Abstract
Biosynthesis of the highly biologically active long-chain polyunsaturated fatty acids, arachidonic (ARA), eicosapentaenoic (EPA), and docosahexaenoic (DHA) acids, in vertebrates requires the introduction of up to three double bonds catalyzed by fatty acyl desaturases (Fad). Synthesis of ARA is achieved by Δ6 desaturation of 18∶2n - 6 to produce 18∶3n - 6 that is elongated to 20∶3n - 6 followed by Δ5 desaturation. Synthesis of EPA from 18∶3n - 3 requires the same enzymes and pathway as for ARA, but DHA synthesis reportedly requires two further elongations, a second Δ6 desaturation and a peroxisomal chain shortening step. This paper describes cDNAs, fad1 and fad2, isolated from the herbivorous, marine teleost fish (Siganus canaliculatus) with high similarity to mammalian Fad proteins. Functional characterization of the cDNAs by heterologous expression in the yeast Saccharomyces cerevisiae showed that Fad1 was a bifunctional Δ6/Δ5 Fad. Previously, functional dual specificity in vertebrates had been demonstrated for a zebrafish Danio rerio Fad and baboon Fad, so the present report suggests bifunctionality may be more widespread in vertebrates. However, Fad2 conferred on the yeast the ability to convert 22∶5n - 3 to DHA indicating that this S. canaliculatus gene encoded an enzyme having Δ4 Fad activity. This is a unique report of a Fad with Δ4 activity in any vertebrate species and indicates that there are two possible mechanisms for DHA biosynthesis, a direct route involving elongation of EPA to 22∶5n - 3 followed by Δ4 desaturation, as well as the more complicated pathway as described above.
Keywords: Δ4 desaturase, bifunctional Δ6/Δ5 desaturase, polyunsaturated fatty acid biosynthesis, Siganus canaliculatus, teleost
Polyunsaturated fatty acids (PUFA) cannot be synthesized de novo by vertebrates and so must be obtained in the diet. However, the most biologically active essential fatty acids (EFA) including arachidonic (ARA; 20∶4n - 6), eicosapentaenoic (EPA; 20∶5n - 3) and docosahexaenoic (DHA; 22∶6n - 3) acids are long-chain PUFA (LC-PUFA) that can be synthesized in vertebrates through sequential desaturation and elongation of C18 PUFA, 18∶2n - 6 and 18∶3n - 3. Synthesis of ARA is achieved by Δ6 desaturation of 18∶2n - 6 to produce 18∶3n - 6 that is elongated to 20∶3n - 6 followed by Δ5 desaturation (1). Synthesis of EPA from 18∶3n - 3 requires the same enzymes and pathway as for ARA, but DHA synthesis reportedly requires two further elongation steps, a second Δ6 desaturation and a peroxisomal chain shortening step (2). The extent to which any species can convert C18 PUFA to LC-PUFA varies, associated with their complement of fatty acyl desaturase (Fad) and elongase (Elovl) enzymes. Some animals, notably extreme carnivores, have very limited ability to synthesize LC-PUFA, and consequently have a dietary requirement for preformed C20 and C22 PUFA (3–6).
As with all vertebrates, PUFA are essential nutrients in fish, but requirements vary with C18 PUFA being the EFA for freshwater and diadromous (migratory fish that travel between salt and fresh water during their lifecycle) species, whereas marine fish have a dietary requirement for LC-PUFA (7). The molecular basis of LC-PUFA synthesis is understood in fish as well as in any vertebrate, driven by the crucial role fish play as the primary source of n - 3 LC-PUFA in the human diet (8, 9). Evidence suggests that the dependence of marine fish on dietary LC-PUFA is due to deficiency in one or more enzymes required for their biosynthesis (7–10). Like mammals (11), Atlantic salmon possesses separate genes for Δ5 and Δ6 Fads (12–14). Distinct Δ6 Fad cDNAs have been isolated from all fish species studied to date including freshwater and marine species (15–18). Other than salmon, the only other fish Fad with Δ5 activity is the bifunctional Δ6/Δ5 Fad previously isolated from zebrafish (19). Thus the inability of some species to produce DHA might be explained by the lack of a gene encoding Δ5 activity, a deficiency that has no significant consequence in the DHA-rich marine ecosystem. In contrast, the bifunctional zebrafish gene and multiple subfunctionalized salmon genes have enabled these species that spend all, or a significant part, of their lifecycles in relatively nutrient-poor freshwater environments, to produce essential LC-PUFA (10). However, the influence of trophic level (the position of an organism in the food chain) on LC-PUFA biosynthesis capability has never been fully explored. Thus, all the marine species studied to date consume mainly animals, often other fish (carnivores/piscivores) with trophic levels greater than 2.8 (20). In contrast, the freshwater species so far examined consume either mainly plant/detritus (herbivores) or plants/detritus plus animals (omnivores) with trophic levels between 2.0 and 2.79 (20).
