Abstract
The NSD (nuclear receptor-binding SET domain protein) family encodes methyltransferases that are important in multiple aspects of development and disease. Perturbations in NSD family members can lead to Sotos syndrome and Wolf–Hirschhorn syndrome as well as cancers such as acute myeloid leukemia. Previous studies have implicated NSD1 (KMT3B) in transcription and methylation of histone H3 at lysine 36 (H3-K36), but its molecular mechanism in these processes remains largely unknown. Here we describe an NSD1 regulatory network in human cells. We show that NSD1 binds near various promoter elements and regulates multiple genes that appear to have a concerted role in various processes, such as cell growth/cancer, keratin biology, and bone morphogenesis. In particular, we show that NSD1 binding is concentrated upstream of gene targets such as the bone morphogenetic protein 4 (BMP4) and zinc finger protein 36 C3H type-like 1 (ZFP36L1/TPP). NSD1 regulates the levels of the various forms of methylation at H3-K36 primarily, but not exclusively, within the promoter proximal region occupied by NSD1. At BMP4 we find that this reduces the levels of RNAP II recruited to the promoter, suggesting a role for NSD1-dependent methylation in initiation. Interestingly, we also observe that the RNAP II molecules that lie within BMP4 have inappropriate persistence of serine-5 phosphorylation and reduced levels of serine-2 phosphorylation within the C-terminal domain (CTD) of the large subunit of RNAP II. Our findings indicate that NSD1 regulates RNAP II recruitment to BMP4, and failure to do so leads to reduced gene expression and abrogated levels of H3K36Me and CTD phosphorylation.
Keywords: elongation, initiation, C-terminal domain, histone code, ChIP on chip
NSD1 (nuclear receptor-binding SET domain protein 1 or KMT3B) belongs to a family of mammalian histone lysine methyltransferases (NSD1, NSD2/WHSC1, and NSD3/WHSC1L1) that are important in multiple aspects of development and disease (1–7). Human NSD1 is a large protein composed of 2,696 residues and contains several motifs in the C-terminal half that are undoubtedly critical for its proposed functions in signaling and chromatin regulation. This includes a catalytic lysine methyltransferase SET domain and four zinc-binding PHD fingers. NSD1 was originally discovered in a two-hybrid screen through its ability to bind to nuclear receptors such as retinoic acid (8). Mice deficient in NSD1 exhibit an embryonic lethal phenotype due to apoptosis at E10.5 (9). Nearly 5% of all human acute myeloid leukemia (AML) patients harbor a translocation in the NSD1 gene at chromosome 5 that encodes for a chimeric protein encompassing the FG-repeat domain of NUP98 fused to the carboxy-terminus of NSD1 (10). This fusion protein has been shown to promote HOX gene activation by antagonizing repressive chromatin structure in a manner that depends on the methyltransferase activity of the SET domain (10). Furthermore, translocations involving NSD1 have been found in breast cancer (7), and epigenetic inactivation of the NSD1 promoter through CpG hypermethylation has been shown to be important in neuroblastomas and glioblastomas (3). Therefore, translocation-driven NSD1 fusion proteins behave as oncogenes in AML, but inactivation of NSD1 in neuroblastomas behaves as a tumor suppressor (3). In addition to its role in development and cancer, NSD1 is haploinsufficient in Sotos syndrome (11), a childhood overgrowth disease characterized by a broad set of phenotypes, including macrocephaly, advanced bone age, facial dymorphism, learning disabilities, and seizures (12).
Histone side chains undergo a plethora of posttranslational modifications (PTMs) that formulate a “histone code” (13–15). Histone PTMs initiate signaling events by recruiting “reader” proteins or through inducing structural changes in chromatin (13). Lysine residues, including H3-K36, can exist in up to four different forms: nonmethylated (Me0) and the mono-, di-, and trimethylated forms (Me1, Me2, and Me3, respectively). Although NSD1 was originally shown to methylate histones on both H4-K20 and H3-K36 in vitro (9), more recent experiments suggest that the enzyme, as well as its related family members from humans, worms, and flies, is specific for H3-K36 (16–19). Importantly, Li et al. (18) used defined nucleosomal substrates chemically modified at histone H3 to contain the various methylated forms of lysine 36, and showed that human NSD1 is a dimethylase specific for H3-K36. In addition to histones, NSD1 has recently been shown to activate the p65 subunit of NF-κB through both mono- and dimethylation of lysine residues K218 and K221, respectively (20).
The biological significance of the various forms of H3-K36Me is poorly understood. In worms and humans, H3-K36Me3 has been shown to link transcription with splicing (21–23). In flies, H3-K36Me3 functions in dosage compensation (24). In yeast, H3-K36Me2 and H3-K36Me3 have both been implicated in transcription elongation, and this appears to be coupled to histone acetylation through recruitment of the Rpd3S deacetylase complex (14, 25,M–31), which maintains hypoacetylated chromatin in the wake of RNAP II function and prevents cryptic initiation. Rpd3S preferentially binds H3-K36Me2 and H3-K36Me3, but not H3-K36Me1 (30), indicating that the various methylated forms of H3-K36 specify distinct biological signals. In Arabidopsis, the di- and trimethylated states mark actively transcribed chromatin (32), and the levels of H3-K36Me2 and H3-K36Me3 have been shown to peak near the 3′ end of active chicken genes (33). In yeast, H3-K36Me3 correlates with transcriptional frequency, and H3-K36Me2 has been linked to the on/off states of transcription (14). In humans, there is a slight preference for H3-K36Me1 at active promoters, and this mark has also been detected in transcriptionally active regions of the β-globin locus (34). At the human globin genes, H3-K36Me1 is broadly distributed, but the di- and trimethylation marks correlate with transcription in opposite ways (34).
