Summary
Escherichia coli strains displaying hyper-accurate (restrictive) and ribosomal ambiguity (ram) phenotypes have long been associated with alterations in rpsL and rpsD/rpsE, respectively. Crystallographic evidence shows the ribosomal proteins S12 and S4/S5 (corresponding to these genes) to be located in separate regions of the small ribosomal subunit that are important for domain-closure thought to take place during tRNA selection. Mechanistically, the process of tRNA selection is separated into two distinct phases, initial selection and proofreading. Here, we explore the effects of mutations in rpsL and rpsD on these steps. Surprisingly, both restrictive and ram ribosomes primarily affect tRNA selection through alteration of the off-rates of the near-cognate tRNA species, but during distinct phases of the overall process (proofreading and initial selection, respectively). These observations suggest that closure interfaces (S12/h27/h44 versus S4/S5) in two distinct regions of the small ribosomal subunit function independently to promote high-fidelity tRNA selection.
Introduction
The faithful decoding of the genetic code into functional protein sequences is an intricate process that involves several biological complexes. Chief among them is the ribosome, which has evolved to complete the task of decoding mRNA sequences into peptides with both high fidelity (1 error in 103-104 incorporation events) and speed (a rate of 20 s-1), (reviewed in (Zaher and Green, 2009a)). During each cycle of elongation, the ribosome must select the cognate aminoacyl-tRNA (aa-tRNA), in a ternary complex with elongation factor Tu (EFTu) and GTP, that matches the codon occupying the A site from a large-pool of competing aa-tRNAs. This tRNA selection process is divided into two main phases, initial selection and proofreading, separated by the nearly irreversible step of GTP hydrolysis by EFTu (Figure 1). As a result, the ribosome is given two opportunities to inspect the codon-anticodon pairing and discard incorrect aa-tRNAs, a strategy commonly referred to as “Kinetic Proofreading” (Hopfield, 1974; Ninio, 1975). While this thermodynamically-based discrimination, despite its name, can in principle be sufficient to yield a high level of accuracy (Johansson et al., 2008; Nierhaus, 2006; Ninio, 2006; Thompson, 1988), it has been argued that such an equilibrium-based process can not easily account for the observed rapid high fidelity protein synthesis (Gromadski et al., 2006). Instead, the ribosome apparently circumvents this hurdle by utilizing induced-fit mechanisms, where conformational changes stabilized only by correct codon-anticodon interactions accelerate two key forward rates: activation of EFTu for GTP hydrolysis and accommodation of the aa-tRNA into the A site of the large subunit where it participates in the peptidyl-transfer reaction (Gromadski and Rodnina, 2004; Ogle et al., 2002; Pape et al., 1999).
Figure 1. Simplified kinetic scheme of the tRNA selection pathway.
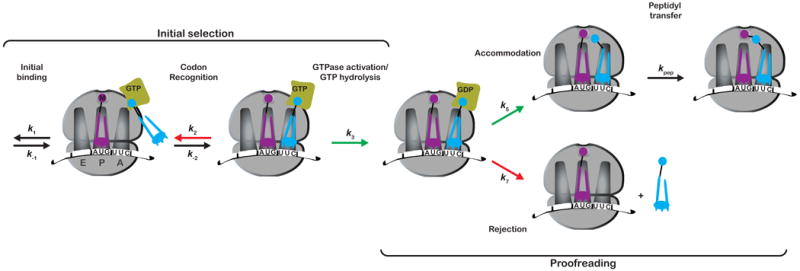
Only the steps that govern discrimination are shown. The pathway is divided into two phases, initial selection and proofreading, separated by the GTPase activation step. Steps accelerated for cognate tRNAs are depicted by green arrows, whereas steps that are slowed down for near-cognate tRNAs are depicted by red arrows.
Recent high-resolution crystal structures of the ribosome have provided important molecular details about this kinetically driven mechanism. Upon binding cognate aa-tRNAs, three key universally conserved nucleotides (A1492, A1493, and G530) in the decoding center of the small ribosomal subunit undergo substantial conformational rearrangements to engage the minor groove of the codon-anticodon helix (Carter et al., 2001; Ogle et al., 2002; Wimberly et al., 2000). Further interactions specific to cognate tRNAs are also made by the small ribosomal protein S12 to help orient several nucleotides for contact with the codon-anticodon helix. These local changes, in turn, are thought to stabilize global conformational changes in the small subunit referred to as “domain closure” (Ogle et al., 2002), which have been also seen in recent high-resolution structures of the whole 70S ribosome (Schmeing et al., 2009). These conformational changes involve rotation of the head of the small subunit toward the subunit interface and shoulder and ultimately towards the h44/h27/platform region (Figure 2). In this closed conformation of the small subunit, ternary complex makes contacts with both the 50S and 30S subunits, which are thought to activate EFTu for GTPase activation through interactions with the GTPase activation center (GAC) (Ogle et al., 2003). The transition from the open to the closed conformation involves distinct hydrogen bonding networks between conserved lysine residues in ribosomal protein S12 and backbone phosphate groups in h27 and h44. Streptomycin binds in a pocket at the interface between S12 and h27 (Carter et al., 2001), and hence is thought to stabilize the closed form and induce miscoding. In contrast to these contacts, polar interactions between ribosomal proteins S4 and S5, which lie on the opposite side of the shoulder domain relative to the S12/h44/h27 interface (on the solvent face of the subunit), are disrupted as a result of domain-closure.
Figure 2. Global conformational change (“domain closure”) in the small ribosomal subunit.
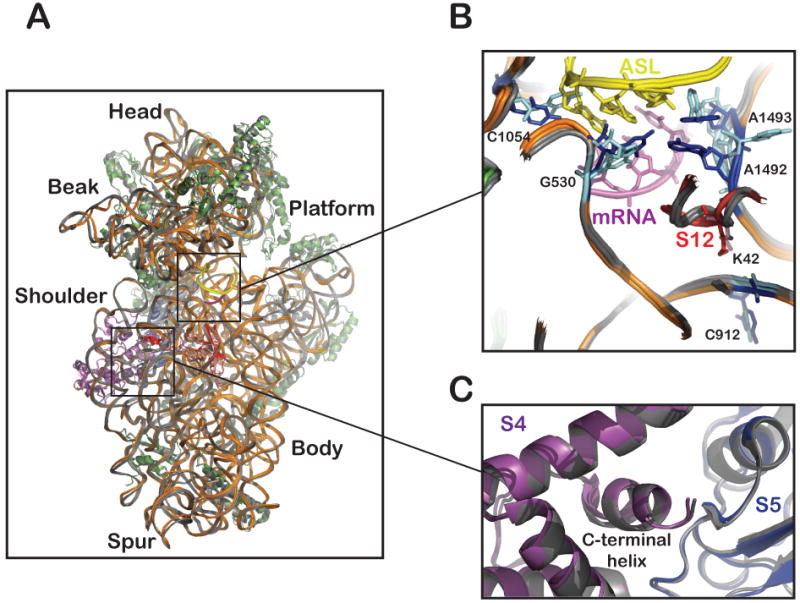
(A) The crystal structure of cognate anticodon stem loop (ASL)-bound 30S subunit (PDB 1IBM, r-proteins and rRNA are shown in green and orange, respectively) was superimposed on a near-cognate ASL-bound 30S subunit (PDB 1N34, shown in light gray) (Ogle et al., 2002). ASL, mRNA, S4, S5 and S12 of the cognate ASL-bound structure are depicted in yellow, pink, magenta, blue and red, respectively. As a result of cognate ASL binding, the shoulder and head domains of the subunit rotate towards the center of the subunit, where decoding takes place. For instance, regions of the 16S rRNA in the beak domain move closer to the shoulder domain (gray to orange) when the A-site is occupied with a cognate ASL.
(B) Close-up of the S12/h44/h27 interface is shown, with emphasis on the decoding center, where key universally conserved nucleotides undergo conformational changes (Light blue in the presence of near-cognate ASL to dark blue in the presence of cognate ASL). Ribosomal protein S12 also moves toward the decoding center upon cognate ASL-binding. Note the proximity of the rpsL141 mutation (K42N) to the ASL-mRNA helix and to residue C912 of 16S rRNA.
