Abstract
The catalytic domain of the F-ATPase in mitochondria protrudes into the matrix of the organelle, and is attached to the membrane domain by central and peripheral stalks. Energy for the synthesis of ATP from ADP and phosphate is provided by the transmembrane proton-motive-force across the inner membrane, generated by respiration. The proton-motive force is coupled mechanically to ATP synthesis by the rotation at about 100 times per second of the central stalk and an attached ring of c-subunits in the membrane domain. Each c-subunit carries a glutamate exposed around the midpoint of the membrane on the external surface of the ring. The rotation is generated by protonation and deprotonation successively of each glutamate. Each 360° rotation produces three ATP molecules, and requires the translocation of one proton per glutamate by each c-subunit in the ring. In fungi, eubacteria, and plant chloroplasts, ring sizes of c10–c15 subunits have been observed, implying that these enzymes need 3.3–5 protons to make each ATP, but until now no higher eukaryote has been examined. As shown here in the structure of the bovine F1-c-ring complex, the c-ring has eight c-subunits. As the sequences of c-subunits are identical throughout almost all vertebrates and are highly conserved in invertebrates, their F-ATPases probably contain c8-rings also. Therefore, in about 50,000 vertebrate species, and probably in many or all of the two million invertebrate species, 2.7 protons are required by the F-ATPase to make each ATP molecule.
Keywords: ATP synthase, rotational catalysis, c-ring structure, protons per ATP, vertebrates
Almost all ATP in respiring cells is made by the membrane bound enzyme F-ATPase (F-ATP synthase). In the F-ATPase in the inner membranes of mitochondria, the energy of the transmembrane proton-motive-force, generated by respiration, is coupled mechanically to the synthesis of ATP from ADP and phosphate in its membrane extrinsic catalytic domain by rotating the asymmetrical central stalk in a clockwise direction (as viewed from the membrane) at about 100 times per second (1–4). The spherical catalytic domain, which protrudes into the matrix of the organelle, has three catalytic sites in β-subunits at interfaces with α-subunits (5). The rotational torque is resisted by the peripheral stalk which links the surface of the catalytic domain to subunit a (ATPase-6) in the membrane domain; together they constitute the stator (6). The asymmetry of the central stalk imposes different conformations on the three catalytic sites. In a ground state structure of the catalytic domain, two of them, the βDP and the βTP sites, have similar but significantly different closed conformations. Both bind nucleotides, but catalysis occurs at the βDP site. The third, or βE site, has a different open conformation with low nucleotide affinity (5). These three catalytic conformations correspond to “tight,” “loose,” and “open” states in a binding change mechanism of ATP hydrolysis and synthesis (7). Each 360° rotation of the central stalk takes each catalytic site through these conformations in which substrates bind, and three ATP molecules are made and released. The turning of the rotor is impelled by protons, driven across the inner membrane into the mitochondrial matrix by the transmembrane proton-motive force. The transmembrane pathway for protons in the a-subunit has not been defined structurally. This pathway allows protons in the intermembrane space to access an essential ionized carboxylate of a glutamate residue, midmembrane on the C-terminal α-helix of subunit c. Once protonated, this carboxylate moves to a more hydrophobic environment by Brownian motion generating a rotation of the ring. As succeeding c-subunits become protonated, each neutralized carboxylate reaches an environment in subunit a where it reionises, releasing the proton into the mitochondrial matrix (8). According to current models based on structures, the number of translocated protons for generation of each 360° rotation is the same as the number of c-subunits in the ring, as each c-subunit carries a carboxylate involved in protonation and deprotonation events. In the yeast F-ATPase, the ring has ten c-subunits, and so ten protons are translocated per three ATP molecules made during a 360° cycle; therefore, the bioenergetic cost to the enzyme is 3.3 protons per ATP (9). However, the c-ring sizes differ between species; c10–c15 rings have been found in yeast, eubacterial, and plant chloroplast F-ATPases (10–13). Therefore, the bioenergetic cost of these F-ATPases making an ATP molecule ranges from 3.3–5 protons per ATP.
Until now, the c-ring symmetry and the bioenergetic cost of making an ATP in a mammalian F-ATPase has been unknown. As described here, we have determined the ring size in the structure of the bovine F1-c-ring complex at 3.5 Å resolution.
Results and Discussion
Isolation of the Bovine F1-c-ring Complex.
The complex was prepared from the purified bovine ATP synthase by dissociation of the peripheral stalk, subunit a, and other minor membrane subunits (subunits A6L, e, f, and g) with detergents. The purified complex consisted of the α-, β-, γ-, δ- and ϵ-subunits that constitute the catalytic F1-domain plus the membrane protein, subunit c. All of these subunits were present in crystals of the complex (Fig. S1).