Despite the fact that NSD1 targets H3-K36, little is known about its role in transcriptional regulation. To further delineate the role of NSD1 in transcription and chromatin regulation, we identified and validated an NSD1 transcriptional network in multiple human cell lines. We performed a chromatin immunoprecipitation/microarray (“ChIP on chip”) assay and determined a set of over 300 candidate target genes in HCT116 cells for NSD1. We found that NSD1 binds to the promoter regions of genes implicated in various processes, including those consistent with its known role in diseases such as cancer and Sotos. In particular, we show that NSD1 regulates transcription of bone morphogenetic protein 4 (BMP4). Here, NSD1 binding is concentrated in a region ≈1,200 bp upstream (−1,200) of the BMP4 promoter and enforces H3-K36Me levels within this region. Depletion of NSD1 reduces the levels of H3-K36Me1, 2, and 3, suggesting that NSD1 is a mono/dimethylase and that this modification serves as a potential substrate for trimethylation. A similar profile was found for ZFP36L1/TPP. We further show that in the absence of NSD1, the abrogated H3-K36Me levels at −1,200 reduces RNAP II occupancy at the BMP4 promoter. We find that the RNAP II that does elongate through the BMP4 gene is enriched in serine-5 phosphorylation and diminished in serine 2-phosphorylation. These data suggest that NSD1 is involved in the regulation of gene expression through stimulating the transition of RNAP II from an initiation to fully elongation-competent state.
Results
Predominant Expression of the Short Isoform of NSD1 in Multiple Cell Types.
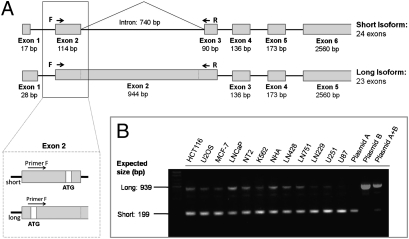
To begin to understand the role of NSD1 in chromatin regulation, we sought to determine the relative ratios of the short and long isoforms in various cell types. Through an examination of the NSD1 gene on Ensembl, we deduced that the short isoform of NSD1 is differentiated from the long isoform by an inclusion of an additional 740 nucleotides that result from an intron-retention mechanism. In this particular case there is an mRNA splicing event that removes an intron from within exon 2 (Fig. 1A). This exon contains coding information that is part of the long form. To determine the extent of intron retention and hence the relative levels of the short and long isoforms of NSD1, we designed a PCR-based assay to differentiate between the long and short isoforms in various cell types. Using PCR conditions that could amplify both the long (939 bp) and short (199 bp) isoforms (with the same primer set), we reproducibly observed that the short isoform was predominantly expressed in a variety of cell types (Fig. 1B). Partial NSD1 clones were used as positive control for the short (exons 1–5) and long isoforms (exons 1–2).
Fig. 1.
Predominance of expression of the short NSD1 isoform in multiple cell types. (A) Intron-exon structure (not drawn to scale) of NSD1 gene. The diagrams show the intron retention mechanism that results in two isoforms. In detail: localization of the forward primer and ATG sites in exon 2 of both isoforms. (B) PCR analysis from cDNA using one primer set that recognizes sequences common to both isoforms of NSD1 in different cell lines: HCT116 (colorectal cancer cells), U2OS (osteosarcoma), MCF-7 (breast cancer), LNCaP (prostate cancer), NT-2 (teratocarcinoma), K562 (leukemia), NHA (normal human astrocytes), LN428, LN751, LN229, U251, U87 (glioblastomas). The PCR products were sequenced to confirm the amplification of the NSD1 short isoform. Sequences: primer F: TGATGCCGGCCAGGATGGA and primer R: TGGCAATTCCTGTGAAGTAGATGATGATG. NSD1 partial clones for the short and long isoforms (plasmids A and B, respectively) were used as control [clone IDs 2117843 (A) and 3908832 (B); Open Biosystems].
Establishment of an NSD1 Transcriptional Network.
Very little is known about the number and types of endogenous targets of NSD1. One study showed that NSD1 regulates MEIS1 in cells of neuronal lineages (3). An earlier report showed that the HOXA7, 9, 10, and MEIS1 genes are regulated by the NUP98-NSD1 oncoprotein that contains a fusion between Nup98 and the C-terminal half of NSD1 (10). However, it is not clear if NSD1 by itself or the fusion protein regulates these targets under “normal” conditions (10). To determine the natural genomic targets that NSD1 associates with, we first used homologous recombination (HR) to generate an epitope-tagged fusion at the 3′ end of the endogenous NSD1 gene on chromosome 5 in human colorectal cancer HCT116 cells (Fig. S1). Because the C terminus of NSD1 was previously shown to be dispensable for its methyltransferase activity and its interaction with the HOX locus (10), we were confident that manipulating the 3′ end of NSD1 would not interfere with the resulting activity of the protein in our subsequent analysis. Using this HCT116 NSD1-FLAG cell line, we performed a chromatin immunoprecipitation/microarray (ChIP on chip, or ChIP microarray) experiment with two-color tiling arrays. HCT116 cells expressing NSD1-FLAG were subjected to a ChIP protocol with anti-FLAG antibodies (Materials and Methods). As NSD1 has been implicated in transcriptional events, we examined the ability of NSD1-FLAG to associate with any of 24,659 promoter regions throughout the human genome. Here, the promoter regions were covered by probes ranging from 50 to 75 bp in length with a median spacing of 50 bp. To identify significant peaks and to assign P values using a t test, we used the recently described MA2C algorithm that incorporates a normalization method based upon the GC content of the probes (35). Using P values ranging from 0.001 to 0.0005, we determined that there were over 300 candidate NSD1 targets in HCT116 cells. From this, we identified a spectrum of candidate targets for NSD1 in HCT116 cells (Table S1). This includes those genes implicated in cancer: Bone morphogenetic protein 4 (BMP4), Cathepsin D (CTSD), and Kruppel-like factor 6 (KLF6), von Hippel tumor suppressor (VHL), and Kallikreins KLK6 and KLK14. Additionally, several cell cycle-related genes were identified: CDK4, CDC6, CHEK1, CCNE1 (cyclin E), CDKN2A, and the cyclin B interacting protein CCNB1IP1. We found that NSD1 binds to the promoter regions of 20 different keratin genes, including two (KRT3 and KRT6B) known to be regulated by the p65 subunit of NF-κB. Moreover, we identified the p65 targets SLC16A1, IGF2BP2 as candidate NSD1-regulated genes. We also identified the development and mental retardation candidate gene (ZMYM3) and the early response gene Tristetraprolin (TTP or ZFP36L1) that encodes an RNA binding protein as potential targets of NSD1 (36–39) (Table S1).