(C) Close-up of the S4/S5 interface, in particular the location of the S4 deletion (C-terminal helix) found in the rpsD12 mutant.
The relationship between an “open” to a “closed” transition of the small subunit and tRNA selection seemed to be further supported by mutations in the ribosome known to affect translational fidelity. Streptomycin-resistance (SmR) mutations are typically associated with alterations in S12, and are typically found to exhibit a hyperaccurate phenotype (Gorini and Kataja, 1964; Ozaki et al., 1969). Because these mutations restrict the miscoding action of streptomycin, they are referred to as restrictive. Most of these mutations in S12 are located at the interface with h27/h44 of the 16S rRNA, likely destabilizing interactions that are important for closure (and the completion of tRNA selection). Revertants of the streptomycin-dependent (SmD) phenotype (typically displayed by the most restrictive variants) were found to carry compensatory mutations in either protein S4 or S5 (Birge and Kurland, 1970; Brownstein and Lewandowein, 1967; Stoffler et al., 1971). On their own, these mutations typically confer an error-prone phenotype and are referred to as ribosomal ambiguity (ram) mutants (Rosset and Gorini, 1969). Most of the ram mutations are located at the interface between S4 and S5 on the solvent face of the small subunit where they likely disrupt mutual interactions. As this interface is disrupted during the transition from the open to the closed form, it is speculated that the ram mutants more readily facilitate this structural transition.
While elegant, the generality of the closure model in explaining translational fidelity has been recently called into question. A number of studies have argued that the “closure model” is too simplistic, and fails to explain some of the genetic data. In particular, these discussions cite ribosomal mutations that alter the fidelity of decoding without affecting the S4/S5 and S12/h44/h27 interfaces (Bjorkman et al., 1999; Kirthi et al., 2006; Vallabhaneni and Farabaugh, 2009).
While there is much known genetically about these ram and restrictive strains, we lack a complete biochemical and kinetic framework describing their mode of action. A priori one might predict that these mutations exploit the same steps in the tRNA selection pathway, thus resulting in their divergent phenotypes. Indeed, earlier steady-state studies examining misincorporation of tRNALeu on a polyU transcript suggested that while ram ribosomes are poor proofreaders (Andersson and Kurland, 1983), restrictive ribosomes are aggressive proofreaders (Bohman et al., 1984). Later pre-steady state studies by Ehrenberg and colleagues correlated this enhanced proofreading by restrictive ribosomes with slow rates of GTP hydrolysis on EFTu (Bilgin et al., 1992). However, as these latter studies were only performed with cognate aa-tRNAs, their implications for miscoding are unknown. Moreover, equivalent studies with ram ribosomes have not been conducted.
In this study, we conducted a kinetic study on an example of each ribosome miscoding phenotype, restrictive and ram, using a well-defined system to define the effects of these mutations on various steps of the tRNA selection pathway. While restrictive ribosomes were found to modestly affect the forward rates of the selection process, elevated discrimination was to a large extent the result of an increase in the rejection rate of near-cognate aa-tRNAs during the proofreading step. On the other hand, ram ribosomes were found to exploit only the initial phase of the selection to reduce discrimination against near-cognate aa-tRNAs. Surprisingly, neither the restrictive nor ram mutant ribosomes had any direct effects on the observed maximal rates of peptide release. These distinctive kinetic profiles will be discussed in the context of ribosome structure.
Results
Experimental Approach
We chose the genetically well-studied SmR rpsL141 mutant as a model for the restrictive phenotype (henceforth referred to as rpsL(res)), while the rpsD12 mutant (Andersson et al., 1982) served as a model for the ram phenotype (henceforth referred to as rpsD(ram)). rpsL141 carries an Asn for Lys substitution at position 42 in protein S12; this site makes contacts with a decoding center nucleotide (C912) close to the codon-anticodon helix in the A site (Figure 2B). rpsD12 carries a deletion of 5 nucleotides (C528-A532) resulting in a truncated S4 protein (due to a frameshift); the deleted portion of S4 directly interacts with the S5 protein in the wild-type (WT) ribosome (Figure 2C).
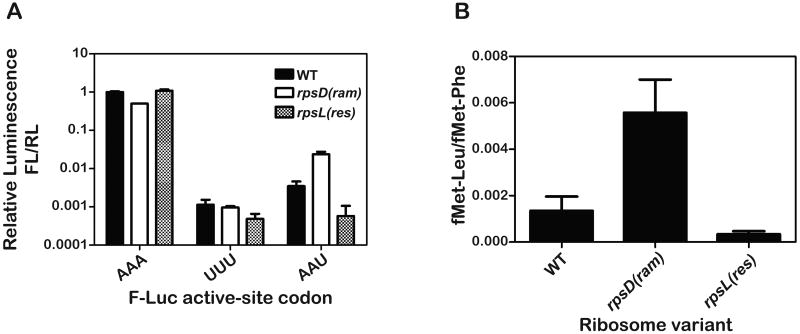
To establish that the previously characterized fidelity phenotypes are exhibited by these two strains, we used a dual-luciferase reporter system where the identity of a key active site residue (lysine 529) dictates the observed level of luciferase activity (Kramer and Farabaugh, 2007). As expected, the rpsD(ram) mutation increases misreading of the AAU asparagine codon by the near-cognate Lys-tRNALys by almost one order of magnitude, while the rpsL(res) mutation reduces the same error-rate to background levels (at least 6-fold) (Figure 3A).
Figure 3. The rpsD(ram) and rpsL(res) variants exhibit the expected phenotype both in vivo and in vitro.
(A) The relative luminescence of firefly luciferase (F-Luc) active site (K529) variants to that of control renilla-luciferase (R-Luc) expressed in the indicated strains is plotted (normalized to K529 F-Luc value in the WT strain) (Kramer and Farabaugh, 2007). When the K529 codon (AAA) is replaced by the non-cognate phe codon (UUU), only background activity of F-Luc is observed in all strains. In contrast, when this codon is replaced by the near-cognate codon (AAU), activity associated with the error frequency of tRNALys reading the substituted AAU codon is observed.
(B) The error frequency of WT, rpsL(res) and rpsD(ram) ribosome variants in our in vitro system. The error frequency was determined on UUC programmed ribosomes with excess ternary complex containing equimolar concentrations of Phe-tRNAPhe and Leu-tRNA2Leu where the relative amounts of fMet-Leu and fMet-Phe dipeptide were determined. In both graphs, the mean of three independent experiments is plotted and the error bars represent standard deviations from the mean.
We next set out to determine the effect of these ribosomal mutations on the individual steps of the tRNA selection process. For our studies, we first used ribosomal initiation complexes programmed with an mRNA that displays the Phe codon UUC in the A site, and ternary complexes of EFTu:GTP with either the cognate Phe-tRNAPhe (anticodon GAA, from yeast) or near-cognate Leu-tRNA2Leu (anticodon GAG, from E. coli). Discrimination during tRNA selection is governed by the rate constants of dissociation of the ternary complex (k-2), and GTPase activation (k3) during the initial phase of the pathway, and the rejection rate (k7), and accommodation (k5) during the proofreading stage (Figure 1). Codon recognition during the initial phase was monitored using a stopped-flow approach originally developed by Rodnina and colleagues (Rodnina et al., 1995) with proflavin-fluorescent derivatives of Phe and Leu tRNAs (Prf16/17 tRNAPhe and Prf16/17/20 tRNA2Leu). The chemical steps of GTP hydrolysis, which reports on k3, and peptide bond formation, which reports on k5 and k7 (Pape et al., 1998), were measured by quench-flow techniques using radio-labeled substrates (Cochella et al., 2007) (using ternary complexes containing [γ-32P]-GTP and ribosomes programmed with f[35S]-Met-tRNAfMet, respectively). The results that we describe with the initiation complexes were corroborated using dipeptidyl-tRNA programmed “elongation” complexes (see end of Results section).
