Abstract
Profound chromatin changes occur during mitosis to allow for gene silencing and chromosome segregation followed by re-activation of memorized transcription states in daughter cells. Using genome-wide sequencing, we found H2A.Z containing +1 nucleosomes of active genes shift upstream to occupy TSSs during mitosis, significantly reducing nucleosome-depleted regions. Single molecule analysis confirmed nucleosome shifting and demonstrated that mitotic shifting is specific to active genes that are silenced during mitosis and thus is not seen on promoters, which are silenced by methylation or mitotically expressed genes. Using the GRP78 promoter as a model, we found H3K4 tri-methylation is also maintained while other indicators of active chromatin are lost and expression is decreased. These key changes provide a potential mechanism for rapid silencing and re-activation of genes during the cell cycle.
Highlights.
Global H2A.Z patterns are maintained during mitosis
+1 Nucleosomes shift during mitosis reducing nucleosome-depleted regions
+1 Nucleosome shifting during mitosis is restricted to transiently silenced promoters
Introduction
Eukaryotic genomes are organized into repeating arrays of nucleosomes, which consist of 146 base pairs (bp) of DNA wrapped around a histone core comprised of two copies of H3, H4, H2A and H2B proteins. DNA methylation, histone variants and modifications and nucleosome positioning work together to define the epigenetic landscape of a cell. For example, nucleosome occupancy at transcriptional start sites (TSSs) of genes make DNA inaccessible to transcription machinery and must be remodeled for transcription activation (Li et al., 2007; Mellor, 2005). The region immediately upstream of the TSSs of active genes is depleted of stable nucleosomes, generating a nucleosome-depleted region (NDR), which allows transcription factor binding and formation of the pre-initiation complex. Thus the presence of an NDR is critical for active transcription (Gal-Yam et al., 2006; Jiang and Pugh, 2009; Lee et al., 2004; Lin et al., 2007; Mito et al., 2005). This NDR is bordered by nucleosomes containing the H2A variant, H2A.Z (Guillemette and Gaudreau, 2006), which preferentially marks active genes and genes that are poised for activation (Barski et al., 2007; Creyghton et al., 2008) and is not present on promoters that are silenced by DNA methylation in plants (Zilberman et al., 2008). The presence of H2A.Z can stabilize or destabilize nucleosomes depending on which other histone proteins are present in the nucleosome (Jin and Felsenfeld, 2007). Thus DNA methylation, histone variants and modifications and nucleosome positioning work together and generate active or inactive chromatin configurations.
Cell division requires choreographed processes of DNA replication (S-phase) followed by chromatin condensation and distribution of duplicated DNA into two daughter cells (M-phase or mitosis). During mitosis, the nuclear envelope breaks down and there is global phosphorylation of serine residues on histone 3, which enables chromatin condensation (Hans and Dimitrov, 2001; Nowak and Corces, 2004; Van Hooser et al., 1998) and histone de-acetylation, which allows for stabilization of higher order chromatin structure (Horn and Peterson, 2002). Ultimately, chromatin condenses in a multistep process involving condensin complexes and topoisomerase 2, resulting in two sister chromatids that are joined at a centromere from which the duplicated DNA will separate (Belmont, 1997; Belmont, 2006). While the global changes to chromatin structure are well established, a detailed understanding of the alterations that occur at specific chromatin regulatory regions during mitosis is still unclear.
As a result of this profound condensation and global phosphorylation of transcription factors, both of which prevent DNA binding, almost all gene transcription is shut off during mitosis, except for those that are necessary for cell cycle progression (Gottesfeld and Forbes, 1997; Prescott and Bender, 1962). Upon mitotic exit, memorized transcription states must be re-established in daughter cells and for some genes, rapid re-expression is necessary for proper cellular function and survival. The mechanisms by which gene expression patterns are re-established after mitotic exit are still under investigation. For several years, it has been known that subsets of genes can be marked for rapid re-expression following division, by continued TATA binding protein (TBP) and transcription factor binding (Verdeguer et al., 2010; Xing et al., 2008; Xing et al., 2005; Young et al., 2007). However, recently the role of specific histone proteins and their modifications have received significant attention and been shown to impact reactivation of gene expression following mitotic exit (Blobel et al., 2009; Dressler, 2010; Verdeguer et al., 2010).
Studies of individual gene promoters have demonstrated the variable maintenance of H3K4 methylation and H3 and H4 acetylation along with increases in H3K79 dimethylation at gene promoters and coding regions during mitosis (Kouskouti and Talianidis, 2005; Valls et al., 2005). However, more recent studies have reported inconsistent results when examining broader patterns of H3K4 tri-methylation and MLL (a histone methyltransferase which catalyzes H3K4 tri-methylation) maintenance during mitosis. For example, Mishra et al., reported H3K4 tri-methylation maintenance on mitotic chromatin, while MLL binding was lost (Mishra et al., 2009). In contrast, Blobel et al., demonstrated that MLL binding was maintained during mitosis and furthermore, it was re-distributed to genes that were highly expressed in interphase and contributed to reactivation following mitotic exit (Blobel et al., 2009).
This previous work has established a role for maintenance of transcription factor binding and histone modifications in mitotic inheritance of gene expression patterns. The importance of these two mechanisms in retaining gene expression patterns was recently highlighted by Dressler, which also pointed out the potential for locus specificity of such marks (Dressler, 2010). Since epigenetic mechanisms work together with each other and transcriptional machinery to regulate gene expression, it is important to examine the role of additional epigenetic mechanisms in regulating cellular memory of transcription states. However, the role of histone variants and nucleosome positioning in maintaining gene expression patterns through cell divisions has largely been overlooked.