The present study was initiated to study a marine species, Siganus canaliculatus (Park, 1797) (white-spotted spinefoot or rabbitfish), which is truly herbivorous consuming algae and seagrasses with a trophic level below 2.8 (21). Previously, the cDNA for a Fad had been cloned (fad1) but not functionally characterized (22). Here we report the cloning of the cDNA for a further S. canaliculatus gene (fad2) and the results of heterologous expression of both fad cDNAs in the yeast Saccharomyces cerevisiae. The results show the two S. canaliculatus Fads display bifunctional Δ6/Δ5 and Δ4/Δ5 activities, respectively. This is a unique report of a vertebrate Fad with Δ4 activity and suggests that there is potentially more than one possible pathway for the synthesis of DHA in vertebrates.
Results
Sequence and Phylogenetic Analysis of Fad2.
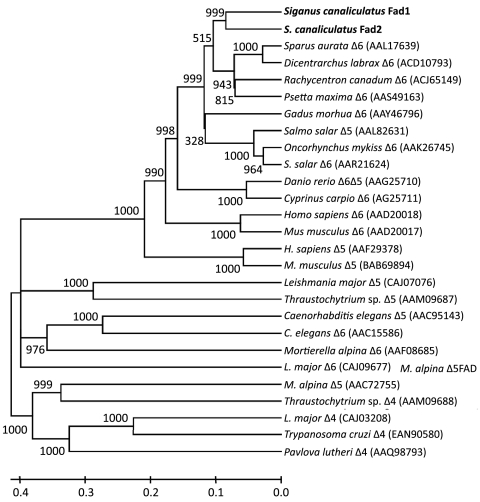
The Fad2 cDNA was 1831 bp in length (excluding polyA tail) and contains a 1338 bp open-reading frame (ORF) (GenBank accession number GU594278). The deduced protein has 445 amino acids and is 82.7% identical to the previously isolated S. canaliculatus Fad1 (EF424276), and 67.8 %, 57.8 % and 63.6 % identical to Danio rerio Δ6/Δ5 (AF309556), Homo sapiens Δ5 (AF199596) and Δ6 (AF126799), respectively. The deduced Fad2 polypeptide sequence has a number of characteristic features of microsomal Fad proteins, including three histidine boxes, an N-terminal cytochrome b5 domain containing the heme-binding motif, and two transmembrane regions. Phylogenetic analysis of Fad1 and Fad2 with a variety of Fads of other species shows that S. canaliculatus desaturases are most closely related to marine teleost Δ6 Fads, and more distantly from lower eukaryotes Δ4 and Δ5 Fads (Fig. 1).
Fig. 1.
Phylogenetic tree comparing the deduced aa sequences of S. canaliculatus Fad1 and Fad2 with desaturase proteins from fish and other organisms. The tree was constructed using the neighbor joining method (47) with MEGA4. The horizontal branch length is proportional to aa substitution rate per site. The numbers represent the frequencies with which the tree topology presented was replicated after 1,000 iterations.
Functional Characterization.
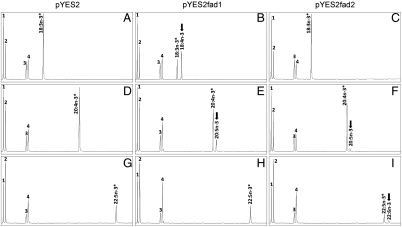
The S. canaliculatus Fad proteins were functionally characterized by determining the fatty acid (FA) profiles of yeast transformed with either the pYES2 vector alone or the vector with the putative Fad cDNA inserts (pYES2fad1 and pYES2fad2), and grown in the presence of potential FA substrates including those for Δ6 (18∶3n - 3 and 18∶2n - 6), Δ5 (20∶4n - 3 and 20∶3n - 6) and Δ4 (22∶5n - 3 and 22∶4n - 6). Yeast transformed with pYES2 vector alone showed the main FA normally found in S. cerevisiae, namely 16∶0, 16∶1 isomers, 18∶0 and 18∶1n - 9, together with the exogenously added FA. This is consistent with S. cerevisiae not possessing Δ4, Δ5, or Δ6 desaturase activities (19). Additional peaks were observed in the FA profiles of pYES2fad1 grown in the presence of Δ6, 18∶3n - 3, and 18∶2n - 6, and Δ5, 20∶4n - 3, and 20∶3n - 6, substrates. Similarly, the FA profile of yeast transformed with pYES2fad2 also showed additional peaks when grown in presence of Δ4 substrates, 22∶5n - 3 and 22∶4n - 6, and Δ5 substrates. The GC traces obtained with n - 3 FAs are shown in Fig. 2A–I. Based on GC retention times, the additional peaks observed with the presence of the S. canaliculatus cDNAs were identified as 18∶4n - 3 (Fig. 2B), 20∶5n - 3 (Fig. 2 E and F) and 22∶6n - 3 (Fig. 2I).
Fig. 2.
Functional characterization of the S. canaliculatus putative fatty acyl desaturases in transgenic yeast (S. cerevisiae). FAME were extracted from yeast transformed with pYES2 vector alone (A, D, G) or the constructs pYES2fad1 (B, E, H) and pYES2fad2 (C, F, I), and grown in the presence of FA substrates (*) 18∶3n - 3 (A–C), 20∶4n - 3 (D–F), and 22∶5n - 3 (G–I). Peaks 1–4 represent the main endogenous FAs of S. cerevisiae, namely 16∶0, 16∶1 isomers, 18∶0 and 18∶1n - 9, respectively. Based in retention times, additional peaks (arrowed) were identified as 18∶4n - 3 (B), 20∶5n - 3 (E and F) and 22∶6n - 3 (I). Vertical axis, FID response; horizontal axis, retention time.