Before carrying out the detailed kinetic analysis, we set out to determine whether the fidelity phenotypes observed in vivo could be recapitulated in our in vitro system. This was accomplished by conducting a competition experiment wherein equal amounts of ternary complexes carrying either the cognate Phe-tRNAPhe or the near-cognate Leu-tRNA2Leu were incubated with the f-Met-tRNAfMet-programmed ribosomes. We measured the overall error-frequency by determining the amount of dipeptide fMet-Leu relative to fMet-Phe. Consistent with previous in vivo analysis, the error-frequencies on the WT, rpsD(ram), and rpsL(res) ribosomes were measured to be 1.6 ×10-3, 5.5 × 10-3, and 2.3 × 10-4 respectively (Figure 3B).
Stability of the Codon-Recognition Complex, k2 and k-2
Two distinct changes in fluorescence signal are observed as a result of mixing the ternary complex containing the proflavin-labeled aa-tRNA with the programmed ribosomes: an increase corresponding to the codon recognition step (k2), and a decrease corresponding to accommodation and/or rejection of aa-tRNA (k5 and k7, respectively) (Pape et al., 1998). The observed rate of codon recognition for the cognate aa-tRNA was determined to be quite similar for the WT, rpsD(ram) and rpsL(res) ribosomes (ranging from 25 to 40 s-1) (Table 1, Figure 4A and Supplemental Figure 1). We next observed that the apparent k2 for the near-cognate aa-tRNA with the WT ribosome was similar to that of cognate aa-tRNA (27 s-1), consistent with previous reports (Gromadski et al., 2006). Similarly, for both rpsD(ram) and rpsL(res), the observed k2 values for near-cognate aa-tRNA were not significantly different from those measured for the cognate aa-tRNA. We note that the second phase of the fluorescence profile (the observed decrease) revealed interesting information about the three ribosomal variants; these data will be discussed later as they pertain to the proofreading phase of tRNA selection.
Table 1. Observed rates for cognate and near-cognate aa-tRNAs on variant ribosomes.
| App. rate (s-1) | WT | rpsD(ram) | rpsL(res) | |||
|---|---|---|---|---|---|---|
| Cognate | Near Cognate | Cognate | Near Cognate | Cognate | Near Cognate | |
| k2 | 40 ± 6.5 | 27 ± 8.6 | 37 ± 2.1 | 36 ± 2.9 | 27 ± 6.5 | 25 ± 2.3 |
| k-2 | 0.50 ± 0.0070 | 47 ± 2.1 | 0.43 ± 0.0020 | 4 ± 0.1 | 0.41 ± 0.0080 | 46 ± 1.9 |
| kGTP | 25 ± 1.8 | 1.2 ± 0.12 | 31 ± 1.5 | 3.9 ± 0.25 | 14 ± 0.60 | 0.49 ± 0.029 |
| kpep | 8.5 ± 0.85 | 0.71 ± 0.13 | 7.7 ± 0.87 | 0.62 ± 0.19 | 4.8 ± 0.52 | > 0.49* |
| Fp | > 0.9 | 0.05 ± 0.002 | > 0.9 | 0.05 ± 0.004 | > 0.9 | 0.007 ± 0.005 |
| k5 + k7** | 4.6 ± 0.57 | 1.4 ± 1.0 | 5.8 ± 0.87 | 1.4 ± 0.4 | 1.6 ± 0.4 | 0.34 ± 0.16 |
Rates were determined at 1 μM initiation complex in polymix buffer at 20°C.
Limited by kGTP.
As determined from fluorescence stopped-flow experiments.
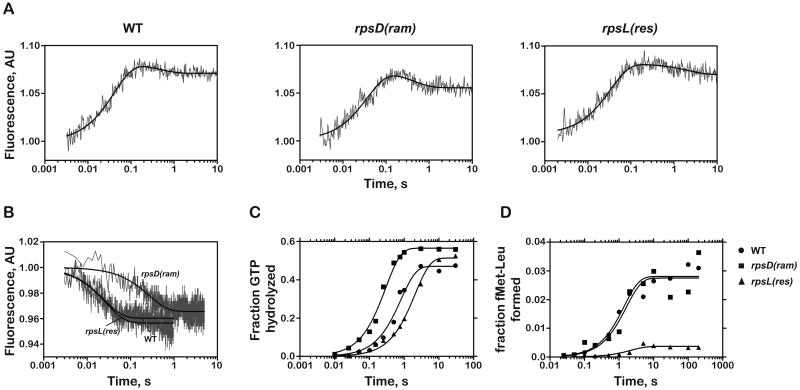
Figure 4. Kinetic parameters for near-cognate aa-tRNAs on variant ribosomes.
(A) Determination of apparent k2 and relative amount of aa-tRNA that goes through initial selection by monitoring fluorescence changes in Leu-tRNA2Leu (Prf16/17/20) on each of three ribosomal variants. Ternary complex containing labeled aa-tRNA (0.15 μM after mixing) was rapidly mixed with ICs (1 μM after mixing) displaying the Phe UUC codon in the A site. The resulting change in fluorescence signal was fit to double exponential kinetics (smooth lines).
(B) Determination of k-2. The time courses were obtained by first generating codon-recognition complexes with the GTPase-deficient EFTu H84A (0.3 μM after mixing) and then rapidly mixing it with a 10-fold excess of ternary complex containing unlabeled Phe-tRNAPhe.
(C) Time courses of GTP hydrolysis with the ternary complex EFTu:GTP:Leu-tRNA2Leu with 1 μM ICs.
(D) Time courses of dipeptide formation. Ternary complex was added to a final concentration of 0.5 μM (thus the maximal end-point is 0.5).
Next we set out to assess the effects of the ribosome mutations on the dissociation rate of the ternary complex from the codon-recognition complex (k-2). To measure this rate, the selection process was halted at the codon recognition step (with the fluorescently labeled aa-tRNA) by preventing subsequent steps from taking place through the utilization of a GTPase deficient EFTu (H84A) (Daviter et al., 2003). The dissociation reaction was initiated by rapidly mixing the complex with unlabeled cognate ternary complex in a stopped-flow spectrophotometer where the decrease in fluorescence signal could be monitored. As previously observed, WT ribosomes discriminated effectively against near-cognate aa-tRNA at this step (Gromadski and Rodnina, 2004; Pape et al., 1999); the measured rates for cognate and near cognate aa-tRNAs were 0.50 s-1, and 47 s-1, respectively (Figure 4B, Table 1 and Supplemental Figure 1). While the same extent of discrimination at the codon recognition step (k-2) was observed for the rpsL(res) ribosomes (0.41 s-1 vs. 46 s-1 for cognate and near-cognate aa-tRNAs, respectively), the rpsD(ram) ribosomes were significantly less effective at discriminating against near-cognate aa-tRNAs (0.43 s-1 vs. 4.0 s-1 for cognate and near-cognate aa-tRNAs, respectively). These data provide our first indication of how the rpsD(ram) ribosomes decrease the overall fidelity of the tRNA selection process.
GTPase Activation, k3
We next determined the effects of the ribosome mutations on the rate-limiting GTPase activation step k3 by measuring the rate of GTP hydrolysis (Gromadski and Rodnina, 2004; Pape et al., 1999). Previous studies with WT ribosomes showed that conformational changes induced by cognate aa-tRNA stimulate the rate of k3 by a factor of 50-200 fold over those observed with near-cognate aa-tRNA (Gromadski et al., 2006). The observed rate of GTPase activation (k3) for the cognate Phe-tRNAPhe ternary complex (25 s-1) was ∼20-fold faster than that for the near-cognate Leu-tRNA2Leu complex (1.2 s-1) (Figure 4C and Table 1), consistent with earlier studies performed at similar concentrations (Gromadski and Rodnina, 2004).
While the hyperaccurate ribosomes exhibited slightly reduced rates of GTP hydrolysis for both cognate and near-cognate ternary complexes (14 s-1 and 0.5 s-1, respectively), the difference in these rates is about 30-fold which is quite similar to the differences observed with WT ribosomes. The reduced rate for cognate ternary complex was previously reported by Ehrenberg and colleagues (Bilgin et al., 1992), but the rate for near-cognate aa-tRNA had not been measured. These data thus indicate that the rpsL(res) ribosomes do not modulate the GTPase activation step (k3) to discriminate against near-cognate aa-tRNAs.
