Abstract
Pacidamycins are a family of uridyl tetra/pentapeptide antibiotics that act on the translocase MraY to block bacterial cell wall assembly. To elucidate the biosynthetic logic of pacidamcyins, a putative gene cluster was identified by 454 shotgun genome sequencing of the producer Streptomyces coeruleorubidus NRRL 18370. The 31-kb gene cluster encodes 22 proteins (PacA-V), including highly dissociated nonribosomal peptide synthetase (NRPS) modules and a variety of tailoring enzymes. Gene deletions confirmed that two NRPSs, PacP and PacO, are required for the biosynthesis of pacidamycins. Heterologous expression and in vitro assays of PacL, PacO, and PacP established reversible formation of m-Tyr-AMP, l-Ala-AMP, and diaminopropionyl-AMP, respectively, consistent with the amino acids found in pacidamycin scaffolds. The unusual Ala4-Phe5 dipeptidyl ureido linkage was formed during in vitro assays containing purified PacL, PacJ, PacN, and PacO. Both the genetic and enzymatic studies validate identification of the biosynthetic genes for this subclass of uridyl peptide antibiotics and provide the basis for future mechanistic study of their biosynthesis.
Keywords: adenylation, ureido-bond, uridine
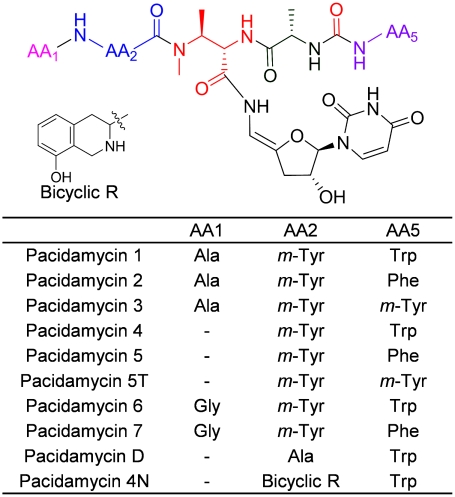
Pacidamycins are a family of uridyl tetra/pentapeptide antibiotics isolated from Streptomyces coeruleorubidus. Since their discovery in 1989, at least 10 related compounds have been reported (1, 2), which share a common structural skeleton with a 3′-deoxyuridine nucleoside attached to an N-methyl 2,3-diaminobutyric acid (DABA) residue via a 4′,5′-enamide linkage. Attached to the α-amino of DABA is l-Ala, which is linked to the C-terminal aromatic amino acid via a ureido linkage. An N-terminal amino acid or dipeptide is attached to the β-amino of DABA to give the tetra/pentapeptide framework (Fig. 1). Similar to many other uridyl peptides and uridyl lipopeptides, including liposidomycins (3), tunicamycins (4), capuramycins (5, 6), muraymycins (7), and more closely related mureidomycins (8) and napsamycins (9), pacidamycins exhibited antimicrobial activity targeting the translocase MraY to block formation of lipid I from UDP-N-acetylmuramoyl-pentapeptide and undecaprenyl phosphate during bacterial cell wall assembly (Fig. S1) (10, 11). The uracil-ribose moiety is a key determinant of binding to the MraY target (10).
Fig. 1.
Structures of pacidamycins.
We have begun to focus on the biosynthesis of pacidamycin family uridyl peptide antibiotics due to their three unusual structural features (Fig. 1). The first one is the presence of the nonproteinogenic amino acids l-meta-tyrosine (l-m-Tyr) and DABA, which suggests that nonribosomal peptide synthetase (NRPS) modules might be involved in the tetra/pentapeptide framework formation. The second feature is that the peptide chain direction reverses twice during assembly. As exemplified in pacidamycin 1, the peptide bond between m-Tyr2 and DABA3 is a β- rather than an α-peptide linkage. Consequently, the DABA3-Ala4 peptide bond uses the α-amino of DABA moiety and the chain is reversed at Ala4. Next, the attachment of that Ala4 to Trp5 involves a ureido linkage, which constitutes a second chain direction reversal such that Tyr5 has a free C-terminal carboxylate. Our laboratory has recently established that an NRPS module in syringolin biosynthesis catalyzes such a ureido-bond formation (12). The third remarkable feature of the pacidamycin scaffold is the nature and attachment mode of the uracil-based nucleoside to the peptide chain. The carboxyl moiety of DABA, rather than being utilized for a normal peptide linkage in chain extension (for example, linked to the amino group of Ala4), is tethered as an amide but a special one: the cis-enamide of a 3′-deoxy-4′,5′-enamino-uridine. Thus the DABA residue serves both as a branching element and a connection point for the unusual uridine moiety.