In addition to playing a structural role, nucleosomes can enhance or inhibit gene transcription. Nucleosomes can bring distal regulatory regions in close proximity to promoters to initiate or enhance transcription, yet they can also block access of DNA binding proteins and transcriptional machinery, thereby inhibiting expression. A previous study which examined nucleosome positioning between the proximal promoters and enhancers of c-FOS and U6, found that the well positioning of the nucleosome within this region was lost during mitosis leading to the conclusion that nucleosome occupancy did not play a role in regulating gene expression during the cell cycle (Komura and Ono, 2005). However, well-positioned nucleosomes are also found after the TSS of actively transcribed genes (Fatemi et al., 2005; Gal-Yam et al., 2006; Lin et al., 2007), whether there are changes in positioning of these +1 nucleosomes is unknown. Furthermore, it is unclear whether the NDRs upstream of TSSs of mitotically silent genes are maintained in the context of condensed chromatin that occurs during mitosis, potentially allowing for rapid re-activation following mitotic exit.
In this study, we used genome-wide ChIP-seq combined with high-resolution single molecule analysis to examine changes in nucleosome composition and positioning at TSSs during mitosis. We found that H2A.Z is maintained during mitosis and marks the +1 nucleosome of active genes, which shifts during mitosis resulting in occupancy at the TSS and a reduced NDR. These key changes provide a potential mechanism for rapid silencing and re-activation of genes during the cell cycle.
Results
H2A.Z Containing Nucleosomes Surrounding the TSSs of Active Genes Shift During Mitosis
In mammals, the H2A variant, H2A.Z, is preferentially localized to nucleosomes near the TSSs of active genes (Barski et al., 2007) and is maintained during mitosis in murine L929 cells (Bruce et al., 2005). Therefore we probed promoter architecture during G0/G1 and M-phase using chromatin immunoprecipitation (ChIP) for H2A.Z followed by Solexa sequencing, deriving good sequence coverage around TSSs, which allowed for accurate comparative genome-wide analysis of the H2A.Z patterns. We obtained 12,830,000 alignable reads from M-phase cells and 8,630,000 alignable reads from G0/G1 cells. Global H2A.Z patterns were similar in G0/G1 and M-phase, chromosome region 9q33.3 shown as an example (Fig. 1a). In this region, H2A.Z was localized to the TSSs of active but not inactive genes, with additional peaks in H2A.Z signal, which do not correlate with known TSSs, potentially marking chromatin regulatory regions, like enhancers and CTCF binding sites (Jin et al., 2009). The mitotic maintenance of H2A.Z within nucleosomes near TSSs of active genes enables analysis of changes in chromatin structure that occur specifically at TSSs of active genes that are temporarily silenced during mitosis.
Figure 1. Genome-wide analysis by ChIP-seq shows that H2A.Z localization is maintained during mitosis and preferentially marks +1 nucleosomes of active genes, which shift upstream to occupy the TSS during mitosis, shrinking the NDR.

H2A.Z ChIP-seq reads from G0/G1 and M-phase cells were aligned to the Human Genome. (A) Low resolution example demonstrates that global H2A.Z localization is similar during G0/G1 and M-phase. Significant H2A.Z signal is found at the TSSs of expressed (*), but less so at silent (§) genes. In addition, H2A.Z is also present at regions that do not correspond to known TSS (†). (B) Well-positioned +1 nucleosomes of active genes shift upstream to occupy the TSS during mitosis. The size of the shift is promoter specific resulting in a broad peak during M-phase (dashed ovals). (C) Genes, which are not expressed, or expressed at low levels contain little H2A.Z during both M-phase and G0/G1. Forward and reverse reads (solid and dashed lines, respectively) correspond to orientation to the genome. Reads were normalized to the number of promoters analyzed. TSS in b & c are indicated by yellow lines.
In humans, -2 and +1 nucleosomes border the NDR while the -1 nucleosome refers to the unstable H3.3/H2A.Z nucleosome that is seldom-present within the NDR (Jiang and Pugh, 2009; Jin et al., 2009; Schones et al., 2008). H2A.Z is present in both the -2 and +1 nucleosomes of active promoters (Fig. 1b). Sequencing reads were aligned based on distance from the TSS revealing that the +1 nucleosome positioned after the TSS in G0/G1 cells slides upstream to occupy the TSS during mitosis (Fig. 1b). The 3′ end of the -2 nucleosome also shifts during mitosis, shrinking the NDR. Thus, during mitosis, localization of the -2 and +1 nucleosomes changes resulting in a shortening of the NDR. The size of the +1 nucleosome shift is promoter specific resulting in a broad peak corresponding to its localization during mitosis (Fig. 1b). In contrast, little H2A.Z was present surrounding TSSs of genes which have little to no expression (Fig. 1c) consistent with preferential localization of H2A.Z to active promoters (Barski et al., 2007). While global H2A.Z patterns are maintained at active promoters during mitosis the specific localization of H2A.Z containing nucleosomes near the TSS is altered resulting in a shortened NDR and nucleosome occupancy at the TSS.
