Abstract
In facultative photoperiodic flowering plants, noninductive photoperiods result in a delay in flowering, but such plants eventually flower, illustrating plasticity in an important developmental transition, flowering. The model plant, Arabidopsis, has a facultative photoperiod response. Although the inductive flowering promotion pathway has been extensively studied, the pathway to flowering in noninductive photoperiods is not well understood. Here, we show that a Plant Homeo Domain finger-containing protein, VIN3-LIKE 2 (VIL2), is necessary to maintain the epigenetically repressed state of MAF5 and permit more rapid flowering in noninductive photoperiods in Arabidopsis. Levels of both VIL2 mRNA and protein are under diurnal fluctuation and maintain the repressed state at MAF5 chromatin in a photoperiod-specific manner. VIL2 binds preferentially to dimethylated histone H3 Lys-9 (H3K9me2) peptides in vitro and VIL2 is required for the maintenance of H3K9me2 at MAF5 chromatin in vivo. Furthermore, VIL2 is required for the maintenance of trimethylated histone H3 Lys-27 at MAF5 through the physical association with a component of polycomb repression complex 2. Thus, the repression of MAF5 by VIL2 provides a mechanism to promote flowering in noninductive photoperiods, which contributes to the facultative nature of the Arabidopsis photoperiodic response.
Keywords: chromatin, flowering, diurnal
Like all organisms, plants have evolved to maximize their reproductive success, and optimizing the timing of flowering plays a critical role in this success. Flowering time is influenced by environmental cues, including temperature and day length. Floral regulatory cues ultimately act to regulate a group of genes including FLOWERING LOCUS T (FT), SUPPRESSOR OF OVER-EXPRESSION OF CONSTANS1 (SOC1) and LEAFY (LFY) (1, 2). These genes are often referred to as “floral integrators” because their expression is in turn regulated by flowering pathways that sense environmental cues as well as developmental states (1, 2). FLOWERING LOCUS C (FLC) and its related genes (hereafter referred to as the FLC clade) are major floral repressors that act to repress the floral integrators FT and SOC1 (3–7). The FLC clade includes FLOWERING LOCUS M (FLM)/MADS AFFECTING FLOWERING 1 (MAF1) and MAF2–5 (3–7). Polymorphisms in FLM/MAF1 are responsible for natural variations in flowering time under short days (SD) in certain ecotypes (8).
VERNALIZATION INSENSITIVE 3 (VIN3) acts to repress FLC during exposure to prolonged periods of cold, through a process known as vernalization (9–11). VIN3 is induced by vernalizing cold and thus acts to trigger epigenetic repression in response to environmental changes at its targets, including FLC and some FLC clade members (9, 11–14). VIN3 exists as a small gene family, known as the VIN3/VERNALIZATION LIKE (VEL) gene family (13, 15). Another member of the gene family, VIN3-LIKE 1 (VIL1)/VERNALIZATION 5 (VRN5), is also involved in the vernalization-mediated repression of FLC and another member of the FLC clade, FLM/MAF1 (13, 15). The VIN3/VEL gene family includes three additional members, VIL2/VEL1, VIL3/VEL2, and VIL4/VEL3, although VIL4/VEL3 appears to be a pseudogene (13, 15).
Arabidopsis is a facultative long-day (LD) plant; i.e., it flowers rapidly in LD and flowers much later in SD. The inductive photoperiod pathway (also known as the LD pathway) has been well characterized (16, 17). Perception of inductive day length appears to be a function of the coincidence between light and expression of the circadian-regulated gene, CO (16, 17). Once CO is activated by light, it directly activates floral integrators (i.e., FT and SOC1), which in turn initiate flowering. On the other hand, flowering promotion under SD is poorly understood. A phytohormone, gibberellic acid (GA), plays a role in the promotion of flowering under noninductive SD photoperiods. The flowering effects of GA are exerted through the floral integrators, LFY and SOC1 (2), likely via a DELLA-dependent mechanism (18). Lack of flowering in the absence of GA under SD is likely the combinatorial consequence of a lack of inductive photoperiod stimulation and the lack of activation of floral integrators by GA, thus creating a situation in which no floral promoters are present.
Interestingly, VIL1/VRN5 promotes flowering specifically under SD via the repression of FLM/MAF1 through chromatin modifications (13). Although FLC is the major floral repressor, other FLC-related genes act as floral repressors as well (5–7). FLM/MAF1 is a repressor of flowering and repression of FLM/MAF1 contributes to flowering in SD (5, 6, 8, 13). Other MAF genes, including MAF2–5, also share common characteristics as floral repressors (5, 14). The induced nature of VIN3 by vernalization (11) and that of VIL1 by SD conditions (13) demonstrate that members of the VIN3/VEL gene family are involved in the control flowering time in response to environmental cues.
Here, we show that VIL2/VEL1 is required for the repression of MAF5 under SD. VIL2 binds preferentially to dimethylated histone H3 Lys-9 (H3K9me2) in vitro and is necessary for the establishment and/or maintenance of H3K9me2 at MAF5 chromatin. Furthermore, VIL2 coprecipitates with a component of PRC2 and is required for the establishment and/or maintenance of trimethylated histone H3 K27 (H3K27me3) at MAF5 chromatin. The pattern of VIL2 expression is part of the system for SD-specific promotion of flowering in a facultative LD plant, Arabidopsis.
Results and Discussion
VIL2 Is Necessary for the Floral Promotion Under Short Days.