In contrast, the rpsD(ram) ribosomes do appear to utilize the GTPase activation step (k3) to increase miscoding. The observed rate of GTPase activation for cognate aa-tRNA on the rpsD(ram) ribosomes was similar to that of WT (31 s-1 vs. 25 s-1, respectively), while the rate for near-cognate aa-tRNA was significantly accelerated (4 s-1 for WT vs. 1.2 s-1 for rpsD(ram)). This observed 3-fold increase in k3 is expected to contribute to the miscoding seen with the rpsD(ram) ribosomes. These observations, together with the earlier finding that the error-prone ribosomes alter the dissociation rate of the codon recognition complex (k-2), indicate that the rpsD(ram) ribosomes utilize the initial phase of the selection process for reducing discrimination against near-cognate aa-tRNAs.
Peptide-Bond Formation and Proofreading
We next assessed the effects of the rpsL(res) and rpsD(ram) ribosome mutations on the proofreading phase of tRNA selection. For this we followed the rate and extent of dipeptide formation with cognate and near-cognate aminoacyl-tRNAs on our standard initiation complexes. In this assay, the rate of peptide-bond formations (kpep) reports on the sum of the forward rate of accommodation (k5) and the rate of rejection of aa-tRNA (k7), while the endpoint of the reaction reveals the effectiveness of proofreading Fp (equal to k5/(k5+k7)) (Figure 1 and (Gromadski and Rodnina, 2004)). Normally during tRNA selection, the ribosome utilizes differences in both kpep and Fp to efficiently discriminate against near-cognate aa-tRNAs.
To measure these parameters in the context of our variant ribosomes, ribosome initiation complexes (1 μM final concentration) carrying radio-labeled f-[35S]-Met-tRNAfMet in the P site, were reacted with limiting concentrations (0.5 μM final concentration) of either cognate or near-cognate ternary complexes in a quench-flow apparatus. For each ribosome variant (WT, rpsL(res) and rpsD(ram)), the reaction proceeded to completion with the cognate aa-tRNA (Supplemental Figure 1), indicating that the mutations do not affect the rate of cognate aa-tRNA drop off (k7) during the proofreading phase. In addition, the rate of the reaction (kpep) for all three variants was similar, ranging from 5 to 8 s-1. These values are generally consistent with those previously reported (Gromadski and Rodnina, 2004).
Equivalent reactions with near-cognate aa-tRNAs exhibited interesting behaviors that help to explain the fidelity phenotype of the rpsL(res), but not the rpsD(ram), ribosomes. In their interaction with near-cognate aminoacyl-tRNAs, the rpsD(ram) ribosomes display very similar rates of peptidyl transfer (kpep of ∼0.7 s-1) and Fp (∼0.05) (Figure 4D). These findings indicate that contributions from proofreading to the overall selectivity are the same for the WT and rpsD(ram) ribosomes (∼ a 20-fold contribution). We note that the Fp value of 0.05 is identical to the one reported by Rodnina and colleagues under their different high-fidelity conditions (Gromadski and Rodnina, 2004).
In contrast, the rpsL(res) ribosomes were found to be aggressive proofreaders, as previously reported (Bohman et al., 1984). Whereas the rate of peptide-bond formation (kpep) appeared to be limited by that of GTPase activation (∼0.5 s-1) and did not differ significantly from the WT rate, the end-point was almost one order of magnitude lower than the value determined for both the WT and rpsD(ram) ribosomes (0.007 vs. 0.05, Table 1). These findings indicate that rpsL(res) ribosomes rely heavily on the proofreading stage (∼ a 150-fold contribution) to increase the accuracy of tRNA selection.
These findings were corroborated by similar experiments using different tRNAs (Supplemental Figure 2) and by the fluorescence stopped-flow experiment we described earlier that evaluated the codon-recognition step of tRNA selection (Figure 4A). In these experiments, the initial increase in fluorescence signal corresponds to the formation of codon recognition complex, while the subsequent decrease in signal reflects the accommodation and/or dissociation of aa-tRNA during the proofreading stage. For the WT and rpsD(ram) ribosomes with near-cognate ternary complexes, similar rates were observed during the second fluorescence phase (∼ 1.5 s-1), agreeing roughly with the measured rate of peptide-bond formation (∼0.8 s-1); agreement between the two sets of experiments was also seen with the rpsL(res) mutant (0.4 s-1 and 0.5 s-1 for the stopped-flow and quench data, respectively). Furthermore, the amplitude change for this phase with the near-cognate aa-tRNA on the rpsD(ram) mutant was observed to be higher than on the WT or rpsL(res) ribosomes (Figure 4A).
Kinetic analysis with elongation ribosome complexes yields similar results
To address concerns that initiation complexes might not be optimal for the analysis of tRNA-selection parameters, we assembled ribosome “elongation” complexes with dipeptidyl-tRNA bound in the P site (referred to as ribosomal nascent chains, or RNCs) and deacylated tRNAMet bound in the E site (Remme et al., 1989). We then evaluated the various tRNA-selection parameters for the WT, rpsD(ram) and rpsL(res) ribosomes and compared them with those determined for the initiation complexes above. Dipeptidyl RNCs were prepared by reacting initiation complexes carrying an AAA codon in the A site with Lys-tRNALys ternary complex (EFTu:GTP:Lys-tRNALys) to form f-Met-Lys-tRNALys-programmed ribosomal complexes. Following translocation with elongation factor G (EFG), the dipeptidyl-tRNA ribosomes complexes displaying different codons in the A site - cognate UUU (MKF) or near-cognate UUG (MKL) - were reacted with EFTu:GTP:Phe-tRNAPhe ternary complex.
The rates for the WT and rpsD(ram) RNCs displaying a cognate A-site codon (MKF) were almost identical (Table 2). Furthermore, similar to the effects seen with initiation complexes, the rate of ternary complex dissociation from the rpsD(ram) near-cognate codon recognition RNC (MKL) decreased by more than 20-fold relative to that observed with the WT RNC (1.4 s-1 vs. 34 s-1, respectively). Additionally, the rate of GTP hydrolysis was slightly increased in the rpsD(ram) RNC (MKL) with near-cognate tRNA (from 0.3 s-1 for WT to 0.7 s-1 for the rpsD(ram)). The rpsD(ram) mutation, as previously observed, had little to no effect on the proofreading phase of tRNA selection (Table 2). Also consistent with our earlier observations, the rpsL(res) ribosomes appear not to modulate the initial selection phase, but instead modulate the proofreading phase of tRNA selection (see Table 2).
Table 2. Observed rates for cognate and near-cognate RNCs.
| App. rate (s-1) | WT | rpsD(ram) | rpsL(res) | |||
|---|---|---|---|---|---|---|
| Cognate | Near Cognate | Cognate | Near Cognate | Cognate | Near Cognate | |
| k2 | 32 ± 4.6 | 26 ± 5.4 | 28 ± 4.9 | 30 ± 5.9 | 27 ± 4.2 | 32 ± 4.4 |
| k-2 | 0.50 ± 0.013 | 34 ± 1.5 | 0.47 ± 0.05 | 1.4 ± 0.065 | 0.78 ± 0.010 | 18 ± 0.9 |
| kGTP | 18 ± 1.6 | 0.29 ± 0.022 | 18 ± 1.5 | 0.71 ± 0.084 | 13 ± 0.88 | 0.06 ± 0.01 |
| kpep | 6.9 ± 0.47 | 0.2 ± 0.004* | 9.2 ± 1.0 | 0.3 ± 0.01 | 4.6 ± 0.54 | ND** |
| Fp | > 0.9 | 0.04 ± 0.004 | > 0.9 | 0.1 ± 0.02 | 0.65 ± 0.02 | ND** |
| k5 + k7*** | 5.0 ± 1.1 | 0.16 ± 0.060 | 4.4 ± 0.61 | 0.21 ± 0.090 | 1.8 ± 0.4 | 0.20 ± 0.14 |
Rates were determined at 1 μM initiation complex in polymix buffer at 20°C.
Limited by kGTP.
ND indicates that the values were too low to be determined.