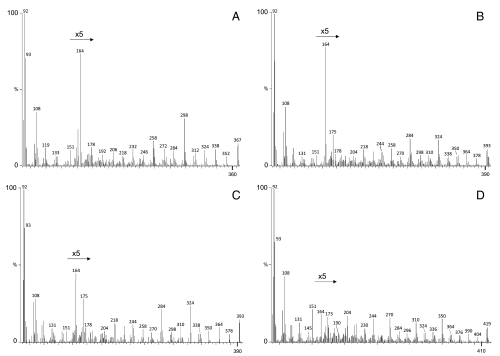
FA methyl esters (FAME) from transgenic yeast incubated with PUFA were derivatized to picolinyl esters and subjected to electron ionization (EI) GC-MS to confirm the structures of PUFA produced in the presence of fad constructs (Fig. 3). The samples all showed prominent ions at m/z = 92, 108, 151, and 164, which are characteristic of picolinyl derivatives, representing pyridine ring fragments (23). The EI spectra of the additional peak in pYES2fad1-transformed yeast incubated with 18∶3n - 3 showed a fragmentation pattern with a mass ion of 367 m/z and prominent peaks at 352, 338, 312, 298, 272, 258, 232, 218, and 192 (Fig. 3A). The initial interval of 15 represented the terminal methyl and was followed by an interval of 14, indicating one methylene group. The subsequent intervals of 26 denoted the positions of four double bonds indicating that this FA is Δ15,12,9,618∶4 = 18∶4n - 3 (Fig. 3A). The EI spectra of the additional FA from both pYES2fad1- and pYES2fad2-transformed yeast incubated with 20∶4n - 3 showed mass ions of 393 m/z with prominent ions at intervals of 26 (364–338, 324–298, 284–258, 244–218, and 204–178 m/z), confirming that the product FAs are Δ17,14,11,8,520∶5 = 20∶5n - 3 (Fig. 3 B and C). The spectra of the additional peak observed in yeast transformed with pYES2fad2 and incubated with 22∶5n - 3 showed a mass ion of 419 m/z, with prominent ions at intervals of 26 (390–364, 350–324, 310–284, 270–244, 230–204, and 190–164 m/z), confirming that this FA is Δ19,16,13,10,7,422∶6 = 22∶6n - 3 (Fig. 3D). The GC-MS data confirmed that the S. canaliculatus Fad1 cDNA is a Fad that introduces double bonds into 18∶3n - 3 at the Δ6 position and also into 20∶4n - 3 at the Δ5 position. Additionally, Fad2 cDNA encodes a Fad that introduces double bonds at the Δ4 position of 22∶5n - 3 and, to a lesser extent, the Δ5 position of 20∶4n - 3.
Fig. 3.
Mass spectra of the arrowed peaks in Fig. 2. Picolinyl esters were prepared from FAME extracted from yeast transformed with pYES2fad1 and grown in the presence of 18∶3n - 3 (A) and 20∶4n - 3 (B), or pYES2fad2 and grown in the presence of 20∶4n - 3 (C) and 22∶5n - 3 (D). Fatty acid picolinyl ester derivatives were analyzed by GC-MS as described in Methods. The identities of the peaks were confirmed as 18∶4n - 3 (A), 20∶5n - 3 (B and C) and 22∶6n - 3 (D). Vertical axis, % abundance; horizontal axis, mass-to-charge (m/z) ratio.
The analyses indicated that the yeast cells transformed with pYES2fad1 acquired functional Δ6 and Δ5 desaturation activity, whereas cells transformed with pYES2fad2 had functional Δ4 desaturation and Δ5 desaturation capability. On the basis of the percentages of substrate FA converted to product, the S. canaliculatus Fad1 cDNA is more active on Δ6 than on Δ5 substrates, with Fad2 showing higher specificity on Δ4 than Δ5 substrates (Table 1). Both Fads preferentially converted n - 3 rather than n - 6 FAs.
Table 1.