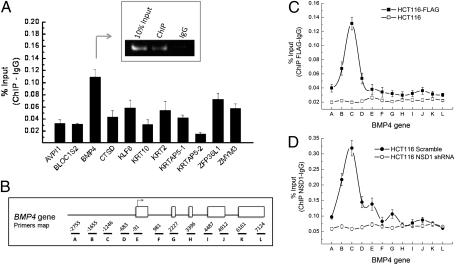
We used ChIP followed by real-time PCR (qPCR) to validate our ChIP microarray results. We confirmed the occupancy of NSD1 at multiple promoter elements, including BMP4 (Fig. 2A). We chose to analyze the BMP4 gene more thoroughly, because bone morphogenesis is perturbed in Sotos syndrome and because of the amenable size of the gene (7,100 bp). Because the microarrays contained probes that represented promoter regions, it remained possible that we would not have detected NSD1 binding events outside of this region. We therefore analyzed the distribution of NSD1 across the length of the entire BMP4 gene in HCT116 cells. To examine this, we performed ChIP analysis with primer sets at multiple sites throughout the BMP4 gene. Primers were designed covering the region from −3,000 bp to 7,100 bp from the TSS site of BMP4 gene (Fig. 2B). We found that NSD1 localization was concentrated to a region 1,200 bp upstream of the +1 site of the BMP4 promoter (Fig. 2C). Importantly, we failed to observe a ChIP signal in control HCT116 cells that did not harbor the FLAG-tagged allele, demonstrating the specificity for the interaction of NSD1 at the −1,200 region (Fig. 2C, open squares). We also confirmed this with an independent antibody specific to NSD1 (Fig. 2D). This second antibody gave slightly elevated NSD1 signals within the body of the gene but were absent in HCT116 cells depleted of NSD1 through shRNA (Fig. 2D, open circles). Therefore, the association between NSD1-FLAG and the −1,200 region of BMP4 is conclusively a function of NSD1 and is not due to any possible FLAG-associated artifacts. Additionally, we performed an analogous set of experiments for the ZFP36L1 gene and found that the binding profile of NSD1 to this target was also concentrated near 5′ promoter elements (Fig. S2 A–C).
Fig. 2.
(A) Validation of ChIP-on-chip data. Individual ChIP (n = 4) was performed for NSD1 targets in HCT116 NSD1-FLAG cells: AVPI1 (arginine vasopressin induced 1), BLOC1S2 (biogenesis of lysosomal organelles complex1 subunit 2), BMP4 (bone morphogenetic protein 4), CTSD (cathepsin D), KLF6 (Kruppel-like factor 6), KRT10 (keratin 10), KRT2 (keratin 2), KRTAP5-1 and 2 (keratin-associated protein 5: 1 and 2), ZFP36L1 (zinc finger protein CH3-like), and ZMYM3 (zinc finger MYM-type 3). Primer sequences are listed in Table S2. (Inset) ChIP at the BMP4 promoter as a representative of the other individual ChIP experiments. (B) Diagram of BMP4 intron/exon gene structure (not drawn to scale) showing the location of amplicons used to verify NSD1 binding profile. Numbers represent the first base of forward primer. Average sizes of amplicons are between 150 and 200 bp. Negative numbers indicate the region upstream +1 site (promoter region). Primer sequences are listed in Table S3. (C and D) NSD1 binding is concentrated upstream of the BMP4 promoter (near −1,200). HCT116 NSD1-FLAG were processed for ChIP using FLAG (C) or NSD1 (D) antibodies. All primer sets were validated by DNA sequencing and through their ability to amplify one band. Control cells (parental cells void of the FLAG-tag and cells depleted of NSD1 by shRNA) were used with FLAG and NSD1 antibody, respectively, to show specificity of NSD1 binding by ChIP. Error bars in A, C, and D represent SD from the mean (n = 3).
NSD1 Depletion Reduces Expression of Target Genes in a Cell Type-Specific Manner.
