Abstract
Recent data indicate that a variety of regulatory molecules active in embryonic development may also play a role in the regulation of early hematopoiesis. Here we report that the human Vent-like homeobox gene VENTX, a putative homolog of the Xenopus xvent2 gene, is a unique regulatory hematopoietic gene that is aberrantly expressed in CD34+ leukemic stem-cell candidates in human acute myeloid leukemia (AML). Quantitative RT–PCR documented expression of the gene in lineage positive hematopoietic subpopulations, with the highest expression in CD33+ myeloid cells. Notably, expression levels of VENTX were negligible in normal CD34+/CD38− or CD34+ human progenitor cells. In contrast to this, leukemic CD34+/CD38− cells from AML patients with translocation t(8,21) and normal karyotype displayed aberrantly high expression of VENTX. Gene expression and pathway analysis demonstrated that in normal CD34+ cells enforced expression of VENTX initiates genes associated with myeloid development and down-regulates genes involved in early lymphoid development. Functional analyses confirmed that aberrant expression of VENTX in normal CD34+ human progenitor cells perturbs normal hematopoietic development, promoting generation of myeloid cells and impairing generation of lymphoid cells in vitro and in vivo. Stable knockdown of VENTX expression inhibited the proliferation of human AML cell lines. Taken together, these data extend our insights into the function of embryonic mesodermal factors in human postnatal hematopoiesis and indicate a role for VENTX in normal and malignant myelopoiesis.
Keywords: human, myelopoiesis, Xvent, VENTX2
Hematopoiesis is a finely orchestrated process of cell proliferation and differentiation. Much progress has been made in understanding the molecular network that controls fate decisions of hematopoietic cells. Interestingly, factors playing an important role in embryonic body development were shown to play a key role in postnatal hematopoietic development. In this respect, clustered homeobox (HB) genes (HOX genes), pivotal regulatory transcription factors of body development during embryogenesis, were characterized as “master genes” of adult murine and human hematopoietic development and as key players in postnatal leukemogenesis (1–3). Clustered HB genes such as HOXB4 and HOXA9/10 are highly expressed in normal hematopoietic stem cells (HSC), but sharply down-regulated in more mature cells. This stem-cell–associated expression pattern as well as gain- or loss-of-function studies have provided support for the hypothesis that HOX genes play a significant role in the development of normal hematopoietic progenitor cells (1, 2, 4–6). Notably, aberrant expression of HOX genes is one of the molecular hallmarks in human AML, characterizing more than every third patient with this disease (7). Dysregulated expression is not restricted to clustered HB genes but includes nonclustered HB genes such as ParaHox genes, composing the Caudal related homeobox gene 2 (CDX2), which is not expressed in normal HSC and their progenitors but aberrantly expressed in the majority of patients with acute myeloid leukemia (AML). Functional studies have clearly demonstrated that aberrant expression of members of the HOX gene and ParaHOX gene family are highly leukemogenic in murine model systems (4–6, 8–10).
Another class of nonclustered Hox genes is the Vent HB gene family (11): this HB gene family comprises multiple genes, which are subdivided into two families on the basis of their sequence homology outside the homeodomain (HD): the vent1 and vent2 family. In Xenopus, the vent1 subfamily includes genes such as xvent1, xvent1b, and pvi (12, 13), the vent2 family includes the genes xvent2, xvent2b, vox1, vox15, and xom genes (14). The pivotal role of xvent2 in modulating hematopoietic development was supported by observations that this member of the vent2 gene family can interact with key hematopoietic transcription factors such as GATA2, SMAD1, and the ets family transcription factor ERG and acts in establishing a positive autoregulatory loop of BMP4 activation by binding to its own promoter and the bmp4 promoter (15–18).
The mammalian homolog of the Xenopus xvent gene is the Vent-like Homeobox gene VENTX2 (19), which was later renamed to VENTX to distinguish it from several VENTX pseudogenes in the human genome (20). The predicted protein is 258 residues in length, with an estimated molecular weight of 28 kDa. The VENTX HD shares the highest identity to the HD in Xenopus laevis xvent2b with 65% homology. In addition to the homology within the HD and a small conserved N-terminal motif, there is little homology between VENTX and the other known Vent homeoproteins (12). On the basis of the HD homology and genomic organization VENTX is considered to belong to the same class of HD and is considered to be the first member of the Vent gene family in mammals (19). Importantly, no Xenopus xvent gene homolog could be identified in the mouse, so that studies on the function of the gene in murine experimental models are not possible. On the basis of this and the given key role of the xvent gene family in the formation of primitive hematopoietic cells during embryogenesis, we tested the hypothesis that VENTX is a yet-unidentified regulator of human hematopoietic differentiation. In this report we demonstrate that, in contrast to HOX genes such as HOXB4 or HOXA9, VENTX is highly expressed in normal human myeloid cells with nearly extinct expression in early CD34+/lin− cells. In contrast, high expression of VENTX could be documented not only in the leukemic myeloid bulk population of patients with t(8;21) positive or normal karyotype AML, but also in the CD34+leukemic subpopulation, thought to comprise the leukemic stem-cell pool. Enforced expression of VENTX in normal CD34+ human progenitor cells perturbed normal hematopoietic development with a shift into myeloid development, whereas VENTX depletion in human AML cell lines impaired their growth. Thus, these data characterize VENTX as a unique regulatory factor of normal and leukemic postnatal hematopoietic cells.