The biosynthesis of peptidyl nucleoside antibiotics has been actively pursued recently for the possible development of previously undescribed classes of MraY-targeted antibacterial drugs. In the past 2 years the biosynthetic gene clusters were identified for polyoxins (13), caprazamycins (14) and related liposidomycins (15), and A500359s (a member of the capuramycin family) (16) via PCR screening of the cosmid libraries. However, little is known about the biosynthesis of the subfamily of pacidamycins, mureidomycins, and napsamycins uridyl peptide antibiotics, and none of the above PCR probes would be useful for identifying the gene cluster for this subfamily. A β-replacement reaction has been reported in the conversion of threonine to DABA in mureidomycin A, albeit the dedicated enzyme was not identified (17). To understand what types of reactions and enzyme catalysts are involved in both the construction of the nonproteinogenic amino acids and uridine building blocks and the assembly of these branched deoxyuridine-ureido-peptide scaffolds, we have undertaken the identification of the pacidamycin biosynthetic gene cluster from the producer S. coeruleorubidus NRRL 18370 through genome scanning. The identity of the cluster has been confirmed by both genetic and biochemical characterizations. After completion of this study, a parallel effort in S. coeruleorubidus genome sequencing has independently identified the same 22 gene cluster (18).
Results and Discussion
Identification of Pacidamycin Gene Cluster from S. coeruleorubidus.
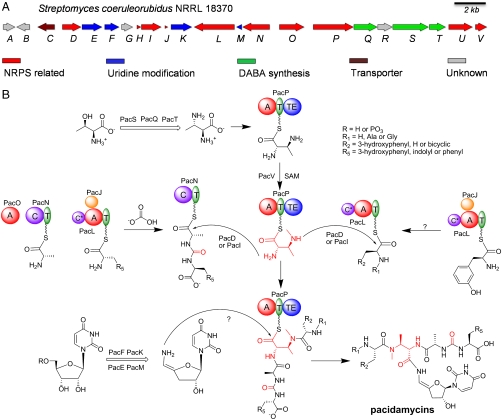
The 454 shotgun sequencing of S. coeruleorubidus NRRL 18370 with the GS FLX Titanium series at University of California at Los Angeles (UCLA) generated a total of ∼208 million bases. Assembly of the unpaired sequence reads resulted in 10,850,001 nonredundant bases distributed over 212 contigs. Using a local BLASTP program queried against a database consisting of all the contigs, more than 20 putative NRPS modules were found. To spot the sequence region potentially responsible for pacidamycin synthesis, sequences of a cysteine synthase and an argininosuccinate lyase were further used as probes for BLASTP. These two enzymes have been demonstrated to be essential for DABA synthesis in the lipopeptide antibiotic friulimicin by gene inactivation and subsequent DABA feeding experiments (19). The bioinformatic search identified one putative gene cluster for pacidamycins, which spans ∼31 kb of genomic DNA on a single contig and consists of 22 open reading frames (ORFs), here designated pacA-V (Fig. 2 and Table 1). Of these ORFs 17 can be assigned roles in pacidamycin biosynthesis: 8 encode dissociated NRPS modules including a total of 4 adenylation (A) domains, 4 thiolation (T) domains, 3 condensation (C) domains, and 1 thioesterase (TE) domain. None of these NRPSs have more than three domains, analogous to the previously described fragmented assembly line of andrimid (20). Thus there are one freestanding T domain and two freestanding A and C domains, raising questions about how functional modules assemble and provide specific flux during peptidyl-chain growth. One MbtH-like protein (PacJ) is encoded in the cluster that might interact with A domains as indicated previously (21, 22). One stand-alone S-adenosylmethionine (SAM)-dependent N-methyltransferase (PacV) was also detected, and putatively assigned to N-methylation of DABA. We propose that all of these NRPS-related ORFs function in the assembly of tetra/pentapeptide framework, with some domains perhaps used iteratively. The highly dissociated nature of these NRPSs, together with nonstringent domain–domain interactions and relaxed amino acid substrate specificity, may account for the production of up to 10 pacidamycins varied in the peptide scaffold.
Fig. 2.
Map of pacidamycin gene cluster and proposed biosynthetic pathway. (A) Organization of pac genes. (B) Proposed pathway for pacidamycin biosynthesis. Domain notation: T, thiolation; A, adenylation; C, condensation; TE, thioesterase.
Table 1.