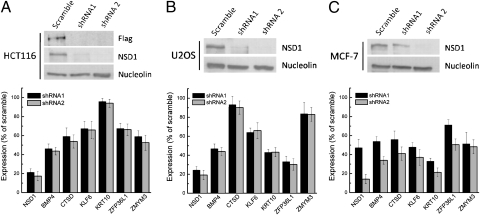
To determine the impact of NSD1 deficiency on target gene function, we used real-time PCR to measure the levels of candidate target gene expression in HCT116 cells depleted of NSD1 by shRNAs. For this, we generated two independent HCT116 cell lines where NSD1 was knocked down through the introduction of lentiviruses that express shRNAs targeting two independent sequences. A third line expressing a scrambled shRNA served as a control. Furthermore, to determine the generality of NSD1 regulation at candidate targets in additional cell types, we also generated U2OS osteosarcoma and MCF-7 breast cancer cells knocked down in NSD1 as well as their companion, scrambled controls. We found that knocking down NSD1 resulted in severely reduced levels of NSD1 protein in all cell types examined (Fig. 3 Upper). Using real-time PCR, we found significant and reproducible reductions in transcript levels for several candidate targets, confirming that NSD1 regulates the expression of various targets identified in the ChIP array (Fig. 3 Lower). Notably, we found that although the levels of the KRT10 transcripts were severely reduced in MCF-7 cells defective in NSD1 function, KRT10 levels were largely independent of NSD1 function in HCT116 cells. Moreover, ZFP36L1/TPP transcript levels were significantly abrogated in U2OS cells knocked down in NSD1, but they were only modestly affected in NSD1-deficient HCT116 cells. Therefore, NSD1 regulates target gene expression in a tissue-specific manner. Lentiviral shRNA1 was highly effective at knocking down NSD1 transcript and protein levels in HCT116 and U2OS cells (Fig. 3 A and B Upper). Although effective in these cell types, shRNA1 reduced NSD1 transcript and protein levels by about a factor of 2 in MCF-7 cells (Fig. 3C). Interestingly, knockdown of NSD1 to these levels was nearly as effective in abrogating target gene expression to those levels seen in the highly effective shRNA2 (Fig. 3C). This finding is reminiscent of the fact that NSD1 is haploinsufficient in Sotos syndrome and suggests that reduced, but not completely abolished, levels of NSD1 transcription and translation are sufficient to creating a mutant state.
Fig. 3.
Knockdown of NSD1 results in reduced expression of targets. Two different shRNAs against NSD1 or a scrambled control were transduced into HCT116 NSD1-FLAG, U2OS, and MCF-7 cells. (Upper) Immunoblotting shows efficacy of NSD1 knockdown in HCT116 (A), U2OS (B), and MCF-7 (C) cells. Nucleolin was used as loading control. (Lower) Real-time PCR for NSD1, BMP4, CTSD, KLF6, KRT10, ZFP36L1, and ZMYM3 expression shows their levels of transcription in the absence of NSD1 in HCT116 (n = 6), U2OS (n = 3), and MCF-7 cells. Primers sequences are listed in Table S2. Error bars represent SD (n = 3).
NSD1 Regulates H3-K36Me Levels at the BMP4 Gene.
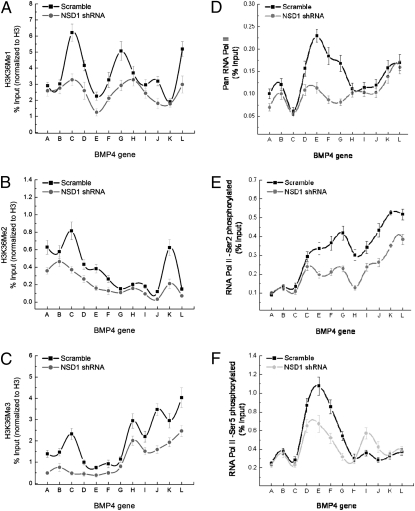
Our in vitro assays with recombinant NSD1 show that the enzyme is specific for nucleosomal histone H3, but not octamers (Fig. S3). This is consistent with a recent report from Li et al. (18) that showed the specificity of NSD1 as a dimethylase (as well as NSD2 and NSD3) toward nucleosomal histone H3. To examine how NSD1 deficiency impacted histone methylation of H3-K36 at the BMP4 locus in vivo, we performed ChIP assays with antibodies directed against the various methylated forms of H3-K36 or total H3 (as described in Materials and Methods). First, we confirmed the specificity of the antibodies in dot blots with various peptides containing the mono-, di-, and trimethylated forms of H3-K36 and several other histone methylated peptides (Fig. S4 A and B). Next, we examined the distribution of the various forms of H3-K36Me across the entire BMP4 gene as a function of NSD1. When normalized to total H3, high levels of H3-K36Me1, 2, and 3 were reproducibly observed near −1,200 and coincided with the peak region of NSD1 binding, suggesting that NSD1 enforces methylation at H3-K36 within this region. Indeed, we consistently detected reduced levels of all three forms of H3-K36Me at this region in NSD1 knockdown cells (Fig. 4 Left). Consistent with earlier studies that have determined that the levels of H3-K36Me2 and H3-K36Me3 peak near the 3′ end of genes (33), we found that this trend is also phenocopied at BMP4. In particular, NSD1 knockdown cells generated reproducible reductions in the di- and trimethylation signals, but not to the same extent as what we found near −1,200 (Fig. 4 Left). Consistent with this, we also observed that NSD1 enforced H3-K36Me levels at ZFP36L1 (Fig. S2 D and E).
Fig. 4.
