Abstract
Double-stranded RNA induces the homology-dependent degradation of cognate mRNA in the cytoplasm via RNA interference (RNAi) but also is a target for adenosine-to-inosine (A-to-I) RNA editing by adenosine deaminases acting on RNA (ADARs). An interaction between the RNAi and the RNA editing pathways in Caenorhabditis elegans has been suggested recently, but the precise mode of interaction remains to be established. In addition, it is unclear whether this interaction is possible in mammalian cells with their somewhat different RNAi pathways. Here we show that ADAR1 and ADAR2, but not ADAR3, avidly bind short interfering RNA (siRNA) without RNA editing. In particular, the cytoplasmic full-length isoform of ADAR1 has the highest affinity among known ADARs, with a subnanomolar dissociation constant. Gene silencing by siRNA is significantly more effective in mouse fibroblasts homozygous for an ADAR1 null mutation than in wild-type cells. In addition, suppression of RNAi effects are detected in fibroblast cells overexpressing functional ADAR1 but not when overexpressing mutant ADAR1 lacking double-stranded RNA-binding domains. These results identify ADAR1 as a cellular factor that limits the efficacy of siRNA in mammalian cells.
Double-stranded RNA (dsRNA)1 is produced within cells not only during viral infection but also by processes that have an important impact on cell and developmental biology (1, 2). RNA interference (RNAi) evolved to deal with these viral and cellular dsRNA molecules (3–5). One of the critical steps involved in the RNAi mechanism is processing of a long targeting dsRNA (trigger) into 21-nt short dsRNA fragments (termed short interfering RNA or siRNA) by the dsRNA-specific ribonuclease DICER (6, 7). These RNAi intermediates contain 2-nt 3′-overhangs in the sense and antisense strands. Each siRNA is used in multiple rounds of degradation of the target mRNA following integration into the RNA-induced silencing complex (4, 5). Introduction of trigger dsRNAs (usually ≥30 bp) into mammalian cells initiates a series of cellular responses including the induction of interferons, which in turn activate a group of interferon-inducible genes such as PKR (interferon-inducible serine/threonine kinase). PKR catalyzes the phosphorylation of the α subunit of protein synthesis initiation factor 2 (eIF-2α), which leads eventually to a global shutdown of protein synthesis (8). Activation of this nonspecific pathway could mask any sequence-specific effects that might result from the RNAi pathway (4, 5). Nevertheless, successful application of RNAi has been reported in many mammalian tissue culture cell lines by using chemically synthesized siRNAs instead of trigger dsRNAs, thus bypassing activation of PKR (9, 10). However, there may also be factors other than PKR that limit RNAi efficacy in mammalian cells.
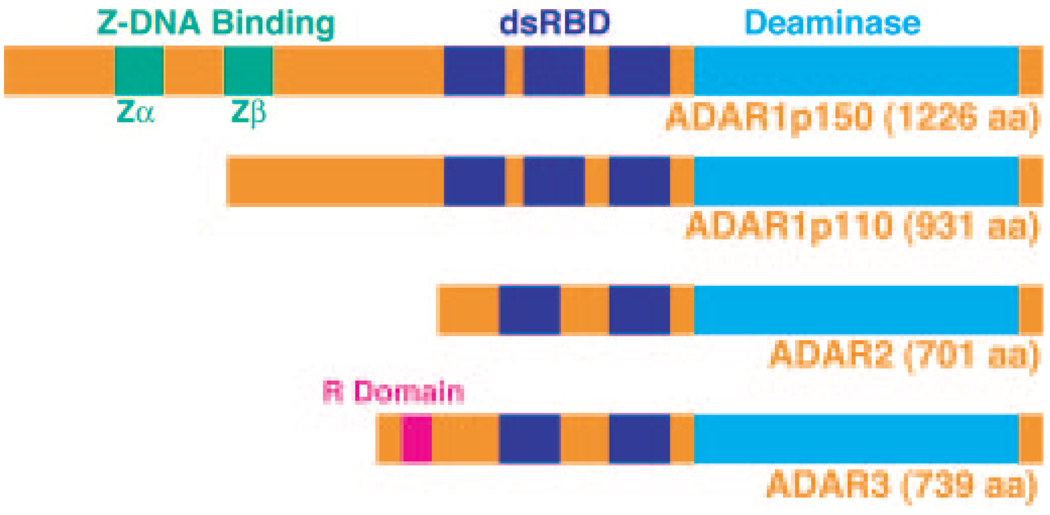
ADAR is responsible for site-specific RNA editing of coding sequences (11, 12) and noncoding intron and untranslated region sequences containing inverted repeats such as Alu and L1 elements (13–15). This family of enzymes convert selected adenosine residues into inosine in substrate RNAs containing a relatively short dsRNA region (1, 2). A-to-I RNA editing of coding regions can lead to functional alterations of the affected genes (11, 12), whereas editing of noncoding regions may affect the splicing rate, the translation efficacy, or the stability of the edited mRNAs (13–16). Three separate ADAR gene family members (ADAR1 to ADAR3) have been identified in mammals (Fig. 1) (17–23). In addition, two forms of ADAR1, a full-length 150-kDa form (p150) and a shorter 110-kDa protein (p110), are synthesized via alternative translation initiation codons (24). The ADAR1 mRNA transcribed from an interferon- and dsRNA-inducible promoter directs translation of p150, whereas two other ADAR1 mRNAs transcribed from constitutive promoters direct synthesis of p110 initiated from a downstream methionine codon (24). The transcriptional regulation of ADAR2 and ADAR3 genes are currently unknown. ADAR1p150 is found mainly in the cytoplasm, whereas both ADAR1p110 and ADAR2 localize to the nucleus and nucleolus (24–26). Recent proteomic analyses have identified ADAR3 among proteins localized within the nucleoli (27). Both ADAR1 and ADAR2 form a stable, enzymatically active homodimer complex, whereas ADAR3 remains as a monomeric, enzymatically inactive form, indicating that oligomerization may play a role in the site-selective editing mechanism (28).
FIG. 1. Three mammalian ADAR gene family members.
Indicated are the Zα and Zβ Z-DNA-binding domains (ADAR1), the dsRNA-binding domains (dsRBD), the arginine-rich (R domain) sense strand of siRNA-binding domain (ADAR3), and the deaminase domain. aa, amino acids.
Because ADARs possess unique enzymatic activity capable of generating multiple I-U mismatched base pairs in dsRNA, it has been suggested that they may suppress RNAi efficacy (6, 29). A-to-I RNA editing of dsRNAs derived from transgenes and endogenous genes indeed appears to prevent their silencing by RNAi in Caenorhabditis elegans, revealing one potential intersection of the RNA editing and RNAi pathways and the antagonistic effects of ADAR in vivo on RNAi (30, 31). In these previous studies, the long trigger dsRNA was proposed to be the target of ADAR (6, 29–31), but potential direct interactions between ADAR and siRNA have never been investigated. In the present study, we examined the interaction of ADAR with siRNAs. We find that ADAR1p150 binds siRNAs with extremely high affinity. In addition, our in vivo RNAi experiments conducted with fibroblasts homozygous for an ADAR1 null mutation revealed ADAR1 suppression of siRNA-based RNAi. We propose cytoplasmic ADAR1p150 as a cellular factor limiting siRNA efficacy in mammalian cells.