Results
Rapid Evolution of VENTX in Mammals.
We and others have not been able to identify a murine Xenopus xvent gene homolog. Strikingly, even though the two genes that flank the VENTX gene in the human genome (UTF1 and ADAM8) are also syntenic in the mouse genome, there is no functional VENTX gene in the mouse. Therefore, we examined the occurrence and divergence of the VENTX genes in different mammalian and other vertebrate and nonvertebrate species. A phylogenetic tree based on the protein sequence of VENTX was constructed. This tree shows that the protein sequence of VENTX has undergone rapid evolution in vertebrates and mammals when compared with a similar tree constructed for the conserved TALE homeobox gene Meis1 (Fig. S1). VENTX shows more sequence divergence among mammalian species (the slightly darker shaded box) than Meis1 in all vertebrates (the lightly shaded box). In several species (e.g., rat and mouse) whose complete genome has been sequenced, no VENTX homolog was found. Thus, it seems that VENTX has assumed very specific functions in various species. Given the known role of xvent2 in hematopoiesis in Xenopus, we analyzed VENTX function in normal and malignant human hematopoiesis.
VENTX Is Highly Expressed in Normal Human Myeloid Cells.
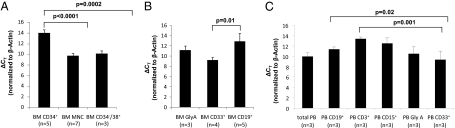
First, we checked the expression levels of VENTX in human hematopoietic stem cells and bone marrow (BM) subpopulations by analyzing previously published gene expression microarrays (n = 3000) (http://www.genesapiens.com) (21). These microarray results suggest that VENTX displays higher expression in circulating myeloid and erythroid cells compared with HSCs (Fig. S2). To confirm these gene expression results, the transcription of VENTX was quantified by TaqMan quantitative reverse transcription polymerase chain reaction (qRT-PCR) in total BM cells, highly purified normal CD34+, CD34+/CD38−, and CD34−/38+ BM subpopulations: VENTX expression was not detectable in the primitive CD34+/CD38− population (45 cycles PCR). In contrast, gene expression was readily detectable in CD34+ human hematopoietic progenitors, but expression was significantly lower (19.4- and 15.1-fold) compared with more mature BM mononuclear cells (MNCs) (P < 0.001) or CD34−/38+ cells (P < 0.001), respectively (Fig. 1A). Among committed progenitors VENTX was significantly more highly expressed in CD33+ BM myeloid cells (ΔCT 9.2 ± 0.61 SEM, n = 4; 4 of 4 positive) compared with CD19+ BM lymphoid cells (ΔCT 12.9 ± 1.6 SEM, n = 7; 5 of 7 positive, P = 0.01) or GlyA+ erythroid subpopulations (ΔCT 11.2 ± 0.8 SEM, n = 4; 3 of 4 positive) (Fig. 1B). In CD3+ lymphoid cells, expression of VENTX was also low to negative (ΔCT 13.2, n = 4; 2 of 4 positive). These results were confirmed for hematopoietic subpopulations purified from normal peripheral blood with a significantly higher expression of VENTX in CD33+ myeloid cells compared with CD19+ B-cells and CD3+ T-cells (4-fold, P = 0.021, and 15.9-fold, P = 0.001, respectively) (Fig. 1C). Thus VENTX expression is most consistent with a role in myeloid differentiation.
Fig. 1.
(A) Expression of VENTX in total BM vs. CD34+ BM progenitors and CD34−/38+ BM cells and in (B) GlyA+ erythroid, CD33+ myeloid, and CD19+ lymphoid cells (C). VENTX expression in hematopoietic subpopulations derived from the peripheral blood. Expression analyses were performed by TaqMan qRT–PCR, and ΔCT values were obtained by normalization to β-actin. Bars represent average expression ±SD; the number of the tested samples is indicated. Note that all ΔCT values are inversely correlated to the expression level.
Malignant Myeloid Cells Show High Expression of VENTX.
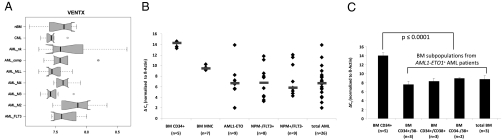
Because normal myeloid cells showed high expression levels of VENTX compared with lymphoid cells, we next asked whether VENTX is also expressed in malignant myeloid and lymphoid cells. To address this question, we quantified the expression of VENTX in the myeloid cell lines NB4, EOL1, OCI-AML3, MV4-11, HL-60, and K562, originally derived from patients with myeloid leukemia, as well as HUT78, NALM6, SKW, HBL2, SD1, Jurkat, and MM6 derived from acute lymphoid leukemia (ALL) patients. Consistent with the data above from primary cell populations, we observed significantly higher VENTX in myeloid compared with lymphoid cell lines (P = 0.03). Furthermore, VENTX expression in myeloid cell lines was significantly higher compared with normal CD34+ progenitor cells (P = 0.002) with the highest expression in the K562, NB4, and EOL1 cell lines (Fig. S3A). In the next step, we extended our analysis to primary AML samples: 10 samples from different AML subtypes and normal BM were analyzed by cDNA microarray analysis for expression of VENTX. The majority of patients expressed VENTX at a low level; however, there was a notable trend toward higher expression of VENTX in FAB M2 t(8;21)-positive AML cases compared, e.g., to FAB M3 t(15;17)-positive cases (Fig. 2A). When we examined VENTX expression in a larger cohort of AML patients with normal karyotype (n = 194), a small proportion (n = 30) of the AML patients exhibited high VENTX expression (Fig. S3B). These results suggest that there might be a distinct subset of human AMLs with normal karyotype that is characterized by increased VENTX expression.