As determined from fluorescence stopped-flow experiments
Peptide Release
We next examined the effects of the ribosome mutations on the related step of peptide release as catalyzed by class 1 release factors (RF). As for tRNA selection, both release factors (RF1 and RF2) have been shown to depend on a kinetic discrimination mechanism, where maximal rates of release (khyd) on authentic stop codons can be as high as 1000 fold faster than those on near-cognate-stop codons (Freistroffer et al., 2000). While both tRNAs and RFs interact with the same functional centers on the ribosome, it remains unclear how conformational changes in the decoding center induced by RFs are capable of causing such dramatic changes in catalysis some 70 Å away. More interestingly perhaps, the rpsL141 and rpsD12 strains were identified through screens involving nonsense suppression, where effects on either RF or tRNA selection might be critical. Here we can evaluate whether the rpsL(res) and rpsD(ram) ribosomes exhibit similar defects in RF selection, and thus can address how similar or different the processes of tRNA and RF selection might be.
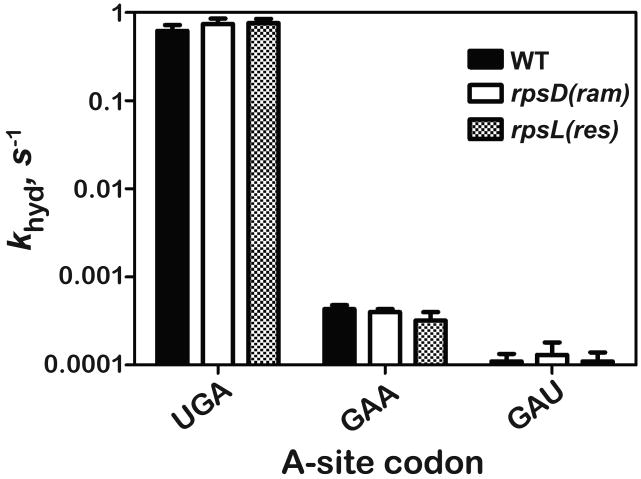
We examined the effects of the rpsD(ram) and rpsL(res) mutations on the maximal rate of peptide release (khyd) by employing a dipeptide-release assay (Brunelle et al., 2008). Briefly, ribosomes were programmed with dipeptidyl-tRNA (f[35S]-Met-Lys-tRNALys) in the P site on their cognate codon (AAA) with either the RF2-cognate-stop codon (UGA), near-cognate (GAA) or non-cognate (GAU) in the A site. As expected, RF2 effectively discriminated against non-stop codons on WT ribosomes; the rate of peptide release on the near-cognate (GAA) codon was more than three orders of magnitude slower than that seen on the stop codon, and that on the non-cognate (GAU) codon was barely detectable (Figure 5). Somewhat surprisingly, the khyd values measured for the rpsL(res) and rpsD(ram) ribosomes were almost indistinguishable from those measured for WT, suggesting that the S4/S5 and S12/h44/h27 interfaces make no discernible contributions to the catalysis of peptide release. Although a role for these interfaces during the binding phase of RF to the ribosome cannot be ruled out, the conformation of the protein once bound to the ribosome, just preceding catalysis, is likely not affected by changes at these interfaces.
Figure 5. Effects of the ribosomal mutations on the rates of peptide release.
Dipeptidyl-tRNA RNCs carrying f-Met-Lys-tRNALys (∼200 nM) in the P site, and displaying the indicated A-site codon were reacted with RF2 - 10 μM for the stop-codon (UGA) complex, and 100 μM for the near-stop (GAA) and non-stop (GAU) complexes. Rates plotted are the average of two independent measurements, obtained from single exponential fitting of time courses. The error bars represent the standard deviation from the mean.
Discussion
Our detailed kinetic analysis of classical restrictive (rpsL141) and ram (rpsD12) ribosomes has uncovered interesting features about the decoding process and its relation to ribosomal mutations proposed to alter the equilibrium between the closed and open states of the ribosome. Because the ram strains were isolated as suppressors of the SmD phenotype (typically exhibited by the most restrictive ribosomes) (Brownstein and Lewandowein, 1967), the initial prediction was that ribosomes from the ram strain would inversely affect the same steps of the tRNA selection pathway as the restrictive ribosomes. In contrast, we observe disparate kinetic effects on the pathway conferred by these mutations in the S4 and S12 proteins. As it turns out, the rpsD(ram) ribosomes characterized here exclusively alter steps involved in the initial phase of the pathway (initial selection, Figure 4B), and in particular the stability of the codon-recognition complex and subsequent step of GTPase activation. As established by earlier work, WT ribosomes discriminate against near-cognate aa-tRNAs through accelerated dissociation rates (k-2) and reduced rates of GTPase activation (k3) relative to cognate aa-tRNA. Here, we show that rpsD12 ram ribosomes (with a mutated S4 protein) exhibit significant differences in these steps; k-2 for near-cognate ternary complex was reduced by more than one order of magnitude (from 47 s-1 to 4 s-1 for WT and rpsD(ram) ribosomes, respectively), while k3 was found to be more than 3-fold higher on rpsD(ram) than on WT ribosomes (3.9 s-1 vs. 1.2 s-1, respectively). While earlier studies suggested that ram ribosomes are “defective proofreaders” (Andersson and Kurland, 1983), our data instead indicate that the rpsD(ram) ribosomes exploit initial selection steps to elicit miscoding. These discrepancies are likely reconciled by differences in the nature of the assays (the earlier studies utilized steady state approaches that depended on a larger number of variables while here we used a pre-steady state approach). By contrast, the rpsL(res) ribosomes characterized here do not exploit the steps of initial selection, but instead exploit the proofreading phase to reduce the rate of misreading (with Fp values of 0.05 and 0.007 for WT and rpsL(res) ribosomes, respectively). Taken at face value, these data immediately indicate that the rpsD12 mutation does not directly suppress the effects of the rpsL141 mutation, but overcomes them through a distinct mechanism.
The idea that rpsD12 is not a “true” suppressor of rpsL141 (at least in terms of molecular mechanism) is corroborated by recent studies indicating that the classic ram mutations are not suppressors of the restrictive phenotype, but only of streptomycin-dependence. In one study, the peculiarities of which codon misreadings were affected by the S4 and S12 variant ribosomes suggested that the read-out was more complicated than might be expected from reciprocal effects on subunit closure (Kramer and Farabaugh, 2007). Furthermore, genetic studies found that SmD-compensatory mutations identified in rpsD in S. typhimirium were themselves restrictive, not ram. These results again argue that effects on miscoding may be secondary to those demanded by the screen (streptomycin-dependence) (Bjorkman et al., 1999).
Interestingly, the effects of the rpsD(ram) ribosomes on the tRNA selection steps are distinct from those seen with nonsense suppressor tRNAs and miscoding agents. For instance, several miscoding tRNA variants (the G24A tRNATrp Hirsh suppressor (Hirsh and Gold, 1971) and certain Ala tRNA variants) increase their acceptance into the A site through the acceleration of the forward rates of GTPase activation and accommodation, without altering the stability of the aa-tRNA on the ribosome (Cochella and Green, 2005; Ledoux et al., 2009; Murakami et al., 2009). More recent biochemical studies suggest that the action of the Hirsh variant tRNA is independent of the action of universally conserved nucleotides in the small subunit decoding center (Cochella et al., 2007), instead functioning to constitutively transduce signals from the decoding center to the GTPase activation center (GAC). On the other hand, the aminoglycoside paromomycin increases error-frequency during tRNA selection by altering all of the steps that govern discrimination during the pathway (k-2, k3, k5 and k7) (Pape et al., 2000). These observations have been reconciled by crystal structures of the 30S subunit bound to paromomycin in the presence of near-cognate tRNA mimics wherein closure of the subunit is seen, even in the absence of cognate codon:anticodon interactions (Ogle et al., 2002).