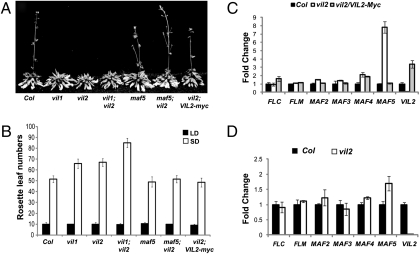
To evaluate whether another member of the VIN3/VEL family, VIL2, has a role in flowering, we determined the flowering behavior of a vil2 mutant under different photoperiods (Fig. S1 and Fig. 1 A and B). Interestingly, vil2 mutants display delayed flowering in SD conditions, but flower similarly to the wild type in LD, similar to vil1 mutants. Because VIN3 and VIL1 are required for the repression of a subset of the FLC clade, we examined mRNA levels of FLC clade genes in vil2 mutants. The level of MAF5 mRNA is elevated in vil2 mutants especially in SD (Fig. 1C). Unlike vil1 mutants (13), however, there is no significant change observed in mRNA levels of FLM in vil2 mutants (Fig. 1 C and D), indicating VIL1 and VIL2 operate independently to regulate flowering time in SD through different members of the FLC clade. Furthermore, vil1/vil2 double mutants flower much later than either single mutant, supporting parallel activities of VIL1 and VIL2 (Fig. 1 A and B). Interestingly, the elevated mRNA level of MAF5 is more apparent in SD than in LD (Fig. 1 C and D), consistent with SD-specific flowering phenotypes in vil2 mutants (Fig. 1 A and B). MAF5 acts as a floral repressor similar to other members of the FLC clade (5, 6). Expression of VIL2 using 35S CaMV promoter can repress MAF5 and rescue vil2 mutants in SD (vil2/VIL2-myc; Fig. 1 B and C). Furthermore, a mutation in MAF5 completely abolishes the late-flowering phenotype of vil2 mutants (Fig. 1B), genetically confirming that the late flowering of vil2 mutants is mainly due to an effect on MAF5.
Fig. 1.
vil2 mutants flower late only under short days. Flowering times are determined by rosette leaf numbers at the time of bolting (A and B). Solid bars indicate leaf numbers under long days (16 h light:8 h dark) and open bars indicate leaf numbers under short days (8 h light:16 h dark) (B). The levels of transcripts of the FLC clade and VIL2 under short days (C) and under long days (D) were determined by using the real-time reverse transcription followed by PCR (RT-PCR) analysis. Relative fold changes are shown compared between wild type (Columbia; WT; solid bars) and vil2 mutants (open bars). The levels of transcripts in complemented lines (VIL2-Myc) are shown as gray bars (C).
VIL2 was biochemically copurified with PRC2-VIN3-VIL1/VRN5, suggesting they are acting together to mediate stable repression of their targets (19). However, we do not observe significant differences in MAF5 mRNA expression in either vin3 or vil1 mutants compared with wild type under SD (Fig. S2), suggesting that the repression of MAF5 by VIL2 under SD is independent of the PRC2-VIN3-VIL1/VRN5–containing complex.
Given the biochemical evidence that VIL2 is part of a PRC2-VIN3-VIL1/VRN5 complex involved in vernalization-mediated FLC repression (19), we evaluated the role of VIL2 in the vernalization response. Accordingly, the vil2 mutation was (i) introgressed into the vernalization-responsive genetic fca mutant background and (ii) a FRIGIDA transgene was transformed into vil2 mutants (Fig. S3). In both cases, however, vil2 mutants exhibit a wild-type vernalization response. It is possible that the lack of phenotype is due to the redundancy of VIL2 with other members of the VIN3 gene family in the complex.
VIL2 Exhibits a Diurnal Pattern of Expression.
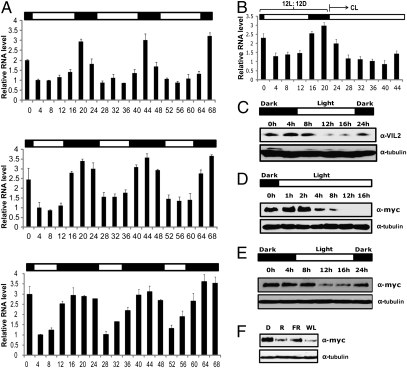
The late flowering of vil2 mutants only under SD prompted us to evaluate the influence of photoperiod on VIL2 expression. We examined VIL2 mRNA levels during various light–dark cycles (Fig. 2A). Interestingly, VIL2 mRNA levels are diurnally regulated: VIL2 transcripts accumulate during the dark period and are reduced in light. VIL2 mRNA is not circadian regulated as the diurnal pattern of VIL2 mRNA quickly disappears in continuous light condition after entrainment in 12 h light/12 h dark for 7 d (Fig. 2B). We obtained polyclonal antibodies against VIL2 to evaluate the expression patterns of endogenous VIL2 protein. Similar to the patterns of VIL2 mRNA expression, VIL2 protein also exhibits a diurnal pattern of expression (Fig. 2C). We also examined the VIL2 protein level in 35S::VIL2 lines in which VIL2 is fused to a c-Myc tag and rescues vil2 mutants (Fig. 1C). Interestingly, the level of VIL2 protein is reduced upon exposure to light (Fig. 2D), despite expression from a constitutive promoter. Furthermore, VIL2 protein is also diurnally regulated in overexpression lines (Fig. 2E). The level of the VIL2 protein is reduced upon exposure to light regardless of the light quality (Fig. 2F). However, the maximum amounts of VIL2 protein under different photoperiods are similar to one another (Fig. S4). Thus, the diurnal nature of VIL2 protein results in a different amount of available VIL2 protein over the course of different photoperiods. As day length shortens, VIL2 protein would become more abundant overall.