Genes in the pacidamycin biosynthetic cluster from S. coeruleorubidus NRRL 18370 contig00048 and deduced roles based on sequence homology
| Gene | Size, aa | Deduced role | Protein homolog* | Accession number | Protein similarity/identity, %/% |
| pacA | 258 | Hypothetical protein | Caci_2672 [Catenulispora acidiphila DSM44928] | YP_003113429 | 39/52 |
| pacB | 285 | Hypothetical protein | SSEG_09055 [Streptomyces sviceus ATCC 29083] | ZP_05017494 | 43/60 |
| pacC | 364 | Major facilitator transporter | Caci_7785 [Catenulispora acidiphila DSM 44928] | YP_003118450 | 54/71 |
| pacD | 398 | NRPS (C) | Pden_3015 [Paracoccusdenitrificans PD1222] | YP_916794 | 28/40 |
| pacE | 426 | PLP-dependent aminotransferase | CetH [Actinomyces sp. Lu 9419] | ACH85568 | 51/65 |
| pacF | 311 | Fe(II)/2-oxoglutarate oxygenase | RegB [Actinoplanes friuliensis] | CAD32906 | 37/51 |
| pacG | 244 | Hypothetical protein | SSCG_02980 [Streptomyces clavuligerus ATCC 27064] | ZP_06771352 | 29/48 |
| pacH | 93 | NRPS (T) | SghaA1_010100031768 [Streptomyces ghanaensis ATCC 14672] | ZP_04689798 | 40/66 |
| pacI | 421 | NRPS (C) | NrpA [Ralstonia eutropha H16] | YP_841202 | 28/42 |
| pacJ | 72 | MbtH domain-containing protein | Bcep1808_1578 [Burkholderia vietnamiensis G4] | YP_001119420 | 58/74 |
| pacK | 443 | FAD-dependent oxidoreductase | SSEG_08816 [Streptomyces sviceus ATCC 29083] | ZP_05016850 | 54/65 |
| pacL | 849 | NRPS (C*-A-T) | PstC [Actinoplanes friuliensis] | CAM56770 | 46/60 |
| pacM | 121 | Cupin-2 domain-containing isomerase | Mbar_A1689 [Methanosarcina barkeri str. Fusaro] | YP_305211 | 32/56 |
| pacN | 574 | NRPS (C-T) | AptA2 [Anabaena sp. 90] | ACZ55942 | 31/50 |
| pacO | 527 | NRPS (A) | bcere0026_56870 [Bacillus cereus AH603] | ZP_04200912 | 40/57 |
| pacP | 848 | NRPS (A-T-TE) | SghaA1_010100001257 [Streptomyces ghanaensis ATCC 14672] | ZP_04683778 | 48/61 |
| pacQ | 498 | Argininosuccinate lyase | orf_R2 [Streptomyces collinus Tu 365] | CAN89657 | 51/62 |
| pacR | 318 | Kinase | PduX [Streptomyces kanamyceticus NBRC 13414] | BAE95583 | 54/65 |
| pacS | 755 | Fusion protein containing a PLP-dependent cysteine synthase and an argininosuccinate lyase | orf_R4 [Streptomyces collinus Tu 365] | CAN89659 | 58/68 |
| pacT | 350 | PLP-dependent threonine aldolase | bcere0007_32170 [Bacillus cereus AH621] | ZP_04295986 | 61/76 |
| pacU | 507 | NRPS (A) | Strop_2774 [Salinispora tropica CNB-440] | YP_001159594 | 58/69 |
| pacV | 250 | SAM-dependent N-methyltransferase | sce7770 [Sorangium cellulosum “So ce 56”] | YP_001618419 | 37/51 |
*Results generated by BLAST analysis, S. roseosporus homologs (best homologs for all ORFs) are excluded.
Three genes predicted to be on the same operon, pacQST, are postulated to be involved in the biosynthesis of the DABA moiety. PacQ displayed significant homology to an argininosuccinate lyase, and PacS was predicted to be a fusion protein containing both a pyridoxal phosphate (PLP)-dependent cysteine synthase and an argininosuccinate lyase domain. Consistent with DABA synthesis in friulimicin (19), PacS is proposed to catalyze the β-replacement of threonine hydroxyl by the α-amino of aspartate, and the intermediate then breaks down to give DABA with the release of fumarate promoted by argininosuccinate lyase PacQ or PacS. The configuration of DABA in pacidamycins was determined to be 2S,3S, different from the 2S,3R configuration of DABA in friulimicin and natural l-Thr (11). PacT, a PLP-dependent threonine aldolase homolog, not encoded in friulimicin gene cluster, is thus proposed to be responsible for the 3S configuration of DABA (Fig. S2). The remaining four tailoring enzymes encoded in the gene cluster are putatively assigned for uridine nucleoside modification, including a Fe(II)/α-ketoglutarate-dependent oxygenase (PacF), a flavin adenine dinucleotide (FAD)-dependent oxidoreductase (PacK), a PLP-dependent aminotransferase (PacE), and a cupin-2 domain-containing isomerase (PacM). Although uridine-5′-aldehyde might be a common intermediate in the nucleoside modification of pacidamycins, caprazamycins, and liposidomycins, the proposed dedicated alcohol dehydrogenase (LpmW/Cpz25) encoded in caprazamycin and liposidomycin gene clusters was not found in the pacidamycin cluster (14, 15). Instead, either PacF or PacK might work on uridine or uridine-5′-monophophate to give uridine-5′-aldehyde, which is then subjected to 3′,4′-dehydration, 5′-transamination possibly catalyzed by PacE, and isomerization presumably catalyzed by PacM to yield the unique building block 3′-deoxy-4′,5′-enamino-uridine (Fig. S3).
PacC showed strong sequence similarity to major facilitator transporters, and probably functions to export pacidamycins out of the cell. There are other ORFs in the gene cluster encoding proteins with no obvious function, some of which might be related to regulation and antibiotic resistance. Interestingly, the whole biosynthetic gene cluster was also found in the published genome of Streptomyces roseosporus NRRL 15998, a known daptomycin producer, although no pacidamycin production has been reported from S. roseosporus. By comparing the sequences around the two clusters, the boundary of the pac cluster was putatively identified (Fig. S4). Notably there is a phenylalanine hydroxylase encoded in the cluster from S. roseosporus, which presumably catalyzes the formation of m-Tyr from Phe. The absence of the corresponding gene in the pac cluster from S. coeruleorubidus indicates that the identified cluster might be incomplete, and the putative gene encoding a phenylalanine (meta)-hydroxylase is located elsewhere on the genome revealed by BLASTP analysis.