The measured values of the individual steps of tRNA selection for the different ribosomes can be used to provide reasonable estimates of the contributions of these mutations to overall error-frequency. The amplitude change in the fluorescence signal of the second phase observed during k2 measurements (Figure 4A) is correlated with the amount of ternary complex that goes through the initial selection process; the amplitude change with the near-cognate aa-tRNA on the rpsD(ram) mutant was observed to be higher than on the WT ribosomes. The error-frequency for the hyperaccurate mutant is readily determined, because it exploits the proofreading stage whose contribution to fidelity is reflected by the inverse of the Fp value. While this contribution was found to be ∼20 for the WT ribosomes, its value increased to ∼150 for the rpsL(res) ribosomes; this can be interpreted as a 7-fold improvement in the overall accuracy. These predicted alterations in the overall fidelity from calculations depending on individual rate constants nicely match the empirically determined values from our competition experiment (Figure 3B).
For the first time we have specifically investigated the effect of ram and restrictive mutations on peptide release. Recent crystallographic and biochemical studies have provided substantial insight into how stop codons are recognized by RFs, and how this process differs from that of sense codon recognition by aa-tRNAs. First, high-resolution structures have shown that in the presence of RF, the decoding center nucleotides adopt conformations that are different than those seen with bound cognate anticodon stem-loops (Korostelev et al., 2008; Laurberg et al., 2008; Weixlbaumer et al., 2008). Second, biochemical studies from our lab have revealed that certain ribosomal mutations and antibiotics, which have been amply documented to alter kinetic parameters in the tRNA selection pathway, have no effect on peptide release (Youngman et al., 2007). The observation that the two ribosomal mutations studied here (rpsL141 and rpsD12) have no effect on the rate of peptide release are consistent with these earlier studies. Moreover, these data suggest that molecular conformational switches on the ribosome might be quite distinct for these two processes of tRNA and RF selection.
Our findings have immediate ramifications for the structural rearrangements that are proposed to happen during the decoding process. The presence of a cognate tRNA in the A site is proposed to stabilize a transition from an “open” to a “closed” state, which is thought to increase the stability of ternary complex binding to the ribosome, and to activate EFTu for GTP hydrolysis and downstream events (Ogle et al., 2003). This “closed” form of the 30S subunit involves disruption of close contacts between S4 and S5, and the creation of new contacts between S12 and the 16S rRNA. While these structural observations come from a snap-shot of the small subunit at the end of the decoding process, we have little information about the sequence of events that lead to the ultimate “closed” state. As such, the simplest model has argued that the two interfaces (S4/S5 and S12/h44/h27) serve as opposite poles of a toggle, whose rearrangements are intimately correlated (Ogle et al., 2003).
Our data suggests that these interfaces may be altered in a temporal manner; mutations affecting the S4/S5 interface modulating only the initial binding phase of the tRNA selection pathway, with those affecting the S12/h44/h27 interface modulating the proofreading phase. We propose that the interface between S4 and S5 is broken during the codon-recognition step, activating EFTu for GTP hydrolysis, whereas the S12/h44/h27 interface forms subsequently, but as a requisite feature for the accommodation of aa-tRNA into the A site. The observed decoupling of GTPase activation and accommodation in our analysis suggest that the ribosome might depend on distinct molecular switches for initial selection and proofreading. Such a model is appealing as it provides distinct checkpoints by which the ribosome can evaluate the codon:anticodon interaction. It will be exciting to visualize such potential intermediates in the decoding process as functional ribosome complexes are resolved with increasing resolution (Schmeing et al., 2009; Vila-Sanjurjo et al., 2003).
Experimental Procedures
Materials and Reagents
Experiments were performed in polymix buffer (Jelenc and Kurland, 1979) [95 mM KCl, 5 mM NH4Cl, 5 mM Mg(OAc)2, 0.5 mM CaCl2, 8 mM putrescine, 1 mM spermidine, 5 mM potassium phosphate pH 7.5, 1 mM DTT].
Bacterial strains used in this study were: Xac (ara Δ[lacproAB] gyrA rpoB argEamber), US157 (Xac rspL141 zcg-174::Tn10) and UD 131 (Xac rpsD12) (Andersson et al., 1982). 70S ribosomes from these strains were prepared as described previously (Moazed and Noller, 1986). Bacterial translational factors were over-expressed and purified according to earlier methods (Zaher and Green, 2009b). Yeast S100 extract used for the charging of yeast tRNAPhe was prepared as described (Sampson and Uhlenbeck, 1988). tRNAGlu, tRNAPhe, tRNAfMet (all from E. coli) and yeast tRNAPhe were purchased from Sigma-Aldrich. tRNA2Leu was obtained from Subriden. Total E. coli tRNA was purchased from Roche. Aminoacylation of the E. coli tRNAs was performed as described earlier (Zaher and Green, 2009b), while that of yeast tRNAPhe is described elsewhere (Sampson and Uhlenbeck, 1988). mRNA templates were prepared from double-stranded DNA templates using run-off transcription by T7 RNA polymerase (Zaher and Unrau, 2004), and purified by PAGE. The resulting mRNA transcripts used for the initiation complexes shared the following sequence: UAAUACGACUCACUAUAGGGU UAACUUUAGAAGGAGGUAAAAAAA AUG X UUUUUCUUU, where X denotes the codon occupying the A site. Those used for dipeptidyl RNCs and peptide release had the following sequence: GGGUGUCUUGCGAGGAUAAGU GCAUU AUG (X) (Y) UUUGCCCUUCUGUA GCCA, where X and Y denote codons occupying the A and P sites, respectively.
Ribosome Complex Formation
Initiation complexes (ICs) were first prepared by incubating 70S ribosomes (2 μM) with IF1, IF2, IF3, f-[35S]-Met-tRNAfMet (3 μM each), and mRNA (6 μM) in polymix buffer in the presence of GTP at 2 mM at 37°C for 45 minutes. Dipeptidyl tRNA ribosomal nascent chains (RNCs), were prepared by mixing equivalent volumes of ICs and a pre-incubated elongation mixture containing EFTu (15 μM), charged tRNA (6 μM), EF-G (6 μM), and GTP (2 mM) and incubating at 37°C for 10 minutes. To purify both ICs and RNCs away from unincorporated tRNAs and factors, the reaction mixture was layered over a 1300 μL sucrose cushion (1.1 M sucrose, 20 mM Tris-HCl pH 7.5, 500 mM NH4Cl, 10 mM MgCl2, 0.5 mM EDTA) and spun at 69,000 rpm in a TLA100.3 rotor for 2 hours. The resulting pellet was resuspended in polymix buffer, aliquoted and stored at -80°C. The typical yield for the dipeptidyl RNCs was > 80% (i.e. amount of fMet that was converted to dipeptide). Additionally, the fractional radioactivity that pelleted was used to determine the concentration of the RNC.
Codon Recognition Assay
Prf16/17 yeast tRNAPhe and Prf16/17/20 E. coli tRNA2Leu were prepared as described earlier (Wintermeyer and Zachau, 1979). The procedure for determining apparent rates of ternary complex binding to ICs and the following steps of accommodation and/or rejection was adapted from ref (Pape et al., 1998), except that proflavin was excited at 465 nm. Briefly, ternary complexes containing; 10 μM EFTu, 0.3 μM labeled aa-tRNA and 2 mM GTP, were mixed with equal volumes of 2 μM ICs at 20°C on an SX-18MV stopped-flow spectrometer (Applied Photophysics). The resulting change in fluorescence signal was fit to double exponential kinetics after smoothing the data by averaging the nearest 4 neighbors. Rates shown in Table 1 are the average of at least four runs on the machine. k-2 was determined by first preparing a codon recognition complex (0.3 μM after mixing), where ternary complexes made with EFTu (H84A) were mixed with ICs on ice (to prevent slow accommodation observed with the EFTu mutant), and rapidly mixing with a 10 fold excess of ternary complex containing unlabeled aa-tRNA. These reactions were carried out in triplicates.
GTP Hydrolysis Assay
The protocol used to make ternary complexes containing [γ-32P]-GTP was similar to the one used in ref (Ledoux and Uhlenbeck, 2008). EFTu at 20 μM was incubated with 5 mCi/mL of [γ-32P]-GTP and 5 μM of unlabeled GTP for 15 minutes, before the addition of an equal volume of 30 μM aa-tRNA. After incubating the mixture for another 15 minutes, unbound GTP and aa-tRNA were purified away by passing the ternary complex twice over P-30 spin column (Biorad). The purified ternary complex was diluted to 1 μM in polymix buffer (0.5 μM final concentration in the reaction) and rapidly mixed with an equal volume of 2 μM ICs (1 μM after mixing) at 20°C in a quench-flow instrument (RQF-3 quench-flow, KinTek Corporation). The reactions were quenched by the addition of 40% formic acid. The products (inorganic phosphate) were resolved from unreacted GTP by PEI cellulose thin-layer chromatography with 0.5 M KH2PO4 pH 3.5 as a mobile phase. Fractional radioactivity corresponding to inorganic phosphate at each time-point was quantified using ImageQuant v5.2 (Molecular Dynamics) and plotted against time. Apparent rate constants were determined by signal exponential fitting.