Fig. 2.
VENTX expression in AML cell lines and primary AML samples. (A) Box plots of mRNA expression levels (microarray signal intensity values) of VENTX in normal bone marrow (nBM) (n = 6), chronic myelocytic leukemia (CML) and AML (10 cases each) with normal karyotype (AML_nk), AML with complex karyotype (AML_comp), AML with mixed lineage leukemia (MLL) rearrangement (AML_MLL), AML with CBFB/MYH11 fusion (AML_M4), AML-M3 with PML/RARA fusion (AML_M3), AML with AML1/ETO fusion (AML_M2), and AML with normal karyotype and FLT3 length mutation (AML_FLT3). The normalized and variance stabilized expression values are shown on a logarithmic scale (log 2). Solid bar represents median, boxes represent the 25–75% quantile range (inter-quantile range, IQR), whiskers represent the 1.5-fold IQR, small circles represent outliers. Ten samples are included in each group. (B) Quantitative expression level of VENTX in AML patients with either AML1-ETO translocation or normal karyotype and NPMc+/FLT3-LM− or NPMc−/FLT3-LM+. Diamonds indicate single patients; bars indicate median expression level. (C) Quantitative VENTX expression in human normal CD34+ enriched BM and sorted BM subpopulations of AML1-ETO positive AML patients. Expression analyses were performed by TaqMan qRT–PCR, and ΔCT values were obtained by normalization to β-actin. Note that all ΔCT values are inversely correlated to the expression level.
High expression of VENTX in primary AML samples was further confirmed by real-time PCR: the majority of AML patients (23 of 26 patients) (Table S1) showed significantly elevated expression of VENTX compared with normal BM cells (n = 7) and CD34+ BM cells. Specifically, patients with normal karyotype and NPM1 mutation (NPM1c+/FLT3-LM−; n = 9) or FLT3-length mutation (NPM1c−/FLT3-LM+; n = 8) or patients with t(8;21) (n = 9) had a >100-fold higher expression of VENTX compared with normal CD34+ BM cells and between 5- and 7.8-fold higher expression compared with BM MNCs (Table 1 and Fig. 2B).
Table 1.
Expression levels of VENTX in AML subtypes compared with normal hematopoietic cells
| BM from healthy controls |
||
| AML patients | BM CD34+ (n = 5) | BM MNCs (n = 7) |
| NPM1c−/FLT3-LM+ (n = 8) | 151-fold, P < 0.0000 | 7.8-fold, P < 0.027 |
| NPM1c+/FLT3-LM− (n = 9) | 101-fold, P < 0.0001 | 5.23-fold, P < 0.05 |
| t(8;21) (n = 9) | 137-fold, P < 0.0000 | 7.09-fold, P < 0.03 |
| Total AML (n = 26) | 129-fold, P < 0.0000 | 6.66-fold, P < 0.02 |
Aberrantly high expression of VENTX in leukemic cells could be confirmed when we analyzed the expression of VENTX in highly purified leukemic CD34+CD38−, CD34+CD38+, and CD34−CD38+ cells isolated from t(8;21) AML patients (n = 3 each): expression levels for VENTX in the different leukemic compartments were significantly elevated compared with expression levels detectable in normal CD34+ BM cells (50-fold higher; P ≤ 0.0001) (Fig. 2C). In particular, in contrast to leukemic CD34+/CD38− cells, normal CD34+/CD38− progenitor cells did not show any VENTX expression as assessed by real-time RT–PCR with up to 45 cycles as mentioned before. Thus, aberrant expression levels of VENTX in AML patients extended to the most primitive progenitor compartment, which is thought to harbor the leukemic stem cell candidates (LSCs).
Taken together, these data demonstrate that VENTX is aberrantly expressed in malignant myeloid cells with particularly high expression in AML cases with t(8;21) or normal karyotype.
Overexpression of VENTX in CD34+ Cord Blood Cells Up-Regulates Genes Associated with Myeloid Development.
As we had seen that VENTX is aberrantly expressed in leukemic CD34+ cells with negligible expression in normal counterparts, we assessed the impact of forced VENTX gene expression in normal CD34+ human progenitor cells on the transcription program: differential regulation of genes was assessed by comparing normal CD34+ cord blood cells transduced with the MSCV-VENTX-IRES-GFP construct to CD34+ cord blood cells containing only the vector backbone 48 h after the end of the transduction. The at least twofold differentially regulated genes are listed in Table S2. Notably, most genes, including CD11b, CD125, CD9, and CD14, that were up-regulated by VENTX in this analysis were linked to myelopoiesis. In contrast, several genes associated with erythroid development, such as EPOR, CD35, and CD36, were down-regulated (Fig. S4 and Fig. S5A).