Fig. 2.
Diurnal patterns of VIL2 expression. VIL2 transcripts are diurnally regulated. (A) Diurnal regulation of VIL2 mRNA was determined by real-time RT-PCR analysis. Relative fold changes are shown under different light (open bar)/dark (black bar) periods. The light/dark cycles were: 16 h light:8 h dark (Top), 12 h light:12 h dark (Middle), and 8 h light:16 h dark (Bottom) photoperiod conditions. Samples were taken every 4 h starting from “hour 0.” Hour 0 is at the end of the dark period. (B) After 12 h light:12 h dark entrainment, seedlings were exposed to continuous light (CL). Samples were taken every 4 h starting from hour 0. Hour 0 is at the end of the dark period as indicated by black bar (dark) and open bar (light). (C) VIL2 protein exhibits diurnal patterns of expression. Seedlings were grown under 12 h light:12 h dark photoperiod conditions. Samples were taken every 4 h starting from hour 0. Hour 4 is at the end of the dark period as indicated by black bar (dark) and open bar (light). (D) VIL2 protein is degraded upon light exposure. Dark grown 35S::VIL2-Myc transgenic seedlings were exposed to continuous light and seedlings were taken during the intervals as indicated for the Western blot analysis. Samples were taken every 4 h starting from hour 0. Hour 0 is at the end of the dark period as indicated by black bar (dark) and open bar (light). (E) VIL2 protein in 35S::VIL2-Myc transgenic lines also undergoes diurnal regulation. Seedlings were grown under 12 h light:12 h dark photoperiod conditions. Samples were taken every 4 h starting from hour 0. Hour 4 is at the end of the dark period as indicated by black bar (dark) and open bar (light). (F) Light quality does not affect the VIL2 degradation upon light exposure. Dark-grown seedlings were illuminated by Red (R; 3 μmol·m−2·s−1), Far-red (FR; 0.5 μmol·m−2·s−1) and white light (WL; ∼70 μmol·m−2·s−1) for 12 h.
VIL2 Binds Preferentially to Dimethylated H3 Lys-9 Peptides in Vitro.
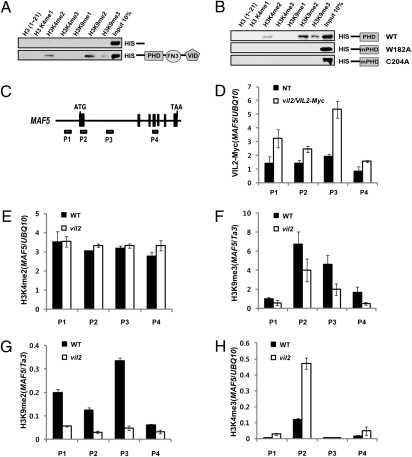
The VIN3 family of proteins contains three distinct domains: a Plant Homeo Domain (PHD) finger, a fibronectin type III (FNIII) domain, and a VIN3-interacting domain (VID) (Fig. 3A). The PHD finger can serve as a “reader” motif for modified histones (20). To determine whether VIL2 binds to modified histones, we performed in vitro histone peptide-binding assays (21–23) using a series of synthetic histone peptides with various modifications (see Table S1 for the full list of peptides tested; Fig. S5 and Fig. 3 A and B, results only with positive binding are shown). Among the tested peptides, VIL2 exhibits binding to dimethylated histone H3 Lys-4 (H3K4me2), trimethylated histone H3 Lys-9 (H3K9me3), as well as H3K9me2 peptides. VIL2 exhibits the strongest binding to H3K9me2 peptides (Fig. 3 A and B). H3K9me2 is a modification that is enriched in repressed chromatin in plants (11, 24–26). We observed binding using full-length proteins as well as the PHD finger motif in isolation (Fig. 3 A and B).
Fig. 3.
VIL2 binds preferentially to H3K9me2 peptides in vitro and is required for the maintenance of H3K9me2 at MAF5 chromatin. (A and B) In vitro histone peptides pull-down assay using recombinant VIL2 proteins. Full-length VIL2 recombinant proteins with His-tag (A) or the PHD motif of VIL2 (amino acid 146–326) with His-tag (B) were used. His-tag–only controls were used as a negative control. Full list of synthesized histone peptides used in this study is described in Table S1. No binding to other histone peptides was observed unless presented here. (C) Schematic representation of MAF5 genomic regions and relative position of primers used in chromatin immunoprecipitation (ChIP) assays followed by quantitative real-time PCR (qPCR). (D) VIL2-Myc epitope-tagged transgenic lines were used to detect the enrichment of VIL2 at MAF5 chromatin. (E–H) Chromatin signatures at MAF5 chromatin were determined using antibodies against H3K4me2 (E), H3K9me3 (F), H3K9me2 (G), and H3K4me3 (H). The relative enrichments were shown compared with indicated controls (D–H). The enrichments of controls and MAF5 regions were first calculated on the basis of the fractions of precipitates compared with each input. Relative enrichments were then calculated on the basis of comparison with each indicated control (D–H).