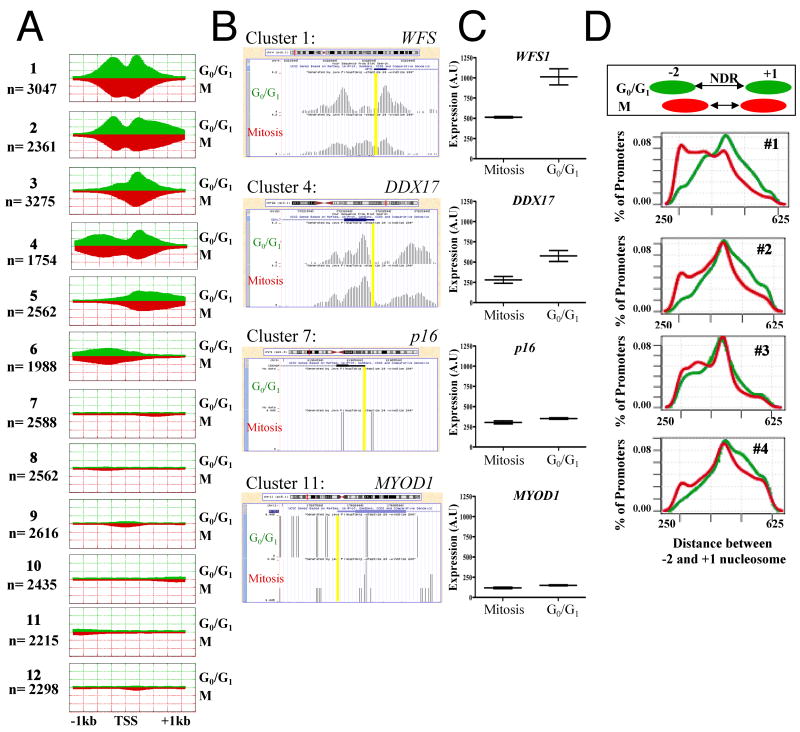
To examine differences in H2A.Z localization in more detail, localization profiles were clustered based on similarity of distribution pattern (Fig. 2a and Fig. S2a). In clusters 1-4, whose genes are actively expressed (Fig. S2b), H2A.Z occupancy is high and we observe an NDR at the TSS in G0/G1. The NDR is however weaker in mitotic cells, reflecting a less organized occupancy pattern (Fig. 2a). High-resolution examples of promoters from clusters 1 (WFS1) and 4 (DDX17), which are highly expressed (Fig 2b,c), show robust H2A.Z signal, while examples of promoters from clusters 7 (p16) and 11 (MYOD1), which are not expressed, do not have H2A.Z localized to their promoters (Fig. 2b,c and Fig. S2b). Total H2A.Z occupancy was globally similar between G0/G1 and M-phase promoters for all clusters, and we did not detect a significant group of promoters losing or gaining overall H2A.Z occupancy. Refined analysis of the cluster patterns, and comparison of the profile in G0/G1 and M-phase revealed a more dynamic picture however, and reflected reduced NDR sizes. The reduced NDR during mitosis could result from an averaging effect of poorly positioned nucleosomes in M-phase or from specific but shifted H2A.Z localization patterns (as shown for individual cases in Fig. 2b). To test the hypothesis of shrinking NDR systematically we used a linear model to infer nucleosome positions at all active TSSs and computed the distances between the -2 and +1 nucleosomes in G0/G1 and M-phase cells. We found a reduction in the gap between the nucleosomes (NDR) during mitosis (Fig. 2d), suggesting that the broad occupancy pattern during mitosis is mainly a consequence of variable displacement of the +1 nucleosome and not of general lack of specific positioning during M-phase. This shortened NDR was found for all clusters in which H2A.Z was present on both the -2 and +1 nucleosomes (Fig. 2d).
Figure 2. H2A.Z containing nucleosomes upstream and downstream of TSSs of active genes generate a NDR that is shortened during mitosis.
(A) Average coverage statistics for each H2A.Z profile covering 1000bp upstream and downstream of known transcription start sites (UCSC). Clusters reveal strong nucleosomal phasing in the G0/G1 data, with nucleosomes detected on one or two sides of a strong NDR at the TSS. The mitotic profiles show a weaker NDR and shifted average position of the +1 and -2 nucleosomes. (B) High resolution examples of nucleosome sliding from cluster 1 (WFS1) and cluster 4 (DDX17) demonstrate promoter specificity in the size of the shift, while the height of the peaks indicate that within an individual promoter the nucleosomes are well-positioned during both G0/G1 and M-phase. p16 (Cluster 7) and MYOD1 (cluster 11), which are silenced by DNA methylation, do not contain H2A.Z at their promoters. (C) Gene expression of representative genes from clusters 1 & 4, WFS1 and DDX17, respectively, show that expression is decreased during mitosis, while genes which do not contain H2A.Z, p16 and MYOD1, are not expressed regardless of cell cycle stage. (D) The size distribution of the NDRs is shown as distance between the -2 and +1 H2A.Z nucleosomes in the G0/G1 (green) and M (red) conditions. The percentage of promoters is plotted along the Y-axis and the distance between the -2 and +1 H2A.Z nucleosomes is on the X-axis. There is clear compaction of the NDR in clusters showing H2A.Z presence during M- phase. (A.U.): Arbitrary units.
We could not use this H2A.Z ChIP-seq approach to adequately examine changes in nucleosome positioning at inactive genes since they do not contain significant H2A.Z enrichment (Barski et al., 2007). The lack of H2A.Z on inactive genes is apparent at both high resolution (Fig. 2b) and genome-wide (Fig. 1c). Furthermore, genes within clusters 7-12 are weakly expressed and correlate with a general lack of H2A.Z, which remains unchanged between G0/G1 and mitotic cells (Fig. 2a and Fig. S2).
To examine changes in nucleosome occupancy in more detail we next used our high-resolution methylation dependent single promoter analysis (M-SPA) to assess nucleosome positioning during G0/G1 and M-phase. M-SPA is uniquely suitable for footprinting unmethylated CpG island promoters and uses the ability of M.SssI to methylate all CpG sites not bound by nucleosomes or tight binding transcription factors in vivo (Fatemi et al., 2005; Gal-Yam et al., 2006; Lin et al., 2007). To generate footprints, nuclei are treated with M.SssI followed by DNA purification, bisulfite conversion, PCR amplification, cloning and sequencing. The resultant product gives single molecule resolution of the region of interested as a functional unit and allows mapping of DNA binding proteins (inaccessible regions less than 146 bp) and nucleosomes (inaccessible regions equal to or larger than 146 bp). We used the GRP78 promoter to probe nucleosome positioning during mitosis, as it is an endogenously unmethylated (Fig. S3a) CpG island promoter with a TATA box and a well-defined TSS. GRP78 is a constitutively expressed ER chaperone protein that has a well-positioned nucleosome just after the TSS (+1) and a NDR in human fibroblasts (Gal-Yam et al., 2006). As expected from the genome-wide data, we found a well-positioned +1 nucleosome, which is inaccessible to M.SssI at GRP78 in G0/G1 cells (Fig 3a). During mitosis this nucleosome shifted to occupy the TSS (Fig. 3b). These data were also confirmed in HCT116 colon cancer cells, demonstrating that changes in nucleosome occupancy were not cell-type specific (Fig. S3b-e). Similar results were obtained when M-phase cells were harvested by shaking after release from serum starvation (without Nocodazole exposure) showing that the results were not due to a drug-induced artifact (Fig. S3f). Overall accessibility of promoter replicas was not altered during the cell cycle (Fig. S3g), supporting our hypothesis that the change in nucleosome occupancy results from nucleosome reorganization rather than insertion of additional nucleosomes. A similar shift in positioning of the +1 nucleosome occurred on the WFS1 promoter (Fig 3d-e). In addition, the -2 nucleosome on the WFS1 promoter shifts downstream during mitosis further demonstrating a shrinkage of the NDR during mitosis (Fig. S3h).