(Left) H3K36 mono-, di-, and trimethylation profile at the BMP4 locus in the presence or absence of NSD1. The levels of mono (A), di (B), and tri (C) H3-K36 methylation were measured by ChIP using specific antibodies in HCT116 cells, followed by real-time PCR. ChIP-specific signal (% input) was calculated using IgG as control as specified in Materials and Methods. The % input values were normalized to total H3 (% input H3K36Me/% input unmodified H3). As the values were normalized by H3 values <1 may be expected. Error bars represent SD from the mean (n = 3). (Right) RNA polymerase II binding profile in the BMP4 gene in the presence or absence of NSD1 function. Levels of pan (D), Ser-2-phosphorylated (E), and Ser-5-phosphorylated (F) forms of RNA Pol II were measured by ChIP using antibodies specific for RNA Pol II and its phosphorylated forms (Bethyl Laboratories) in HCT116 cells, followed by real-time PCR. ChIP specific signal (% input) was calculated using IgG as control, as specified in Materials and Methods. Error bars represent SD from the mean (n = 3). Primers (A–L) were designed along BMP4 gene according to the diagram in Fig. 2B: −2,755 (A), −1,655 (B), −1,246 (C), −683 (D), −91 (E), 981 (F), 2,227 (G), 3,386 (H), 4,487 (I), 4,912 (J), 6,161 (K), and 7,124 (L).
NSD1 Regulates RNAP II Occupancy at the BMP4 Promoter.
Previous findings have linked H3-K36Me with gene activity, but the role of each modification and how it relates to RNAP II function is still unclear. Because NSD1 regulates the levels of H3-K36Me near −1,200, we examined the levels and distribution of RNAP II across the BMP4 gene in the presence and absence of NSD1 function (Fig. 4 Right). Once recruited to promoter elements, the CTD is rapidly converted to hyperphosphorylated forms during the transition from transcriptional initiation to elongation (40). Here, RNAP II “escapes” the promoter region and transcribes along the length of the gene. Phosphorylation at S5 and S2 of the CTD is known to regulate the initiation and elongation properties of RNAP II, respectively. Serine 5 phosphorylation is present at the 5′ end, but as the polymerase moves toward the 3′ end, the serine-2 phosphorylated form predominates. Therefore, the status of serine-2 phosphorylation is indicative of elongation (41).
In HCT116 control cells (scramble shRNAs), we found that RNAP II levels consistently peaked near the +1 site (Fig. 4D). In contrast, in cells depleted of NSD1, we reproducibly observed reduced levels of RNAP II at the BMP4 +1 site (Fig. 4D), suggesting that the lack of NSD1 caused a reduction in initiation. However, the total levels of RNAP II associated with BMP4 chromatin downstream of the +1 site (i.e., nucleotides +3,396 to end) were not affected by the loss of NSD1 function (Fig. 4D). To explain this paradox, we hypothesized that the elongation efficiency of RNAP II was reduced in cells depleted of NSD1 in this region. In this scenario, RNAP II would be stalled at positions around amplicon +3,396, and a buildup of RNAP II at this locus would account for the equivalent levels of RNAP II detected in control and shRNA-treated cells. To examine the nature of elongating RNAP II at BMP4 as a function of NSD1 activity, we determined the extent of phosphorylation at CTD serine residues 2 and 5 throughout BMP4 (Fig. 4 E and F). Phosphospecific RNAP II antibodies were first validated by phosphatase treatment (Fig. S4C). In amplicons with equivalent amounts of total RNAP II (H-L), we found that the CTD possessed reduced levels of serine-2 phosphorylation (i.e., nearly a 70% reduction at +3,396), but slightly increased levels of serine-5 phosphorylation, a mark associated with initiation. This is accompanied by reduced levels of H3-K36Me3 at this locus. Such defects could account for the apparent stalling observed at BMP4. Because the levels of serine-2 and serine-5 phosphorylation increase and decrease, respectively, as the polymerase approaches the 3′ end, our findings show that the absence of NSD1 generates RNAP II complexes that are defective in CTD phosphorylation.
Discussion
Defects in the NSD family cause various diseases, including Sotos syndrome and Wolf–Hirschhorn syndrome, as well as cancers such as AML, neuroblastomas, and glioblastomas. Despite the clinical importance of this family of enzymes, very little is known about their specific targets or mode of action. NSD2 has been shown to regulate cardiac transcription (42), but contradictory information has been reported in the literature with respect to its substrate specificity as well as that of other NSD family members (ref. 18 and references within). Despite such discrepancies, recent work with defined substrates has indicated that the catalytic SET domain of this family appears sensitive to the nature of its substrates (18). With respect to nucleosomal substrates, as opposed to octamers, our results show that NSD1 appears specific for H3-K36, a finding that is consistent with previous experiments from Li and coworkers (18). Additional experiments from Lu et al. (20) have shown that NSD1 is activated in response to cytokines and methylates nonnucleosomal targets such as the p65 subunit of NF-κB. Here, NSD1 was shown to act as a mono- and dimethylase against lysine residues K218 and K221 of p65, respectively.
As NSD1 is an H3-K36-specific methyltransferase, we set out to define its targets in the context of transcription. Using a ChIP-on-chip strategy and a cell line engineered to express an endogenous epitope tag at the 3′ end of the human NSD1 gene, we identified a number of candidate NSD1 target genes in HCT116 cells. However, because we confirmed only a subset of the candidate NSD1 targets, we cannot formally exclude the possibility that some artifacts associated with the ChIP microarray procedure (i.e., cross-hybridization) may change the list of NSD1 targets. We also examined the role of NSD1 in the regulation of candidate targets in U2OS and MCF-7 cells. We found that the extent of NSD1-dependent regulation of its targets was cell-type specific as, for example, NSD1 regulation of the ZFP36L1/TPP gene was highest in U2OS cells but lowest in HCT116 cells. Because of this, and because NSD1 is activated in response to cytokines, it is likely that the NSD1 transcriptional network, including the extent of target gene regulation, will be variable and inducible in multiple cell types.