Structure Determination.
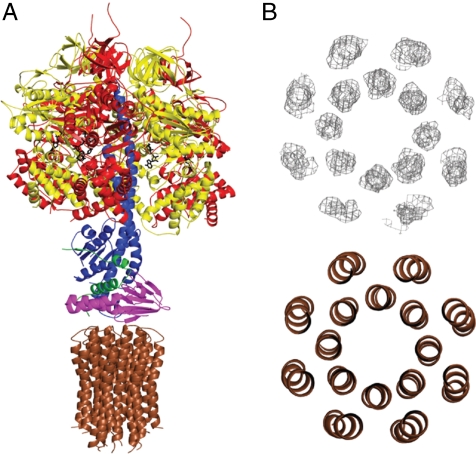
The structure of the bovine F1-c-ring complex (Fig. 1) was solved by molecular replacement with data to 3.5 Å resolution. Data processing and statistics are summarized in Table 1. The final model contains the following residues: 19-510 of the three α-subunits, 9-475 of the three β-subunits, residues 1-61, 67-96, and 101-272 of the γ-subunit, residues 15-145 of the δ-subunit, residues 1-47 of the ϵ-subunit, and residues 2-73 of c-subunits. The c-ring contains eight c-subunits. During refinement, an unsuccessful attempt was made to model a c9-ring into the density (see Fig. S2).
Fig. 1.
The structure of the bovine F1-c-ring complex. Part A. The subunits in the F1-catalytic domain (above) and the c-ring (below) are shown in ribbon representation. The membrane extrinsic region consists of the spherical catalytic domain, made of three α- and three β-subunits (red and yellow, respectively), the central stalk (subunits γ, δ, and ϵ, blue, purple, and green, respectively), The central stalk and the c-ring (brown) together constitute the rotor. Each of the C-terminal α-helices in the c-ring has a glutamate at residue 58, at the midpoint of the lipid bilayer. Part B. Cross-section of the c-ring taken at the midpoint of the α-helices. Above is shown the electron density with the eight N-terminal α-helices in the inner ring, and the eight C-terminal α-helices in the outer ring, and below the structural interpretation of the density.
Table 1.
Data collection and refinement statistics for the F1-c-ring complex from bovine ATP synthase
| Space group | P212121 |
| Unit cell dimensions a, b, c, Å | 155.7, 157.1, 247.3 |
| Resolution range Å | 50.48–3.50 |
| No. unique reflections | 72,959 |
| Multiplicity | 3.6 (3.5)* |
| Completeness, % | 99.8 (99.6)* |
| Rmerge | 0.263 (0.817)* |
| 〈I/σ(I)〉 | 3.7 (1.6)* |
| B factor, from Wilson plot, Å2 | 72.64 |
| R factor, % | 26.13 |
| Free R factor, % | 30.40† |
| rmsd of bonds, Å | 0.006 |
| rmsd of angles, ° | 0.973 |
*Values in parentheses are for the highest resolution bin (3.50–3.69 Å)
†The free R-factor was calculated for 3,855 reflections excluded from refinement.
Structure of the F1-c-ring Complex.
In the structure (Fig. 1A), the F1-domain has a “ground state” structure and the catalytic sites in subunits βTP and βDP contain a bound molecule of the ATP analogue, AMP-PNP, together with Mg2+, and the third catalytic subunit, βE, is devoid of nucleotide. Each noncatalytic α-subunit also contains a bound nucleotide and a magnesium ion, but they do not participate directly in ATP synthesis. A cross-section of the c-ring shows two concentric rings each containing eight regions of density (Fig. 1B). The eight regions in the inner and outer ring correspond to the N- and C-terminal α-helices of the c-subunit, respectively. The latter are significantly longer than the former (52 and 41 Å, respectively), as in other species. The central hole has a diameter of 47 Å at the top and bottom, and 42 Å at the “neck” in middle of the membrane. The hole is probably occupied by phospholipids as in other rings from F- and V-type ATPases (10, 14). The ring of eight c-subunits interacts extensively with the foot of the central stalk via subunits γ, δ, and ϵ and loop regions linking the α-helices of each c-subunit in the ring (Fig. S3). The buried surface area in this contact region is 790 Å2, whereas 600 Å2 was estimated to be buried in the structure of the yeast F1-c10. In both structures, side chains were not built in c-subunits, and so the actual values are higher. Superimposition of the bovine and yeast F1-c-ring structures shows that the rings do not coincide (Fig. S4). The position of the yeast c-ring was influenced by crystal contacts, and the bovine structure, where the c-ring is more symmetrically situated, probably provides a more accurate representation of the domain in the intact F-ATPase.