Figure 3. The +1 nucleosomes on the GRP78 and WFS1 promoters shift upstream to occupy the TSS during mitosis, coinciding with decreased gene expression.
Black and white circles indicate methylated (accessible) and unmethylated (inaccessible) CpG sites, respectively. Orange highlights M.SssI inaccessible regions that can accommodate a nucleosome (>146 bp). The well-positioned nucleosome after the TSS of GRP78 (A) and WFS1 (D) during G0/G1 shifts upstream to occupy the TSS during mitosis (B, E) in T24 cells. (C, F) Nucleosome occupancy at the TSS is anti-correlated with expression. The percentage of promoter replicas that contained a nucleosome at the TSS (gold bar) is plotted along with GRP78 expression (maroon bar). Expression bars are relative to GAPDH expression and represent the mean + SEM of 3 independent experiments.
Nucleosome occupancy at TSSs is incompatible with expression (Pazin et al., 1994). To determine whether the change in nucleosome positioning on the GRP78 promoter during mitosis coincided with decreased expression we calculated the percentage of GRP78 and WFS1 promoter replicas in which a nucleosome blocked M.SssI accessibility before and after the TSS and correlated it with expression level (Fig. 3c,f). During G0/G1, nucleosome occupancy at the TSS was low and expression was high, in contrast, nucleosomes occupied the TSS during mitosis and expression was low. The change in nucleosome localization during mitosis and subsequent decrease in expression points to a potential mechanism of gene silencing during mitosis.
Mitotic Sliding of the +1 Nucleosome Does Not Occur on Mitotically Expressed Genes or Methylated Promoters
We next asked whether similar nucleosome shifting occurred on promoters of genes that are expressed during mitosis and silenced during G0/G1, such that the +1 nucleosome is located after the TSS during M-phase when expression is high, and occupies the TSS during G0/G1 when expression is low. Using the polo like kinase (PLK1) promoter as a model, we found no change in nucleosome occupancy at the TSS of PLK1 during G0/G1, and M-phase (Fig. 4a, b). Instead, the nucleosome remains after the TSS, creating a NDR during both G0/G1 and M-phase, and there is reduced M.SssI accessibility during mitosis likely reflecting the binding of transcription factors (E2F and TBP) (Fig. 4b). Thus, while nucleosome occupancy at the TSS of the PLK1 promoter does not change during the cell cycle, expression is restricted to M-phase (Fig. 4c).
Figure 4. Nucleosome occupancy at TSSs of the mitotically expressed PLK1 promoter does not change during the cell cycle.
Black and white circles indicate methylated (accessible) and unmethylated (inaccessible) CpG sites. Orange highlights M.SssI inaccessible regions that can accommodate a nucleosome (>146 bp). Green and blue highlight M.SssI inaccessible regions that mark E2F and TBP binding sites, respectively. The +1 nucleosome of the PLK1 promoter is positioned after the TSS during G0/G1 (A) and M-phase (B). (C) While nucleosome occupancy at the TSS of PLK1 is similar during G0/G1 and M-phase, expression is restricted to M-phase. The percentage of promoter replicas that contained a nucleosome at the TSS (gold bar) is plotted along with PLK1 expression (maroon bar). Expression bars are relative to GAPDH expression and represent the mean + SEM of 3 independent experiments.
Since H2A.Z is not present on promoters silenced by DNA methylation (Zilberman et al., 2008), our genome-wide sequencing approach did not allow us to assess nucleosome positioning on these promoters. To determine whether +1 nucleosomes of silenced genes also shift during mitosis, we modified the M-SPA protocol to use the M.CviPI methyltransferase, which methylates cytosines in GpC dinucleotides (GM-SPA). Using the p16 promoter as a model, which is methylated in T24 cells (Fig 5a,b), we found that methylated promoters were relatively inaccessible to M.CviPI during both G0/G1 and M-phase (Fig. 5c,d), suggesting that nucleosome occupancy at TSSs of methylated promoters does not change during mitosis consistent with a lack of expression during both G0/G1 and M-phase (Fig 5e). Therefore +1 nucleosome sliding and occupancy at the TSS during mitosis is not a common silencing mechanism for all promoters during mitosis- rather it is specific to gene promoters that are expressed during G0/G1 and silenced during mitosis.
Figure 5. Nucleosome occupancy at TSSs of promoters silenced by methylation does not change during the cell cycle.
(A, B) p16 is methylated in T24 cells. Black and white circles indicate endogenously methylated and unmethylated CpG sites. (C, D) Black and white circles indicate M.CviPI accessible (methylated) and inaccessible (unmethylated) GpC sites. Pink highlights M.CviPI inaccessible regions that can accommodate a nucleosome (>146 bp). The p16 promoter is inaccessible during G0/G1 (C) and M-phase (D). (E) Nucleosome occupancy is high at the TSS of p16 during G0/G1 and M-phase and expression is not detected. The percentage of promoter replicas that contained a nucleosome at the TSS (gold bar) is plotted along with p16 expression (maroon bar). Expression bars are relative to GAPDH expression and represent the mean + SEM of 3 independent experiments.
Histone Modifications on the +1 Nucleosome are Altered During Mitosis
We confirmed the change in nucleosome positioning during mitosis using ChIP for H3 (Fig. 6a) then asked which histone modifications and variants are associated with the shifting nucleosome during mitosis (Fig. 6b). Using primers specifically designed to regions with differential nucleosome occupancy (i.e. region R5.2 in M-phase and region R3.3 in G0/G1) we were able to detect the shift in nucleosome positioning demonstrating that our ChIP methods are sensitive enough to detect shifts at least as small as 80 bp. Thus, these ChIP data combined with the M-SPA data demonstrate that the +1 nucleosome on the GRP78 promoter shifts upstream during mitosis using two independent methods of visualizing nucleosome positioning.