To address the specificity of the binding, we introduced point mutations in the PHD finger motif of VIL2 (Fig. 3B). Not surprisingly, disruption of the zinc finger itself by the substitution of one of the cysteine residues critical for finger formation with alanine completely abolishes the binding of VIL2 to modified histone peptides (Fig. 3B; C204A). Binding specificity depends on a “binding pocket” in the PHD finger (27). In the case of the dimethyl group, the binding cage needs two hydrophobic groups and one charged group to neutralize the positive charge of the epsilon amine (27). We identified candidate hydrophobic residues from the VIL2 protein and changed them to alanines, one by one. Interestingly, the change of a tryptophan (Fig. 3B; W182) to alanine completely abolished the histone peptide binding of the PHD finger of the VIL2 protein, suggesting W182 is a part of the binding pocket for dimethyl histone binding of VIL2 (Fig. S6).
VIL2 Directly Associates with MAF5 Chromatin and Is Required for the Enrichment of H3K9me2 at MAF5 Chromatin.
To examine whether VIL2 directly associates with MAF5 chromatin, we used a c-Myc–tagged VIL2 complemented line (Fig. 1 A and B). Chromatin immunoprecipitation (ChIP) assays using the anti-c-Myc antibody revealed enrichment of VIL2 at MAF5 chromatin, especially in the P3 region (Fig. 3 C and D). We further examined the enrichment of H3K4me2, H3K9me2, and H3K9me3 (modifications that VIL2 binds in vitro) on MAF5 chromatin in wild type and in vil2 mutants to address the in vivo implications of the in vitro histone binding preference by VIL2 (Fig. 3 A and B). We did not observe significant difference at the level of enrichment of H3K4me2 at MAF5 chromatin between wild type and vil2 mutants (Fig. 3E). We did observe above background levels of enrichment of H3K9me3 at MAF5 chromatin and the enrichment of H3K9me3 at MAF5 chromatin is mildly reduced in vil2 mutants (Fig. 3F). Interestingly, H3K9me2 is most highly enriched in P3 region of MAF5 where the VIL2 protein is highly enriched in wild type (Fig. 3 D and G). Furthermore, the enrichment is greatly reduced in vil2 mutants (Fig. 3G), suggesting that VIL2 is required for the enrichment of H3K9me2 at MAF5 chromatin through its direct association with H3K9me2 at MAF5 in a positive autoregulatory loop. The level of H3K4me3 is increased at 5′ regions of MAF5 in vil2 mutants, consistent with the activation of MAF5 in vil2 mutants (Fig. 3H).
VIL2 Mediates a Photoperiod-Dependent Epigenetic Repression of MAF5.
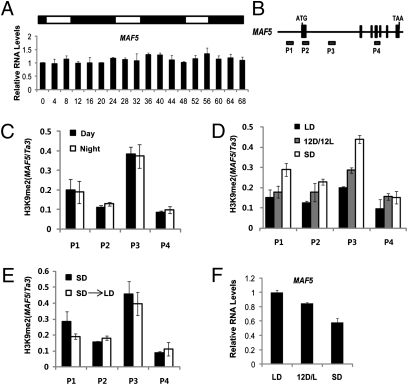
Diurnal patterns of VIL2 expression and the requirement for VIL2 in the enrichment of H3K9me2 at MAF5 chromatin prompted us to test whether MAF5 mRNA expression also undergoes diurnal changes. Unlike VIL2 mRNA, however, MAF5 transcripts do not show obvious diurnal patterns (Fig. 4A). Furthermore, the enrichment of H3K9me2 at MAF5 chromatin does not change diurnally (Fig. 4C). Given the diurnal changes at the level of VIL2 protein, it is likely that the overall abundance of VIL2 under different photoperiods results in different levels of enrichment of H3K9me2 at MAF5 chromatin. If this is the case, the H3K9me2 enrichment at MAF5 chromatin would increase as day length shortens. To address this issue, we examined the level of H3K9me2 under different photoperiods (Fig. 4D). The level of enrichment of H3K9me2 at MAF5 chromatin is most abundant under SD (Fig. 4D). The level of H3K9me2 enrichment at MAF5 chromatin is correlated with the relative levels of MAF5 mRNA expression (Fig. 4F). This enrichment of H3K9me2 at MAF5 chromatin differs according to photoperiod conditions and does not quickly change when plants are moved to different photoperiods (Fig. 4 C and E). Thus, it appears that a shorter day length (i.e., SD) triggers the repression of MAF5 through H3K9me2 enrichment and represses the level of MAF5 mRNA expression. This is consistent with VIL2 being a mediator of H3K9me2 levels at MAF5; shorter days would result in greater overall abundance of VIL2 and thus higher levels of H3K9me2 at MAF5 resulting in decreased expression.
Fig. 4.