We found that the levels of BMP4 were consistently reduced in all three examined cell types. We further examined the role of NSD1 in the regulation of BMP4 transcription. Using FLAG antibodies with the NSD1-FLAG HCT116 cells (or control non–FLAG-tagged cells) as well as a second polyclonal antibody specific to NSD1, we reproducibly demonstrated that NSD1 associates primarily within a region ≈1,200 bp upstream of the BMP4 start site. In contrast, relative NSD1 levels were largely reduced within the body of the gene. A similar NSD1 binding profile was observed for the ZFP36L1 target, suggesting that the localization of NSD1 to 5′ elements is a general, but not necessarily exclusive, feature of the molecule.
We found that depletion of NSD1 abrogated the levels of H3-K36Me1, 2, and 3 at −1,200 and within the BMP4 gene, indicating that NSD1 enforces an activating H3-K36Me signal at the BMP4 locus. This was most pronounced at −1,200 and was accompanied by reduced levels of total RNAP II near +1. This is consistent with a role for H3-K36Me at −1,200 of BMP4 in an initiation event and is interesting because most results have implicated H3-K36Me in elongation (24, 28). Therefore, those reader proteins specific for −1,200 of BMP4 might be expected to be different from RpdS3, a complex that binds to H3-K36Me during elongation (28). Our data raises the question of how NSD1, located primarily near −1,200, can influence downstream H3-K36 methylation events. Because NSD1 occupancy is largely reduced inside the BMP4 gene, we suggest that NSD1 directly methylates nucleosomes around the −1,200 region, but indirectly influences H3-K36Me levels far downstream of the BMP4 start site. In this scenario, reduced and/or faulty initiation events due to NSD1 deficiency could indirectly alter elongation efficiency. Those RNA Pol II molecules that elongate in the context of NSD1 deficiency would be expected to show alterations in CTD phosphorylation (Fig. 5 B and C) that would be expected to be accompanied by defects in those H3-K36Me events that are catalyzed in a cotranscriptional manner.
We advocate a model where NSD1 deposits mono- and/or dimethyl groups onto H3-K36, and that these serve as potential substrates for the trimethylase. As we observe, in the absence of NSD1, the levels of all three forms of H3-K36Me are affected, but determining which form(s) are responsible for recruiting RNAP II to promoters will require further experimentation. However, such a function of NSD1 may not be conserved by other NSD family members. For example, knockdown of the NSD2 methyltransferase in HeLa cells led to reduced levels of H3-K36Me2, but not of H3-K36Me1 or H3-K36Me3 (18). Though informative, it is important to note that this study analyzed global H3-K36Me2 and 3 levels and was not conducted at the resolution of an individual target gene. Our knockdown cells show little, if any, differences in global levels of H3-K36Me. Moreover, a conflicting result identified NSD2 as an H3-K36 specific trimethylase in vivo that represses cardiac-specific transcription (42). This finding is curious in light of reports describing the role of HYPB/SET2 as the only H3-K36-specific trimethylase in murine and fly cells (43). We found no evidence for a repressive role of NSD1 at its gene targets in the three cell types examined here. Despite this, NSD1 has also been described to repress MEIS1 in neuroblastoma cells (3) as well as in reporter assays through its interaction with the NIZP1 zinc finger protein (44). We did not find that MEIS1 was expressed in our cells. Therefore, NSD1, and by extension its other family members, likely performs tissue-specific roles in chromatin function.
We routinely observed that RNAP II levels peaked at the +1 site of the BMP4 promoter. As abrogation of H3-K36Me at −1,200 resulted in a reduction in RNAP II occupancy at +1 of the BMP4 promoter, optimal RNAP II promoter occupancy depends on the status of NSD1-dependent methylation at −1,200. By itself, RNAP II cannot accurately recognize promoters to initiate transcription (45). Rather, transcriptional activators and/or chromatin modifiers can increase transcription initiation by improving the efficiency of RNAP II loading onto promoters. Our data suggest that the NSD1-dependent methyltransferase activity concentrated near −1,200 promotes RNAP II recruitment to the +1 site. This indicates that NSD1 functions in initiation at BMP4 by acting as a transcriptional coactivator in HCT116 cells through maintaining the levels of H3-K36Me at this region. Although we failed to detect immunoprecipitates indicative of physical interactions between RNAP II and NSD1, we speculate that the −1,200 region could be in physical contact with the +1 site through bridging factors. Here, NSD1-dependent methyl marks could recruit effectors (i.e., acetylases) to provide RNAP II with access to the promoter. Whether the decreased levels of RNAP II observed in NSD1-deficient cells reflects the lack of recruitment of factors known to be required for RNAP II-dependent initiation (i.e., TFIIB, D, E, F, and H) remains to be determined. In summary, our data link NSD1 activity with RNAP II loading through H3-K36Me, and suggests that this could, at least indirectly, impact elongation. How NSD1 operates in initiation (i.e., promoter escape and pausing) and what the role of the individual methylation marks are in this process remains the subject of future studies.
Materials and Methods
Cell Lines, Reagents, and Treatment.