Figure 6. The GRP78 promoter has differential nucleosome positioning, unique complements of DNA binding proteins and histone modifications in mitotic and G0/G1 chromatin.
(A) H3 ChIP confirms that there is a change in nucleosome occupancy during mitosis. (B) Acetylation of H3 and H2A.Z are decreased during M-phase, relative to G0/G1, while H3K4me3 and H2A.Z are maintained and H3S10 phosphorylation is increased. The extremely low level of H3K27me3 (data not shown) is unchanged during the cell cycle. Bars in (B) indicate ChIP signal from M-phase chromatin (R5.2) expressed as fold change relative to G0/G1 chromatin (R3.3). (C) Pol II, NF-Y and TBP binding is significantly reduced during mitosis. Paired t-test was performed (*= p-value<0.05). Bars represent mean +SEM of at least 6 ChIPs. Arrows in schematic indicate NF-Y and TBP binding sites.
In order to accurately measure histone modification changes that occur on the shifting nucleosome we compared the signal from GRP78 promoter regions, which correlated to nucleosome occupancy during the specific cell cycle stage (i.e. region R5.2 in M-phase and region R3.3 in G0/G1). As expected, we found increased levels of H3S10 phosphorylation and decreased H3 acetylation during mitosis (Fig. 6b). The overall level of H3K27me3 remains unchanged during the cell cycle (ratio= 1), however, it is 30 fold less than a known polycomb silenced gene (FAM84a) (data not shown), demonstrating that mitosis specific silencing of GRP78 is not due to polycomb repression. Consistent with the genome-wide data (Fig. 1), H2A.Z was present at the GRP78 promoter in both M-phase and G0/G1 cells, while H2A.Z was acetylated during G0/G1 only. In addition H3K4me3 was also maintained on the +1 nucleosome during mitosis. Thus, during mitosis, H3K4me3 and H2A.Z seem to mark chromatin regions that are poised for activation (Henikoff et al., 2009). We next examined DNA binding ability and found that Pol II, TBP and NF-Y were minimally bound to the GRP78 promoter during M-phase (Fig. 6c), consistent with reports that transcription is generally shut down during mitosis (Gottesfeld and Forbes, 1997; Kouskouti and Talianidis, 2005). Thus, in addition to changes in nucleosome occupancy, transcription factor and Pol II binding is altered on the GRP78 promoter during mitosis.
Discussion
Our data show that during mitosis, the only characteristics of active TSSs that we examined which are maintained during mitosis are a reduced NDR, H2A.Z and H3K4me3 (Fig. 7). The maintenance of the NDR, H2A.Z and H3K4me3 during mitosis may keep the +1 nucleosome in an active configuration as it occupies the TSS, while the presence of H3S10 phosphorylation prevents lysine 9 methylation, potentially enabling condensed promoters to remain poised for activation (Nowak and Corces, 2004 and references therein). Thus, the presence of this combination of markers may allow the GRP78 promoter to remain poised for activation in the context of condensed chromatin, and mark the +1 nucleosome so that it can be quickly shifted, allowing for rapid reactivation following the completion of mitosis.
Figure 7. Depiction of Nucleosome Sliding on the GRP78 promoter.
During G0/G1, the +1 nucleosome is positioned just after the TSS allowing for TBP, NF-Y and Pol II binding at the TSS. This nucleosome contains the H2A.Z variant, which is acetylated. H3 is acetylated and lysine 4 is tri-methylated. During mitosis the +1 nucleosome shifts to cover the TSS and transcription factor and Pol II binding are lost. The nucleosome loses H3 and H2A.Z acetylation while maintaining the H2A.Z variant and H3K4me3 and acquiring the mitosis specific phosphorylation of serine 10 of H3.
H2A.Z containing nucleosomes are preferentially localized to regions surrounding the TSSs of active genes (Barski et al., 2007). When paired with H3.3, which is also found at TSSs, H2A.Z containing nucleosomes are less stable than nucleosomes containing the canonical H2A (Jiang and Pugh, 2009; Jin and Felsenfeld, 2007; Mito et al., 2005). Thus, the presence of H2A.Z may act to destabilize these nucleosomes, allowing them to slide upon entering and exiting mitosis. Consistent with this, our data demonstrate that H2A.Z containing nucleosomes are not present at promoters silenced by DNA methylation as has been previously reported (Zilberman et al., 2008), and there is an absence of nucleosome sliding at these promoters during mitosis.
A subset of genes is required for cell cycle progression and expressed during mitosis. Nucleosome sliding also did not occur on these promoters that are silenced during G0/G1 and active during M-phase. Since nucleosome occupancy at the TSS is anti-correlated with expression, one possibility is that promoters of mitotically expressed genes would contain a nucleosome at the TSS during G0/G1 when genes were off, which would shift downstream to occupy the region after the TSS upon activation during M-phase. However, this is not the case with the PLK1 promoter where there was no change in nucleosome positioning during the cell cycle, rather there was a lack of transcription factors during G0/G1 when PLK1 was not expressed. This is consistent with previous work demonstrating that regulation of mitotically expressed genes occurs through active repression during G0/G1 rather than specific activation during mitosis (Martin and Strebhardt, 2006). Thus, nucleosome shifting is not common to all promoters, rather it is specific to promoters which are silenced during mitosis. The shifting identified in our study is a potentially novel mechanism for mitotic gene-silencing whereby H2A.Z containing +1 nucleosomes shift to occupy TSSs, while maintaining a shortened NDR and H3K4me3.
The maintenance of histone modifications can mark subsets of genes for rapid reeactivation following mitotic exit. H3K4me3 is maintained at specific loci during mitosis (Mishra et al., 2009) and MLL gets re-distributed to genes with high expression levels in interphase enabling rapid re-activation following mitotic exit (Blobel et al., 2009). Since GRP78 is an ER stress response protein, which is constitutively and highly expressed, it is not surprising that it maintains H3K4me3 at its promoter during mitosis, allowing for rapid re-activation upon mitotic exit. The maintenance of H3K4me3 can act to keep the +1 nucleosome in an active configuration, while its localization covering the TSS can inhibit transcription.