Photoperiod-mediated epigenetic repression of MAF5. (A) The levels of MAF5 mRNA were determined by real-time RT-PCR under SD. Relative levels of MAF5 mRNA expression were shown compared with PP2A under 8 h light (open bar):16 h dark (black bar) periods. (B) Schematic representation of MAF5 genomic regions and relative position of primers used in ChIP assays (P1–P4). (C) The level of enrichment of H3K9me2 at MAF5 is determined under light and dark conditions at MAF5 chromatin. Samples were taken after 4 h of light exposure (day) and after 4 h of dark (night) under SD. (D) The level of H3K9me2 enrichment at MAF5 chromatin under different photoperiods. Samples were taken at the end of the dark (night) under each photoperiod indicated. (E) The levels of H3K9me2 enrichment at MAF5 chromatin are not changed by photoperiod shifts. For SD ChIP assays, seedlings were germinated under SD and taken for the analysis 7 d after germination under SD. For SD→LD ChIP, seedlings were germinated and grown for 5 d under SD and moved to LD for an additional 2 d. (C–E) The relative enrichments were shown compared with indicated controls. The enrichment of controls and MAF5 regions was first calculated on the basis of the fractions of precipitates compared with each input. Relative enrichments were then calculated on the basis of comparison with each indicated control. (F) mRNA levels of MAF5 under different photoperiods were determined by real-time RT-PCR. Seedlings were taken 5 d after germination under different photoperiods as indicated by LD (16 h light:8 h dark), 12 D/L (12 h light:12 h dark), and SD (8 h light:16 h dark).
VIL2 Coprecipitates with CLF and the Enrichment of CLF at MAF5 Chromatin Increases in SD.
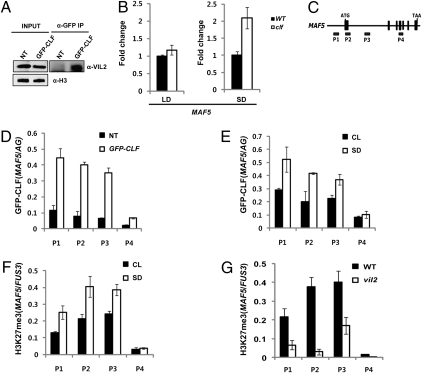
During vernalization, VIN3/VIL1/VIL2 associates with PRC2 to mediate the repression of FLC (19, 28). Although the level of MAF5 mRNA is not altered either in vin3 or in vil1 mutants in SD (Fig. S2), it is possible that VIL2 forms an alternative complex with PRC2 to mediate the repression of MAF5. It was also reported that MAF5 is up-regulated in clf mutants and that CLF associates with MAF5 chromatin (29). To directly test whether VIL2 forms a complex with PRC2, we performed coimmunoprecipitation assays using GFP::CLF transgenic lines (Fig. 5A). Indeed, VIL2 protein was detected from precipitates using an antibody against GFP (Fig. 5A), suggesting that VIL2 associates with a PRC2 complex. Consistent with the physical association of VIL2 with CLF, we observed that the level of MAF5 mRNA in clf mutants is higher in SD than in LD (Fig. 5B). Furthermore, CLF directly associates with MAF5 chromatin (Fig. 5D), as previously reported (29). Interestingly, the enrichment of CLF at MAF5 increases under SD compared with continuous light (Fig. 5E).
Fig. 5.
VIL2 coprecipitates with CLF and is required for the enrichment of H3K27me3 at MAF5 chromatin. (A) Coimmunoprecipitation assay using CLF::GFP transgenic lines. Ten percent of input proteins were visualized by immunoblot using anti-VIL2 antibody and histone H3 antibody (Left). VIL2 coprecipitates with CLF as detected by anti-VIL2 antibody from precipitates using anti-GFP antibody (Right). Nontrangenic plants (NT) were compared for the specificity of precipitation. (B) The level of MAF5 mRNA expression is up-regulated in clf mutants in SD. Relative fold changes compared with the wild type (WT; Col) in each photoperiod condition are shown. (C) Schematic representation of MAF5 genomic regions and relative position of primers used in ChIP assays (P1–P4). (D) ChIP assay using GFP::CLF transgenic plants to address the enrichment of CLF at MAF5 chromatin. Nontrangenic plants (NT) were compared for the specificity of precipitation. Relative enrichments were shown compared with an AGAMOUS (AG) region, a previously reported CLF target (33). (E) The enrichment of CLF at MAF5 chromatin increases in SD. Seedlings of GFP::CLF transgenic plants grown under continuous light (CL) and SD were used for ChIP assays. (F) ChIP assays to address the relative enrichment of H3K27me3 at MAF5 chromatin under different photoperiods. Seedlings of GFP::CLF transgenic plants grown under continuous light (CL) and SD were used for ChIP assays. (G) ChIP assays to address the relative enrichment of H3K27me3 at MAF5 chromatin in vil2 mutants. Seedlings of the wild type (WT; Col) and vil2 mutants grown in SD were used. The relative enrichments were shown compared with indicated controls (D–G). The enrichments of controls and MAF5 regions were first calculated on the basis of the fractions of precipitates compared with input. Relative enrichments were then calculated on the basis of comparison with each indicated control (D–G). For H3K27me3 ChIP analysis, FUS3 was used as a positive control as FUS3 chromatin was previously reported to be enriched with H3K27me3 (34).
PRC2 mediates methylation at H3K27 (30). Consistent with the increased association of PRC2 at MAF5 chromatin in SD, the level of H3K27me3 at MAF5 chromatin also increases in SD (Fig. 5F). Interestingly, the level of H3K27me3 enrichment at MAF5 chromatin is significantly reduced in vil2 mutants (Fig. 6G) and thus VIL2 is required for the enrichment of H3K27me3 at MAF5 chromatin under SD. VIL2 preferentially binds to H3K9me2 (Fig. 3 A and B) but not to H3K27me3 (Fig. S4) in vitro. Thus, VIL2 is likely to contribute to a positive feedback loop between two repressive marks, H3K9me2 and H3K27me3, through interaction with PRC2.