In Vivo Gene Disruption Analysis.
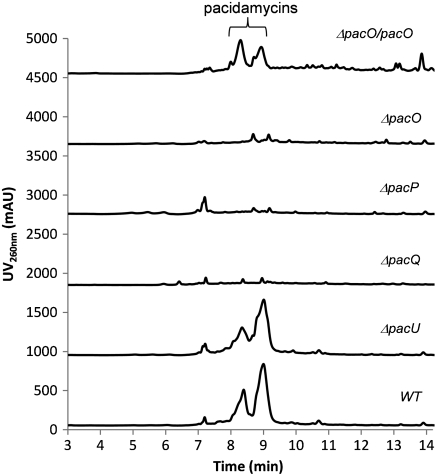
A set of gene disruption experiments was carried out to test the necessity of various genes in the biosynthesis of pacidamycins. The knockout targets include PacO, a single A domain, PacP, an A-T-TE tridomain NRPS, PacU, another single A domain and PacQ, the argininosuccinate lyase homolog. All of the genes were deleted in-frame through double crossover according to standard methods (23), and the resulting mutants were confirmed by PCR (Fig. S5). Following the reported growth condition and extraction technique (24), all 10 known pacidamycins could be detected from the culture of wild-type strain as major products by LC-HRMS analysis (Fig. 3 and Fig. S6). The production of pacidamycins was completely abolished in ΔpacO and ΔpacP mutants, demonstrating that these two genes are essential for the biosynthesis of pacidamycins (Fig. 3 and Figs. S7 and S8). In addition, when pacO was introduced back into the ΔpacO mutant, the transconjugant was found to have resumed pacidamycin production (Fig. 3 and Fig. S9). The disruption of pacQ significantly reduced the yield of all pacidamycins in comparison with that of the wild type (reduced to < 1%), with only trace amount of pacidamycins produced indicated by mass ion extraction (Fig. S10). This result suggested that PacQ plays an important role in the biosynthesis; however, its function might be complemented by other enzymes, such as PacS, the fusion protein with an argininosuccinate lyase domain. Surprisingly, the deletion of pacU had no impact on the production of nine pacidamycin compounds, but did abolish pacidamycin D production in the extract of ΔpacU mutant shown by mass ion extraction (Fig. 3 and Fig. S11). Pacidamycin D is a uridyl tetrapeptide with a single Ala1 attached to the β-amino group of DABA, whereas all other pacidamycins have a m-Tyr2 or its derivative at the corresponding position. The abolishment of pacidamycin D to make the uridyltetrapeptide framework indicated that PacU may be specifically related to activation of the alternative N-terminal Ala. In summary, the in vivo gene disruption experiments unambiguously verified that the identified gene cluster from S. coeruleorubidus genome is directly involved in the biosynthesis of pacidamycins. Independently, Rackham et al. have shown that heterologous expression of this gene cluster in Streptomyces lividans yielded pacidamycin D and a related variant, providing a parallel in vivo identification of the cluster, but they did not report any in vitro studies (18). In contrast, we studied the activity of seven encoded enzymes that allowed the assignment of their functions in the biosynthesis of pacidamycins.
Fig. 3.
UV traces (260 nm) during LC-HRMS analysis of metabolites produced by wild-type and mutant S. coeruleorubidus strains. See SI Figs. S6–S11 for detailed HRMS analysis.
Adenylation Domain Activity Assays.
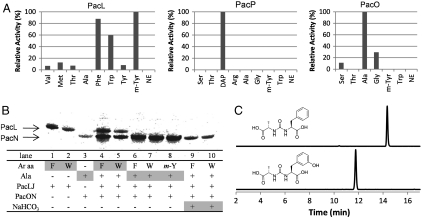
Adenylation domains are gate-keeping enzymes in NRPSs that are responsible for the building block selection in the peptide scaffold assembly. Based on pacidamycin structures, at least three different A domain activities are needed: for l-Ala, DABA, and aromatic amino acids (l-m-Tyr, l-Phe, and l-Trp) activation, respectively. All four A domain-containing proteins, PacL, PacO, PacP, and PacU were overexpressed and purified from Escherichia coli (Fig. S12), and their ability to reversibly adenylate various amino acids was tested using the classical ATP-[32P]PPi exchange assay. PacL is a C*-A-T tridomain protein (C* indicates a truncated and presumably nonfunctional C domain) that was predicted to activate l-Val/l-Leu-like hydrophobic branched amino acids based on the 10-residue specificity sequence (10AA code) (Table S1). As the prediction was inconsistent with amino acid components in pacidamycins, a broad range of substrates was tested. Although PacL alone showed no obvious activity toward all of the amino acids tested, it preferentially activated aromatic amino acids in the presence of PacJ, the small MbtH-like protein (Fig. 4A). MbtH domain-containing proteins have been found in many NRPS-encoding gene clusters and have been shown to be important for metabolite assembly in vivo (22). The PacJ activation of PacL will be explored in detail elsewhere. The preferential activation of m-Tyr, l-Phe, and l-Trp but not l-Tyr executed by PacL/PacJ is in good agreement with the aromatic amino acids found at the C terminus of pacidamycins. PacP is a A-T-TE tridomain protein predicted to activate l-Ser/l-Arg by bioinformatic analysis. It is encoded in the same operon with pacQST (genes related to DABA synthesis) and is therefore postulated to be involved in DABA activation. Because 2S,3S-DABA was not readily available, its analog l-2,3-diaminopropionate (DAP) was used in the ATP-PPi exchange assay. PacP exhibited a strong preference for activation of DAP over all other amino acids, strongly suggesting that it is a DABA activation enzyme (Fig. 4A). The remaining two stand-alone A domain proteins, PacO and PacU, were predicted to activate l-Ala and l-Pro/l-p-hydroxyphenylglycine, respectively, based on the 10AA code. Indeed, PacO demonstrated a preference toward reversible formation of l-Ala-AMP, with modest activation of l-Gly (Fig. 4A). However, no substantial activation of any amino acids was detected in PacU assays, with or without the presence of PacJ. The amino acid specificities of PacP, PacO, and PacU correlate well with the in vivo gene disruption results, in which pacP and pacO were essential for pacidamycins synthesis, whereas pacU was not needed for the synthesis of 9 out of 10 pacidamycins (Fig. 3). It is unclear, given the in vivo deletion result noted above on loss of pacidamycin D, why PacU did not activate l-Ala in the in vitro assay; it is possible that PacU was purified from E. coli in an inactive form. It can also be deduced that Ala4 in pacidamycins is selected by PacO.