Despite the dramatic changes in chromatin structure that occur during the cell cycle, few studies have examined changes in nucleosome positioning during mitosis. One such study found a loss of well positioned nucleosomes between proximal promoters and upstream regulatory elements concluding that nucleosome occupancy does not play a role in regulating gene expression during the cell cycle (Komura and Ono, 2005). However this study examined nucleosomes located upstream of the proximal promoter, which often act to enhance gene expression by bringing distal regulatory elements in close proximity to promoters, rather than nucleosomes located immediately surrounding the TSS where the transcriptional machinery is assembled.
Assembly of the transcriptional machinery at TSSs is necessary for gene activation. The continued binding of transcription factors or TBP has been shown to mark some genes for rapid re-expression following mitotic exit (Denissov et al., 2007; Xing et al., 2008; Sarge and Park-Sarge, 2009; Verdeguer et al., 2010). However, the majority of transcription factors are displaced during mitosis due to both chromatin condensation as well as their phosphorylation (Delcuve et al., 2008; Gottesfeld and Forbes, 1997; Martinez-Balbas et al., 1995) thus making it unlikely that transcription factor binding is a global mechanism for marking genes for rapid re-activation following mitotic exit.
We show that NF-YA and TBP binding are lost during mitosis and the TBP binding site on the GRP78 promoter becomes occupied by a nucleosome. Thus, it is possible that once transcription factors and components of the transcriptional machinery are removed from the GRP78 promoter during mitosis, the +1 nucleosome passively shifts to occupy the TSS and TATA box thereby silencing gene expression until mitotic exit, after which the nucleosome shifts downstream of the TSS. Upon viral infection the IFN-β promoter is remodeled by the SWI/SNF complex followed by TBP binding which causes the nucleosome which normally occupies the TSS to shift downstream of the TSS and transcription activation (Lomvardas and Thanos, 2001). Like transcription factors, several components of chromatin remodeling complexes are phosphorylated and excluded from chromatin during mitosis (Muchardt et al., 1996; Sif et al., 1998), however some of the catalytic subunit BRM remains in the insoluble fraction during mitosis and may remain bound to mitotic chromatin (Sif et al., 1998). Thus, chromatin remodeling enzymes may regulate both the upstream and downstream shifting of nucleosomes during the cell cycle, and hence gene silencing and re-activation.
It is possible that subsets of genes are marked for rapid re-expression following mitotic exit using different mechanisms. Which mechanism is used may be determined by a variety of promoter features, including CpG content, TATA box presence, multiple start sites, expression pattern and level and time course of re-expression. The extent to which genes are bookmarked during mitosis using a combination of these mechanisms would be an interesting question for future study.
Methods
Cell Culture
Confluent T24 bladder cancer cells were serum starved for 48 hrs to obtain a relatively pure population of G0/G1 cells or treated with Nocodazole for 20 hours and harvested by shaking to obtain mitotic cells (Fig. S1).
ChIP & ChIP-Seq
ChIP was performed as previously described using 2×106 cells per IP (Gal-Yam et al., 2006; Lin et al., 2007). Briefly, following formaldehyde fixation, chromatin was sonicated to generate a majority of fragments between 200-600bp. H3 (ab1791), H3S10-phosphorylation (ab14955), H2A.Z (ab4174), Pol II (ab5408) and acetylated H2A.Z (ab18262) antibodies were purchased from Abcam. Acetylated H3 (06-599) and H3K27 trimethylation (17-622) were purchased from Millipore. H3K4 trimethylation antibody (39159) was purchased from Active Motif. TBP (sc-273) and NF-Y (sc-17753) were purchased from Santa Cruz Biotechnology. ChIP-seq samples were generated using 1×108 cells using an antibody directed towards H2A.Z. 20ng of ChIP'd DNA was used to generate libraries using previously described methods (Ku et al., 2008; Mikkelsen et al., 2007). Amplicons between 250 and 700 bp (after 92 bp adaptor addition) were gel purified and used for sequencing on an Illumina GA-II machine. Reads were mapped using the mapq software obtaining 12,830,000 aligned reads from M- phase cells and 8,630,000 from G0/G1 cells. Robustly annotated transcription start sites were taken from the UCSC known genes resource. We compared coverage in the forward and reverse strands, by pooling 36bp reads into 20bp bins according to their position relative to the nearest TSS (oriented according to the strand). We combined reads from both strands by extending them to 150bp and computing overall coverage in bins of 25bps relative to the nearest TSS. We concatenated the profiles for the two conditions, and clustered the TSSs using simple K-means with Euclidian distance. To assess the degree of NDR shrinking and verify it statistically we identified the offset with maximum coverage in the ranges [-400, -200] (-2 nucleosome) and [+50, +250] (+1 nucleosome) NFR for each TSS in each condition. We filtered only cases for which the maxima were at least 4 for both ranges and for both conditions. We then computed the distribution of differences between the heuristically inferred -2 and +1 nucleosome positions for each cluster of TSSs (Fig 2d).
M-SPA/GM-SPA
Methylase-based single promoter assay was performed as previously described (Fatemi et al., 2005; Gal-Yam et al., 2006; Lin et al., 2007; Miranda et al. 2010, http://www.epigenome-noe.net/) with minor modifications. Mitotic cells were harvested by shaking and washes were performed in conical tubes. After washes cells were re-suspended in hypotonic lysis buffer (20 mM K+ Hepes, pH 7.8, 5 mM Potassium Acetate, 0.5 mM MgCl2, 0.5 mM DTT) and incubated on ice for 10 minutes. Cells were centrifuged and re-suspended in lysis buffer and homogenized 20 times using a 1 ml dounce homogenizer. M.SssI (M-SPA) or M.CviPI (GM-SPA) reactions were done according to manufacturer's recommendations (NEB). Nucleosome localization was defined as a region ≥ 146 bp that was inaccessible to M.SssI (M-SPA) or M.CviPI (GM-SPA). M.SssI or M.CviPI treatment was followed by sodium bisulfite conversion, PCR amplification of a region of interest, cloning and sequencing of individual clones to reveal the structure of single promoter replicas as functional units. ChIP-seq data has been deposited in the NCBI GEO database under the series accession number GSE19568.
