackay M, Pickworth J, Trueblood KN, White JG. Structure of vitamin B12. Nature. 1956;178:64–6. doi: 10.1038/178064a0. [DOI] [PubMed] [Google Scholar]
- Hromas R, Collins SJ, Hickstein D, Raskind W, Deaven LL, O’Hara P, Hagen FS, Kaushansky K. A retinoic acid-responsive human zinc finger gene, MZF-1, preferentially expressed in myeloid cells. J Biol Chem. 1991;266:14183–7. [PubMed] [Google Scholar]
- Lindemans J, Kroes AC, van Geel J, van Kapel J, Schoester M, Abels J. Uptake of transcobalamin II-bound cobalamin by HL-60 cells: effects of differentiation induction. Exp Cell Res. 1989;184:449–60. doi: 10.1016/0014-4827(89)90343-1. [DOI] [PubMed] [Google Scholar]
- McLean GR, Quadros EV, Rothenberg SP, Morgan AC, Schrader JW, Ziltener HJ. Antibodies to transcobalamin II block in vitro proliferation of leukemic cells. Blood. 1997;89:235–42. [PubMed] [Google Scholar]
- Morris JF, Hromas R, Rauscher FJ., 3rd Characterization of the DNA-binding properties of the myeloid zinc finger protein MZF1: two independent DNA-binding domains recognize two DNA consensus sequences with a common G-rich core. Mol Cell Biol. 1994;14:1786–95. doi: 10.1128/mcb.14.3.1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paranchych W, Cooper BA. Factors influencing the uptake of cyanocobalamin (vitamin B12) by Ehrlich ascites carcinoma cells. Biochim Biophys Acta. 1962;60:393–403. doi: 10.1016/0006-3002(62)90415-8. [DOI] [PubMed] [Google Scholar]
- Quadros EV, Nakayama Y, Sequeira JM. The protein and the gene encoding the receptor for the cellular uptake of transcobalamin-bound cobalamin. Blood. 2009;113:186–92. doi: 10.1182/blood-2008-05-158949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quadros EV, Regec AL, Khan KM, Quadros E, Rothenberg SP. Transcobalamin II synthesized in the intestinal villi facilitates transfer of cobalamin to the portal blood. Am J Physiol. 1999;277:G161–6. doi: 10.1152/ajpgi.1999.277.1.G161. [DOI] [PubMed] [Google Scholar]
- Quadros EV, Rothenberg SP, Jaffe EA. Endothelial cells from human umbilical vein secrete functional transcobalamin II. Am J Physiol. 1989;256:C296–303. doi: 10.1152/ajpcell.1989.256.2.C296. [DOI] [PubMed] [Google Scholar]
- Ramji DP, Foka P. CCAAT/enhancer-binding proteins: structure, function and regulation. Biochem J. 2002;365:561–75. doi: 10.1042/BJ20020508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakami W, Welch AD, Lafaye JM. Synthesis of labile methyl groups by the rat in vivo and in vitro. J Biol Chem. 1950;187:379–84. [PubMed] [Google Scholar]
- Shaulian E, Karin M. AP-1 in cell proliferation and survival. Oncogene. 2001;20:2390–400. doi: 10.1038/sj.onc.1204383. [DOI] [PubMed] [Google Scholar]
- Taylor RT, Hanna LM. Folate-dependent enzymes in cultured Chinese hamster ovary cells: induction of 5-methyltetrahydrofolate homocysteine cobalamin methyltransferase by folate and methionine. Arch Biochem Biophys. 1975;171:507–20. doi: 10.1016/0003-9861(75)90060-0. [DOI] [PubMed] [Google Scholar]
- Thiagalingam A, De Bustros A, Borges M, Jasti R, Compton D, Diamond L, Mabry M, Ball DW, Baylin SB, Nelkin BD. RREB-1, a novel zinc finger protein, is involved in the differentiation response to Ras in human medullary thyroid carcinomas. Mol Cell Biol. 1996;16:5335–45. doi: 10.1128/mcb.16.10.5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker PR, Smith B, Carson C, LeBlanc J, Sikorska M, Woodhouse CS, Morgan AC. Induction of apoptosis in neoplastic cells by depletion of vitamin B12. Cell Death Differ. 1997;4:233–41. doi: 10.1038/sj.cdd.4400225. [DOI] [PubMed] [Google Scholar]
- Wang H, Gauthier BR, Hagenfeldt-Johansson KA, Iezzi M, Wollheim CB. Foxa2 (HNF3beta) controls multiple genes implicated in metabolism-secretion coupling of glucose-induced insulin release. J Biol Chem. 2002;277:17564–70. doi: 10.1074/jbc.M111037200. [DOI] [PubMed] [Google Scholar]
- Weir DG, Scott JM. The biochemical basis of the neuropathy in cobalamin deficiency. Baillieres Clin Haematol. 1995;8:479–97. doi: 10.1016/s0950-3536(05)80217-3. [DOI] [PubMed] [Google Scholar]
- Weissbach H, Taylor R. Role of vitamin B12 in methionine synthesis. Fed Proc. 1966;25:1649–56. [PubMed] [Google Scholar]
- Wu KL, Gannon M, Peshavaria M, Offield MF, Henderson E, Ray M, Marks A, Gamer LW, Wright CV, Stein R. Hepatocyte nuclear factor 3beta is involved in pancreatic beta-cell-specific transcription of the pdx-1 gene. Mol Cell Biol. 1997;17:6002–13. doi: 10.1128/mcb.17.10.6002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngdahl-Turner P, Mellman IS, Allen RH, Rosenberg LE. Protein mediated vitamin uptake. Adsorptive endocytosis of the transcobalamin II-cobalamin complex by cultured human fibroblasts. Exp Cell Res. 1979;118:127–34. doi: 10.1016/0014-4827(79)90590-1. [DOI] [PubMed] [Google Scholar]
- Youngdahl-Turner P, Rosenberg LE, Allen RH. Binding and uptake of transcobalamin II by human fibroblasts. J Clin Invest. 1978;61:133–41. doi: 10.1172/JCI108911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Zhao J, Edenberg HJ. A human Raf-responsive zinc-finger protein that binds to divergent sequences. Nucleic Acids Res. 1999;27:2947–56. doi: 10.1093/nar/27.14.2947. [DOI] [PMC free article] [PubMed] [Google Scholar]