EXPERIMENTAL PROCEDURES
Oligonucleotides and Synthetic siRNAs
Synthetic RNAs for preparation of 36- and 29-bp siRNAs or varying sizes of EGFP siRNAs corresponding to part of the sense or antisense EGFP-C1 sequence (Clontech) and nonspecific control siRNA (NS siRNA) were obtained from Dharmacon (Lafayette, CO): 38RNAS1, 5′-AAUUAACCAAGGAAAAUAACAAGGACAGGGACCAGGUU-3′; 38RNAA1, 5′-CCUGGUCCCUGUCCUUGUUAUUUUCCUUGGUUAAUUUU-3′; 31RNAS1, 5′-AAUUAACCAAGGAAAAUAACAAGGACAGGUU-3′; 31RNAA1, 5′-CCUGUCCUUGUUAUUUUCCUUGGUUAAUUUU-3′; 25EGFPS1, 5′-GCAGCACGACUUCUUCAAGUCCGUU-3′; 25EGFPA1, 5′-CGGACUUGAAGAAGUCGUGCUGCUU-3′; 21EGFPS1, 5′-GCAGCACGACUUCUUCAAGUU-3′; 21EGFPA1, 5′-CUUGAAGAAGUCGUGCUGCUU-3′; 19EGFPS1, 5′-GCAGCACGACUUCUUCAAG-3′; 19EGFPA1, 5′-CUUGAAGAAGUCGUGCUGC-3′; 17EGFPS1, 5′-GCAGCACGACUUCUUUU-3′; 17EGFPA1, 5′-AAGAAGUCGUGCUGCUU-3′; 21NSS1, 5′-AUUGUAUGCGAUCGCAGACUU-3′; and 21NSA1, 5′-GUCUGCGAUCGCAUACAAUUU-3′. The following sense and antisense DNA strands corresponding to 19-bp EGFP siRNA were synthesized at the University of Pennsylvania Cancer Center Nucleic Acid Facility: 21DNAS1, 5′-GCAGCACGACTTCTTCAAGTT-3′; 21DNAA1, 5′-CTTGAAGAAGTCGTGCTGCTT-3′.
siRNA or siDNA single strands were phosphorylated by T4 polynucleotide kinase (New England Biolabs, Beverly, MA) in the presence of 50 µCi of [γ-32P]ATP (3000 Ci/mmol) (Amersham Biosciences). For some experiments, RNA and DNA single strands were phosphorylated in the presence of nonradioactive ATP. The combinations of sense and antisense strand RNAs or DNAs used for preparation of specific siRNAs or siDNAs were as follows: 36-bp siRNA, 38RNAS1 and 38RNAA1; 29-bp siRNA, 31RNAS1 and 31RNAA1; 23-bp siRNA, 25EGFPS1 and 25EGFPA1; 19-bp siRNA, 21EGFPS1 and 21EGFPA1; 19-bp blunt end dsRNA, 19EGFPS1 and 19EGFPA1; 15-bp siRNA, 17EGFPS1 and 17EGFPA1; 19-bp nonspecific control siRNA, 21NSS1 and 21NSA1; and 19-bp siDNA, 21DNAS1 and 21DNAA1.
Recombinant ADAR Proteins
FLAG epitope-tagged ADAR proteins (ADAR1p150 and p110, ADAR2, ADAR3, ADAR1p150E912A, and ADAR2E396A) were ectopically expressed in Sf9 cells and purified by affinity column chromatography using anti-FLAG M2 mAb agarose gel (Sigma) as described previously (32). The purity of recombinant proteins (>95% pure) was monitored by electrophoresis on a 8% SDS-polyacrylamide gel followed by silver staining. If necessary, purified ADAR proteins were concentrated using Centriplus spin columns (Millipore, Bedford, MA). A-to-I RNA editing by wild-type ADAR1 and ADAR2 was monitored by using a long synthetic dsRNA substrate to ensure that these recombinant ADAR proteins are fully functional for their deaminase activities.
RNA Binding Assays
Electrophoretic mobility shift assays were conducted in the presence of 0.1 m NaCl and 1 mm Mg2+, i.e. physiological ionic strength. Unless otherwise indicated, a 20-µl reaction mixture containing 10 mm Tris-HCl (pH 8), 0.1 m NaCl, 1 mm MgCl2, 0.5 mm dithiothreitol, 10% glycerol, and various concentrations of purified recombinant ADAR protein was incubated with 10 pm 32P-labeled siRNA at 20 °C for 20 min. Different binding conditions (temperature, 30 °C; duration, 60 min; EDTA, 50 mm; Mg2+, 5 or 10 mm; and NaCl, 0.2–1.0 m) were tested, but none of these conditions except the salt concentration (see “Results”) affected formation of ADAR-siRNA complexes. For competitive inhibition experiments, cold competitors (5–15× excess) were preincubated with ADAR for 5 min prior to addition of 32P-labeled siRNA. Reaction mixtures were then loaded onto nondenaturing 4% polyacrylamide gels (40:1; acrylamide:bisacrylamide), pre-run at 4 °C for 30 min, in 1× TBE (0.1 m Tris, 83 mm H3BO3, 1 mm EDTA). Gels were electrophoresed further at 500 V for 3 h. Radioactivity in the dried gels was quantitated using a PhosphorImager and the ImageQuant program (Amersham Biosciences). The fraction of siRNA bound to ADARs was determined by dividing the radioactivity measured in the siRNA-ADAR complex band by the radioactivity in the free siRNA band. The radioactivity of the smeared material between the siRNA-ADAR complex and the free siRNA bands was included in the bound fraction. Kd was defined as the protein concentration required for 50% binding (32).
Two-dimensional TLC Analysis for siRNA
For analysis of inosine modification, RNA samples subjected to complex formation with ADAR1p150 (10 nm) or ADAR2 (25 nm) at 30 °C for 60 min were first digested to 3′ NMPs with RNase T2, phenol/chloroform-, chloroform-, and ether-extracted, and dried. The 5′-ends of lyophilized 3′ NMPs were then labeled with polynucleotide kinase and 10 µCi of [γ-32P]ATP at 37 °C for 30 min; polynucleotide kinase was heat-inactivated at 65 °C for 20 min. The resulting 32P-labeled 5′-,3′-phospho-mononucleosides were then digested to 5′ [32P]NMPs with P1 nuclease (5′-phosphomononucleoside analysis). As a control, c-myc dsRNA, which had been subjected to A-to-I modification by ADAR1p150 (10 nm) at 30 °C for 60 min, was analyzed in parallel. The digests, 5′ NMPs, were analyzed by two-dimensional TLC. The solvent system used for the first dimension contained isobutyric acid/NH4OH/water, 66:1:33 (by volume), and that used for the second dimension contained 0.1 m sodium phosphate (pH 6.8)/ammonium sulfate/1-propanol, 100:60:2 (v/w/v) (33).
Preparation of ADAR2−/− and PKR−/− MEF Cells
Mutant mouse line ADAR2−/−/GluR-BR/R is an ADAR2−/− mouse line rescued by the introduction of a GluR-BR allele containing an arginine codon for the Q/R site (34). MEF cells were prepared with E14.0 embryos derived from inter se crosses of ADAR2−/−/GluR-BR/R or PKR−/− mice (35) and cultured in Dulbecco’s modified Eagle’s medium with 10% fetal calf serum. All procedures involving mice were approved by the Wistar Institutional Animal Care and Use Committee.
In Vivo Silencing of EGFP by siRNA in MEF Cells
In vivo effects of ADAR1 on the siRNA-based RNA silencing of EGFP by 19-bp EGFP siRNAs were assayed in wild-type, ADAR1−/− (36), ADAR2−/−/GluR-BR/R, and PKR−/− MEF cells (see above), by using a dual fluorescence reporter system (37). MEF cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum, trypsinized, and washed twice with phosphate-buffered saline prior to transfection. A transfection mixture of 1.5 µg each of pEGFP-C1 and pCMV-DsRed-Express reporter plasmids (Clontech) and a combination of EGFP siRNA and nonspecific control siRNA (total concentration adjusted to be equal) was electroporated into 2 × 106 MEF cells using 100 µl of an electroporation buffer optimized for MEF cells and Nucleofector™ (Amaxa Biosystems, Cologne, Germany). For the rescue experiments, 3 µg of pCMV-F-ADAR1p150, pCMV-F-ADAR1ΔMIM2M3, or pRC/CMV (vector-only control) was also included in the transfection mixture. The dsRNA-binding domains mutant ΔMIM2M2 lacks all three dsRNA-binding domains, as described previously (32). The entire transfection mixture including siRNA was added to 10 ml of the complete media. The specific siRNA concentration was indicated as the final concentrations in 10 ml of culture media (0–25 nm). The transient transfection efficiency was comparable between pEGFP-C1 and pCMV-DsRed-Express plasmids (70–90%). The MEF cells transfected and continuously cultured with siRNA were harvested 44 h post-electroporation, trypsinized, washed with phosphate-buffered saline, suspended in chilled phosphate-buffered saline, and subjected to fluorescence-activated cell sorting (FACS) analysis using a FACS-flow cytometer (Dakocytomation, Carpinteria, CA). The relative levels of green (EGFP) and red fluorescence (RFP) for different samples were quantified by using the geometric means, and the normalized ratio of target (EGFP) to control (RFP) expression was determined at various concentrations of specific siRNA in the culture media. Data were analyzed using Summit Software (DakoCytomation).