We then used the Gene Ontology approach to test whether genes differentially regulated by VENTX fell into biological processes/pathways. Interestingly, the top 10 cellular processes that were annotated included cytokine-cytokine receptor interaction, hematopoietic cell lineage, MAPK signaling pathway, Wnt signaling pathway, and JAK-STAT signaling pathway.
When KegArray analysis was performed for pathway analysis, one of the top five most enriched pathways identified in this analysis was the “hematopoietic cell lineage” (KEGG hsa04640) (Fig. S5A). In line with this, SCF and M-CSF were found to be up-regulated. In contrast, both IL-7 and IL-9R were down-regulated. Furthermore, when we used the KegArray analysis to investigate the impact of VENTX on the LEF1/TCF pathway known to be important for lymphoid development, the analysis identified LEF1/TCF and one of the downstream targets of LEF1 C-JUN as the genes being down-regulated after VENTX expression (Fig. S5B). Thus, gene expression data and pathway analysis indicated that enforced expression of VENTX in CD34+ early human progenitor cells alters normal expression programs, promoting gene expression with myeloid development and suppressing genes associated with lymphoid and erythroid development.
VENTX Expression Promotes the Formation of Myeloid Colonies and Reduces the Number of Erythroid Colonies in Vitro.
We then tested whether enforced expression of VENTX in CD34+ cells is able to alter the hematopoietic development of early human progenitors as indicated by gene expression and pathway analyses: for this we retrovirally transduced human CD34+ cord blood cells with either VENTX or the empty GFP control vector. Protein expression in transduced cells was demonstrated by Western blot (Fig. S6).
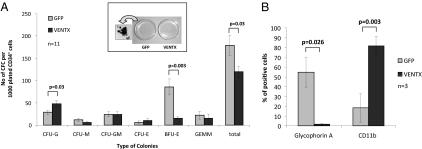
Constitutive expression of VENTX induced a significant increase in the number of myeloid colonies compared with the GFP control with 48 ± 6.5 compared with 28.9 ± 4.8 CFU-G per initially plated 1,000 CD34+ cells (n = 11; P = 0.03; Fig. 3A). Strikingly, constitutive expression of VENTX resulted in an almost complete block in erythroid colony formation with an 81% reduction of the number of BFU-E compared with the control (15.5 ± 3.9 and 81.9 ± 17.9 for VENTX and GFP control, respectively; n = 11; P < 0.003; Fig. 3 A and B), resulting in an overall reduction of the total colony number by 33% (119.5 ± 22.5 and 179.2 ± 12.2 for VENTX and GFP control, respectively; n = 11; P = 0.03). This effect of VENTX was confirmed when the colonies were stained for the erythroid marker glycophorin A and the myeloid marker CD11b: 63% of the cells in the control arm were positive for glycophorin A, whereas only 2% of the cells in the VENTX arm stained positive for this antigen (Fig. 3C). In replating experiments, VENTX expression resulted in an increase in the number of myeloid colonies paralleled by a reduced number of erythroid colonies compared with the control, although the difference was not significant. In total, the number of secondary colonies were comparable in both experimental groups [n = 6; VENTX mean = 124.4 colonies; GFP mean = 131.2 colonies/1,000 initially CD34+ cells plated in the primary colony-forming cells (CFCs); P = not significant (n.s.)]. The impact of VENTX on erythroid cells was confirmed in liquid expansion cultures, which were initiated with highly purified CD34+ cord blood cells: after 1 wk in culture, 22.9% of the cells in the control arm stained positive for glycophorin A compared with 7.9% in the VENTX arm (n = 4; P < 0.025). These data demonstrate the ability of VENTX to promote a myeloid fate on early hematopoietic progenitor cells while suppressing erythroid differentiation.
Fig. 3.
(A) Morphology of colonies and colony numbers derived from CB cells transduced with VENTX compared with the GFP control. Bars represent the average colony number of 11 independent experiments ±SEM. A macroscopic picture of primary colonies and magnification of one representative GFP+ colony are shown. (B) Immunophenotypic analysis of primary colonies by glycophorin A and CD11b staining. Bars represent the average percentage of positively stained cells ±SD.
VENTX Promotes Myelopoiesis in a Feeder-Dependent Lymphoid Coculture System.
To evaluate whether VENTX expression affects generation of B-lymphoid cells in vitro, VENTX and GFP-transduced cells were plated in the MS-5 coculture system using B-cell–promoting conditions. In this assay, expression of VENTX reduced the total output of CD19+ B-cells 9.3-fold (P < 0.001; n = 3). At the same time, VENTX induced a striking 6.4-fold increase (P = 0.002) in the number of CD15+ myeloid cells, resulting in a significant 64-fold decrease in the lymphoid/myeloid ratio in the culture system (Fig. S7 and Table S3). Thus, VENTX also strongly promotes the generation of myeloid cells in a feeder-dependent coculture system while impairing the development of B-lymphoid cells.
VENTX Enhances the Proportion of Human Myeloid Cells in Nonobese Diabetic/Severe Combined Immunodeficient Mice.