Substrate conversions of pYES2fad1 and pYES2fad2-transformed yeast grown in presence of Δ6, Δ5, and Δ4 fatty acid (FA) substrates
| FA substrate | Product | Conversion (%) | Activity | |
| pYES2fad1 | pYES2fad2 | |||
| 18∶3n - 3 | 18∶4n - 3 | 59 | 0 | Δ6 |
| 18∶2n - 6 | 18∶3n - 6 | 35 | 0 | Δ6 |
| 20∶4n - 3 | 20∶5n - 3 | 22 | 6 | Δ5 |
| 20∶3n - 6 | 20∶4n - 6 | 12 | 2 | Δ5 |
| 22∶5n - 3 | 22∶6n - 3 | 1 | 23 | Δ4 |
| 22∶4n - 6 | 22∶5n - 6 | 0 | 14 | Δ4 |
Discussion
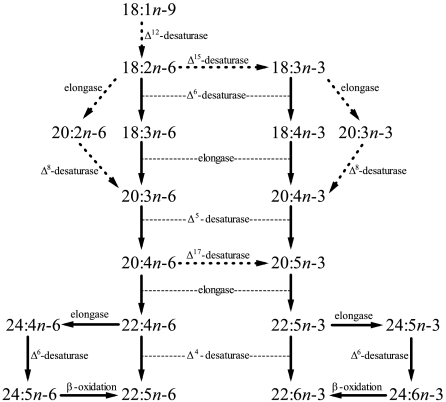
This paper is a unique report of Δ4 Fad activity in a vertebrate. Historically, production of DHA from EPA in animals was thought to occur via an elongation followed by Δ4 desaturation (see refs. 1 and 2). However, in the late 1990’s, Sprecher and coworkers indicated an alternative pathway in rats with sequential elongations of EPA or ARA to C24 substrates followed by desaturation at the Δ6 position (2). Biochemical studies indicated that this pathway also likely operated in rainbow trout (24, 25), and that mammalian Δ6 Fads were capable of desaturating both C18 and C24 substrates (26, 27). Molecular studies showed that the zebrafish Δ6/Δ5 and salmon Δ6 Fad could utilize C24 FA in addition to C18 FA when expressed in S. cerevisiae (28). Furthermore, no Fad with Δ4 activity had been isolated from a vertebrate species and, consistent with this, none of the previously cloned and functionally characterized Fad cDNAs of fish had shown any measurable Δ4 activity (12, 13, 17–19). Thus, it became the paradigm that vertebrates in general produced DHA from EPA via the alternative “Sprecher” pathway and did not possess a Δ4 Fad (1, 2). The present study has clearly demonstrated the presence of a Fad with Δ4 activity in a teleost fish and that an alternative pathway for the production of DHA from EPA, utilizing Δ4 desaturation, is thus possible in at least some vertebrate species (Fig. 4).
Fig. 4.
PUFA biosynthesis pathways. Solid lines indicate pathways confirmed in teleosts, whereas broken lines indicate pathways shown in other organisms but unconfirmed in teleosts.
The rate of DHA synthesis could be faster in the more direct Δ4 pathway that only requires endoplasmic reticulum, whereas the Sprecher pathway also involves peroxisomes, translocation of PUFA intermediates and limited fatty acid oxidation, a catabolic step. Previously, Δ4 Fads have been demonstrated in other organisms including protozoan trypanosomes (29), the photosynthetic freshwater protist Euglena gracilis (30) and marine microalgae Pavlova lutheria and Thraustochytrids (31, 32), the latter of which is particularly interesting as it also has two alternative pathways for DHA synthesis and contains a polyketide pathway not requiring aerobic desaturases (33, 34). However, as expected, the phylogenetic analysis showed that the S. canaliculatus Δ4 Fad was more closely related to other vertebrate Δ6 and Δ5 Fads than the algal and protozoan Δ4 desaturases (35).
The first bifunctional desaturase isolated from a vertebrate was the zebrafish Fad with Δ6/Δ5 activity (19), and subsequently desaturases with dual function have also been described in moths (Δ11 and Δ10, 12) (36), and fungus (Δ12 and Δ15) (37). Recently, a baboon Fad was reported to possess both Δ6 and Δ8 activity (38). Phylogenetic analysis showed that the S. canaliculatus Fads clustered with Fads from taxonomically similar species rather than with the zebrafish bifunctional desaturase a pattern that has been observed previously (17, 18). The multiple desaturase encoding genes in Atlantic salmon shared more than 90% identity to each other but were strictly monofunctional, with three Δ6 Fads and a Δ5 Fad (12–14). Gene duplication and environmental pressure may underpin the diversity of Fad isoforms and functions. Recently, molecular evolution and functional diversification of Fads after recurrent gene duplication was shown in Drosophila (39). Fish possess varied and “plastic” genomes as a result of frequent genomic changes including polyploidy, gene and chromosomal duplications, and gain of introns, making them interesting models for environmental genomics (40). Indeed, the evolution of stearoyl-CoA desaturases in teleost fishes has been described in relation to ancient and modern duplication events (41). Thus it is likely that environmental pressure and genome duplication has led to molecular evolution and functional diversification of other Fads in teleosts.