Western Immunoblot Analysis
The total protein extracted from wild-type, ADAR1−/−, or ADAR1−/− MEF cells transiently transfected with pCMV-F-ADAR1p150, pCMV-F-ADAR1ΔMIM2M3, or pRC/CMV (vector-only control) plasmid DNA for 44 h was fractionated on an SDS-8% polyacrylamide gel and analyzed by Western blotting using mAb M2 specific for FLAG epitope-tagged proteins. Various known amounts of recombinant ADAR1p150 protein loaded onto the gel were used for calibration. Chemiluminescence intensity was recorded on film. The relative amount of ADAR1 protein was estimated by comparing the integrated density of each chemiluminescence spot, measured with a flatbed scanning microdensitometer (model 1010G, PerkinElmer Life Sciences) using a 50 × 400-µm slit aperture.
RESULTS
Specific and High Affinity Binding of siRNA by ADAR1p150
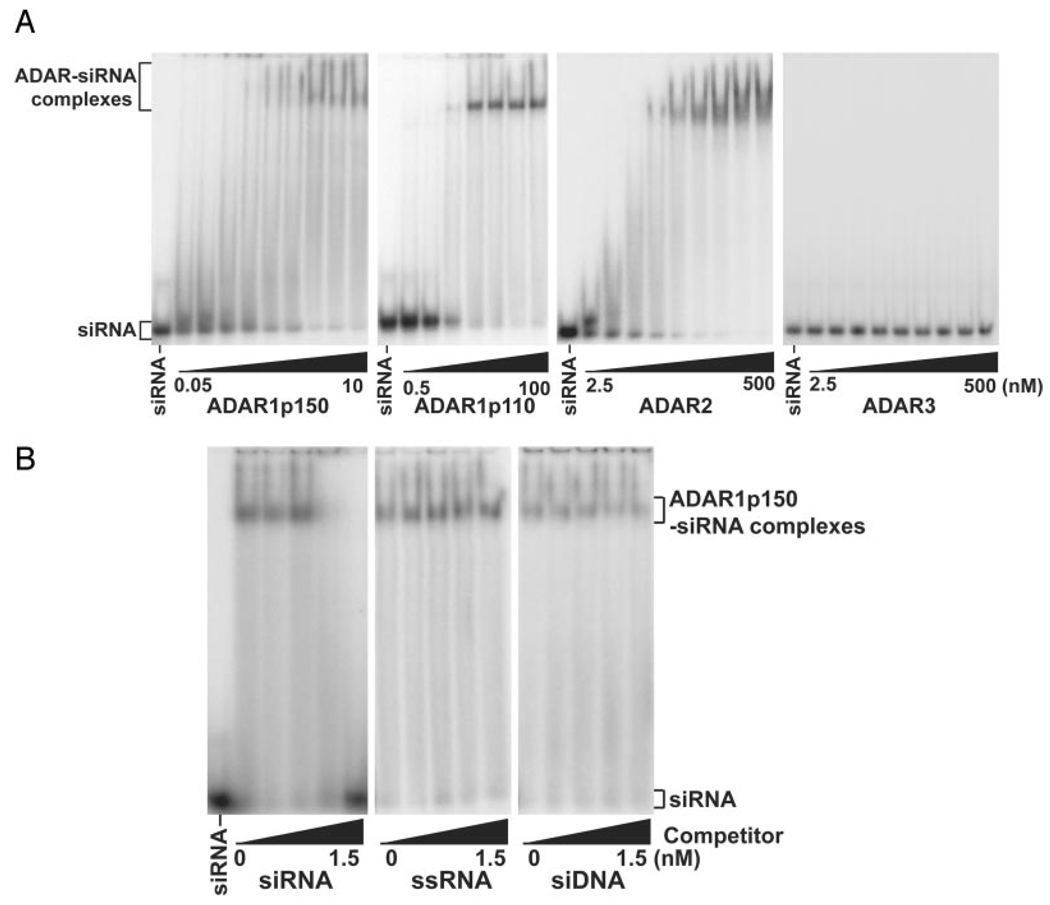
Because RNAi acts in the cytoplasm, the most relevant member of the ADAR gene family is ADAR1p150 (Fig. 1). Therefore, using an electrophoretic mobility shift assay (EMSA), we first tested binding of ADAR1p150 to a synthetic 21-nt siRNA, homologous to EGFP containing a double-stranded region of 19-bp and 2-nt 3′ overhangs on both sense and antisense strands (19-bp EGFP siRNA). ADAR1p150 bound this EGFP siRNA very strongly with an apparent dissociation constant (Kd) of 0.21 nm (Fig. 2A and Table I). This unexpectedly high affinity is comparable with the affinity of p150 for a long, completely complementary dsRNA of 575 bp (Kd = 0.12 nm) (Table I). The Kd value for two additional siRNAs that corresponded to different regions of the EGFP sequence as well as one siRNA unrelated to the EGFP sequence was identical to that for binding of the 19-bp EGFP siRNA, indicating a sequence-independent interaction (data not shown). Competitive inhibition assays indicated that binding of ADAR1p150 to siRNA is specific and is inhibited only by siRNA but not by the sense strand of siRNA or by a short dsDNA matching the sequence of the siRNA (Fig. 2B).
FIG. 2. High affinity binding of ADARs.
A, binding of homodimeric ADAR1 and ADAR2 to siRNA. Varying concentrations of FLAG-tagged ADAR1p150 (0.05, 0.075, 0.1, 0.15, 0.2, 0,5, 1, 5, and 10 nm), ADAR1p110 (0.5, 1, 2, 10, 20, 50, and 100 nm), ADAR2 and ADAR3 (2.5, 5, 10, 15, 25, 50, 100, 250, and 500 nm) were examined by EMSA using a native 4% polyacrylamide gel. The 5′-end of the sense strand of 19-bp EGFP siRNA (10 pm) was labeled with 32P. The 5′-end of the antisense strand was also phosphorylated but without 32P. B, specificity of ADAR1 binding to siRNA. Competitive inhibition experiments were conducted by preincubating ADAR1p150 (5 nm) with varying concentrations of cold competitors (0, 0.05, 0.25, 0.75, and 1.5 nm) for 5 min prior to addition of 32P-labeled 19-bp siRNA probe (10 pm). siRNA, 19-bp EGFP siRNA; ssRNA, single-stranded RNA, the sense strand of the 19-bp EGFP siRNA; siDNA, dsDNA corresponding to the 19-bp EGFP siRNA.
TABLE I.
Difference in siRNA binding affinity among ADAR gene family members
| Binding affinity (Kd, nm)a | ||||||
|---|---|---|---|---|---|---|
| ADARp150 | ADAR1E912A p150 | ADAR1p110 | ADAR2 | ADAR2E396A | ADAR3 | |
| 575-bp dsRNAb | 0.12 ± 0.01 | 0.14 ± 0.02 | 0.24 ± 0.03 | 0.29 ± 0.06 | 0.34 ± 0.08 | 0.43 ± 0.03 |
| 36-bp siRNAc | 0.11 ± 0.01 (~3)d | 0.32 ± 0.02 (~5)d | 2.6 ± 0.2 (~25d, ~50e) | 3.1 ± 0.5 (~50)d | ||
| 29-bp siRNAc | 0.11 ± 0.01 | 0.69 ± 0.10 | 3.1 ± 0.3 (~25d, ~50e) | 3.3 ± 0.3 (~50)d | ||
| 23-bp siRNA | 0.12 ± 0.01 | 1.9 ± 0.1 | 7.5 ± 0.3 | |||
| 19-bp siRNA | 0.21 ± 0.02 | 2.2 ± 0.2 | 3.0 ± 0.1 | 10 ± 1 | 20 ± 1 | NBf |
| 15-bp siRNA | 0.55 ± 0.03 | 4.1 ± 0.2 | 31 ± 2 | |||
Kd values for siRNA (means ± S.E.) were estimated from the results of five independent EMSA experiments and are expressed in terms of the concentration of ADAR monomer, although all ADARs examined in the present study except ADAR3 are predominantly homodimer complexes.
Kd values for this long dsRNA were obtained using the filter binding assay.
Kd values for these two longer siRNAs represent an apparent dissociation constant for the first complex (fast migrating on EMSA gels).
Approximate Kd values for the second complex (slow migrating) are indicated.
Approximate Kd values for the third complex (slow migrating) are indicated.