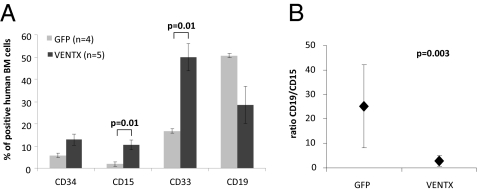
We next examined the effect of forced VENTX expression on hematopoietic differentiation in vivo. Irradiated nonobese diabetic/severe combined immunodeficient (NOD/SCID) mice were transplanted with the unselected progeny of 3–5 × 105 CD34+ cells 4–6 h after ending the transduction with VENTX-GFP or the GFP control. Mice were killed and assessed for engraftment and differentiation 9 wk post transplantation. Multilineage engraftment of transduced human CD19+ B-lymphoid and CD15+ myeloid cells was detected in five of seven mice transplanted with VENTX-GFP–transduced human CD34+ cells and in four of four mice in the control arm. As observed in the in vitro assays, VENTX expression promoted generation of myeloid cells with a >5- and 2.5-fold increase in the proportion of human CD15+ and CD33+ primitive myeloid cells, respectively, as compared with the GFP control (10.6 ± 2.3% vs. 2.0 ± 0.6% and 50.0 ± 6.1% vs. 16.7 ± 8.6% within the transduced compartment; P = 0.01) (Fig. 4A). This increase in the proportion of CD15+ myeloid cells also translated into a 1.5-fold higher absolute number of engrafted CD15+-transduced cells with 5.1 × 104 ± 1.5 × 104 SEM cells for VENTX mice compared with 3.3 × 104 ± 2 × 104 SEM cells for the control mice. In line with its promoting effect on myeloid cells, VENTX also increased the proportion of CD33+ myeloid cells significantly. There was a substantially lower proportion of transduced CD19+ B-cells in the VENTX arm compared with the control (mean 28.5% vs. 50.7%, respectively) (Fig. 4A), which also resulted in a lower number of engrafted CD19+ cells in the VENTX mice with 8.8 × 104 ± 3.4 × 104 cells vs. 85 × 104 ± 3.8 × 105 cells in the controls, respectively. This resulted in a striking inversion of the lympho-myeloid ratio from 25.1 in the control to 2.7 in the VENTX arm (P = 0.003) (Fig. 4B).
Fig. 4.
Overexpression of VENTX in human CB cells increases myeloid engraftment in mice. (A) Immunophenotypic analysis of human CB cells 9–12 wk after transplantation in NOD/SCID mice transduced with either GFP or VENTX. Bars represent the average percentage of cells positive for the respective marker ±SEM. (B) Lymphoid/ myeloid (CD19/CD15) ratio of the same mice. Whiskers indicate SD.
To analyze whether forced VENTX expression would have any impact on the properties of primitive human hematopoietic cells, we performed in vitro “limiting dilution” long-term culture-initiating cell (LTC-IC) assays (n = 2) from CD34+-transduced cord blood (CB) cells to determine the frequency of long-term culture-initiating cells and the average number of CFCs produced per LTC-IC: no significant differences were detected between VENTX and the GFP control with regard to the frequency of LTC-IC (453 LTC-IC vs. 801 LTC-IC/1× 106 cells, respectively; P = n.s.). Furthermore, the number of colonies generated per LTC-IC did not significantly differ between the two arms (175.5 ± 13 CFCs/LTC-IC for VENTX and 170 ± 18 CFCs/LTC-IC for the control) (P = n.s.).
In vivo VENTX engrafted mice showed a 2.3-fold increase in the proportion of engrafted transduced human CD34+ cells (VENTX: 13.0% ± 2.5% SEM, n = 5; GFP: 5.8% ± 1.2% SEM, n = 4; absolute cell numbers 7.1 × 104 ± 3 × 104 SEM vs. 1.3 × 105 ± 3.8 × 105 SEM, respectively). However, this difference was not statistically significant.
To determine if VENTX would affect the frequency of long-term repopulating cells, limiting dilution competitive repopulating unit (CRU) assays were performed (n = 4). In this assay, constitutive expression of VENTX failed to induce a significant change in the CRU frequency (Table S4). In conclusion, these data demonstrate that VENTX expression promotes myeloid development but has no major impact at the hematopoietic stem cell level in vitro and in vivo.
Stable Knockdown of VENTX Reduces the Proliferative Potential of AML Cell Lines.
To determine the functional significance of aberrant expression of VENTX in myeloid leukemias, we used lentivirus-mediated short hairpin RNA (shRNA) delivery for long-term silencing of VENTX in human AML cell lines.
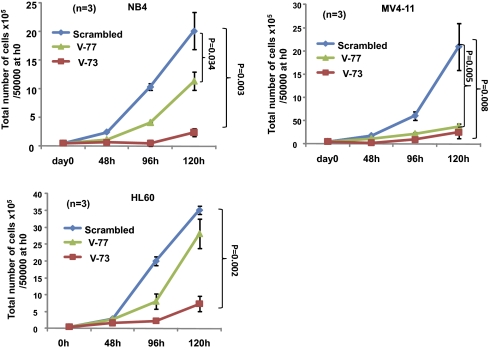
We first validated the VENTX shRNA constructs in NB4 and MV4-11 cell lines, transducing them with four individual shRNA constructs: the shRNA TRCN15973 (V73) (56% and 66% mRNA knockdown, respectively, in NB4 and MV4-11 cell lines) and the TRCN15977 (V77) (62% and 62% mRNA knockdown, respectively, in NB4 and MV4-11 cell lines) exhibited the most potent effect as compared with the nonsilencing control construct (scrambled) (Fig. S8A). Western blot showed that shRNA-mediated suppression of VENTX mRNA was associated with greatly diminished levels of VENTX protein (Fig. S8B). We then analyzed the effect of VENTX silencing in three VENTX-expressing AML cell lines: MV4-11, NB4, and HL60 [expression level: ΔCT 4.47, 5.30, and 9.30, respectively, when normalized to TATA binding protein (TBP)]. In these cell lines, silencing of VENTX expression by shRNA V-73 (mRNA knockdown between 58% and 75% and undetectable until 45 cycles, respectively) led to a significant reduction in cell number compared with the nonsilencing control construct (>79% after 120 h; Fig. 5). Similarly, shRNA V-77 induced a significant reduction in cell number compared with the nonsilencing control construct (56–79% after 120 h; Fig. 5). These results indicate that growth of human leukemic cell lines depends on VENTX expression in vitro.