The c8-ring is the smallest c-ring yet observed among F-ATPases. Models of c8 and c9-rings were constructed with the bovine diameter using the structure of a yeast c-subunit from a model of the yeast F1-c10 complex. In both models, there were serious stereo-chemical clashes between the side chains of residues Ile-13, Leu-19, and Ile-23 with the equivalent side chains of an adjacent protomer in the neck of the hour-glass shaped c-ring. However, in the bovine c-subunit, these residues are all alanines. In the bovine c8-ring, there are no side chain clashes in the neck of the ring, and in a model of a c9-ring using the bovine structure and sequence, they were greatly reduced relative to the yeast c9- and c8-rings. Replacement of these alanine residues with amino acids with larger side chains, such as valine, leads to side chain clashes and destabilizes the ring. Replacement of the alanines by glycines abolishes hydrophobic packing interactions that contribute to the ring’s stability. Only in one invertebrate species, the tube-worm, Hydroides elegans, is alanine-13 evidently replaced by cysteine. Therefore, the presence of alanine residues at positions 13, 19, and 23 (bovine numbering) appears to be essential for the formation of the c8-ring.
Mosaic Structure of Bovine ATP Synthase.
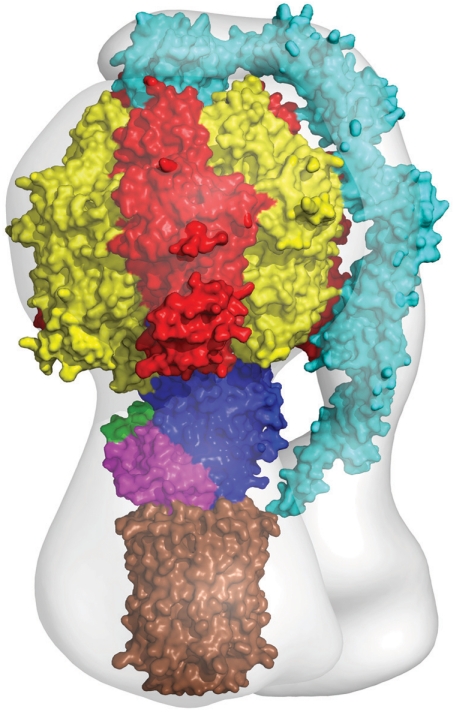
The structure of the bovine F1-c-ring provides an important contribution to the overall structure of bovine ATP synthase. In Fig. 2, a mosaic structure has been built by docking high resolution component structures into a structure of the intact enzyme determined by electron cryomicroscopy at 32 Å resolution (pale gray outline in Fig. 2) (15). The membrane extrinsic part of the structure is derived from a structure of bovine F1-ATPase with most of the peripheral stalk attached to it (16), augmented by information from a structure of an overlapping fragment of the peripheral stalk (17). Many structures have been determined of bovine F1-ATPase inhibited in various ways. The most accurate at 1.9 Å resolution is a structure representing the ground state in the catalytic cycle of ATP hydrolysis in which the enzyme was inhibited with the nonhydrolysable ATP analogue, AMP-PNP (18). The empty gray region on the right of Fig. 2 represents the part of the structure in the membrane domain of the enzyme where detailed structural information is lacking for subunit a (ATPase-6), which sits close to the surface of the c-ring and provides a transmembrane path for protons. Also, it contains the membrane intrinsic region of subunit b (two transmembrane α-helices), and for subunits A6L, e, f, and g (each with a single transmembrane α-helix), which are not involved directly in the synthesis of ATP. It is likely that, as in the structure of the V-ATPase from Thermus thermophilus at 16 Å resolution, the gray area in the membrane domain will contain an annular belt of detergent (19).
Fig. 2.