Cell lines used in this study were obtained from American Type Culture Collection (ATCC). Cells were cultured in DMEM containing 10% FBS, and maintained in a humidified incubator (5% CO2) at 37 °C. To detect FLAG signal on Western blotting or in ChIP experiments we used FLAG M2 (Sigma) as per recommendation of the manufacturer. H3 antibody was obtained from Abcam (ab1791); H3K36Me1, Me2, and Me3 antibodies were purchased from Abcam (9048), Upstate (369), and Abcam (9050), respectively. An NSD1-specific antibody was obtained from Bethyl Laboratories (BL715). RNA polymerase II antibodies (pan, Ser2-P, and Ser5-P) were also purchased from Bethyl Laboratories as AbVantage Pack (A310-190A). Additional experimental details are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. Pierre Chambon (Strasbourg, France) for the gift of the murine NSD1 cDNA; Dr. Z. Wang (Case Western, Cleveland, OH) for plasmids and protocols for generating FLAG knockin cells; Dr. Joseph Alcorn (University of Texas, Houston, TX) for adenoviruses; and Dr. Shelley Barton (University of Texas-M. D. Anderson Cancer Center, Houston, TX) and Dr. Eric Wagner (University of Texas, Houston, TX) for other reagents. J.C.R. is a Pew Scholar in the Biomedical Sciences and is partially supported by National Institutes of Health Grant GM075094. This work was supported by Welch Foundation Grant AU-1569 (to P.B.C.) and National Institutes of Health Grant R56GM065812 (to P.B.C.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1002653107/-/DCSupplemental.
References
- 1.Angrand PO, et al. NSD3, a new SET domain-containing gene, maps to 8p12 and is amplified in human breast cancer cell lines. Genomics. 2001;74:79–88. doi: 10.1006/geno.2001.6524. [DOI] [PubMed] [Google Scholar]
- 2.Battaglia A, Filippi T, Carey JC. Update on the clinical features and natural history of Wolf–Hirschhorn (4p-) syndrome: Experience with 87 patients and recommendations for routine health supervision. Am J Med Genet C Semin Med Genet. 2008;148C:246–251. doi: 10.1002/ajmg.c.30187. [DOI] [PubMed] [Google Scholar]
- 3.Berdasco M, et al. Epigenetic inactivation of the Sotos overgrowth syndrome gene histone methyltransferase NSD1 in human neuroblastoma and glioma. Proc Natl Acad Sci USA. 2009;106:21830–21835. doi: 10.1073/pnas.0906831106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bergemann AD, Cole F, Hirschhorn K. The etiology of Wolf–Hirschhorn syndrome. Trends Genet. 2005;21:188–195. doi: 10.1016/j.tig.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 5.Keats JJ, et al. Overexpression of transcripts originating from the MMSET locus characterizes all t(4;14)(p16;q32)-positive multiple myeloma patients. Blood. 2005;105:4060–4069. doi: 10.1182/blood-2004-09-3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stec I, et al. WHSC1, a 90 kb SET domain-containing gene, expressed in early development and homologous to a Drosophila dysmorphy gene maps in the Wolf–Hirschhorn syndrome critical region and is fused to IgH in t(4;14) multiple myeloma. Hum Mol Genet. 1998;7:1071–1082. doi: 10.1093/hmg/7.7.1071. [DOI] [PubMed] [Google Scholar]
- 7.Zhao Q, et al. Transcriptome-guided characterization of genomic rearrangements in a breast cancer cell line. Proc Natl Acad Sci USA. 2009;106:1886–1891. doi: 10.1073/pnas.0812945106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang N, et al. Two distinct nuclear receptor interaction domains in NSD1, a novel SET protein that exhibits characteristics of both corepressors and coactivators. EMBO J. 1998;17:3398–3412. doi: 10.1093/emboj/17.12.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rayasam GV, et al. NSD1 is essential for early post-implantation development and has a catalytically active SET domain. EMBO J. 2003;22:3153–3163. doi: 10.1093/emboj/cdg288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang GG, Cai L, Pasillas MP, Kamps MP. NUP98-NSD1 links H3K36 methylation to Hox-A gene activation and leukaemogenesis. Nat Cell Biol. 2007;9:804–812. doi: 10.1038/ncb1608. [DOI] [PubMed] [Google Scholar]
- 11.Kurotaki N, et al. Haploinsufficiency of NSD1 causes Sotos syndrome. Nat Genet. 2002;30:365–366. doi: 10.1038/ng863. [DOI] [PubMed] [Google Scholar]
- 12.Leventopoulos G, et al. A clinical study of Sotos syndrome patients with review of the literature. Pediatr Neurol. 2009;40:357–364. doi: 10.1016/j.pediatrneurol.2008.11.013. [DOI] [PubMed] [Google Scholar]
- 13.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 14.Shilatifard A. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu Rev Biochem. 2006;75:243–269. doi: 10.1146/annurev.biochem.75.103004.142422. [DOI] [PubMed] [Google Scholar]
- 15.Turner BM. Cellular memory and the histone code. Cell. 2002;111:285–291. doi: 10.1016/s0092-8674(02)01080-2. [DOI] [PubMed] [Google Scholar]
- 16.Bell O, et al. Localized H3K36 methylation states define histone H4K16 acetylation during transcriptional elongation in Drosophila. EMBO J. 2007;26:4974–4984. doi: 10.1038/sj.emboj.7601926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bender LB, et al. MES-4: An autosome-associated histone methyltransferase that participates in silencing the X chromosomes in the C. elegans germ line. Development. 2006;133:3907–3917. doi: 10.1242/dev.02584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Y, et al. The target of the NSD family of histone lysine methyltransferases depends on the nature of the substrate. J Biol Chem. 2009;284:34283–34295. doi: 10.1074/jbc.M109.034462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stabell M, Larsson J, Aalen RB, Lambertsson A. Drosophila dSet2 functions in H3-K36 methylation and is required for development. Biochem Biophys Res Commun. 2007;359:784–789. doi: 10.1016/j.bbrc.2007.05.189. [DOI] [PubMed] [Google Scholar]