Peptidyl Transferase (PT) assay
EFTu at 100 μM was first incubated with 2 mM GTP in polymix buffer for 15 minutes before diluting to 20 μM in polymix buffer containing E. coli Phe-tRNAPhe or Leu-tRNA2Leu (0.5 μM) and 2 mM GTP, followed by another 15 minute incubation. The mixture was rapidly mixed with an equal volume of complex (ICs or dipeptidyl RNCs, final concentration 1 μM) at 20°C in the quench-flow instrument. Reaction was stopped by the addition of KOH to a concentration of 500 mM. Dipeptide or tripetide reaction products were separated from unreacted fMet or dipeptides using cellulose TLC plates that were electrophoresed in pyridine-acetate at pH 2.8 (Youngman et al., 2004). Fractional radioactivity corresponding to the product at each time point was quantified and analyzed as described earlier.
Release Assay
Dipeptidyl RNCs at ∼200 nM, containing a cognate-stop codon in the P site, were incubated with RF2 at 10 μM (determined to be saturating) at 37°C in the quench-flow instrument. In reactions where the A site did not contain a cognate-stop codon, the concentration of RF2 was increased to 100 μM. These latter reactions were slow enough to be carried out on the bench. Time-points were obtained by taking aliquots at different time intervals and stopping the reaction with 5% formic acid. Released dipeptides were separated from unreacted peptidyl-tRNA by electrophoretic TLC as above, and analyzed similarly. In all cases, greater than 70% of the dipeptidyl-tRNA was hydrolyzed at the end of the reaction.
Supplementary Material
Acknowledgments
We thank P. Farabaugh for supplying bacterial strains and plasmids. We thank Nicholas R. Guydosh for reading the manuscript and members of the lab for useful discussions. The work was supported by a grant from the NIH and HHMI salary support to R.G. H.S.Z. is supported by NSERC.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andersson DI, Bohman K, Isaksson LA, Kurland CG. Translation rates and misreading characteristics of rpsD mutants in Escherichia coli. Mol Gen Genet. 1982;187:467–472. doi: 10.1007/BF00332630. [DOI] [PubMed] [Google Scholar]
- Andersson DI, Kurland CG. Ram ribosomes are defective proofreaders. Mol Gen Genet. 1983;191:378–381. doi: 10.1007/BF00425749. [DOI] [PubMed] [Google Scholar]
- Bilgin N, Claesens F, Pahverk H, Ehrenberg M. Kinetic properties of Escherichia coli ribosomes with altered forms of S12. J Mol Biol. 1992;224:1011–1027. doi: 10.1016/0022-2836(92)90466-w. [DOI] [PubMed] [Google Scholar]
- Birge EA, Kurland CG. Reversion of a streptomycin-dependent strain of Escherichia coli. Mol Gen Genet. 1970;109:356–369. doi: 10.1007/BF00267704. [DOI] [PubMed] [Google Scholar]
- Bjorkman J, Samuelsson P, Andersson DI, Hughes D. Novel ribosomal mutations affecting translational accuracy, antibiotic resistance and virulence of Salmonella typhimurium. Mol Microbiol. 1999;31:53–58. doi: 10.1046/j.1365-2958.1999.01142.x. [DOI] [PubMed] [Google Scholar]
- Bohman K, Ruusala T, Jelenc PC, Kurland CG. Kinetic impairment of restrictive streptomycin-resistant ribosomes. Mol Gen Genet. 1984;198:90–99. doi: 10.1007/BF00328706. [DOI] [PubMed] [Google Scholar]
- Brownstein Bl, Lewandowein LJ. A mutation suppressing streptomycin dependence. I. An effect on ribosome function. J Mol Biol. 1967;25:99–109. doi: 10.1016/0022-2836(67)90281-1. [DOI] [PubMed] [Google Scholar]
- Brunelle JL, Shaw JJ, Youngman EM, Green R. Peptide release on the ribosome depends critically on the 2′ OH of the peptidyl-tRNA substrate. RNA. 2008;14:1526–1531. doi: 10.1261/rna.1057908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter AP, Clemons WM, Brodersen DE, Morgan-Warren RJ, Hartsch T, Wimberly BT, Ramakrishnan V. Crystal structure of an initiation factor bound to the 30S ribosomal subunit. Science. 2001;291:498–501. doi: 10.1126/science.1057766. [DOI] [PubMed] [Google Scholar]
- Cochella L, Brunelle JL, Green R. Mutational analysis reveals two independent molecular requirements during transfer RNA selection on the ribosome. Nat Struct Mol Biol. 2007;14:30–36. doi: 10.1038/nsmb1183. [DOI] [PubMed] [Google Scholar]
- Cochella L, Green R. An active role for tRNA in decoding beyond codon:anticodon pairing. Science. 2005;308:1178–1180. doi: 10.1126/science.1111408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daviter T, Wieden HJ, Rodnina MV. Essential role of histidine 84 in elongation factor Tu for the chemical step of GTP hydrolysis on the ribosome. J Mol Biol. 2003;332:689–699. doi: 10.1016/s0022-2836(03)00947-1. [DOI] [PubMed] [Google Scholar]
- Freistroffer DV, Kwiatkowski M, Buckingham RH, Ehrenberg M. The accuracy of codon recognition by polypeptide release factors. Proc Natl Acad Sci USA. 2000;97:2046–2051. doi: 10.1073/pnas.030541097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorini L, Kataja E. Phenotypic repair by streptomycin of defective genotypes in E Coli. Proc Natl Acad Sci USA. 1964;51:487–493. doi: 10.1073/pnas.51.3.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromadski KB, Daviter T, Rodnina MV. A uniform response to mismatches in codon-anticodon complexes ensures ribosomal fidelity. Mol Cell. 2006;21:369–377. doi: 10.1016/j.molcel.2005.12.018. [DOI] [PubMed] [Google Scholar]
- Gromadski KB, Rodnina MV. Kinetic determinants of high-fidelity tRNA discrimination on the ribosome. Mol Cell. 2004;13:191–200. doi: 10.1016/s1097-2765(04)00005-x. [DOI] [PubMed] [Google Scholar]
- Hirsh D, Gold L. Translation of UGA triplet in-vitro by tryptophan transfer RNAs. J Mol Biol. 1971;58:459–468. doi: 10.1016/0022-2836(71)90363-9. [DOI] [PubMed] [Google Scholar]
- Hopfield JJ. Kinetic proofreading: a new mechanism for reducing errors in biosynthetic processes requiring high specificity. Proc Natl Acad Sci USA. 1974;71:4135–4139. doi: 10.1073/pnas.71.10.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelenc PC, Kurland CG. Nucleoside triphosphate regeneration decreases the frequency of translation errors. Proc Natl Acad Sci USA. 1979;76:3174–3178. doi: 10.1073/pnas.76.7.3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson M, Lovmar M, Ehrenberg M. Rate and accuracy of bacterial protein synthesis revisited. Curr Opin Microbiol. 2008;11:141–147. doi: 10.1016/j.mib.2008.02.015. [DOI] [PubMed] [Google Scholar]
- Kirthi N, Roy-Chaudhuri B, Kelley T, Culver GM. A novel single amino acid change in small subunit ribosomal protein S5 has profound effects on translational fidelity. RNA. 2006;12:2080–2091. doi: 10.1261/rna.302006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korostelev A, Asahara H, Lancaster L, Laurberg M, Hirschi A, Zhu J, Trakhanov S, Scott WG, Noller HF. Crystal structure of a translation termination complex formed with release factor RF2. Proc Natl Acad Sci USA. 2008;105:19684–19689. doi: 10.1073/pnas.0810953105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer EB, Farabaugh PJ. The frequency of translational misreading errors in E. coli is largely determined by tRNA competition. RNA. 2007;13:87–96. doi: 10.1261/rna.294907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurberg M, Asahara H, Korostelev A, Zhu J, Trakhanov S, Noller HF. Structural basis for translation termination on the 70S ribosome. Nature. 2008;454:852–857. doi: 10.1038/nature07115. [DOI] [PubMed] [Google Scholar]