Supplementary Material
Acknowledgments
We would like to thank Phillippa Taberlay, Shinwu Jeong, and Erika Wolff and Daniel DeCarvalho for helpful discussions and careful reading of the manuscript. We would like to thank Bradley Bernstein for helpful conversations, Joseph Aman for technical assistance. The ChIP-seq experiments were performed in conjunction with the USC Epigenome center. We would like to thank Mark Miranda for creating the CpG Bubble Chart Generator Program.
Footnotes
Author Contributions T.K.K, T.B.M and J.C.L performed the experiments, T.K.K., G.L. and P.A.J. designed the experiments, B.P.B and A.T. performed bioinformatics analysis, and all authors discussed the results and commented on the manuscript. T.K.K., G.L. and P.A.J. wrote the paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Belmont AS. Large-scale chromatin organization. Dordrecht: Kluwer Academic Publishers; 1997. [Google Scholar]
- Belmont AS. Mitotic chromosome structure and condensation. Curr Opin Cell Biol. 2006;18:632–638. doi: 10.1016/j.ceb.2006.09.007. [DOI] [PubMed] [Google Scholar]
- Blobel GA, Kadauke S, Wang E, Lau AW, Zuber J, Chou MM, Vakoc CR. A reconfigured pattern of MLL occupancy within mitotic chromatin promotes rapid transcriptional reactivation following mitotic exit. Mol Cell. 2009;36:970–983. doi: 10.1016/j.molcel.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce K, Myers FA, Mantouvalou E, Lefevre P, Greaves I, Bonifer C, Tremethick DJ, Thorne AW, Crane-Robinson C. The replacement histone H2A.Z in a hyperacetylated form is a feature of active genes in the chicken. Nucleic Acids Res. 2005;33:5633–5639. doi: 10.1093/nar/gki874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Markoulaki S, Levine SS, Hanna J, Lodato MA, Sha K, Young RA, Jaenisch R, Boyer LA. H2AZ is enriched at polycomb complex target genes in ES cells and is necessary for lineage commitment. Cell. 2008;135:649–661. doi: 10.1016/j.cell.2008.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delcuve GP, He S, Davie JR. Mitotic partitioning of transcription factors. J Cell Biochem. 2008;105:1–8. doi: 10.1002/jcb.21806. [DOI] [PubMed] [Google Scholar]
- Denissov S, van Driel M, Voit R, Hekkelman M, Hulsen T, Hernandez N, Grummt I, Wehrens R, Stunnenberg H. Identification of novel functional TBP-binding sites and general factor repertoires. Embo J. 2007;26:944–954. doi: 10.1038/sj.emboj.7601550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dressler GR. Turning the Page on Epigenetic Bookmarks. Developmental Cell. 2010;18:4–5. doi: 10.1016/j.devcel.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatemi M, Pao MM, Jeong S, Gal-Yam EN, Egger G, Weisenberger DJ, Jones PA. Footprinting of mammalian promoters: use of a CpG DNA methyltransferase revealing nucleosome positions at a single molecule level. Nucleic Acids Res. 2005;33:e176. doi: 10.1093/nar/gni180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal-Yam EN, Jeong S, Tanay A, Egger G, Lee AS, Jones PA. Constitutive nucleosome depletion and ordered factor assembly at the GRP78 promoter revealed by single molecule footprinting. PLoS Genet. 2006;2:e160. doi: 10.1371/journal.pgen.0020160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesfeld JM, Forbes DJ. Mitotic repression of the transcriptional machinery. Trends Biochem Sci. 1997;22:197–202. doi: 10.1016/s0968-0004(97)01045-1. [DOI] [PubMed] [Google Scholar]
- Guillemette B, Gaudreau L. Reuniting the contrasting functions of H2A.Z. Biochem Cell Biol. 2006;84:528–535. doi: 10.1139/o06-077. [DOI] [PubMed] [Google Scholar]
- Hans F, Dimitrov S. Histone H3 phosphorylation and cell division. Oncogene. 2001;20:3021–3027. doi: 10.1038/sj.onc.1204326. [DOI] [PubMed] [Google Scholar]
- Henikoff S, Henikoff JG, Sakai A, Loeb GB, Ahmad K. Genome-wide profiling of salt fractions maps physical properties of chromatin. Genome Res. 2009;19:460–469. doi: 10.1101/gr.087619.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn PJ, Peterson CL. Molecular biology. Chromatin higher order folding--wrapping up transcription. Science. 2002;297:1824–1827. doi: 10.1126/science.1074200. [DOI] [PubMed] [Google Scholar]
- Jiang C, Pugh BF. Nucleosome positioning and gene regulation: advances through genomics. Nat Rev Genet. 2009;10:161–172. doi: 10.1038/nrg2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C, Felsenfeld G. Nucleosome stability mediated by histone variants H3.3 and H2A.Z. Genes Dev. 2007;21:1519–1529. doi: 10.1101/gad.1547707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C, Zang C, Wei G, Cui K, Peng W, Zhao K, Felsenfeld G. H3.3/H2A.Z double variant-containing nucleosomes mark ‘nucleosome-free regions’ of active promoters and other regulatory regions. Nat Genet. 2009;41:941–945. doi: 10.1038/ng.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komura J, Ono T. Disappearance of nucleosome positioning in mitotic chromatin in vivo. J Biol Chem. 2005;280:14530–14535. doi: 10.1074/jbc.M500637200. [DOI] [PubMed] [Google Scholar]