- 20.Lu T, et al. Regulation of NF-kappaB by NSD1/FBXL11-dependent reversible lysine methylation of p65. Proc Natl Acad Sci USA. 2010;107:46–51. doi: 10.1073/pnas.0912493107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kolasinska-Zwierz P, et al. Differential chromatin marking of introns and expressed exons by H3K36me3. Nat Genet. 2009;41:376–381. doi: 10.1038/ng.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sims RJ, 3rd, Reinberg D. Processing the H3K36me3 signature. Nat Genet. 2009;41:270–271. doi: 10.1038/ng0309-270. [DOI] [PubMed] [Google Scholar]
- 23.Spies N, Nielsen CB, Padgett RA, Burge CB. Biased chromatin signatures around polyadenylation sites and exons. Mol Cell. 2009;36:245–254. doi: 10.1016/j.molcel.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Larschan E, et al. MSL complex is attracted to genes marked by H3K36 trimethylation using a sequence-independent mechanism. Mol Cell. 2007;28:121–133. doi: 10.1016/j.molcel.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 25.Carrozza MJ, et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 26.Joshi AA, Struhl K. Eaf3 chromodomain interaction with methylated H3-K36 links histone deacetylation to Pol II elongation. Mol Cell. 2005;20:971–978. doi: 10.1016/j.molcel.2005.11.021. [DOI] [PubMed] [Google Scholar]
- 27.Kaplan CD, Laprade L, Winston F. Transcription elongation factors repress transcription initiation from cryptic sites. Science. 2003;301:1096–1099. doi: 10.1126/science.1087374. [DOI] [PubMed] [Google Scholar]
- 28.Kizer KO, et al. A novel domain in Set2 mediates RNA polymerase II interaction and couples histone H3 K36 methylation with transcript elongation. Mol Cell Biol. 2005;25:3305–3316. doi: 10.1128/MCB.25.8.3305-3316.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee JS, Shilatifard A. A site to remember: H3K36 methylation a mark for histone deacetylation. Mutat Res. 2007;618:130–134. doi: 10.1016/j.mrfmmm.2006.08.014. [DOI] [PubMed] [Google Scholar]
- 30.Li B, et al. Histone H3 lysine 36 dimethylation (H3K36me2) is sufficient to recruit the Rpd3s histone deacetylase complex and to repress spurious transcription. J Biol Chem. 2009;284:7970–7976. doi: 10.1074/jbc.M808220200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morris SA, et al. Histone H3 K36 methylation is associated with transcription elongation in Schizosaccharomyces pombe. Eukaryot Cell. 2005;4:1446–1454. doi: 10.1128/EC.4.8.1446-1454.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu L, et al. Di- and tri- but not monomethylation on histone H3 lysine 36 marks active transcription of genes involved in flowering time regulation and other processes in Arabidopsis thaliana. Mol Cell Biol. 2008;28:1348–1360. doi: 10.1128/MCB.01607-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bannister AJ, et al. Spatial distribution of di- and tri-methyl lysine 36 of histone H3 at active genes. J Biol Chem. 2005;280:17732–17736. doi: 10.1074/jbc.M500796200. [DOI] [PubMed] [Google Scholar]
- 34.Kim A, Kiefer CM, Dean A. Distinctive signatures of histone methylation in transcribed coding and noncoding human beta-globin sequences. Mol Cell Biol. 2007;27:1271–1279. doi: 10.1128/MCB.01684-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Song JS, et al. Model-based analysis of two-color arrays (MA2C) Genome Biol. 2007;8:R178. doi: 10.1186/gb-2007-8-8-r178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bell SE, et al. The RNA binding protein Zfp36l1 is required for normal vascularisation and post-transcriptionally regulates VEGF expression. Dev Dyn. 2006;235:3144–3155. doi: 10.1002/dvdy.20949. [DOI] [PubMed] [Google Scholar]
- 37.Chen D, Zhao M, Mundy GR. Bone morphogenetic proteins. Growth Factors. 2004;22:233–241. doi: 10.1080/08977190412331279890. [DOI] [PubMed] [Google Scholar]
- 38.Simmen RC, et al. The emerging role of Krüppel-like factors in endocrine-responsive cancers of female reproductive tissues. J Endocrinol. 2010;204:223–231. doi: 10.1677/JOE-09-0329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van der Maarel SM, et al. Cloning and characterization of DXS6673E, a candidate gene for X-linked mental retardation in Xq13.1. Hum Mol Genet. 1996;5:887–897. doi: 10.1093/hmg/5.7.887. [DOI] [PubMed] [Google Scholar]
- 40.Komarnitsky P, Cho EJ, Buratowski S. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 2000;14:2452–2460. doi: 10.1101/gad.824700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Buratowski S. Progression through the RNA polymerase II CTD cycle. Mol Cell. 2009;36:541–546. doi: 10.1016/j.molcel.2009.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nimura K, et al. A histone H3 lysine 36 trimethyltransferase links Nkx2-5 to Wolf-Hirschhorn syndrome. Nature. 2009;460:287–291. doi: 10.1038/nature08086. [DOI] [PubMed] [Google Scholar]
- 43.Edmunds JW, Mahadevan LC, Clayton AL. Dynamic histone H3 methylation during gene induction: HYPB/Setd2 mediates all H3K36 trimethylation. EMBO J. 2008;27:406–420. doi: 10.1038/sj.emboj.7601967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nielsen AL, et al. Nizp1, a novel multitype zinc finger protein that interacts with the NSD1 histone lysine methyltransferase through a unique C2HR motif. Mol Cell Biol. 2004;24:5184–5196. doi: 10.1128/MCB.24.12.5184-5196.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ptashne M, Gann A. Transcriptional activation by recruitment. Nature. 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.