This is also a unique report of a marine fish expressing Δ5 desaturase activity as previously no Δ5 activity or gene had been reported in any marine fish. Indeed lack of Δ5 activity (8, 10) formed the biochemical and molecular basis for the difference in EFA requirements between freshwater/diadromous species (e.g., zebrafish, carp, trout, and salmon) whose EFA requirements can be satisfied by 18∶3n - 3 and 18∶2n - 6, and marine species (e.g., cod, turbot, sea bream, and sea bass) whose EFA requirements are not satisfied by C18 PUFA and instead require preformed LC-PUFA (7). The demonstration of genes encoding enzymes with Δ6/Δ5 and Δ4 Fad activities in the marine species S. canaliculatus is thus interesting in relation to the determinants of LC-PUFA biosynthesis in fish and vertebrates in general. Previously, it was unclear whether the above distinction between marine and freshwater species was related simply to environment. The underpinning hypothesis was actually based on nutrient supply in the two environments (i.e., higher levels of EPA) and, especially, DHA in the marine environment (8), and so could also be related to feeding habits and trophic level (42). The trophic level of the marine fish studied were generally > 3.0 (carnivores) compared to the lower trophic level (< 2.8) of the freshwater species investigated (20). S. canaliculatus consumes exclusively benthic algae and seagrasses and thus is a rare example of an exclusively herbivorous species inhabiting the marine environment (21, 22). This study therefore is evidence that trophic level can prevail over other environmental factors, and that the above distinction between freshwater/marine species is too simplistic (10). The presence of Δ6/Δ5 Fad indicates that S. canaliculatus has the desaturation activities necessary for the production of EPA and ARA from 18∶3n - 3 and 18∶2n - 6, respectively, and furthermore the presence of Δ4 Fad enables the subsequent endogenous production of DHA from EPA.
The above discussion is of far more than simple scientific interest as it is also highly relevant to aquaculture and the provision of n - 3 LC-PUFA to the burgeoning human population. Fish are the major dietary source of n - 3 LC-PUFA (20) and, with declining fisheries worldwide (43), farmed fish constitute an ever-increasing proportion of the fish in the human food basket amounting to one half in 2009 (44). Until now, high n - 3 LC-PUFA levels in flesh of farmed fish have been obtained by the use in the feeds of fish oils, paradoxically themselves derived from marine fisheries, but this is not sustainable and will constrain continuing growth of aquaculture activities (45). Alternatives to fish oil are urgently required but the prime candidates, vegetable oils, are rich in C18 PUFA but devoid of the n - 3 LC-PUFA abundant in fish oil (9). Feeding fish on vegetable oil can thus have important consequences for the human consumer as it lowers the n - 3 LC-PUFA content of the flesh compromising nutritional value (46). The problem is particularly acute in fish species that do not have the capability of endogenous production of LC-PUFA and this has included all marine species investigated to date (7–10). S. canaliculatus, the white-spotted spinefoot or rabbitfish, is a reef-associated, perciform, oceanodromous species that inhabits tropical marine or brackish water, and is a common food fish in its region and a prime candidate for aquaculture (22). As such, it is an example of a marine species that would likely thrive on feeds formulated with C18-rich vegetable oils and endogenously produce the healthful LC-PUFA, ARA, EPA, and DHA.
In conclusion, this work has demonstrated the presence of Δ4 Fad activity in a vertebrate species indicating an alternative, simpler pathway for the production of DHA from EPA (Fig. 4). Furthermore the demonstration of further examples of bifunctional Fads suggests these may be more common among vertebrate species. The isolation and characterization of these Fad activities in an herbivorous, marine teleost demonstrates that trophic level has likely taken precedent over environment in the evolution of LC-PUFA biosynthesis pathways in fish.
Methods
Molecular Cloning of S. canaliculatus Δ4 fad and Sequence Analysis.
As with the previously cloned S. canaliculatus desaturase Fad1 cDNA (gb|EF424276|) (22), the highly conserved sequences of vertebrate Δ6 or Δ5 Fads were used to design primers for the amplification of fad2. A fragment was obtained by polymerase chain reaction (PCR) and further extended by 5′ and 3′ rapid amplification of cDNA ends (GeneRacer™ Kit, Invitrogen) to produce full-length Fad2 cDNA (gb|GU594278|).
The amino acid (aa) sequence deduced from the S. canaliculatus Fad2 cDNA was compared with mammalian and teleost Fad proteins using EMBOSS Pairwise Alignment Algorithms tool (http://www.ebi.ac.uk/Tools/emboss/align/). A phylogenetic tree was constructed on the basis of the aa sequence alignments between the S. canaliculatus Fads and those from other organisms, using the neighbor joining method (47).
Heterologous Expression of fad ORFs.