The mosaic structure of the F-ATPase from bovine mitochondria. The components of the structure were docked into a structure of the intact enzyme determined by electron cryomicroscopy (pale gray outline). The F1-domain and the c-ring are taken from the present work. In addition many, structures of the isolated F1-domain have been determined, the most accurate at 1.9 Å resolution. The peripheral stalk (cyan; made of single copies of subunits OSCP, b, d, and F6) penetrates into the membrane domain and links subunit a to the external surface of the catalytic domain. The structure of the peripheral stalk is derived from a structure of bovine F1-ATPase with most of the peripheral stalk attached to it, augmented by information from a structure of an overlapping fragment of the peripheral stalk. The foot of the central stalk makes extensive contacts with the bovine c-ring (this work). The empty gray region on the right represents the part of the structure in the membrane domain of the enzyme where detailed structural information is lacking for subunit a (ATPase-6), which sits close to the surface of the c-ring and provides a transmembrane path for protons. Also, it contains the membrane intrinsic region of subunit b (two transmembrane α-helices), and subunits A6L, e, f, and g (each with a single transmembrane α-helix), which are not involved directly in the synthesis of ATP. It is likely that, as in the 16 Å resolution structure of the V-ATPase from Thermus thermophilus, the gray area in the membrane domain will contain an annular belt of detergent.
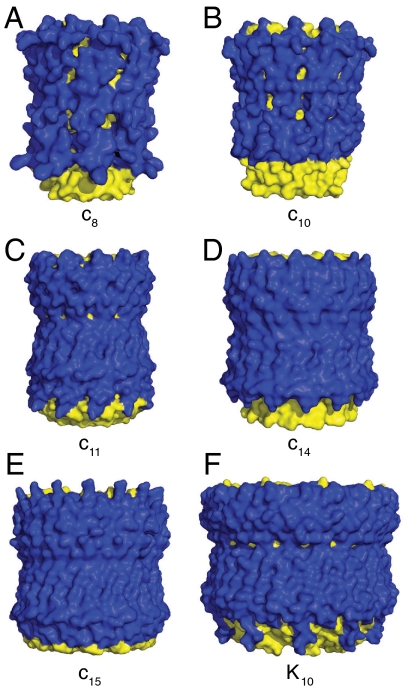
The bovine c-subunit has an unusual feature as the ϵ-amino group of Lys-43, which is at the beginning of the C-terminal α-helix, is completely trimethylated (20). The trimethylated side chain, terminated by a quaternary amino group (which is similar to choline), is exposed to the phospholipid bilayer in the head-group region. In this position, this side chain would clash with the head groups of phospholipids and would probably impede their binding to the ring. Cardiolipin is a characteristic and abundant lipid of the inner membranes of mitochondria, and is an essential component of an active mitochondrial F-ATP synthase fully coupled to the proton-motive force (21). Cardiolipin has no head group, and so the eight trimethyl-lysine residues in the c8-ring probably mark cardiolipin binding sites. Each bound cardiolipin with its covalently joined phosphatidylglycerols, each bearing two acyl side chains could help to strengthen the c8-ring by linking adjacent c-subunits together. Unlike the outer ring of α-helices in c15-rings, which are tightly packed, the c8-outer ring has gaps between α-helices that expose the inner ring to the lipid bilayer (Fig. 3). It is likely that these gaps in the c8-ring are occupied by the acyl groups of cardiolipins. The Lys-43 residues in the human, pig, sheep, and rabbit c-subunits are also completely trimethylated. Lys-43 is conserved throughout the known c-subunit sequences in animalia, and it is likely that the trimethylation of this residue is conserved also as a possible means of stabilizing their c-rings. Lys-43 is also conserved in the c-subunit of the F-ATPase in Saccharomyces cerevisiae, where the ring has ten c-subunits. However, the residue is not trimethylated, and in other fungi it is replaced by arginine. Therefore, the c10 and larger rings may not have such a strong requirement for bound cardiolipins.
Fig. 3.
External surfaces of rotor rings from F- and V-ATPases. The rings are shown in solid representation. The N- and C-terminal α-helices are yellow and blue, respectively. Parts A–E, c-rings from F-ATPases from bovine and yeast mitochondria, from Ilyobacter tartaricus, from Spinacea oleracea, and from Spirulina platensis. Part F, the K-ring from the V-ATPase from Enterococcus hirae.
Bioenergetic Implications.