NB, no measurable binding by EMSA.
We assessed siRNA binding affinity in a separate experiment using a nitrocellulose filter-binding assay previously used to evaluate the binding of long dsRNA (32). The Kd value measured in this assay was identical to the Kd value measured with EMSA, confirming the strong binding of ADAR1p150 to siRNA (data not shown). Dissociation of the ADAR1p150-siRNA complex was then examined with the filter-binding assay. A binding reaction carried out with 5 nm ADAR1p150 was diluted 50-fold by adding binding buffer and continuously incubating for various times before filtering. The amount of ADAR1p150-siRNA complex was unaffected by the rapid reduction in siRNA and protein concentration for up to 2 h after dilution, revealing that the ADAR1p150-siRNA complex, once formed, is very stable (data not shown). These results indicate that the off-rate for dissociation of siRNA from the ADAR1p150-siRNA complex is <10−4 s−1, consistent with binding affinity in the subnanomolar range.
Binding of Other ADARs and Effects of the siRNA Size
ADAR1p110 and ADAR2 also bound the 19-bp siRNA, but their binding affinities were 15 and 50 times lower than that of ADAR1p150, respectively; ADAR3 capable of binding long dsRNA failed to bind the 19-bp siRNA (Fig. 2A and Table I). Both ADAR1 and ADAR2 form a stable, enzymatically active homodimer complex, whereas ADAR3 remains monomeric (28). Our previous studies suggested that homodimerization and interactions of the two monomers of ADAR1 or ADAR2 may play a critical role in the site-selective editing mechanism (28). Most interestingly, formation of the ADAR1- or ADAR2-siRNA complexes was quite sensitive to the salt concentration. For instance, formation of ADAR1p150-siRNA complexes decreased 10-fold when the NaCl concentration was raised from 0.1 to 0.5 m, possibly indicating also the importance of ionic interactions between the two monomers for binding of siRNA. In contrast, ADAR1p150 binding to long dsRNAs was essentially unaffected in the presence of 0.5 m NaCl, although the binding affinity for long dsRNAs diminished slightly in 1 m NaCl (data not shown). We also examined the E912A and E396A mutants of ADAR1 and ADAR2, respectively, which retain intact dsRNA-binding domains but lack a functional deaminase domain (thus catalytically dead) (28, 32) and also lack productive interactions between the two monomers in a homodimer complex (28). These mutants bound siRNA, although their binding affinity was substantially (2–10-fold) lower than that of the wild-type proteins (Table I). Their lower affinity for siRNAs may indicate once again the importance of the functional monomer interactions perturbed in these mutants in the siRNA binding.
We next examined binding to two different siRNAs with shorter or longer dsRNA regions (15 and 23 bp). All ADARs that were capable of binding the 19-bp siRNA (ADAR1p150 and p110 and ADAR2) also bound siRNAs containing either the 15- or 23-bp dsRNA regions, although shortening or lengthening of the dsRNA region in the siRNA affected the binding affinity (Table I). For instance, the binding affinity of ADAR1p150 or ADAR2 for the 15-bp siRNA was 3-fold weaker than that for the 19-bp siRNA (Table I). We also examined binding of ADAR1p150 to the 19-bp siRNA that lacked 2-nt 3′-overhangs (blunt end). ADAR1p150 bound this blunt end siRNA, but the 2-nt 3′-overhangs of siRNA appeared to make an additional contribution to strong binding (e.g. ADAR1p150 binding to the 19-bp siRNA, Kd = 0.21 nm versus the 19-bp blunt end siRNA, Kd = 0.43 nm). A 5′ phosphate group of the antisense strand of siRNA is required for the RNAi mechanism (37, 38). However, high affinity binding of ADAR1p150 to siRNA appeared to be independent of 5′-end phosphorylation, because ADAR1p150 also bound strongly to siRNA consisting of a 5′-phosphorylated sense and nonphosphorylated antisense strand. Furthermore, siRNA with both strands nonphosphorylated at the 5′-end effectively competed with normal siRNA having both 5′-phosphorylated sense and antisense strands in a competitive inhibition assay (data not shown).
A-to-I RNA Editing Independent Binding of siRNA
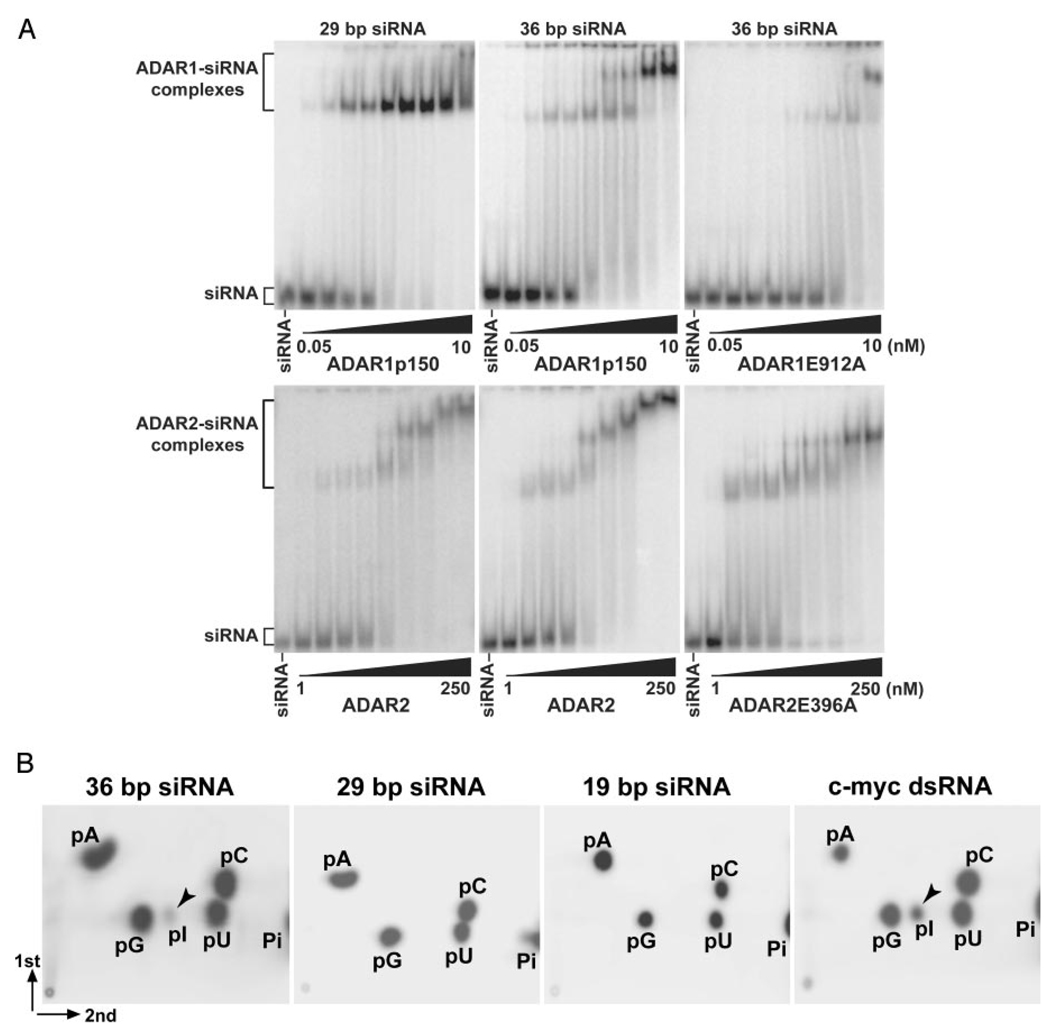
Because the mutants lacking deaminase activity (ADAR1E912A and ADAR2E396A) are capable of binding to siRNAs, complex formation of ADAR1 or ADAR2 with a typical 19-bp siRNA is independent of A-to-I RNA editing. Efficient A-to-I editing occurs with dsRNAs of ≥30 bp in length, although a very limited editing may occur with dsRNAs as short as 23 bp (39). In order to confirm A-to-I RNA editing independent binding of typical siRNAs, we tested the 19-bp EGFP siRNA as well as two additional siRNAs, containing a 29- and a 36-bp dsRNA region, for the A-to-I conversion reaction. Two-dimensional TLC analysis of mononucleotides derived from siRNAs complexed with wild-type ADAR1p150 revealed that low efficiency editing of adenosines (~10%) occurs in the 36- but not the 29- and 19-bp siRNAs (Fig. 3B). Identical results for siRNA bound to ADAR2 were obtained; low efficiency editing was only with 36-bp siRNA (not shown). These results confirm that the length of siRNA determines whether bound siRNA is edited or held in a stable complex without change of sequence; the critical size threshold appears to be ≥30 bp.