Fig. 4.
Characterization of NRPSs. (A) A domain activity of PacL, PacP, and PacO. NE, no enzyme; m-Tyr, meta-tyrosine; DAP, l-2,3-diaminopropionate. All acids are l-configuration except m-Tyr (dl). One hundred percent relative activity for Ala, DAP, and m-Tyr-dependent exchange corresponds to 253 k, 251 k, and 139 k cpm, respectively. (B) Autoradiograph of SDS-PAGE gel illustrating the covalent loading of 14C-labeled substrate (shaded). Enzymes and amino acids used in each lane are indicated in Table 1. (C) Extracted ion chromatograms showing production of ureido dipeptides Ala-CO-Phe and Ala-CO-m-Tyr. The calculated mass with 10-ppm mass error tolerance was used.
Covalent Loading of Holo Forms of Thiolation Domains.
The loading of the adenylated amino acids to the adjacent or dissociated T domain holo forms could then be investigated using 14C-labeled substrate. Particularly significant is the loading of l-Ala activated by the freestanding adenylation domain PacO to identify its in trans loading partner. All four T domain-containing proteins, PacH, PacL, PacN, and PacP, were overexpressed and purified from E. coli BAP1 strain that contains a chromosomal copy of the phosphopantetheinyl transferase Sfp to ensure their posttranslational modification to the pantetheinylated forms (Fig. S12) (25). The 14C-labeled covalent aminoacyl-S-thiolation intermediate was then detected by SDS-PAGE autoradiography. Using [14C]l-Ala, PacN (a C-T didomain protein) was determined to be the cognate loading partner of PacO (Fig. 4B, lane 3); no Ala loading was detected on any of the other three holo T domains. In addition, as expected, PacL was covalently loaded with aromatic amino acid when using [14C]l-Phe and [14C]l-Trp as the substrate, respectively (Fig. 4B, lanes 1–2).
Ureido-Bond Formation Between Covalently Tethered Ala and Phe/Trp/m-Tyr.
When [14C]Phe/Trp-loaded PacL and Ala-S-PacN were combined, 14C-labeled l-Phe or l-Trp was transferred to PacN, whereas [14C]l-Ala remained on PacN (Fig. 4B, lanes 4–8). These results indicated the formation of covalent linkage between Ala tethered on PacN and aromatic amino acyl moiety undergoing transfer, presumably catalyzed by the C domain of PacN. To test if this represents ureido-linked Phe/Trp5-Ala4-S-PacN, because this 4–5 ureido link is observed in all the pacidamycins, [14C]NaHCO3 was fed as the only radioactive substrate to the PacL, PacN assays. Our prior work on a comparable ureido link in syringolin formation has shown that bicarbonate is the carbon source of the carbonyl moiety of that ureido linkage (12). Radioactivity migrated with PacN but not PacL (Fig. 4B, lanes 9–10), suggesting that indeed a Phe/Trp5-Ala4 ureido dipeptyl thioester was formed and tethered on the pantetheinyl arm of PacN. The identification of the ureido dipeptide was confirmed by liquid chromatography–high resolution mass spectrometry after hydrolytic release of the peptidyl-S-T domain intermediate with thioesterase TycF (26). In vitro assay with ATP, l-Ala, l-Phe, PacO, PacN, PacL, and PacJ yielded the ureido dipeptide Ala-CO-Phe (m/z = 303.0951 [M + Na]+, Δ = 0.8 mmu), and its mass was shifted by +1 using [13C]NaHCO3, or by +2 using [2,3-13C2]l-Ala as an alternative substrate (Fig. 4C and Fig. S13). The dipeptide Ala-CO-m-Tyr (m/z = 319.0901[M + Na]+, Δ = -0.2 mmu) was also formed when Phe was replaced by m-Tyr in vitro (Fig. 4C and Fig. S14). Therefore, we have successfully reconstituted the assembly of Ala4 and C-terminal aromatic amino acid residue (Phe/Trp/m-Tyr) with the chain-reversing ureido linkage. Ala4 is selected and activated as the Ala-AMP by PacO and loaded on HS-pantetheinyl-PacN; the C-terminal aromatic amino acid (Phe, Trp, or m-Tyr) is activated by PacL with the help of PacJ and loaded onto HS-pantetheinyl-PacL. PacN may then catalyze the ureido-bond formation between Ala-S-PacN and Phe/Trp/m-Tyr-S-PacL, yielding a ureido dipeptide tethered on PacN (Fig. 2B).