PCR fragments corresponding to the ORFs of S. canaliculatus Fad1 (gb|EF424276|) and Fad2 (gb|GU594278|) cDNAs were amplified from brain cDNA (Pfu Turbo polymerase, Stratagene). Briefly, gene-specific primers 5′-GGAGGATGGGGATGTGAGTA-3′ (forward) and 5′-ATAAAACCATGTGGGCAGGT-3′ (reverse) for fad1, and 5′-GAAGACGGAGGATGAGGATG-3′ (forward) and 5′-TGCTCAGCACAGGATTGAGT-3′ (reverse) designed on the untranslated regions were used in first-round PCR. The isolation of the fad1 and fad2 ORFs was achieved in second-round (nested) PCR using first-round PCR products primed with 5’-CCCAAGCTTAGGATGGGAGGTGGAGGTC-3′ and 5′-CCGTCTAGATCATTTATGGAGATATGC-3′ containing restriction sites (underlined) for HindIII (forward) and XbaI (reverse). The amplified DNA fragments were digested with the corresponding restriction endonucleases (New England BioLabs) and ligated into pYES2 vector (Invitrogen). The resulting plasmid constructs, pYES2fad1 and pYES2fad2, were transformed into S. cerevisiae (strain INVSc1) using the S.C. EasyComp Transformation kit (Invitrogen). Yeast transformed with pYES2 (control), or the constructs pYES2fad1 or pYES2fad2, were grown in S. cerevisiae minimal medium-uracil using galactose induction of gene expression as described previously (19). Recombinant yeast cultures were supplemented with one of the following FA substrates: α-linolenic (18∶3n - 3), linoleic (18∶2n - 6), eicosatetraenoic (20∶4n - 3), dihomo-γ-linolenic (20∶3n - 6), docosapentaenoic (22∶5n - 3), and docosatetraenoic (22∶4n - 6) acids. FA substrates were added to the yeast cultures at final concentrations of 0.5 (C18), 0.75 (C20) and 1.0 (C22) mM as uptake efficiency decreases with increasing chain length. After 2 days, approximately equal amounts of yeast cells were transferred to glass test tubes, and collected by centrifugation (500 × g for 2 min), washed twice with 5 mL Hanks’s balanced salt solution (Invitrogen), and lipid extracted by homogenization in chloroform/methanol (2∶1, v/v) containing 0.01% butylated hydroxytoluene (BHT) as antioxidant, as described previously (19).
GC-MS Analysis.
FAME were prepared, extracted, and purified by thin-layer chromatography (19), and picolinyl esters were prepared from FAME (48). Briefly, FAME samples dissolved in 1 mL dry dichloromethane were added to a mixture of 0.1 mL potassium tert-butoxide in tetrahydrofuran (1.0 M) and 0.2 mL 3-pyridylcarbinol. After incubation at 40 °C for 30 min, the picolinyl esters were extracted by adding 2 mL water and 4 mL isohexane, and the organic phase collected and evaporated under dry nitrogen. Picolinyl derivatives were subjected to electron ionization (EI) GC-MS using a Fisons GC8000 gas chromatograph coupled to an MD800 mass spectrometer (ThermoFisher Scientific). The gas chromatograph was equipped with a fused silica capillary column (30 m × 0.32 mm i.d.) coated with Zebron ZB-Wax (Phenomenex) and used helium as carrier gas. Samples were applied using on-column injection with the oven temperature programmed to rise from 80 to 250 °C at 40 °C/ min. Proportions of substrate FA converted to desaturated FA product were calculated as [product area/(product area + substrate area)] × 100 as described previously (19).
Materials
Eicosatetraenoic, docosapentaenoic, and docosatetraenoic acids (> 98–99% pure) were from Cayman Chemical Co. and the remaining FA substrates (> 99% pure) were from Sigma–Aldrich Co. Chemicals used to prepare the S. cerevisiae minimal medium-uracil, BHT, potassium tert-butoxide in tetrahydrofuran and 3-pyridylcarbinol were from Sigma–Aldrich Co. TLC (20 × 20 cm × 0.25 mm) plates precoated with silica gel 60 (without fluorescent indicator) were from Merck. All solvents were HPLC grade and obtained from Fisher Scientific.
Acknowledgments.
We acknowledge financial support from the National Natural Science Foundation of China (NSFC) (Grants 30972266 and 30671629), as well as an NSFC-Royal Society Joint Grant (31011130156 and JP090748). O.M. was supported by a Marie Curie IntraEuropean Fellowship within the 7th European Community Framework Program (PIEF-GA-2008-220929).
Footnotes
References
- 1.Cook HW, McMaster RCR. In: Biochemistry of Lipids, Lipoproteins, and Membranes. Vance DE, Vance JE, editors. Amsterdam: Elsevier; 2004. pp. 181–204. [Google Scholar]
- 2.Sprecher H. Metabolism of highly unsaturated n - 3 and n - 6 fatty acids. Biochim Biophys Acta. 2000;1486:219–231. doi: 10.1016/s1388-1981(00)00077-9. [DOI] [PubMed] [Google Scholar]
- 3.Rivers JPW, Sinclair AJ, Crawford MA. Inability of the cat to desaturate essential fatty acids. Nature. 1975;258:171–173. doi: 10.1038/258171a0. [DOI] [PubMed] [Google Scholar]