- Kouskouti A, Talianidis I. Histone modifications defining active genes persist after transcriptional and mitotic inactivation. Embo J. 2005;24:347–357. doi: 10.1038/sj.emboj.7600516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, Presser A, Nusbaum C, Xie X, Chi AS, et al. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet. 2008;4:e1000242. doi: 10.1371/journal.pgen.1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CK, Shibata Y, Rao B, Strahl BD, Lieb JD. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat Genet. 2004;36:900–905. doi: 10.1038/ng1400. [DOI] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Lin JC, Jeong S, Liang G, Takai D, Fatemi M, Tsai YC, Egger G, Gal-Yam EN, Jones PA. Role of nucleosomal occupancy in the epigenetic silencing of the MLH1 CpG island. Cancer Cell. 2007;12:432–444. doi: 10.1016/j.ccr.2007.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomvardas S, Thanos D. Nucleosome sliding via TBP DNA binding in vivo. Cell. 2001;106:685–696. doi: 10.1016/s0092-8674(01)00490-1. [DOI] [PubMed] [Google Scholar]
- Martin BT, Strebhardt K. Polo-like kinase 1: target and regulator of transcriptional control. Cell Cycle. 2006;5:2881–2885. doi: 10.4161/cc.5.24.3538. [DOI] [PubMed] [Google Scholar]
- Martinez-Balbas MA, Dey A, Rabindran SK, Ozato K, Wu C. Displacement of sequence-specific transcription factors from mitotic chromatin. Cell. 1995;83:29–38. doi: 10.1016/0092-8674(95)90231-7. [DOI] [PubMed] [Google Scholar]
- Mellor J. The dynamics of chromatin remodeling at promoters. Mol Cell. 2005;19:147–157. doi: 10.1016/j.molcel.2005.06.023. [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda TB, Kelly TK, Bouazoune K, Jones PA. Methylation-sensitive single-molecule analysis of chromatin structure. Curr Protoc Mol Biol. Chapter 21(Unit 21 17):21–16. doi: 10.1002/0471142727.mb2117s89. [DOI] [PubMed] [Google Scholar]
- Mishra BP, Ansari KI, Mandal SS. Dynamic association of MLL1, H3K4 trimethylation with chromatin and Hox gene expression during the cell cycle. Febs J. 2009;276:1629–1640. doi: 10.1111/j.1742-4658.2009.06895.x. [DOI] [PubMed] [Google Scholar]
- Mito Y, Henikoff JG, Henikoff S. Genome-scale profiling of histone H3.3 replacement patterns. Nat Genet. 2005;37:1090–1097. doi: 10.1038/ng1637. [DOI] [PubMed] [Google Scholar]
- Muchardt C, Reyes JC, Bourachot B, Leguoy E, Yaniv M. The hbrm and BRG-1 proteins, components of the human SNF/SWI complex, are phosphorylated and excluded from the condensed chromosomes during mitosis. Embo J. 1996;15:3394–3402. [PMC free article] [PubMed] [Google Scholar]
- Nowak SJ, Corces VG. Phosphorylation of histone H3: a balancing act between chromosome condensation and transcriptional activation. Trends Genet. 2004;20:214–220. doi: 10.1016/j.tig.2004.02.007. [DOI] [PubMed] [Google Scholar]
- Pazin MJ, Kamakaka RT, Kadonaga JT. ATP-dependent nucleosome reconfiguration and transcriptional activation from preassembled chromatin templates. Science. 1994;266:2007–2011. doi: 10.1126/science.7801129. [DOI] [PubMed] [Google Scholar]
- Prescott DM, Bender MA. Synthesis of RNA and protein during mitosis in mammalian tissue culture cells. Exp Cell Res. 1962;26:260–268. doi: 10.1016/0014-4827(62)90176-3. [DOI] [PubMed] [Google Scholar]
- Sarge KD, Park-Sarge OK. Mitotic bookmarking of formerly active genes: keeping epigenetic memories from fading. Cell Cycle. 2009;8:818–823. doi: 10.4161/cc.8.6.7849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schones DE, Cui K, Cuddapah S, Roh TY, Barski A, Wang Z, Wei G, Zhao K. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–898. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sif S, Stukenberg PT, Kirschner MW, Kingston RE. Mitotic inactivation of a human SWI/SNF chromatin remodeling complex. Genes Dev. 1998;12:2842–2851. doi: 10.1101/gad.12.18.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valls E, Sanchez-Molina S, Martinez-Balbas MA. Role of histone modifications in marking and activating genes through mitosis. J Biol Chem. 2005;280:42592–42600. doi: 10.1074/jbc.M507407200. [DOI] [PubMed] [Google Scholar]
- Van Hooser A, Goodrich DW, Allis CD, Brinkley BR, Mancini MA. Histone H3 phosphorylation is required for the initiation, but not maintenance, of mammalian chromosome condensation. J Cell Sci. 1998;111(Pt 23):3497–3506. doi: 10.1242/jcs.111.23.3497. [DOI] [PubMed] [Google Scholar]
- Verdeguer F, Le Corre S, Fischer E, Callens C, Garbay S, Doyen A, Igarashi P, Terzi F, Pontoglio M. A mitotic transcriptional switch in polycystic kidney disease. Nat Med. 2010;16:106–110. doi: 10.1038/nm.2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing H, Vanderford NL, Sarge KD. The TBP-PP2A mitotic complex bookmarks genes by preventing condensin action. Nat Cell Biol. 2008;10:1318–1323. doi: 10.1038/ncb1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing H, Wilkerson DC, Mayhew CN, Lubert EJ, Skaggs HS, Goodson ML, Hong Y, Park-Sarge OK, Sarge KD. Mechanism of hsp70i gene bookmarking. Science. 2005;307:421–423. doi: 10.1126/science.1106478. [DOI] [PubMed] [Google Scholar]
- Young DW, Hassan MQ, Yang XQ, Galindo M, Javed A, Zaidi SK, Furcinitti P, Lapointe D, Montecino M, Lian JB, et al. Mitotic retention of gene expression patterns by the cell fate-determining transcription factor Runx2. Proc Natl Acad Sci U S A. 2007;104:3189–3194. doi: 10.1073/pnas.0611419104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman D, Coleman-Derr D, Ballinger T, Henikoff S. Histone H2A.Z and DNA methylation are mutually antagonistic chromatin marks. Nature. 2008;456:125–129. doi: 10.1038/nature07324. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.