The most important inference from the presence of the c8-ring in bovine F-ATP synthase, is that eight protons are translocated across the inner mitochondrial membranes per 360° rotation of its rotor. As each 360° rotation produces three ATP molecules from the F1-domain, the bioenergetic cost of the enzyme making an ATP is 2.7 protons. Where they are known, the sequences of c-subunits are identical in almost all vertebrates (Fig. 4). Also, they are highly conserved across animal phyla (Fig. 5), and mutations are confined mostly to the N and C termini of the protein, away from the c-ring-central stalk interface, and the ion binding site around Glu-47. With one exception, alanines 13, 19, and 23 are absolutely conserved throughout the animalia sequences, but not throughout lower eukarya and eubacteria (Figs. S5 and S6). Therefore, it appears that F-ATPases in all vertebrates and probably all or most invertebrates, will contain c8-rings. Therefore, the bioenergetic cost of the enzyme making an ATP is 2.7 protons in vertebrates and probably in invertebrates also (exceptions dictated by particular energetic demands of a species may exist). There are estimated to be 48–58,000 vertebrate species and about two million invertebrates. This picture of an evidently universal cost of making an ATP molecule in multicellular animals is very different from the picture in prokarya, chloroplasts, and fungi where, for reasons that are not understood, the F-ATP synthases have evolved a range of ring sizes from c10–c15 with associated higher bioenergetic costs of 3.3–5 protons per ATP (22). Thus, the vertebrate and probably the invertebrate F-ATP synthases are the most efficient that have been found. In mitochondria, it is thought that for each two electrons transferred to oxygen from NADH or succinate, ten or six protons, respectively, are pumped out of the matrix into the space between the outer and inner membranes of the organelle. The combination of the electrogenic exchange of internal ATP for an external ADP by the ADP/ATP translocase and the nonelectrogenic symport of phosphate and a proton by the phosphate carrier protein adds one proton to the total required to provide ATP to the cellular cytoplasm, and so the bioenergetic cost to the vertebrate mitochondrion is 3.7 protons. Therefore, the number of moles of ADP phosphorylated to ATP per two electrons transferred to oxygen, known as the P/O ratio, will be 10/3.7 and 6/3.7, or 2.7 and 1.6 for NADH and succinate, respectively, close to experimental values of 2.5 and 1.5 (23). It is to be expected that as the F-ATPase from S. cerevisiae contains a c10 ring the P/O ratio for succinate in yeast mitochondria (and probably in other fungal mitochondria) will be lower than in animal mitochondria.
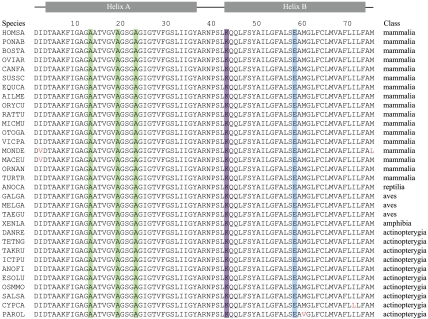
Fig. 4.
Sequences of c-subunits from F-ATP synthase in vertebrates. The sequences have been aligned from the following species with their names indicated on the left, and the animal class to which the species belongs is shown on the right: HOMSA, Homo sapiens (human); PONAB, Pongo albelii (Sumatran orangutan); BOSTA, Bos taurus (cattle); OVIAR; Ovis aries (sheep); CANFA, Canis lupus familiaris (dog); SUSSC, Sus scrofa (pig); EQUCA, Equus caballus (horse); AILME, Ailuropoda melanoleuca (giant panda); ORYCU, Oryctolagus cuniculus (rabbit); RATTU, Rattus norvegicus (Norwegian rat); MICMU, Microcebus murinus (mouse lemur); OTOGA, Otolemur garnettii (small eared galago); VICPA, Vicugna pacos (alpaca); MONDE, Mondelphis domestica (gray short tailed opposum); MACEU, Macropus eugenii (wallaby); ORNAN; Ornithorhynchus anatinus (duckbill platypus); TURTR, Tursiops truncates (bottle nosed dolphin); ANOCA, Anolis carolinesis (green anole lizard); GALGA, Gallus gallus (chicken); MELGA, Meleagris gallopavo (turkey); TAEGU, Taenio guttat (zebra finch); XENLA, Xenopus laevis (clawed toad); DANRE, Danio rerio (zebrafish); TETNG, Tetraodon nigroviridis (green pufferfish); TAKRU, Takifugu rubripes (fugu pufferfish); ICTPU, Ictalurus punctatus (channel catfish); ANOFI, Anoplopoma fimbria (sablefish); ESOLU, Esox lucius (northern pike); OSSMO, Osmerus mordax (rainbow smelt); SALSA, Salmo salar (Atlantic salmon); CYPCA, Cyprinus carpio (common carp); and PAROL, Paralichthys olivaceus (Japanese flounder). The green boxes indicate alanines 13, 19, and 23 that are required for the formation of the c8-ring. The purple box and blue boxes show, respectively, the positions of the lysine residue that is known to be trimethylated in the human, bovine, ovine, porcine, and rabbit enzymes and of the glutamate residue that is involved in proton translocation through the inner membranes of mitochondria
Fig. 5.