In summary, we have identified the gene cluster for pacidamycins that sets the stage for further deciphering of the chemical logic for assembly of this class of peptidyl nucleoside antibiotics. Our in vivo and in vitro experiments allow a first proposal of the pacidamycin biosynthetic pathway (Fig. 2B). The key bifunctional amino acid building block DABA is most probably synthesized from l-Thr catalyzed by PacQST, activated by and loaded onto PacP, and N-methylated by PacV. Promoted by one of the condensation domains (PacD or PacI), the α-amino of DABA nucleophilically attacks the PacN tethered thioester of the C-terminal ureido dipeptide, which is assembled by PacLJON as described above. The remaining condensation domain presumably directs the nucleophilic attack of β-amino of DABA (before or after its N-methylation) on the carbonyl of a thioester tethered N-terminal amino acid or dipeptide. Whether one or more of the above PacLJON also assembles Ala/Gly-m-Tyr as a normal dipeptidyl-S-T intermediate (rather than the ureido-linked dipeptide) for Ala/Gly1-m-Tyr2 is yet to be demonstrated. Finally, the TE domain of PacP is presumed to catalyze the release of tetra/pentapeptidyl-S-PacP by 3′-deoxy-4′,5′-enamino-uridine, a modified nucleoside possibly synthesized by PacEFKM from uridine, to yield uridyl tetra/pentapeptide pacidamycins via a 4′,5′-enamide linkage. Further biochemical study will provide more insight in the timing and mechanisms of the biosynthesis of this intriguing class of antibiotics.
Materials and Methods
454 Sequencing and Bioinformatic Analysis.
Streptomyces coeruleorubidus NRRL 18370 was obtained from U.S. Department of Agriculture Agricultural Research Service Culture Collection. The genomic DNA used for sequencing was prepared using the phenol/chloroform extraction method. The shotgun sequencing was performed at the GenoSeq (UCLA Genotyping and Sequencing Core) with the GS FLX Titanium system (Roche). The 454 sequencing reads were assembled into contigs with the GS De Novo Assembler software (Roche). The assembled data were converted into a local BLAST database for search using stand-alone BLAST software (ver. 2.2.18) downloaded from the National Center for Biotechnology Information Web site. ORFs were detected and analyzed using online program FGENESB (Softberry), and the putative roles of the proteins were assigned using protein–protein BLAST and Pfam analysis. The NRPS A domain specificity was predicted using online program NRPSpredictor (27). The nucleotide sequence of the gene cluster was deposited at GenBank.
Gene Disruptions in S. coeruleorubidus and Mutants Analysis.
In vivo generation of targeted mutations in S. coeruleorubidus was achieved by conjugative transfer of disruption plasmids from E. coli WM6026 and Streptomyces according to general protocols (23). The knockout cassettes were constructed using the ReDirect technology (28). An example for pacP is detailed here. A 6.5-kb fragment containing pacP flanked by 2-kb arms was PCR amplified from genomic DNA and inserted into PCRBlunt vector. This plasmid was introduced into E. coli BW25113/pIJ790 by electroporation. The acc(3)IV-oriT cassette amplified by PCR from pIJ773 was then introduced to replace the entire pacP using PCR targeting and λ-Red-mediated recombination. The resulting knockout cassette was transformed into E. coli WM6026 (a diaminopimelic acid auxotroph) for conjugation with S. coeruleorubidus. The double-crossover strain was obtained from antibiotic selection (ApraRKanS) and confirmed by PCR (Fig. S5). The pacO complementation experiment was performed using plasmid pIJ6902 and confirmed by PCR. Mutant analysis was carried out following the reported procedure (24). See SI Appendix for details. LC-HRMS analysis was performed on an Agilent Technologies 6520 Accurate-Mass QTOF LC-MS instrument with a Luna 3u C18 column (4.6 × 75 mm) from Phenomenex. A linear gradient of 2 to 95% CH3CN (vol/vol) over 15 min in H2O supplemented with 0.1% (vol/vol) formic acid at a flow rate of 0.5 mL/ min was used.
ATP-PPi Exchange Assays.
All proteins were expressed and purified with His6 tag following the general protocol (see SI Appendix, Fig. S12). The assays were preformed in 100 μL of reaction buffer (50 mM Tris-HCl/2 mM MgCl2, pH 7.8) containing 5 mM ATP, 1 mM Na4[32P]PPi (∼4 × 106 cpm/mL), 1 mM (tris(2-carboxyethyl)phosphine) (TCEP), 5 mM substrate, and 1 μM enzyme. Reactions were incubated at 25 °C for 1 h, then quenched by the addition of charcoal suspension (1.6% wt/vol activated charcoal, 0.1 M Na4PPi, 3.5% HClO4). Free [32P]PPi was removed by centrifugation of the sample followed by washing twice with wash solution (0.1 M Na4PPi and 3.5% HClO4). Charcoal-bound radioactivity was measured on a Beckman LS 6500 scintillation counter.