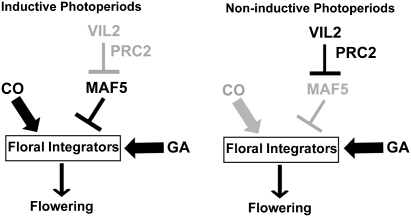
Fig. 6.
A model for the role of VIL2 in noninductive photoperiod-specific repression of MAF5. Under inductive photoperiod, the LD pathway becomes dominant to activate floral integrators, resulting in early flowering (Left). Under noninductive photoperiods, the LD pathway becomes inactive. Instead, VIL2 becomes available to repress a floral repressor, MAF5 (Right). The repression of MAF5 under noninductive condition prevents further delay in flowering in the absence of the LD pathway.
Conclusion
The diurnal nature of VIL2 expression is part of a mechanism to monitor different photoperiods and represses MAF5 specifically under noninductive photoperiods (Fig. 6). This contributes to the molecular system for the facultative nature of Arabidopsis photoperiod-dependent flowering. Under LD, the LD promotion pathway is active (i.e., CO) and the LD pathway can overcome the effect of MAF5. In SD, however, the LD pathway is inactive and there is no active floral promotion pathway except the GA promotion pathway. VIL2 becomes more available to repress MAF5 in SD. The repression of MAF5 in SD ensures that flowering eventually happens in the absence of the LD promotion pathway.
The VIL2/MAF5 component of SD flowering is independent of the GA promotion pathway, which also plays a role in flowering promotion and is indispensable for flowering under noninductive photoperiods. GA treatments do result in earlier flowering either in wild type or vil2 mutants (Fig. S7A). However, the effect of GA is neither through VIL2 nor MAF5, but rather due to direct activation of floral integrators, SOC1 and LFY (Fig. 6 and Fig. S7 B–F), as previously reported (2, 18). Thus, VIL2 functions to prevent further delay in flowering under noninductive photoperiods in a GA-independent manner.
It is interesting to note that all VIN3 family members characterized so far act to monitor certain environmental changes (i.e., temperature and day length), suggesting that the VIN3 family of proteins may have evolved to serve as a gateway between environmental stimuli and developmental programs through chromatin-based mechanisms. VIN3 is induced by vernalizing cold to mediate the repression of FLC by vernalization, whereas the level of VIL2 protein is diurnally regulated to mediate the repression of MAF5 in SD. Furthermore, diurnal patterns of an H3K9me2 binding protein and its requirement for enrichment of H3K9me2 and H3K27me3 at its target in a photoperiod-specific manner provides insight on environmentally induced epigenetic switches that play roles in developmental transitions.
Materials and Methods
Plants Materials and Growth Conditions.
The vil2 mutant (vil2-1) was obtained from the SAIL T-DNA population (SAIL_323_A03). The maf5 mutant (maf5-1) was obtained from the SALK collection (SALK_095092). vil1-1 is previously described (13). Primers used to determine genotypes are in Table S1. Plants were grown at 22 °C in different photoperiods as described in each experiment. Vernalization treatments were as previously described (13).
Antibodies.
The primary antibodies for immunoblot assays used in this study were histone H3 antibody (Abcam; 10799), anti-tubulin monoclonal antibody (Sigma-Aldrich; T9026), anti-myc antibody (Santa Cruz Biotechnology; sc-40), anti-His antibody (Santa Cruz Biotechnology; sc-803), and polyclonal anti-VIL2 antibody (produced by Genscript). Polyclonal anti-VIL2 antibody was obtained using synthesized peptide antigen (473 PKKPSSKNEDNNSP 486; Genscript). Synthesized peptide was injected into rabbit and resulting antisera was further purified by affinity purification (Genscript).
In Vivo Coimmunoprecipitation Assays.
Arabidopsis GFP–CLF transgenic plants were used for coimmunoprecipitation analysis. Using a mortar, Arabidopsis seedling (10 g) grown in dark condition for 4 d after seed germination was frozen and ground in liquid nitrogen. Ground samples were resuspended in the nuclei extraction buffer [0.25 M sucrose, 15 mM Pipes (pH 6.8), 5 mM MgCl2, 60 mM KCl, 15 mM NaCl, 1% Triton X-100, 1 mM PMSF, 1 tablet of proteinase inhibitor mixture (Roche)], and incubated on ice for 15 min. Samples were centrifuged at 14,500 rpm for 10 min at 4 °C and pellets were resuspended with 1 mL lysis buffer [50 mM hepes (pH 7.4), 150 mM NaCl, 1 mM EDTA (pH 8.0), 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS, 1 mM PMSF, 1 tablet of proteinase inhibitor mixture (Roche)]. Resuspended samples were sonicated. It was centrifuged at 4 °C at 14,500 rpm for 10 min. Superantant was transferred to new tubes and incubated overnight with GFP polyclonal antibody (Invitrogen; A11122) at 4 °C with gentle rotation. On the following day, 0.1 volume of protein A/agarose beads was added and incubated at 4 °C for 2 h with gentle rotation. Beads were collected by centrifugation and washed three times with washing buffer [50 mM Tris-HCl (pH 7.5), 1% Triton X-100, 1 mM EDTA (pH 8.0), 150 mM NaCl]. Samples were then resolved in SDS/PAGE gel and transferred for Western blot analysis with VIL2 polyclonal antibody.
In Vitro Histone Peptide Pull-Down Assay.