- 4.Rivers JPW, Hassam AG, Crawford MA, Brambell MR. The inability of the lion (Panthero leo, L.) to desaturate linoleic acid. FEBS Lett. 1976;67:269–270. doi: 10.1016/0014-5793(76)80544-3. [DOI] [PubMed] [Google Scholar]
- 5.Hassam AG, Rivers JPW, Crawford MA. The failure of the cat to desaturate linoleic acid: Its nutritional implications. Nutr Metab. 1977;21:321–328. doi: 10.1159/000176079. [DOI] [PubMed] [Google Scholar]
- 6.Sinclair AJ, McLean JG, Monger EA. Metabolism of linoleic acid in the cat. Lipids. 1979;14:932–936. doi: 10.1007/BF02533508. [DOI] [PubMed] [Google Scholar]
- 7.Tocher DR. Fatty acid requirements in ontogeny of marine and freshwater fish. Aquac Res. 2010;41:717–732. [Google Scholar]
- 8.Bell MV, Tocher DR. In: Lipids in Aquatic Ecosystems. Arts MT, Brett M, Kainz M, editors. New York: Springer; 2009. pp. 211–236. [Google Scholar]
- 9.Tocher DR. Issues surrounding fish as a source of ω3 long-chain polyunsaturated fatty acids. Lipid Technol. 2009;21:13–16. [Google Scholar]
- 10.Leaver MJ, et al. Towards fish lipid nutrigenomics: Current state and prospects for fin-fish aquaculture. Rev Fish Sci. 2008;16:71–92. [Google Scholar]
- 11.Marquardt A, Stohr H, White K, Weber BHF. cDNA cloning, genomic structure, and chromosomal localization of three members of the human fatty acid desaturase family. Genomics. 2000;66:175–183. doi: 10.1006/geno.2000.6196. [DOI] [PubMed] [Google Scholar]
- 12.Hastings N, et al. Molecular cloning and functional characterization of fatty acyl desaturase and elongase cDNAs involved in the production of eicosapentaenoic and docosahexaenoic acids from α-linolenic acid in Atlantic salmon (Salmo salar) Mar Biotechnol. 2005;6:463–474. doi: 10.1007/s10126-004-3002-8. [DOI] [PubMed] [Google Scholar]
- 13.Zheng X, et al. Highly unsaturated fatty acid synthesis in vertebrates: New insights with the cloning and characterization of a Δ6 desaturase of Atlantic salmon. Lipids. 2005;40:13–24. doi: 10.1007/s11745-005-1355-7. [DOI] [PubMed] [Google Scholar]
- 14.Monroig O, et al. Multiple fatty acyl desaturase (FAD) genes in Atlantic salmon: Cloning and functional expression of cDNAs confirm presence of three Δ6 FADs. Biochim Biophys Acta. 2010;1801:1072–1081. doi: 10.1016/j.bbalip.2010.04.007. [DOI] [PubMed] [Google Scholar]
- 15.Seiliez I, Panserat S, Kaushik S, Bergot P. Cloning, tissue distribution and nutritional regulation of a delta 6-desaturase-like enzyme in rainbow trout. Comp Biochem Physiol, Part B: Biochem Mol. 2001;130:83–93. doi: 10.1016/s1096-4959(01)00410-9. [DOI] [PubMed] [Google Scholar]
- 16.Seiliez I, Panserat S, Corraze G, Kaushik S, Bergot P. Cloning and nutritional regulation of a delta 6-desaturase-like enzyme in the marine teleost gilthead seabream (Sparus aurata) Comp Biochem Physiol, Part B: Biochem Mol. 2003;135:449–460. doi: 10.1016/s1096-4959(03)00111-8. [DOI] [PubMed] [Google Scholar]
- 17.Zheng X, et al. Characterization and comparison of fatty acyl Δ6 desaturase cDNAs from freshwater and marine teleost fish species. Comp Biochem Physiol, Part B: Biochem Mol. 2004;139:269–279. doi: 10.1016/j.cbpc.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 18.Tocher DR, et al. Highly unsaturated fatty acid synthesis in marine fish: Cloning, functional characterization, and nutritional regulation of fatty acyl Δ6 desaturase of Atlantic cod (Gadus morhua L.) Lipids. 2006;41:1003–1016. doi: 10.1007/s11745-006-5051-4. [DOI] [PubMed] [Google Scholar]
- 19.Hastings N, et al. A vertebrate fatty acid desaturase with Δ5 and Δ6 activities. Proc Natl Acad Sci USA. 2001;98:14304–14309. doi: 10.1073/pnas.251516598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tacon AGJ, Metian M, Turchini GM, DeSilva SS. Responsible aquaculture and trophic level implications to global fish supply. Rev Fish Sci. 2010;18:94–105. [Google Scholar]
- 21.Woodland DJ. Revision of the fish family Siganidae with descriptions of two new species and comments on distribution and biology. Indo-Pacific Fishes. 1990;19:1–136. [Google Scholar]
- 22.Li Y, et al. The effects of dietary fatty acids on liver fatty acid composition and delta 6-desaturase expression differ with ambient salinities in Siganus canaliculatus. Comp Biochem Physiol, Part B: Biochem Mol. 2008;151:183–190. doi: 10.1016/j.cbpb.2008.06.013. [DOI] [PubMed] [Google Scholar]
- 23.Christie WW. Lipid Analysis. Bridgwater: Oily Press; 2003. pp. 1–289. [Google Scholar]