FIG. 3. A-to-I editing of 36-bp but not 29- or 19-bp siRNA.
A, ADAR1p150 (wild-type and E912A mutant) and ADAR2 (wild-type and E396A mutant) were tested for binding at 30 °C for 60 min to 29- or 36-bp siRNA (10 pm), and the reaction products were analyzed by EMSA. ADAR1p150, 0.05, 0.075, 0.1, 0.15, 0.2, 0.5, 1, 5, and 10 nm. ADAR2, 1, 2, 2.5, 5, 10, 20, 50, 100, and 250 nm. B, TLC analysis of siRNAs bound by ADAR1p150. Varying sizes of siRNA complexed with ADAR1p150 (wild-type) were analyzed for A-to-I editing of internal adenosine residues.
As expected, EMSA analysis revealed that both ADAR1p150 and ADAR2 bound these longer siRNAs. At low concentrations of protein, a single discrete siRNA-ADAR complex was detected (Fig. 3A). Their binding affinity was slightly higher than siRNAs with a shorter dsRNA region (Table I). However, as the ADAR concentration increased, formation of multiple complexes was noted during binding of 36-bp siRNA by ADAR1p150 and during binding of 29- and 36-bp siRNAs by ADAR2 (Fig. 3A). It appears that different siRNA-ADAR complexes were formed as the ratio of siRNA to protein changed. Formation of multiple complexes for these longer siRNAs was detected also with catalytically inactive ADAR1p150E912A and ADAR2 E396A mutants, suggesting that formation of multiple complexes is unlikely to be due to editing of these longer siRNAs (Fig. 3A). A possible explanation for formation of multiple complexes is that they differ in the number of siRNA molecules bound to one ADAR homodimer, but currently available data cannot eliminate other possibilities (see “Discussion”).
Enhanced Silencing by siRNA in ADAR1 Null Mutant Fibroblast Cells
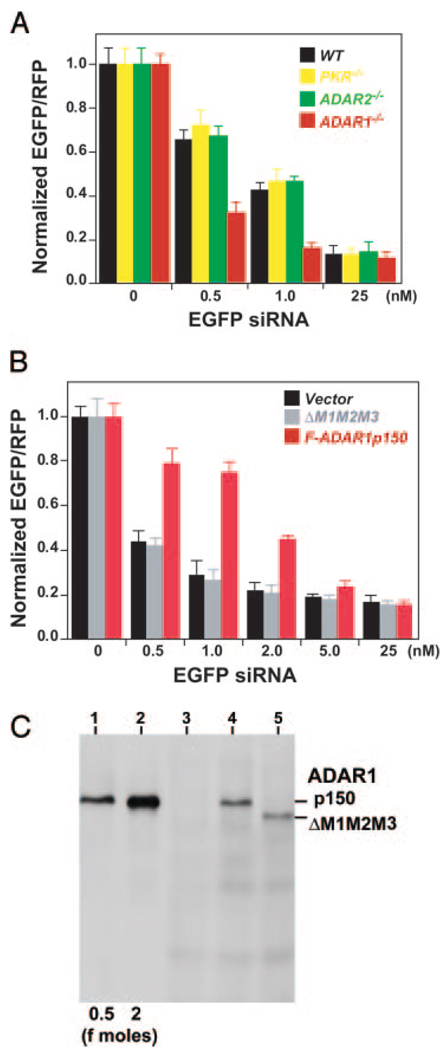
To assess the in vivo effects of siRNA binding by ADARs, we compared the efficacy of siRNA-based gene silencing in mutant mouse embryonic fibroblasts (MEF) null for ADAR1 (35, 40) or ADAR2 (34) to that in wild-type cells (Fig. 4A). As an additional control, PKR null MEF cells (35) were also tested. Expression of EGFP was targeted with 19-bp EGFP siRNA used for the in vitro binding EMSA analysis and assayed with an in vivo dual fluorescence reporter system using pEGFP-C1 (target) and pCMV-DsRed-Express (control) plasmids. A control 19-bp siRNA with sequence unrelated to EGFP or RFP had no effects on the expression of EGFP and RFP at the highest concentration tested, in both wild-type and three separate mutant MEF cells (25 nm control 19-bp siRNA tested: see Fig. 4A, EGFP siRNA 0 nm control experiments). Although suppression of target EGFP expression by ~10-fold was detected in wild-type and all mutant MEF cells with relatively high concentrations, substantially enhanced silencing effects (up to ~3-fold) were detected in ADAR1−/− MEF cells at relatively low concentrations of siRNA. For instance, although EGFP expression was decreased only to 42–46% in wild-type, PKR−/−, or ADAR2−/− MEF cells by 1 nm EGFP siRNA, the expression was down-regulated to 16% in ADAR1−/− cells. The enhanced RNAi effects in ADAR1−/− MEF cells were confirmed with experiments using a different siRNA that corresponded to a different region of the EGFP sequence (data not shown).
FIG. 4. Suppression of siRNA-based RNAi by ADAR1.
A, effects of endogenous ADAR1 proteins. A combination of varying concentrations of 19-bp EGFP siRNA and nonspecific control siRNA together with pEGFP-C1 target (1.5 µg) and pCMV-DsRed-Express control (1.5 µg) plasmid DNA were co-transfected into wild-type, PKR null, ADAR2 null, and ADAR1 null MEF cells. Quantitative measurement of EGFP and RFP expression levels were done by FACS analysis, and the EGFP expression level relative to that of RFP was determined. Results are presented as means ± S.E. (bars) values of five independent experiments performed in triplicate. The reduction in the relative EGFP expression levels in ADAR1 null MEF cells was examined statistically by individual unpaired Student’s t tests in comparison to those in wild-type, PKR null, and ADAR2 null MEF cells. p < 0.001 for both 0.5 and 1 nm concentrations. B, effects of recombinant ADAR1p150 proteins. In addition to pEGFP-C1 and pCMV-DsRed-Express, 3.0 µg of pCMV-F-ADAR1p150 or pCMV-F-ADAR1ΔM1M2M3 were transfected into ADAR1 null MEF cells. The vector-only control experiments were also conducted with pRC/CMV plasmid DNA. Results are presented as the mean ± S.E. (bars) values from three independent experiments performed in triplicate. The difference between the vector-only control or pCMV-F-ADAR1ΔMIM2M3 experiments and the pCMV-F-ADAR1p150 experiments (0.5, 1.0, and 2.0 nm concentrations) were significant (p < 0.001) by unpaired Student’s t tests. C, the total protein extracted from 1 × 105 ADAR1 null MEF cells transfected for 44 h with vector-only (lane 3), pCMV-F-ADAR1p150 (lane 4), or pCMV-F-ADAR1ΔMIM2M3 (lane 5), together with varying amounts of FLAG epitope-tagged p150 standard proteins (0.5 or 2 fmol) was examined by Western blotting analysis.
In order to further demonstrate the in vivo suppressive effects of ADAR1, we examined silencing of the EGFP in ADAR1 null MEF cells transfected with a FLAG-tagged ADAR1p150 expression plasmid. The vector-only control and an additional mutant construct ADAR1ΔMIM2M3, in which all three dsRNA-binding domains were deleted (32), were also tested (Fig. 4B). These rescue experiments confirmed the quenching effects by the wild-type ADAR1 but not by the ΔMIM2M3 mutant or vector alone for siRNA mediated EGFP silencing. These results indicated also that the quenching activity of ADAR1 is likely to be exerted through siRNA binding by its dsRNA-binding domains.
Western analysis using M2 mAb specific for FLAG-tagged proteins detected roughly equal levels of wild-type ADAR1p150 or ΔMIM2M3 (130-kDa) proteins in transfected ADAR1 null MEF cells (Fig. 4C). In light of the quenching effects on siRNA efficacy by ADAR1p150, the concentration of recombinant p150 proteins expressed in these cells was determined based on the MEF volume of 2 pl (41) and the known amount of purified recombinant p150 standard (Fig. 4C). We estimated that transfected ADAR1 null MEF cells contained ~1 nm ADAR1p150, sufficient for quenching 0.5–1 nm siRNA. Taken together, our results indicate that ADAR1 (most likely cytoplasmic p150) reduces siRNA potency in vivo, probably by binding, thus decreasing the effective siRNA concentration.