Fig. 5.
Proliferation of AML cell lines after shRNA-mediated silencing of VENTX expression. Down-regulation of VENTX expression by shRNA V73 and V77-inhibited proliferation of the VENTX-expressing AML cell lines MV4-11, NB4, and HL60. In contrast, treatment with scrambled shRNA had no inhibitory effect (control). Values are represented as ±SD.
Discussion
In this report we functionally characterize the human Vent-like gene VENTX in human hematopoiesis. Our data demonstrate that VENTX has a distinct expression pattern in primary human hematopoietic cells, which is opposite to the expression signature of class I HOX genes. Thus, in contrast to HOX genes such as HOXB4 (22), VENTX is silent in the most immature CD34+/CD38− stem-cell compartment, followed by up-regulation of expression in CD34−/lin+ cells. These data demonstrate that homeobox genes of different classes can be expressed in a complementary way during hematopoiesis. Notably, expression of VENTX was highest in myeloid cells and comparably low in lymphoid cells. This expression pattern suggests that VENTX is more involved in myeloid differentiation pathways than in stem cell or lymphoid cell development. This was supported by gene expression and pathway analyses that demonstrated that VENTX expression in normal human progenitors for 48 h initiated genes mostly associated with myeloid development, whereas genes supporting lymphoid development were down-regulated. Interestingly, the LEF1/TCF pathway, which is critical for early B-cell development (23), was identified as being down-regulated by VENTX. This is in line with a recently published report, which demonstrated that VENTX forms a complex with LEF1 and inhibits LEF1/TCF-mediated transcription (24). Thus, high expression of VENTX in the critical phase of early hematopoiesis may promote myeloid lineage specification and impair B-cell development via down-regulation of LEF1.
Importantly, our experiments, which applied the technique of retroviral gene transfer to induce constitutive expression in normal human CD34+ progenitors, exactly recapitulated this with a marked increase in myeloid development and impaired growth of B-lymphoid cells in vitro and in vivo. One of the striking effects of VENTX in vitro was the impairment of red-cell colony formation, which was in line with down-regulation of the EpoR by VENTX in our gene expression studies. There are no data linking VENTX or its target gene LEF1 to erythroid development. However, studies in Xenopus have shown that Xvent-2 can directly interact with the C-terminal domain of GATA-2, an essential transcription factor for erythroid development (15).
Again in line with the observed expression pattern, enforced VENTX expression did not show any significant impact on the stem cell pool as assayed by LTC-IC assays in vitro and by CRU assays in vivo. This is in clear contrast to stem-cell–associated HOX genes such as HOXB4, which has been shown to expand the human hematopoietic stem-cell pool, using the same methodology as employed in this report (2).
Intriguingly, and in contrast to normal CD34+ cells, which showed absent or very low expression of VENTX, leukemic CD34+ cells isolated from patients with t(8;21)-positive AML displayed aberrantly high expression of this gene. The CD34+ compartment in AML is of particular interest as it was shown that, in the vast majority of cases, it harbors LSCs, which are responsible for the initiation and maintenance of the disease (25, 26). Thus, aberrant expression of VENTX in the LSC candidates might contribute to the development of leukemias with myeloid phenotype. In our in vivo NOD/SCID assays, we observed a perturbation of hematopoietic development with a significant shift of the lymphoid/myeloid ratio toward the myeloid side. In addition, knockdown of VENTX in human AML cell lines inhibited their proliferative potential significantly, indicating that VENTX expression is highly relevant for the proliferative activity of AML cells. Despite this, it is unlikely that aberrant expression of VENTX is leukemogenic on its own, but it is interesting to speculate that VENTX might be among the factors in AML that facilitate the decision of the preleukemic or leukemic clone to develop toward the myeloid lineage. The role of VENTX might differ substantially depending on the cellular context: in malignancies that depend on high LEF1 such as chronic lymphoid leukemia (CLL), VENTX might act as a potential tumor suppressor by controlling LEF1-mediated transcription (24, 27).
In summary, our data further support the concept that factors involved in mesodermal development and blood formation during embryonic development can act as key regulators in adult steady-state hematopoiesis and display distinct effects on proliferation and differentiation of hematopoietic progenitors and stem-cell populations. In addition, the data reported here suggest that aberrant expression of mesodermal factors such as VENTX might contribute to human myeloid leukemogenesis.
Materials and Methods
Human CD34+ Cells and Leukemic Patient Samples.