Full-length and partial VIL2 cDNAs were amplified by RT-PCR and cloned into the pVP13 (31) expression vector. Recombinant full-length and partial VIL2 proteins were purified using a Ni-NTA column. Biotinylated histone peptides were purchased from Millipore; see Table S1). Histone pull-down assays were performed as described (22). In short, 2 μg of biotinylated histone peptides (1 mg/mL) was added into 300 μL of pull-down buffer [50 mM Tris-Cl (pH7.5), 150 mM NaCl, 2 mM DTT, 0.05% Nonidet P-40, proteinase inhibitor mixture] together with about 2 μg of recombinant proteins. The 30 μL of streptavidin Sepharose beads (GE Healthcare; 17-5113-01) were used for each binding assay. The mixture was rotated overnight at 4 °C. Beads were harvested by centrifuging at 3,000 rpm for 1 min at 4 °C. The beads were washed three times with pull-down buffer. The beads were resuspended in 60 μL of 2× SDS sample buffer and then boiled and loaded on 10% SDS/PAGE. For control, 10–20% of recombinant proteins were loaded. Proteins were transferred by using mini Transblot electrophoretic transfer cell apparatus. His-tag antibody (Santa Cruz Biotechnology) was used for Western blot analysis.
ChIP Analysis.
Constructs carrying VIL2::c-Myc were transformed into vil2-1 and homozygous lines were identified at T3 generation and used for ChIP analysis. c-Myc antibodies (monoclonal; 9E10) were purchased from Santa Cruz Biotechnology. Antibodies against modified histones were purchased from Abcam. ChIP assays were performed as previously described (26). For primer sequences, see Table S1. All ChIP assays were performed at least three times from at least two biological replicates and produced similar results. Quantitative real-time PCR (qRT-PCR) was done using SYBR green dye reaction mixture (Applied Biosystems) according to the manufacturer's instructions. Real-time PCR was performed on a 7900HT Fast Real-Time PCR system (Applied Biosystems).
RNA Analysis.
Total RNA was isolated using TRIZOL (Invitrogen) according to the manufacturer's instructions. Before reverse transcription reaction, DNase I (Invitrogen) treatment was performed for 30 min at 37 °C to eliminate contaminated genomic DNA from total RNAs. First strand cDNA synthesis was performed on 2 μg of RNAs using the M-MLV system (Promega). The relative transcript level of each gene was determined by normalization of the resulting expression levels versus that of PP2A, as described (32). Whole sets of primer pairs for real-time PCR analysis were determined using the AtRTPrimer program (http://atrtprimer.kaist.ac.kr/). To ensure the specificity and efficiency of each primer pair, RT-PCR product of each primer pair was tested in 3% agarose gel before real-time PCR analyses. For primer sequences used in real-time PCR analyses, see Table S2.
Supplementary Material
Acknowledgments
We thank Justin Goodrich for providing seeds of the GFP::CLF transgenic lines (University of Edinburgh, UK). We thank Jessica Sisavath and Evelyn Joo for technical assistance. This study was supported by start-up funds from the University of Texas at Austin and by the National Science Foundation to S.S.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1010834107/-/DCSupplemental.
References
- 1.Parcy F. Flowering: A time for integration. Int J Dev Biol. 2005;49:585–593. doi: 10.1387/ijdb.041930fp. [DOI] [PubMed] [Google Scholar]
- 2.Moon J, Lee H, Kim M, Lee I. Analysis of flowering pathway integrators in Arabidopsis. Plant Cell Physiol. 2005;46:292–299. doi: 10.1093/pcp/pci024. [DOI] [PubMed] [Google Scholar]
- 3.Helliwell CA, Wood CC, Robertson M, James Peacock W, Dennis ES. The Arabidopsis FLC protein interacts directly in vivo with SOC1 and FT chromatin and is part of a high-molecular-weight protein complex. Plant J. 2006;46:183–192. doi: 10.1111/j.1365-313X.2006.02686.x. [DOI] [PubMed] [Google Scholar]
- 4.Searle I, et al. The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev. 2006;20:898–912. doi: 10.1101/gad.373506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ratcliffe OJ, Kumimoto RW, Wong BJ, Riechmann JL. Analysis of the Arabidopsis MADS AFFECTING FLOWERING gene family: MAF2 prevents vernalization by short periods of cold. Plant Cell. 2003;15:1159–1169. doi: 10.1105/tpc.009506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ratcliffe OJ, Nadzan GC, Reuber TL, Riechmann JL. Regulation of flowering in Arabidopsis by an FLC homologue. Plant Physiol. 2001;126:122–132. doi: 10.1104/pp.126.1.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scortecci KC, Michaels SD, Amasino RM. Identification of a MADS-box gene, FLOWERING LOCUS M, that represses flowering. Plant J. 2001;26:229–236. doi: 10.1046/j.1365-313x.2001.01024.x. [DOI] [PubMed] [Google Scholar]
- 8.Werner JD, et al. Quantitative trait locus mapping and DNA array hybridization identify an FLM deletion as a cause for natural flowering-time variation. Proc Natl Acad Sci USA. 2005;102:2460–2465. doi: 10.1073/pnas.0409474102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim DH, Doyle MR, Sung S, Amasino RM. Vernalization: Winter and the timing of flowering in plants. Annu Rev Cell Dev Biol. 2009;25:277–299. doi: 10.1146/annurev.cellbio.042308.113411. [DOI] [PubMed] [Google Scholar]