Loading Assays with 14C-Labeled Substrates.
A typical assay contained, in a total volume of 20 μL, 5 mM ATP, 2 mM MgCl2, 1 mM TCEP, 50 μM amino acids, 10 μM enzymes, and 50 mM Hepes, pH 8.0. 14C-labeled substrate was added to each reaction accordingly (l-Ala [0.13 μCi], l-Phe [0.4 μCi], l-Trp [0.052 μCi], or NaHCO3 [0.5 μCi]. After 1-h incubation at 25 °C, samples were quenched by adding 1× SDS sample buffer. Following SDS-PAGE, radiolabeled protein was detected using a BAS-III imaging plate (Fuji Film, 48- to 96-h exposure) and a Typhoon 9400 phosphorimager (GE Healthcare).
LC-HRMS Product Assays.
Assays were performed in 50 μL of 50 mM Hepes (pH 8.0) containing 5 mM ATP, 2 mM MgCl2, 1 mM TCEP, 5 mM l-Ala, 5 mM l-Phe or m-Tyr, 10 μM PacO, 20 μM PacN, 20 μM PacL, and 40 μM PacJ. After 2-h incubation at 25 °C, TycF (5 μM) was added and further incubated for 30 min. The proteins were then removed by 5-kDa MWCO (Molecular Weight Cutoff) filter tubes, and the filtered reaction mixture was subjected to LC-HRMS analysis. A linear gradient of 2 to 40% CH3CN (vol/vol) over 15 min in H2O supplemented with 0.1% (vol/vol) formic acid at a flow rate of 0.5 mL/ min was used.
Supplementary Material
Acknowledgments.
We thank Dr. Chris Neumann for providing the purified TycF and Dr. Heidi Imker for helpful discussion and proofreading of the manuscript. This work was supported by NIH Grant GM49338 (C.T.W.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. HM855229).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1011557107/-/DCSupplemental.
References
- 1.Fronko RM, et al. New pacidamycins produced by Streptomyces coeruleorubidus, NRRL 18370. J Antibiot (Tokyo) 2000;53:1405–1410. doi: 10.7164/antibiotics.53.1405. [DOI] [PubMed] [Google Scholar]
- 2.Chen RH, Buko AM, Whittern DN, McAlpine JB. Pacidamycins, a novel series of antibiotics with anti-Pseudomonas aeruginosa activity. II. Isolation and structural elucidation. J Antibiot (Tokyo) 1989;42:512–520. doi: 10.7164/antibiotics.42.512. [DOI] [PubMed] [Google Scholar]
- 3.Muroi M, Kimura K, Osada H, Inukai M, Takatsuki A. Liposidomycin B inhibits in vitro formation of polyprenyl (pyro)phosphate N-acetylglucosamine, an intermediate in glycoconjugate biosynthesis. J Antibiot (Tokyo) 1997;50:103–104. doi: 10.7164/antibiotics.50.103. [DOI] [PubMed] [Google Scholar]
- 4.Tamura G, Sasaki T, Matsuhashi M, Takatsuki A, Yamasaki M. Tunicamycin inhibits the formation of lipid intermediate in cell-free peptidoglycan synthesis of bacteria. Agric Biol Chem. 1976;40:447–449. [Google Scholar]
- 5.Hotoda H, et al. Synthesis and antimycobacterial activity of capuramycin analogues. Part 1: Substitution of the azepan-2-one moiety of capuramycin. Bioorg Med Chem Lett. 2003;13:2829–2832. doi: 10.1016/s0960-894x(03)00596-1. [DOI] [PubMed] [Google Scholar]
- 6.Hotoda H, et al. Synthesis and antimycobacterial activity of capuramycin analogues. Part 2: Acylated derivatives of capuramycin-related compounds. Bioorg Med Chem Lett. 2003;13:2833–2836. doi: 10.1016/s0960-894x(03)00597-3. [DOI] [PubMed] [Google Scholar]
- 7.McDonald LA, et al. Structures of the muraymycins, novel peptidoglycan biosynthesis inhibitors. J Am Chem Soc. 2002;124:10260–10261. doi: 10.1021/ja017748h. [DOI] [PubMed] [Google Scholar]
- 8.Isono F, Inukai M. Mureidomycin A, a new inhibitor of bacterial peptidoglycan synthesis. Antimicrob Agents Chemother. 1991;35:234–236. doi: 10.1128/aac.35.2.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chatterjee S, et al. Napsamycins, new Pseudomonas active antibiotics of the mureidomycin family from Streptomyces sp. HIL Y-82,11372. J Antibiot (Tokyo) 1994;47:595–598. doi: 10.7164/antibiotics.47.595. [DOI] [PubMed] [Google Scholar]
- 10.Winn M, Goss RJ, Kimura K, Bugg TD. Antimicrobial nucleoside antibiotics targeting cell wall assembly: Recent advances in structure-function studies and nucleoside biosynthesis. Nat Prod Rep. 2010;27:279–304. doi: 10.1039/b816215h. [DOI] [PubMed] [Google Scholar]
- 11.Boojamra CG, et al. Stereochemical elucidation and total synthesis of dihydropacidamycin D, a semisynthetic pacidamycin. J Am Chem Soc. 2001;123:870–874. doi: 10.1021/ja003292c. [DOI] [PubMed] [Google Scholar]