- Ledoux S, Olejniczak M, Uhlenbeck OC. A sequence element that tunes Escherichia coli tRNA(Ala)(GGC) to ensure accurate decoding. Nat Struct Mol Biol. 2009;16:359–364. doi: 10.1038/nsmb.1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledoux S, Uhlenbeck OC. Different aa-tRNAs are selected uniformly on the ribosome. Mol Cell. 2008;31:114–123. doi: 10.1016/j.molcel.2008.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed D, Noller HF. Transfer RNA shields specific nucleotides in 16S ribosomal RNA from attack by chemical probes. Cell. 1986;47:985–994. doi: 10.1016/0092-8674(86)90813-5. [DOI] [PubMed] [Google Scholar]
- Murakami H, Ohta A, Suga H. Bases in the anticodon loop of tRNA(Ala)(GGC) prevent misreading. Nat Struct Mol Biol. 2009;16:353–358. doi: 10.1038/nsmb.1580. [DOI] [PubMed] [Google Scholar]
- Nierhaus KH. Decoding errors and the involvement of the E-site. Biochimie. 2006;88:1013–1019. doi: 10.1016/j.biochi.2006.02.009. [DOI] [PubMed] [Google Scholar]
- Ninio J. Multiple stages in codon-anticodon recognition: double-trigger mechanisms and geometric constraints. Biochimie. 2006;88:963–992. doi: 10.1016/j.biochi.2006.06.002. [DOI] [PubMed] [Google Scholar]
- Ninio J. Kinetic amplification of enzyme discrimination. Biochimie. 1975;57:587–595. doi: 10.1016/s0300-9084(75)80139-8. [DOI] [PubMed] [Google Scholar]
- Ogle JM, Carter AP, Ramakrishnan V. Insights into the decoding mechanism from recent ribosome structures. Trends Biochem Sci. 2003;28:259–266. doi: 10.1016/S0968-0004(03)00066-5. [DOI] [PubMed] [Google Scholar]
- Ogle JM, Murphy FV, Tarry MJ, Ramakrishnan V. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell. 2002;111:721–732. doi: 10.1016/s0092-8674(02)01086-3. [DOI] [PubMed] [Google Scholar]
- Ozaki M, Mizushim S, Nomura M. Identification and functional characterization of the protein controlled by the streptomycin-resistant locus in E. coli. Nature. 1969;222:333–339. doi: 10.1038/222333a0. [DOI] [PubMed] [Google Scholar]
- Pape T, Wintermeyer W, Rodnina M. Induced fit in initial selection and proofreading of aminoacyl-tRNA on the ribosome. EMBO J. 1999;18:3800–3807. doi: 10.1093/emboj/18.13.3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pape T, Wintermeyer W, Rodnina MV. Complete kinetic mechanism of elongation factor Tu-dependent binding of aminoacyl-tRNA to the A site of the E-coli ribosome. EMBO J. 1998;17:7490–7497. doi: 10.1093/emboj/17.24.7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pape T, Wintermeyer W, Rodnina MV. Conformational switch in the decoding region of 16S rRNA during aminoacyl-tRNA selection on the ribosome. Nat Struct Biol. 2000;7:104–107. doi: 10.1038/72364. [DOI] [PubMed] [Google Scholar]
- Remme J, Margus T, Villems R, Nierhaus KH. The third ribosomal tRNA-binding site, the E site, is occupied in native polysomes. Eur J Biochem. 1989;183:281–284. doi: 10.1111/j.1432-1033.1989.tb14925.x. [DOI] [PubMed] [Google Scholar]
- Rodnina MV, Fricke R, Kuhn L, Wintermeyer W. Codon-dependent conformational change of elongation factor Tu preceding GTP hydrolysis on the ribosome. EMBO J. 1995;14:2613–2619. doi: 10.1002/j.1460-2075.1995.tb07259.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosset R, Gorini L. A ribosomal ambiguity mutation. J Mol Biol. 1969;39:95–112. doi: 10.1016/0022-2836(69)90336-2. [DOI] [PubMed] [Google Scholar]
- Sampson JR, Uhlenbeck OC. Biochemical and physical characterization of an unmodified yeast phenylalanine transfer RNA transcribed in vitro. Proc Natl Acad Sci USA. 1988;85:1033–1037. doi: 10.1073/pnas.85.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmeing TM, Voorhees RM, Kelley AC, Gao YG, Murphy FV, 4th, Weir JR, Ramakrishnan V. The crystal structure of the ribosome bound to EF-Tu and aminoacyl-tRNA. Science. 2009;326:688–694. doi: 10.1126/science.1179700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoffler G, Deusser E, Wittmann HG, Apirion D. Ribosomal proteins. XIX. Altered S5 ribosomal protein in an Escherichia coli revertant from streptomycin dependence to independence. Mol Gen Genet. 1971;111:334–341. doi: 10.1007/BF00569785. [DOI] [PubMed] [Google Scholar]
- Thompson RC. EFTu provides an internal kinetic standard for translational accuracy. Trends Biochem Sci. 1988;13:91–93. doi: 10.1016/0968-0004(88)90047-3. [DOI] [PubMed] [Google Scholar]
- Vallabhaneni H, Farabaugh PJ. Accuracy modulating mutations of the ribosomal protein S4-S5 interface do not necessarily destabilize the rps4-rps5 protein-protein interaction. RNA. 2009;15:1100–1109. doi: 10.1261/rna.1530509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vila-Sanjurjo A, Ridgeway WK, Seymaner V, Zhang W, Santoso S, Yu K, Cate JH. X-ray crystal structures of the WT and a hyper-accurate ribosome from Escherichia coli. Proc Natl Acad Sci USA. 2003;100:8682–8687. doi: 10.1073/pnas.1133380100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weixlbaumer A, Jin H, Neubauer C, Voorhees RM, Petry S, Kelley AC, Ramakrishnan V. Insights into translational termination from the structure of RF2 bound to the ribosome. Science. 2008;322:953–956. doi: 10.1126/science.1164840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimberly BT, Brodersen DE, Clemons WM, Morgan-Warren RJ, Carter AP, Vonrhein C, Hartsch T, Ramakrishnan V. Structure of the 30S ribosomal subunit. Nature. 2000;407:327–339. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- Wintermeyer W, Zachau HG. Fluorescent derivatives of yeast tRNAPhe. Eur J Biochem / FEBS. 1979;98:465–475. doi: 10.1111/j.1432-1033.1979.tb13207.x. [DOI] [PubMed] [Google Scholar]
- Youngman EM, Brunelle JL, Kochaniak AB, Green R. The active site of the ribosome is composed of two layers of conserved nucleotides with distinct roles in peptide bond formation and peptide release. Cell. 2004;117:589–599. doi: 10.1016/s0092-8674(04)00411-8. [DOI] [PubMed] [Google Scholar]
- Youngman EM, He SL, Nikstad LJ, Green R. Stop codon recognition by release factors induces structural rearrangement of the ribosomal decoding center that is productive for peptide release. Mol Cell. 2007;28:533–543. doi: 10.1016/j.molcel.2007.09.015. [DOI] [PubMed] [Google Scholar]
- Zaher HS, Green R. Fidelity at the molecular level: lessons from protein synthesis. Cell. 2009a;136:746–762. doi: 10.1016/j.cell.2009.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaher HS, Green R. Quality control by the ribosome following peptide bond formation. Nature. 2009b;457:161–166. doi: 10.1038/nature07582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaher HS, Unrau PJ. T7 RNA polymerase mediates fast promoter-independent extension of unstable nucleic acid complexes. Biochemistry. 2004;43:7873–7880. doi: 10.1021/bi0497300. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.