- 10.Dennis ES, Peacock WJ. Epigenetic regulation of flowering. Curr Opin Plant Biol. 2007;10:520–527. doi: 10.1016/j.pbi.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 11.Sung S, Amasino RM. Vernalization in Arabidopsis thaliana is mediated by the PHD finger protein VIN3. Nature. 2004;427:159–164. doi: 10.1038/nature02195. [DOI] [PubMed] [Google Scholar]
- 12.Henderson IR, Dean C. Control of Arabidopsis flowering: The chill before the bloom. Development. 2004;131:3829–3838. doi: 10.1242/dev.01294. [DOI] [PubMed] [Google Scholar]
- 13.Sung S, Schmitz RJ, Amasino RM. A PHD finger protein involved in both the vernalization and photoperiod pathways in Arabidopsis. Genes Dev. 2006;20:3244–3248. doi: 10.1101/gad.1493306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sheldon CC, Finnegan EJ, Peacock WJ, Dennis ES. Mechanisms of gene repression by vernalization in Arabidopsis. Plant J. 2009;59:488–498. doi: 10.1111/j.1365-313X.2009.03883.x. [DOI] [PubMed] [Google Scholar]
- 15.Greb T, et al. The PHD finger protein VRN5 functions in the epigenetic silencing of Arabidopsis FLC. Curr Biol. 2007;17:73–78. doi: 10.1016/j.cub.2006.11.052. [DOI] [PubMed] [Google Scholar]
- 16.Turck F, Fornara F, Coupland G. Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu Rev Plant Biol. 2008;59:573–594. doi: 10.1146/annurev.arplant.59.032607.092755. [DOI] [PubMed] [Google Scholar]
- 17.Kobayashi Y, Weigel D. Move on up, it's time for change—mobile signals controlling photoperiod-dependent flowering. Genes Dev. 2007;21:2371–2384. doi: 10.1101/gad.1589007. [DOI] [PubMed] [Google Scholar]
- 18.Achard P, Herr A, Baulcombe DC, Harberd NP. Modulation of floral development by a gibberellin-regulated microRNA. Development. 2004;131:3357–3365. doi: 10.1242/dev.01206. [DOI] [PubMed] [Google Scholar]
- 19.De Lucia F, Crevillen P, Jones AM, Greb T, Dean C. A PHD-polycomb repressive complex 2 triggers the epigenetic silencing of FLC during vernalization. Proc Natl Acad Sci USA. 2008;105:16831–16836. doi: 10.1073/pnas.0808687105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mellor J. It takes a PHD to read the histone code. Cell. 2006;126:22–24. doi: 10.1016/j.cell.2006.06.028. [DOI] [PubMed] [Google Scholar]
- 21.Li H, et al. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature. 2006;442:91–95. doi: 10.1038/nature04802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shi X, et al. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature. 2006;442:96–99. doi: 10.1038/nature04835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wysocka J, et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- 24.Bastow R, et al. Vernalization requires epigenetic silencing of FLC by histone methylation. Nature. 2004;427:164–167. doi: 10.1038/nature02269. [DOI] [PubMed] [Google Scholar]
- 25.Lippman Z, et al. Role of transposable elements in heterochromatin and epigenetic control. Nature. 2004;430:471–476. doi: 10.1038/nature02651. [DOI] [PubMed] [Google Scholar]
- 26.Johnson L, Cao X, Jacobsen S. Interplay between two epigenetic marks. DNA methylation and histone H3 lysine 9 methylation. Curr Biol. 2002;12:1360–1367. doi: 10.1016/s0960-9822(02)00976-4. [DOI] [PubMed] [Google Scholar]
- 27.Li H, et al. Structural basis for lower lysine methylation state-specific readout by MBT repeats of L3MBTL1 and an engineered PHD finger. Mol Cell. 2007;28:677–691. doi: 10.1016/j.molcel.2007.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wood CC, et al. The Arabidopsis thaliana vernalization response requires a polycomb-like protein complex that also includes VERNALIZATION INSENSITIVE 3. Proc Natl Acad Sci USA. 2006;103:14631–14636. doi: 10.1073/pnas.0606385103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang D, Wang Y, Wang Y, He Y. Repression of FLOWERING LOCUS C and FLOWERING LOCUS T by the Arabidopsis Polycomb repressive complex 2 components. PLoS ONE. 2008;3:e3404. doi: 10.1371/journal.pone.0003404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schubert D, Clarenz O, Goodrich J. Epigenetic control of plant development by Polycomb-group proteins. Curr Opin Plant Biol. 2005;8:553–561. doi: 10.1016/j.pbi.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 31.Thao S, et al. Results from high-throughput DNA cloning of Arabidopsis thaliana target genes using site-specific recombination. J Struct Funct Genomics. 2004;5:267–276. doi: 10.1007/s10969-004-7148-4. [DOI] [PubMed] [Google Scholar]
- 32.Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 2005;139:5–17. doi: 10.1104/pp.105.063743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schubert D, et al. Silencing by plant Polycomb-group genes requires dispersed trimethylation of histone H3 at lysine 27. EMBO J. 2006;25:4638–4649. doi: 10.1038/sj.emboj.7601311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Makarevich G, et al. Different Polycomb group complexes regulate common target genes in Arabidopsis. EMBO Rep. 2006;7:947–952. doi: 10.1038/sj.embor.7400760. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.