- 12.Imker HJ, Walsh CT, Wuest WM. SylC catalyzes ureido-bond formation during biosynthesis of the proteasome inhibitor syringolin A. J Am Chem Soc. 2009;131:18263–18265. doi: 10.1021/ja909170u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen W, et al. Characterization of the polyoxin biosynthetic gene cluster from Streptomyces cacaoi and engineered production of polyoxin H. J Biol Chem. 2009;284:10627–10638. doi: 10.1074/jbc.M807534200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaysser L, et al. Identification and manipulation of the caprazamycin gene cluster lead to new simplified liponucleoside antibiotics and give insights into the biosynthetic pathway. J Biol Chem. 2009;284:14987–14996. doi: 10.1074/jbc.M901258200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaysser L, Siebenberg S, Kammerer B, Gust B. Analysis of the liposidomycin gene cluster leads to the identification of new caprazamycin derivatives. ChemBioChem. 2010;11:191–196. doi: 10.1002/cbic.200900637. [DOI] [PubMed] [Google Scholar]
- 16.Funabashi M, et al. Identification of the biosynthetic gene cluster of A-500359s in Streptomyces griseus SANK60196. J Antibiot (Tokyo) 2009;62:325–332. doi: 10.1038/ja.2009.38. [DOI] [PubMed] [Google Scholar]
- 17.Lam WH, Rychli K, Bugg TD. Identification of a novel beta-replacement reaction in the biosynthesis of 2,3-diaminobutyric acid in peptidylnucleoside mureidomycin A. Org Biomol Chem. 2008;6:1912–1917. doi: 10.1039/b802585a. [DOI] [PubMed] [Google Scholar]
- 18.Rackham EJ, Gruschow S, Ragab AE, Dickens S, Goss RJ. Pacidamycin biosynthesis: Identification and heterologous expression of the first uridyl peptide antibiotic gene cluster. ChemBioChem. 2010;11:1700–1709. doi: 10.1002/cbic.201000200. [DOI] [PubMed] [Google Scholar]
- 19.Muller C, et al. Sequencing and analysis of the biosynthetic gene cluster of the lipopeptide antibiotic Friulimicin in Actinoplanes friuliensis. Antimicrob Agents Chemother. 2007;51:1028–1037. doi: 10.1128/AAC.00942-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jin M, Fischbach MA, Clardy J. A biosynthetic gene cluster for the acetyl-CoA carboxylase inhibitor andrimid. J Am Chem Soc. 2006;128:10660–10661. doi: 10.1021/ja063194c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heemstra JR, Jr, Walsh CT, Sattely ES. Enzymatic tailoring of ornithine in the biosynthesis of the Rhizobium cyclic trihydroxamate siderophore vicibactin. J Am Chem Soc. 2009;131:15317–15329. doi: 10.1021/ja9056008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lautru S, Oves-Costales D, Pernodet JL, Challis GL. MbtH-like protein-mediated cross-talk between non-ribosomal peptide antibiotic and siderophore biosynthetic pathways in Streptomyces coelicolor M145. Microbiology. 2007;153:1405–1412. doi: 10.1099/mic.0.2006/003145-0. [DOI] [PubMed] [Google Scholar]
- 23.Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. Practical Streptomyces Genetics. Norwich, UK: John Innes Foundation; 2000. [Google Scholar]
- 24.Gruschow S, et al. New pacidamycin antibiotics through precursor-directed biosynthesis. ChemBioChem. 2009;10:355–360. doi: 10.1002/cbic.200800575. [DOI] [PubMed] [Google Scholar]
- 25.Pfeifer BA, Admiraal SJ, Gramajo H, Cane DE, Khosla C. Biosynthesis of complex polyketides in a metabolically engineered strain of E. coli. Science. 2001;291:1790–1792. doi: 10.1126/science.1058092. [DOI] [PubMed] [Google Scholar]
- 26.Yeh E, Kohli RM, Bruner SD, Walsh CT. Type II thioesterase restores activity of a NRPS module stalled with an aminoacyl-S-enzyme that cannot be elongated. ChemBioChem. 2004;5:1290–1293. doi: 10.1002/cbic.200400077. [DOI] [PubMed] [Google Scholar]
- 27.Rausch C, Weber T, Kohlbacher O, Wohlleben W, Huson DH. Specificity prediction of adenylation domains in nonribosomal peptide synthetases (NRPS) using transductive support vector machines (TSVMs) Nucleic Acids Res. 2005;33:5799–5808. doi: 10.1093/nar/gki885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gust B, Challis GL, Fowler K, Kieser T, Chater KF. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc Natl Acad Sci USA. 2003;100:1541–1546. doi: 10.1073/pnas.0337542100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.