Cord blood was obtained from mothers undergoing cesarean delivery of normal, full-term infants and collected in heparin-coated syringes. Approved institutional procedures were followed to obtain informed consent from the mothers. Mononuclear cells prepared from diagnostic bone marrow or peripheral blood samples from adult patients with AML were analyzed as described before (9). The AML cases were classified according to the French–American–British criteria and the World Health Organization classification. The study abided by the rules of the local internal review board and the tenets of the revised Helsinki protocol (http://www.wma.net/e/policy/b3.htm).
Quantification of VENTX Expression.
Expression of VENTX was assayed by the TaqMan qRT–PCR method in total human CB and BM cells as well as in highly purified CB and BM subpopulations. VENTX primer and probes were selected in a way that prevented amplification of known VENTX pseudogenes (Applied Biosystems). For further information, see SI Materials and Methods.
Retroviral Vectors and Transduction.
For further information, see SI Materials and Methods.
In Vitro Progenitor Assays.
For further information, see SI Materials and Methods.
Transplantation of Human Cells in NOD/SCID Mice.
Eight-week-old NOD/SCID mice (Taconic) were sublethally irradiated with 275 cGy from a 137Cs source 1 day before injection. For transplantation, transduced cells were washed, counted, resuspended in PBS, and injected into the lateral tail vein of irradiated mice (300–400 μl/mouse: unselected progeny of 3–5 × 105 CD34+ cells in each arm). The mice were killed 6–8 wk after transplantation according to institutional guidelines. BM from the femurs and tibiae were flushed using Iscove's Modified Dulbecco's Medium (IMDM) containing 10% FCS and 0.2 mM EDTA and collected for analysis of human engraftment.
Flow Cytometric Analyses.
Cells isolated from colonies plated in methylcellulose or harvested NOD/SCID BM cells were stained according to standard protocols (SI Materials and Methods).
RNAi-Mediated Silencing of VENTX Expression.
Four pLKO.1-based lentiviral vectors that contain stem-loop cassettes encoding shRNAs targeted to the coding sequence (TRC15973 to exon 3, TRCN15977 and TRCN15975 to exon 2, and TRCN15976 to exon 1) of the human VENTX mRNA (GenBank accession no. NM_014468.2–320s1c1) were obtained from Sigma-Aldrich. For further information, see SI Materials and Methods.
Statistical Analysis.
Differences between two groups were assessed by Student's t test (Microsoft Excel). CRU frequencies were determined using Poisson statistics and the method of maximum likelihood with the assistance of the L-Calc software (Stem Cell Technologies).
Supplementary Material
Acknowledgments
We thank Shiva Bamezai, Nadine Olk, Nicole Behm, and Bianka Ksienzyk for cell sorting and technical support. We also thank the Department of Obstetrics and Gynecology of the Klinikum Grosshadern, Munich, for supplying umbilical cord blood samples. This work was supported by Grant 70-2968 from the German Cancer Foundation and by Grant SFB 684 A7 PD (to M.F.-B) from the Deutsche Forschungsgemeinschaft. V.P.S.R. and M.F.-B. are supported by Grant DFG No RA 1998/1-1 from the German Research Foundation; M.F.-B was supported by Deutsche Forschungsgemeinschaft Grant SFB 684 project A8; C.B. was supported by Deutsche Forschungsgemeinschaft Grant SFB 684 project A7; C.B. and M.F.B. are supported by the Bundesministerium für Bildung und Forschung (NGFN2 Grant 01GS0448); S.K.B. is supported by SFB684 project A6; and M.A.M. is a recipient of a German Academic Exchange Service (Deutscher Akademischer Austauschdienst) scholarship.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1001878107/-/DCSupplemental.
References
- 1.Antonchuk J, Sauvageau G, Humphries RK. HOXB4-induced expansion of adult hematopoietic stem cells ex vivo. Cell. 2002;109:39–45. doi: 10.1016/s0092-8674(02)00697-9. [DOI] [PubMed] [Google Scholar]
- 2.Buske C, et al. Deregulated expression of HOXB4 enhances the primitive growth activity of human hematopoietic cells. Blood. 2002;100:862–868. doi: 10.1182/blood-2002-01-0220. [DOI] [PubMed] [Google Scholar]
- 3.Buske C, Humphries RK. Homeobox genes in leukemogenesis. Int J Hematol. 2000;71:301–308. [PubMed] [Google Scholar]
- 4.Thorsteinsdottir U, Kroon E, Jerome L, Blasi F, Sauvageau G. Defining roles for HOX and MEIS1 genes in induction of acute myeloid leukemia. Mol Cell Biol. 2001;21:224–234. doi: 10.1128/MCB.21.1.224-234.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thorsteinsdottir U, et al. Overexpression of HOXA10 in murine hematopoietic cells perturbs both myeloid and lymphoid differentiation and leads to acute myeloid leukemia. Mol Cell Biol. 1997;17:495–505. doi: 10.1128/mcb.17.1.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buske C, et al. Overexpression of HOXA10 perturbs human lymphomyelopoiesis in vitro and in vivo. Blood. 2001;97:2286–2292. doi: 10.1182/blood.v97.8.2286. [DOI] [PubMed] [Google Scholar]