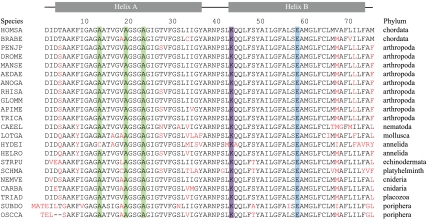
Sequences of c-subunits from F-ATP synthase in invertebrates compared with the human sequence. The sequences have been aligned from the following species with their names indicated on the left, and the phylum to which the species belongs is shown on the right: BRABE, Branchiostoma belcheri (amphioxus or Japanese lancelet); PENJP, Peneus japonica (Kuruma prawn); DROME, Drosophila melanogaster (fruit fly); MANSE, Manduca sexta (tobacco hawkmoth); AEDAE, Aedes aegypti (yellow fever mosquito); ANOGA, Anopheles gambiae (African malarial mosquito); RHISA, Rhicephalus sanguineus (brown dog tick); GLOMM, Glossina morsitans (savannah tsetse fly); APIME, Apis mellifera (honey bee); TRICA, Tribolium castaneum (red flour beetle); CAEEL, Caenorhabditis elegans; LOTGA, Lottia gigantea (sea snail); HYDEI, Hydroides elegans (tube-worm; the C-terminal sequence continues for 17 more residues); HELRO, Helobdella robusta (leech); STRPU, Stronglycentrotus purpuratus (purple sea urchin); SCHMA, Schistosoma mansoni (parasitic worm); NEMVE, Nematostella vectensis (starlet sea anenome); CARBA, Carukia barnesi (Irukandji jellyfish); TRIAD, Trichoplax adhaerans (presponge, simplest nonparasitic invertebrate); SUBDO, Suberites domuncula (sponge); and OSSCA, Oscarella carmela (sponge). Amino acid substitutions are shown in red. For the explanation of the colored boxes, see the legend to Fig. 4.
Materials and Methods
Protein Purification.
Bovine F1Fo-ATPase was purified from bovine heart mitochondria by affinity chromatography with residues 1–60 of the F1-ATPase inhibitor protein, IF1. The F1-c-ring subcomplex was prepared by binding F1Fo-ATPase to a HiTrap Q HP column (5 mL; GE Healthcare; Fig. S1). The bound enzyme was washed at a flow rate of 1 mL/ min, first for 60 min with purification buffer consisting of 20 mM Tris-HCl, pH 8.0, 10% glycerol (v/v), 1 mM ADP, 2 mM MgSO4, and 0.02% NaN3 containing 20 mM N-dodecyl-N,N-(dimethylammonio)butyrate, and then for 60 min with the same buffer containing 20 mM 3-(3-butyl-3-phenylheptanamido)-N,N-dimethylpropan-1-amine oxide. Then the column was washed for 50 min at the same flow rate with the same buffer containing 0.95 mM n-tridecyl-β-maltoside (TDM), and the bound protein was eluted with a NaCl gradient from 0–1 M in the same buffer. The F1-c-ring complex eluted from 0.35–0.40 mM NaCl. The purity of the protein complex was assessed by SDS-PAGE analysis and the peak fractions were pooled and dialyzed against the purification buffer containing 0.95 mM TDM. The F1-c-ring complex was concentrated to 22 mg/mL on a Vivaspin Q mini H spin column and eluted with purification buffer containing 0.95 mM TDM and 1 M NaCl. The concentrated complex was inhibited with 1 mM AMP-PNP, supplemented with TDM to a final concentration of 5.7 mM TDM. The protein concentration was adjusted to 10 mg/mL and the solution was ultracentrifuged at 163,000 x g for 30 min.
Crystallization of the Bovine F1-c-ring Complex.
Crystals were grown by microbatch under light paraffin oil in 72-well microbatch plates (Nalgene, Thermo Fisher Scientific, DK-4000). The drops contained a solution (2 μL) of 14%–16.5% (w/v) polyethylene glycol 4600, 100 mM Hepes, pH 7.0 (adjusted with KOH), 50 mM K2HPO4 (pH 7.0), and an equal volume of protein solution. Plates were covered with filtered liquid paraffin (6 mL; BDH Laboratory Supplies) and were kept at 23 °C for 42 d. Four crystals were harvested, washed three times in the buffer, and then dissolved in water (5 μL) and sample loading buffer (5 μL) containing lithium dodecyl sulphate. The crystals were analyzed by SDS-PAGE. All of the subunits of the F1-c-ring complex were present and undegraded in the crystals. Crystals were cryoprotected by the addition to the well of a solution consisting of 50 mM Hepes, pH 7.0, 25 mM K2HPO4, and 11.5% (w/v) polyethylene glycol 4600 with increasing amounts of glycerol from 5%–25% (w/v). The crystals were left to equilibrate for 60 min at each step. Then the crystals were harvested with micro mounts (MiTeGen) and vitrified in liquid nitrogen.