DISCUSSION
Although RNAi is an extremely powerful gene silencing mechanism, certain cellular and viral factors appear to suppress its efficacy. For instance, ERI-1 is a ribonuclease that limits the efficacy of the endogenous RNAi mechanism by degrading specifically siRNAs (42). In contrast, a 19-kDa protein (p19) homodimer synthesized by tombusvirus, although incapable of binding long dsRNA, binds specifically siRNAs of a defined length (optimal binding at 19–21 bp) with a subnanomolar dissociation constant, thereby suppressing the host plant defense RNAi mechanism (43, 44). It is widely accepted that there are distinctive features of the mammalian RNAi pathway, such as a requirement for higher siRNA concentrations and relatively short lasting effects, possibly due to the lack of RNA-directed RNA polymerase-mediated amplification of siRNA (4, 5, 45). In addition, there may be certain cellular factors similar to ERI-1 and p19 that limit RNAi efficacy in mammals.
Double-strandedness of the trigger RNA is critical for RNAi. Thus, the potential for ADAR-mediated A-to-I editing of trigger dsRNAs and consequent dampening of RNAi efficacy has been pointed out previously (6, 31). As expected, A-to-I editing of a trigger dsRNA in vitro by recombinant ADAR2 greatly reduces the amount of siRNA produced and consequently antagonizes RNAi effects (29). In C. elegans homozygous for null mutations of both c.e.ADAR1 and c.e.ADAR2, transgenes are very effectively silenced through the RNAi mechanism. However, in wild-type worms, transgenes escape silencing because of A-to-I editing of their dsRNA transcripts, presumably derived from simultaneous transcription of the sense and antisense strands (30). These previous studies indicate that ADAR certainly can affect RNAi efficacy through A-to-I editing of trigger dsRNA (29, 30). In the present study, we investigated the interaction between ADAR gene family members and a series of synthetic siRNAs. Our studies revealed that ADAR1p150 bound to 19-bp siRNA, despite its short dsRNA region, with extremely high affinity. Furthermore, siRNA-based RNAi was substantially more effective in mouse fibroblasts homozygous for an ADAR1 null mutation than in wild-type cells. The suppressive effects of ADAR1 were confirmed in ADAR1 null cells when transfected with the ADAR1p150 expression plasmid but not when transfected with a mutant construct lacking dsRNA-binding domains. Our results identify ADAR1 as a cellular factor that limits the efficacy of siRNA in mammalian cells through an unexpected direct interaction with siRNA, in addition to the known effects on trigger dsRNA.
Importance of ADAR Homodimerization for Its Interaction with siRNA
Although the binding of most proteins recognizing A-form dsRNA is sequence-independent, their affinities and the minimum required lengths of target dsRNA appear to differ substantially. For instance, DICER binds long dsRNAs efficiently but not the resulting cleavage product siRNAs (46), whereas the dsRNA-binding domains of PKR (induced by interferons in the cytoplasm) bind 20-bp dsRNA but with relatively low affinity, Kd value of 170 nm (47). Although the co-crystal structure of a monomeric dsRNA-binding domain complexed with dsRNA suggested that a minimum of 16 bp of dsRNA may be sufficient for interaction (48), very little, if any, A-to-I editing of the 23-bp blunt end dsRNA by ADAR1 was detected in our previous studies, indicating that the shortest dsRNA recognized as a substrate for editing by ADAR1 might be ≥30 bp (39). Therefore, it is surprising to find that both ADAR1 and ADAR2 can bind typical siRNAs containing a 19-bp dsRNA region, as well as siRNAs containing an even shorter 15-bp dsRNA region. Most interestingly, the binding affinity of ADAR1p150 for siRNAs with 2-nt 3′-overhangs is slightly higher (~2-fold) than that of blunt end dsRNAs, indicating an additional contribution of the overhangs to the high affinity binding.
ADAR1 and ADAR2 are homodimers, whereas ADAR3, which is incapable of binding siRNAs, is a monomer (28). All three ADARs do bind long dsRNAs equally well (32). Furthermore, the siRNA binding affinity of ADAR1p150E912A and ADAR2E396A, two catalytically inactive mutants, is substantially lower than that of wild-type proteins. The decreased binding affinity of these mutant homodimeric forms is perhaps due to diminished functional interactions between their two monomers, a consequence of the role played by the mutated glutamate residue in formation of the dimer interface (28). Thus, one possible reason that ADAR1 and ADAR2 bind to short dsRNAs, such as siRNAs, is that their homodimeric configuration allows short dsRNAs to interact with more than one dsRNA-binding domain of the two monomers (Fig. 5A).
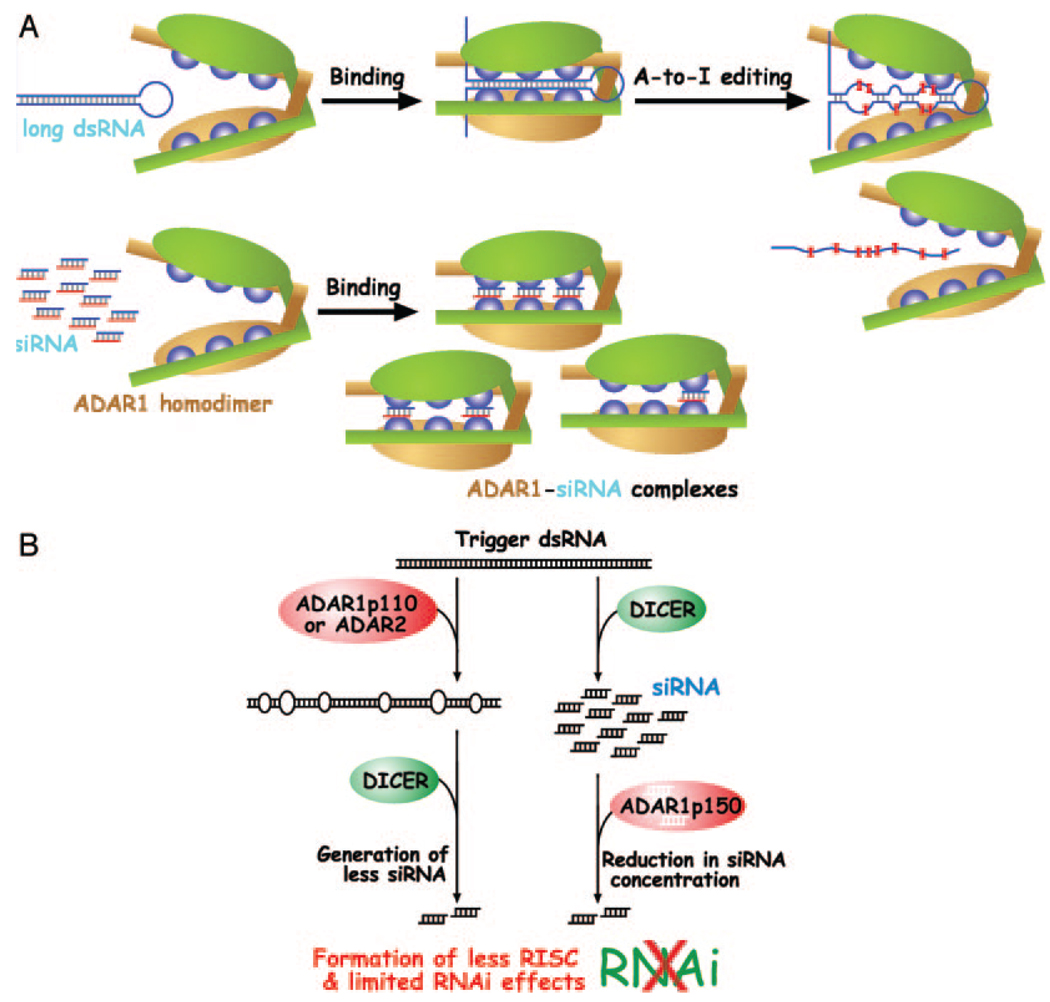
FIG. 5. Model for formation of siRNA-ADAR1p150 complexes and suppression of siRNA efficacy.