- 7.Golub TR, et al. Molecular classification of cancer: Class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 8.Rawat VP, et al. Ectopic expression of the homeobox gene Cdx2 is the transforming event in a mouse model of t(12;13)(p13;q12) acute myeloid leukemia. Proc Natl Acad Sci USA. 2004;101:817–822. doi: 10.1073/pnas.0305555101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rawat VP, et al. Overexpression of CDX2 perturbs HOX gene expression in murine progenitors depending on its N-terminal domain and is closely correlated with deregulated HOX gene expression in human acute myeloid leukemia. Blood. 2008;111:309–319. doi: 10.1182/blood-2007-04-085407. [DOI] [PubMed] [Google Scholar]
- 10.Scholl C, et al. The homeobox gene CDX2 is aberrantly expressed in most cases of acute myeloid leukemia and promotes leukemogenesis. J Clin Invest. 2007;117:1037–1048. doi: 10.1172/JCI30182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davidson AJ, Zon LI. Turning mesoderm into blood: The formation of hematopoietic stem cells during embryogenesis. Curr Top Dev Biol. 2000;50:45–60. doi: 10.1016/s0070-2153(00)50003-9. [DOI] [PubMed] [Google Scholar]
- 12.Rastegar S, Friedle H, Frommer G, Knöchel W. Transcriptional regulation of Xvent homeobox genes. Mech Dev. 1999;81:139–149. doi: 10.1016/s0925-4773(98)00239-1. [DOI] [PubMed] [Google Scholar]
- 13.Ault KT, Dirksen ML, Jamrich M. A novel homeobox gene PV.1 mediates induction of ventral mesoderm in Xenopus embryos. Proc Natl Acad Sci USA. 1996;93:6415–6420. doi: 10.1073/pnas.93.13.6415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huber TL, Zon LI. Transcriptional regulation of blood formation during Xenopus development. Semin Immunol. 1998;10:103–109. doi: 10.1006/smim.1998.0111. [DOI] [PubMed] [Google Scholar]
- 15.Friedle H, Knöchel W. Cooperative interaction of Xvent-2 and GATA-2 in the activation of the ventral homeobox gene Xvent-1B. J Biol Chem. 2002;277:23872–23881. doi: 10.1074/jbc.M201831200. [DOI] [PubMed] [Google Scholar]
- 16.Henningfeld KA, Friedle H, Rastegar S, Knöchel W. Autoregulation of Xvent-2B: Direct interaction and functional cooperation of Xvent-2 and Smad1. J Biol Chem. 2002;277:2097–2103. doi: 10.1074/jbc.M108524200. [DOI] [PubMed] [Google Scholar]
- 17.Deramaudt TB, Remy P, Stiegler P. Identification of interaction partners for two closely-related members of the ETS protein family, FLI and ERG. Gene. 2001;274:169–177. doi: 10.1016/s0378-1119(01)00610-2. [DOI] [PubMed] [Google Scholar]
- 18.Schuler-Metz A, Knöchel S, Kaufmann E, Knöchel W. The homeodomain transcription factor Xvent-2 mediates autocatalytic regulation of BMP-4 expression in Xenopus embryos. J Biol Chem. 2000;275:34365–34374. doi: 10.1074/jbc.M003915200. [DOI] [PubMed] [Google Scholar]
- 19.Moretti PA, et al. Molecular cloning of a human Vent-like homeobox gene. Genomics. 2001;76:21–29. doi: 10.1006/geno.2001.6574. [DOI] [PubMed] [Google Scholar]
- 20.Holland PW, Booth HA, Bruford EA. Classification and nomenclature of all human homeobox genes. BMC Biol. 2007;5:47. doi: 10.1186/1741-7007-5-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kilpinen S, et al. Systematic bioinformatic analysis of expression levels of 17,330 human genes across 9,783 samples from 175 types of healthy and pathological tissues. Genome Biol. 2008;9:R139. doi: 10.1186/gb-2008-9-9-r139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sauvageau G, et al. Differential expression of homeobox genes in functionally distinct CD34+ subpopulations of human bone marrow cells. Proc Natl Acad Sci USA. 1994;91:12223–12227. doi: 10.1073/pnas.91.25.12223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reya T, et al. Wnt signaling regulates B lymphocyte proliferation through a LEF-1 dependent mechanism. Immunity. 2000;13:15–24. doi: 10.1016/s1074-7613(00)00004-2. [DOI] [PubMed] [Google Scholar]
- 24.Gao H, et al. VentX, a novel lymphoid-enhancing factor/T-cell factor-associated transcription repressor, is a putative tumor suppressor. Cancer Res. 2010;70:202–211. doi: 10.1158/0008-5472.CAN-09-2668. [DOI] [PubMed] [Google Scholar]
- 25.Dick JE. Stem cell concepts renew cancer research. Blood. 2008;112:4793–4807. doi: 10.1182/blood-2008-08-077941. [DOI] [PubMed] [Google Scholar]
- 26.Taussig DC, et al. Anti-CD38 antibody-mediated clearance of human repopulating cells masks the heterogeneity of leukemia-initiating cells. Blood. 2008;112:568–575. doi: 10.1182/blood-2007-10-118331. [DOI] [PubMed] [Google Scholar]
- 27.Howe D, Bromidge T. Variation of LEF-1 mRNA expression in low-grade B-cell non-Hodgkin's lymphoma. Leuk Res. 2006;30:29–32. doi: 10.1016/j.leukres.2005.06.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.