Data Collection and Structure Determination.
X-ray diffraction data were collected from the flash-frozen cryoprotected crystals at the Swiss Light Source in the Paul Scherrer Institut, Villigen, Switzerland. The data were collected at a wavelength of 1.0007 Å on beamline X06SA with a micro-diffractometer and a MAR225 mosaic charge coupled detector (Rayonix). Diffraction images were integrated with MOSFLM (24), and data were reduced with SCALA (25) and CTRUNCATE (26). Molecular replacement with bovine F1-ATPase covalently inhibited by reaction of glutamate-199 in one of the three β-subunits with dicyclohexylcarbodiimide (PDB ID 1E79) (27) was carried out with Phaser (28). The data were subjected to a round of rigid body refinement in REFMAC (29) with each the following domains specified: chains A–C 1–95, 96–379, and 600–602, 380–510; chains D–F, 1–82, 83–363, and 600–602, 364–474; chain G 1–272; chain H 1–146; chain I 1–50; and chains J–Q 2–73. Excluding the c8-ring from the model increased the Rfree by 1.0% at this stage, while replacing the c8-ring with a c9-ring increased the Rfree by 2.6%. A further round of restrained refinement was applied. This processing was performed for c-subunits (chains J–Q) using both the bovine c sequence and with a chain of poly alanines. The Rfree did not increase for the poly alanine model. Because the temperature factors for the c-ring model are very high (> 100 Å2), the side chain density is poorly defined, and therefore all nonglycine residues have been truncated to the β-carbon in the final model. Model building was carried out using COOT (30). Figures were prepared using PyMol (31).
Supplementary Material
Acknowledgments.
We thank the staff at beamline X06SA, Swiss Light Source, Villigen; and Dr I. M. Fearnley for his help with the bioinformatic analyses. This work was funded by the Medical Research Council.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1011099107/-/DCSupplemental.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2xnd).
See Commentary on page 16755.
References
- 1.Walker JE. ATP synthesis by rotary catalysis (Nobel lecture) Angewandte Chemie International Edition. 1998;37:5000–5011. doi: 10.1002/(SICI)1521-3773(19980918)37:17<2308::AID-ANIE2308>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 2.von Ballmoos C, Wiedenmann A, Dimroth P. Essentials for ATP synthesis by F1F0 ATP synthases. Annu Rev Biochem. 2009;78:649–672. doi: 10.1146/annurev.biochem.78.081307.104803. [DOI] [PubMed] [Google Scholar]
- 3.Junge W, Sielaff H, Engelbrecht S. Torque generation and elastic power transmission in the rotary FOF1-ATPase. Nature. 2009;459:364–370. doi: 10.1038/nature08145. [DOI] [PubMed] [Google Scholar]
- 4.von Ballmoos C, Cook GM, Dimroth P. Unique rotary ATP synthase and its biological diversity. Annu Rev Biophys. 2008;37:43–64. doi: 10.1146/annurev.biophys.37.032807.130018. [DOI] [PubMed] [Google Scholar]
- 5.Abrahams JP, Leslie AGW, Lutter R, Walker JE. Structure at 2.8 Å resolution of F1-ATPase from bovine heart mitochondria. Nature. 1994;370:621–668. doi: 10.1038/370621a0. [DOI] [PubMed] [Google Scholar]
- 6.Walker JE, Dickson VK. The peripheral stalk of the mitochondrial ATP synthase. Biochim Biophys Acta. 2006;1757:286–296. doi: 10.1016/j.bbabio.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 7.Boyer PD. The binding change mechanism for ATP synthase—some probabilities and possibilities. Biochim Biophys Acta. 1993;1140:215–250. doi: 10.1016/0005-2728(93)90063-l. [DOI] [PubMed] [Google Scholar]
- 8.Junge W. Protons, proteins, and ATP. Photosynth Res. 2004;80:197–221. doi: 10.1023/B:PRES.0000030677.98474.74. [DOI] [PubMed] [Google Scholar]
- 9.Stock D, Leslie AGW, Walker JE. Molecular architecture of the rotary motor in ATP synthase. Science. 1999;286:1700–1705. doi: 10.1126/science.286.5445.1700. [DOI] [PubMed] [Google Scholar]
- 10.Meier T, Polzer P, Diederichs K, Welte W, Dimroth P. Structure of the rotor ring of F-Type Na+-ATPase from Ilyobacter tartaricus. Science. 2005;308:659–662. doi: 10.1126/science.1111199. [DOI] [PubMed] [Google Scholar]