A, two modes of ADAR1 interaction with dsRNA, A-to-I editing of long dsRNA and high affinity binding of siRNA, are indicated. The number of siRNA molecules binding to the three separate dsRNA-binding domains of ADAR1p150 is not known. It is also not known whether binding of one siRNA molecule involves dsRNA-binding domains from one or both monomers. Blue spheres represent individual dsRNA-binding domains. Large ovals represent catalytic domains. The two monomers are indicated with different colors (green and orange) for clarity. B, suppression of RNAi efficacy by ADAR. The introduction of many I-U mismatched base pairs and alteration of trigger dsRNA structure by ADAR1p110 and ADAR2 in the nucleus, as well as the binding of siRNAs by ADAR1p150 in the cytoplasm, reduces the effective siRNA concentration and thus RNAi effects. RISC, RNA-induced silencing complex.
The siRNA binding curve (ADAR-siRNA complex versus ADAR concentration) is sigmoidal and steeper than a normal single-site saturation curve at low ADAR concentrations, indicating that multiple binding sites present on a single molecule interact with positive cooperativity to give the observed high affinity binding. The curve is consistent with all sites being equivalent (i.e. having the same intrinsic binding constant), but of course more complicated models would also fit the data. Formation of multiple complexes is detected for binding of longer siRNAs (29 or 36 bp). A single discrete band is seen on the gel at low concentrations of ADAR. As the protein concentration increases, other bands appear, representing complexes with lower affinity. Once again, a reasonable interpretation is that the lower apparent affinity is the result of less cooperative enhancement of the intrinsic single-site affinity due to under-saturation of the multiple sites on each ADAR homodimer molecule (Fig. 5A). Obviously, a complete analysis of the equilibrium among multiple interacting sites requires much more precise data than can be obtained from our EMSA gels.
Effects of Z-DNA Binding Domains on siRNA Binding
In view of their similar affinity for long dsRNAs, the substantial differences in siRNA binding affinity between ADAR1p150, ADAR1p110, and ADAR2 are surprising. The binding affinity of ADAR1p150 to the 19-bp siRNA was 50-fold greater than that of ADAR2. The difference detected in the binding affinities of ADAR1p150 and p110 is also large (15-fold). The difference may be due to the presence of the N-terminal region present in p150; this region includes two Z-DNA-binding domains, Zα and Zβ. Although a large portion of the Zβ domain is contained within p110 (Fig. 1), efficient binding of ADAR1 to Z-DNA requires both Zα and Zβ (49). The function of these two Z-DNA-binding domains, which are apparently dispensable for A-to-I editing of long dsRNA substrates (50), has been a subject of speculation (49). Our results suggest that one function of the Zα domain of ADAR1p150 may be to mediate high affinity binding to siRNAs. Because ADAR2 can bind siRNAs, despite lacking the Z-DNA-binding domains, direct binding of siRNAs to these domains is unlikely. However, the Zα domain may contribute to the extremely strong binding of ADAR1p150 to siRNAs via its effects on the neighboring dsRNA-binding domains and/or on interactions between monomers. The nuclear export of ADAR1p150 into the cytoplasm, mediated by the nuclear export receptor CMR1 which forms an export-competent complex together with RanGTP, has been reported to be dependent on the Zα domain (51). It is of special interest that the Zα domain is also essential for the high affinity binding of ADAR1p150 to siRNAs. Its presence in ADAR1p150 may be indicative of the specific function of ADAR1p150 in dealing with this type of short dsRNA substrate that localizes to the cytoplasm.
Implication of the ADAR1p150-siRNA Interaction for RNAi Efficacy
Fragmentation of long trigger dsRNAs to siRNAs is carried out by DICER in the cytoplasm (4, 5). Moreover, recent studies (52, 53) using DNA microarrays indicate that not only long dsRNA but also siRNA activate the interferon pathway in contrast to widely accepted assumptions. Thus, the interaction of siRNAs with ADAR1p150, the only interferon-inducible and cytoplasmically localized ADAR gene family member (24, 25), is likely to have the most significant effects on siRNA-based RNAi. Indeed in vivo targeting of EGFP by siRNA is more efficient in fibroblast cells homozygous for an ADAR1 null mutation in comparison to wild-type cells. Quantitative analysis of recombinant ADAR1 protein, derived from an ADAR1 expression plasmid, confirmed their siRNA quenching effects in vivo. Substantial differences in the siRNA-based RNAi efficacy reported with different mammalian cell lines (9, 10) may well be related at least in part to levels of ADAR1p150 synthesized in those cell lines (54). Thus, the efficacy of RNAi may be lessened, not only through introduction of many mismatched I-U base pairs within the trigger dsRNA structure by nuclear localized ADAR1p110 and ADAR2, as suggested previously (6, 29), but also via formation of a stable cytoplasmic ADAR1p150-siRNA complex, which decreases the concentration of siRNA available for RNA-induced silencing complex formation (Fig. 5B). A role for small dsRNA molecules in regulating critical developmental processes is becoming increasingly clear (55). The apoptosis-prone, embryonic lethal, phenotype of ADAR1−/− mice reported recently (36, 40) may be a manifestation of deficiencies in this particular ADAR1 function of buffering endogenous RNAi.
Acknowledgments
We thank Drs. J. Pavlovic and C. Weissmann for PKR−/− mice, Drs. M. Higuchi and P. H. Seeburg for ADAR2−/−/GluR-BR/R mice, Dr. F. Lai for construction of pCMV-F-ADAR1ΔMIM2M3, and Dr. Z. Cao for preparation of ADAR2−/− MEF cells. We also thank the Wistar Expression Vector-Recombinant Protein Production and Flow Cytometry Facilities for technical services and the Wistar Editorial Services Department for preparing the manuscript.
Footnotes
This work was supported in part by grants from the National Institutes of Health, the Doris Duke Charitable Foundation, the March of Dimes, and the Commonwealth Universal Research Enhancement Program, Pennsylvania Department of Health.
The abbreviations used are: dsRNA, double-stranded RNA; ADAR, adenosine deaminases acting on RNA; A-to-I, adenosine-to-inosine; GluR, glutamate receptor; mAb, monoclonal antibody; MEF, mouse embryonic fibroblasts; PKR, interferon inducible serine/threonine kinase; RNAi, RNA interference; siRNA, short interfering RNA; EGFP, enhanced green fluorescent protein; FACS, fluorescence-activated cell sorter; RFP, red fluorescence protein; EMSA, electrophoretic mobility shift assay; nt, nucleotide.
REFERENCES
- 1.Bass BL. Annu. Rev. Biochem. 2002;71:817–846. doi: 10.1146/annurev.biochem.71.110601.135501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maas S, Rich A, Nishikura K. J. Biol. Chem. 2003;278:1391–1394. doi: 10.1074/jbc.R200025200. [DOI] [PubMed] [Google Scholar]
- 3.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 4.Denli AM, Hannon GJ. Trends Biochem. 2003;28:196–201. doi: 10.1016/S0968-0004(03)00058-6. [DOI] [PubMed] [Google Scholar]
- 5.Zamore PD. Science. 2002;296:1265–1269. doi: 10.1126/science.1072457. [DOI] [PubMed] [Google Scholar]
- 6.Bass BL. Cell. 2000;101:235–238. doi: 10.1016/s0092-8674(02)71133-1. [DOI] [PubMed] [Google Scholar]
- 7.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 8.Samuel CE. Clin. Microbiol. Rev. 2001;14:778–809. doi: 10.1128/CMR.14.4.778-809.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caplen NJ, Parrish S, Imani F, Fire A, Morgan RA. Proc. Natl. Acad. Sci. U. S. A. 2001;98:9742–9747. doi: 10.1073/pnas.171251798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 11.Seeburg PH. Neuron. 2002;35:17–20. doi: 10.1016/s0896-6273(02)00760-2. [DOI] [PubMed] [Google Scholar]
- 12.Hoopengardner B, Bhalla T, Staber C, Reenan R. Science. 2003;301:832–836. doi: 10.1126/science.1086763. [DOI] [PubMed] [Google Scholar]
- 13.Morse DP, Aruscavage PJ, Bass BL. Proc. Natl. Acad. Sci. U. S. A. 2002;99:7906–7911. doi: 10.1073/pnas.112704299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levanon EY, Eisenberg E, Yelin R, Nemzer S, Hallegger M, Shemesh R, Fligelman ZY, Shoshan A, Pollock SR, Sztybel D, Olshansky M, Rechavi G, Jantsch MF. Nat. Biotechnol. 2004;22:1001–1005. doi: 10.1038/nbt996. [DOI] [PubMed] [Google Scholar]
- 15.Kim DD, Kim TT, Walsh T, Kobayashi Y, Matise TC, Buyske S, Gabriel A. Genome Res. 2004;14:1719–1725. doi: 10.1101/gr.2855504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nishikura K. Nat. Biotechnol. 2004;22:962–963. doi: 10.1038/nbt0804-962. [DOI] [PubMed] [Google Scholar]
- 17.Kim U, Garner TL, Sanford T, Speicher D, Murray JM, Nishikura K. J. Biol. Chem. 1994;269:13480–13489. [PubMed] [Google Scholar]
- 18.O’Connell MA, Krause S, Higuchi M, Hsuan JJ, Totty N, Jenny A, Keller W. Mol. Cell. Biol. 1995;15:1389–1397. doi: 10.1128/mcb.15.3.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Melcher T, Maas S, Herb A, Sprengel R, Seeburg PH, Higuchi M. Nature. 1996;379:460–464. doi: 10.1038/379460a0. [DOI] [PubMed] [Google Scholar]
- 20.Lai F, Chen C-X, Carter KC, Nishikura K. Mol. Cell. Biol. 1997;17:2413–2424. doi: 10.1128/mcb.17.5.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gerber A, O’Connell MA, Keller W. RNA. 1997;3:453–463. [PMC free article] [PubMed] [Google Scholar]
- 22.Melcher T, Maas S, Herb A, Sprengel R, Higuchi M, Seeburg PH. J. Biol. Chem. 1996;271:31795–31798. doi: 10.1074/jbc.271.50.31795. [DOI] [PubMed] [Google Scholar]
- 23.Chen C-X, Cho D-S, Wang Q, Lai F, Carter KC, Nishikura K. RNA. 2000;6:755–767. doi: 10.1017/s1355838200000170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patterson JB, Samuel CE. Mol. Cell. Biol. 1995;15:5376–5388. doi: 10.1128/mcb.15.10.5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Desterro JMP, Keegan LP, Lafarga M, Berciano MT, O’Connell M, Carmo-Fonseca M. J. Cell Sci. 2003;116:1805–1818. doi: 10.1242/jcs.00371. [DOI] [PubMed] [Google Scholar]
- 26.Sansam CL, Wells KS, Emeson RB. Proc. Natl. Acad. Sci. U. S. A. 2003;100:14018–14023. doi: 10.1073/pnas.2336131100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scherl A, Couté Y, Déon C, Callé A, Kindbeiter K, Sanchez J-C, Greco A, Hochstrasser D, Diaz J-J. Mol. Biol. Cell. 2002;13:4100–4109. doi: 10.1091/mbc.E02-05-0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cho D-S, Yang W, Lee JT, Shiekhattar R, Murray JM, Nishikura K. J. Biol. Chem. 2003;278:17093–17102. doi: 10.1074/jbc.M213127200. [DOI] [PubMed] [Google Scholar]
- 29.Scadden AD, Smith CW. EMBO Rep. 2001;2:1107–1111. doi: 10.1093/embo-reports/kve244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Knight SW, Bass BL. Mol. Cell. 2002;10:809–817. doi: 10.1016/s1097-2765(02)00649-4. [DOI] [PubMed] [Google Scholar]
- 31.Tonkin LA, Bass BL. Science. 2003;302:1725. doi: 10.1126/science.1091340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lai F, Drakas R, Nishikura K. J. Biol. Chem. 1995;270:17098–17105. doi: 10.1074/jbc.270.29.17098. [DOI] [PubMed] [Google Scholar]
- 33.Wagner RW, Smith JE, Cooperman BS, Nishikura K. Proc. Natl. Acad. Sci. U. S. A. 1989;86:2647–2651. doi: 10.1073/pnas.86.8.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Higuchi M, Maas S, Single FN, Hartner J, Rozov A, Burnashev N, Feldmeyer D, Sprengel R, Seeburg PH. Nature. 2000;406:78–81. doi: 10.1038/35017558. [DOI] [PubMed] [Google Scholar]
- 35.Yang YL, Reis LF, Pavlovic J, Aguzzi A, Schäfer R, Kumar A, Williams BR, Aguet M, Weissmann C. EMBO J. 1995;14:6095–6106. doi: 10.1002/j.1460-2075.1995.tb00300.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Q, Miyakoda M, Yang W, Khillan J, Stachura DL, Weiss MJ, Nishikura K. J. Biol. Chem. 2004;279:4952–4961. doi: 10.1074/jbc.M310162200. [DOI] [PubMed] [Google Scholar]
- 37.Chiu Y-L, Rana TM. Mol. Cell. 2002;10:549–561. doi: 10.1016/s1097-2765(02)00652-4. [DOI] [PubMed] [Google Scholar]
- 38.Nykänen A, Haley B, Zamore PD. Cell. 2001;107:309–321. doi: 10.1016/s0092-8674(01)00547-5. [DOI] [PubMed] [Google Scholar]
- 39.Nishikura K, Yoo C, Kim U, Murray JM, Estes PA, Cash FE, Liebhaber SA. EMBO J. 1991;10:3523–3532. doi: 10.1002/j.1460-2075.1991.tb04916.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hartner JC, Schmittwolf C, Kispert A, Miller AM, Higuchi M, Seeburg PH. J. Biol. Chem. 2004;279:4894–4902. doi: 10.1074/jbc.M311347200. [DOI] [PubMed] [Google Scholar]
- 41.Vasquez B, Ishibashi F, Howard BV. In Vitro. 1982;18:643–649. doi: 10.1007/BF02796397. [DOI] [PubMed] [Google Scholar]
- 42.Kennedy S, Wang D, Ruvkun G. Nature. 2004;427:645–649. doi: 10.1038/nature02302. [DOI] [PubMed] [Google Scholar]
- 43.Ye K, Malinina L, Patel DJ. Nature. 2003;426:874–878. doi: 10.1038/nature02213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vargason JM, Szittya G, Burgyán J, Tanaka Hall TM. Cell. 2003;115:799–811. doi: 10.1016/s0092-8674(03)00984-x. [DOI] [PubMed] [Google Scholar]
- 45.Nishikura K. Cell. 2001;107:415–418. doi: 10.1016/s0092-8674(01)00581-5. [DOI] [PubMed] [Google Scholar]
- 46.Provost P, Dishart D, Doucet J, Frendewey D, Samuelsson B, Rådmark O. EMBO J. 2002;21:5864–5874. doi: 10.1093/emboj/cdf578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bevilacqua PC, Cech TR. Biochemistry. 1996;35:9983–9994. doi: 10.1021/bi9607259. [DOI] [PubMed] [Google Scholar]
- 48.Ryter JM, Schultz SC. EMBO J. 1998;17:7505–7513. doi: 10.1093/emboj/17.24.7505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Herbert A, Alfken J, Kim Y-G, Mian IS, Nishikura K, Rich A. Proc. Natl. Acad. Sci. U. S. A. 1997;94:8421–8426. doi: 10.1073/pnas.94.16.8421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maas S, Melcher T, Herb A, Seeburg PH, Keller W, Krause S, Higuchi M, O’Connell MA. J. Biol. Chem. 1996;271:12221–12226. doi: 10.1074/jbc.271.21.12221. [DOI] [PubMed] [Google Scholar]
- 51.Poulsen H, Nilsson J, Damgaard CK, Egebjerg J, Kjems J. Mol. Cell. Biol. 2001;21:7862–7871. doi: 10.1128/MCB.21.22.7862-7871.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sledz CA, Holko M, de Vees MJ, Silverman RH, Williams BR. Nat. Cell Biol. 2003;5:834–839. doi: 10.1038/ncb1038. [DOI] [PubMed] [Google Scholar]
- 53.Bridge AJ, Pebernard S, Ducraux A, Nicoulaz AL, Iggo R. Nat. Genet. 2003;34:263–264. doi: 10.1038/ng1173. [DOI] [PubMed] [Google Scholar]
- 54.Wagner RW, Yoo C, Wrabetz L, Kamholz J, Buchhalter J, Hassan NF, Khalili K, Kim SU, Perussia B, McMorris FA, Nishikura K. Mol. Cell. Biol. 1990;10:5586–5590. doi: 10.1128/mcb.10.10.5586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bartel DP. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]