- 11.Matthies D, et al. The c13 ring from a thermoalkaliphilic ATP synthase reveals an extended diameter due to a special structural region. J Mol Biol. 2009;388:611–618. doi: 10.1016/j.jmb.2009.03.052. [DOI] [PubMed] [Google Scholar]
- 12.Vollmar M, Schlieper D, Winn M, Buchner C, Groth G. Structure of the c14 rotor ring of the proton translocating chloroplast ATP synthase. J Biol Chem. 2009;284:18228–18235. doi: 10.1074/jbc.M109.006916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pogoryelov D, Yildiz O, Faraldo-Gomez JD, Meier T. High-resolution structure of the rotor ring of a proton-dependent ATP synthase. Nat Struct Mol Biol. 2009;16:1068–1073. doi: 10.1038/nsmb.1678. [DOI] [PubMed] [Google Scholar]
- 14.Murata T, Yamato I, Kakinuma Y, Leslie AGW, Walker JE. Structure of the rotor of the V-Type Na+-ATPase from Enterococcus hirae. Science. 2005;308:654–659. doi: 10.1126/science.1110064. [DOI] [PubMed] [Google Scholar]
- 15.Rubinstein JL, Walker JE, Henderson R. Structure of the mitochondrial ATP synthase by electron cryomicroscopy. EMBO J. 2003;22:6182–6192. doi: 10.1093/emboj/cdg608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rees DM, Leslie AGW, Walker JE. The structure of the membrane extrinsic region of bovine ATP synthase. Proc Natl Acad Sci USA. 2009;106:21597–21601. doi: 10.1073/pnas.0910365106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dickson VK, Silvester JA, Fearnley IM, Leslie AGW, Walker JE. On the structure of the stator of the mitochondrial ATP synthase. EMBO J. 2006;25:2911–2918. doi: 10.1038/sj.emboj.7601177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bowler MW, Montgomery MG, Leslie AGW, Walker JE. Ground state structure of F1-ATPase from bovine heart mitochondria at 1.9 Å resolution. J Biol Chem. 2007;282:14238–14242. doi: 10.1074/jbc.M700203200. [DOI] [PubMed] [Google Scholar]
- 19.Lau WC, Rubinstein JL. Structure of intact Thermus thermophilus V-ATPase by cryo-EM reveals organization of the membrane-bound VO motor. Proc Natl Acad Sci USA. 2010;107:1367–1372. doi: 10.1073/pnas.0911085107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen R, Fearnley IM, Palmer DN, Walker JE. Lysine 43 is trimethylated in subunit C from bovine mitochondrial ATP synthase and in storage bodies associated with Batten disease. J Biol Chem. 2004;279:21883–21887. doi: 10.1074/jbc.M402074200. [DOI] [PubMed] [Google Scholar]
- 21.Eble KS, Coleman WB, Hantgan RR, Cunningham CC. Tightly associated cardiolipin in the bovine heart mitochondrial ATP synthase as analyzed by 31P nuclear magnetic resonance spectroscopy. J Biol Chem. 1990;265:19434–19440. [PubMed] [Google Scholar]
- 22.Ferguson SJ. ATP synthase: what dictates the size of a ring? Curr Biol. 2000;10:R804–808. doi: 10.1016/s0960-9822(00)00765-x. [DOI] [PubMed] [Google Scholar]
- 23.Hinkle PC. P/O ratios of mitochondrial oxidative phosphorylation. Biochim Biophys Acta. 2005;1706:1–11. doi: 10.1016/j.bbabio.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 24.Leslie AGW. The integration of macromolecular diffraction data. Acta Crystallogr. 2006;D62:48–57. doi: 10.1107/S0907444905039107. [DOI] [PubMed] [Google Scholar]
- 25.Evans PRE. Scaling and assessment of data quality. Acta Crystallogr. 2006;D62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 26.French G, Wilson K. On the treatment of negative intensity observations. Acta Crystallogr. 1978;A34:517–525. [Google Scholar]
- 27.Gibbons C, Montgomery MG, Leslie AGW, Walker JE. The structure of the central stalk in bovine F1-ATPase at 2.4 Å resolution. Nat Struct Biol. 2000;7:1055–1061. doi: 10.1038/80981. [DOI] [PubMed] [Google Scholar]
- 28.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D . 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 30.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of COOT. Acta Crystallogr D . 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.DeLano WL. The PyMol molecular graphics system. DeLano Scientific USA. 2002 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.