- 24.Buzzi M, Henderson RJ, Sargent JR. The desaturation and elongation of linolenic acid and eicosapentaenoic acid by hepatocytes and liver microsomes from rainbow trout (Oncorhynchus mykiss) fed diets containing fish oil or olive oil. Biochim Biophys Acta. 1996;1299:235–244. doi: 10.1016/0005-2760(95)00211-1. [DOI] [PubMed] [Google Scholar]
- 25.Buzzi M, Henderson RJ, Sargent JR. Biosynthesis of docosahexaenoic acid in trout hepatocytes proceeds via 24-carbon intermediates. Comp Biochem Physiol, Part B: Biochem Mol. 1997;116:263–267. doi: 10.1016/s0305-0491(96)00210-6. [DOI] [PubMed] [Google Scholar]
- 26.D’Andrea S, et al. The same rat Δ6-desaturase not only acts on 18- but also on 24-carbon fatty acids in very-long-chain polyunsaturated fatty acid biosynthesis. Biochem J. 2002;364:49–55. doi: 10.1042/bj3640049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Antueno RJ, et al. Activity of human Δ5 and Δ6 desaturases on multiple n - 3 and n - 6 polyunsaturated fatty acids. FEBS Lett. 2001;509:77–80. doi: 10.1016/s0014-5793(01)03135-0. [DOI] [PubMed] [Google Scholar]
- 28.Tocher DR, Agaba M, Hastings N, Teale AJ. In: Browman HI, Skiftesvik AB, editors. The Big Fish Bang—Proceedings of the 26th Annual Larval Fish Conference; Bergen: Institute of Marine Nutrition; 2003. pp. 211–227. [Google Scholar]
- 29.Tripodi KEJ, Buttigliero LV, Altabe SG, Uttaro AD. Functional characterization of front-end desaturases from trypanosomatids depicts the first polyunsaturated fatty acid biosynthetic pathway from a parasitic protozoan. FEBS J. 2006;273:271–280. doi: 10.1111/j.1742-4658.2005.05049.x. [DOI] [PubMed] [Google Scholar]
- 30.Meyer A, et al. Biosynthesis of docosahexaenoic acid in Euglena gracilis: Biochemical and molecular evidence for the involvement of a Δ4-fatty acyl group desaturase. Biochemistry. 2003;42:9779–9788. doi: 10.1021/bi034731y. [DOI] [PubMed] [Google Scholar]
- 31.Tonon T, Harvey D, Larson TR, Graham IA. Identification of a very long chain polyunsaturated fatty acid Δ4-desaturase from the microalga Pavlova lutheri. FEBS Lett. 2003;553:440–444. doi: 10.1016/s0014-5793(03)01078-0. [DOI] [PubMed] [Google Scholar]
- 32.Qiu X, Hong H, MacKenzie SL. Identification of a Δ4 fatty acid desaturase from Thraustochytrium sp involved in the biosynthesis of docosahexanoic acid by heterologous expression in Saccharomyces cerevisiae and Brassica juncea. J Biol Chem. 2001;276:31561–31566. doi: 10.1074/jbc.M102971200. [DOI] [PubMed] [Google Scholar]
- 33.Metz JG, et al. Production of polyunsaturated fatty acids by polyketide synthases in both prokaryotes and eukaryotes. Science. 2001;293:290–293. doi: 10.1126/science.1059593. [DOI] [PubMed] [Google Scholar]
- 34.Lippmeier JC, et al. Characterization of both polyunsaturated fatty acid biosynthetic pathways in Schizochytrium sp. Lipids. 2009;44:621–630. doi: 10.1007/s11745-009-3311-9. [DOI] [PubMed] [Google Scholar]
- 35.Sperling P, Ternes P, Zank TK, Heinz E. The evolution of desaturases. Prostag Leukotr Ess. 2003;68:73–95. doi: 10.1016/s0952-3278(02)00258-2. [DOI] [PubMed] [Google Scholar]
- 36.Serra M, Gauthier LT, Fabrias G, Buist PH. Δ11 desaturases of Trichoplusia ni and Spodoptera littoralis exhibit dual catalytic behaviour. Insect Biochem Mol Biol. 2006;36:822–825. doi: 10.1016/j.ibmb.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 37.Zhang S, Sakuradani E, Ito K, Shimizu S. Identification of a novel bifunctional Δ12/Δ15 fatty acid desaturase from a basidiomycete, Coprinus cinereus TD#822-2. FEBS Lett. 2007;581:315–319. doi: 10.1016/j.febslet.2006.12.031. [DOI] [PubMed] [Google Scholar]
- 38.Park WJ, Kothapalli KSD, Lawrence P, Tyburczy C, Brenna JT. An alternate pathway to long-chain polyunsaturates: The FADS2 gene product Δ8-desaturates 20∶2n - 6 and 20∶3n - 3. J Lipid Res. 2009;50:1195–1202. doi: 10.1194/jlr.M800630-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fang S, et al. Molecular evolution and functional diversification of fatty acid desaturases after recurrent gene duplication in Drosophila. Mol Biol Evol. 2009;26:1447–1456. doi: 10.1093/molbev/msp057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cossins AR, Crawford DL. Fish as models for environmental genomics. Nat Rev Genet. 2005;6:324–333. doi: 10.1038/nrg1590. [DOI] [PubMed] [Google Scholar]
- 41.Evans H, et al. Ancient and modern duplication events and the evolution of stearoyl-CoA desaturases in teleost fishes. Physiol Genomics. 2008;35:18–29. doi: 10.1152/physiolgenomics.90266.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sargent JR, Tocher DR, Bell JG. In: Fish Nutrition. Halver JE, Hardy RW, editors. San Diego: Academic; 2002. pp. 181–257. [Google Scholar]
- 43.Worms B, et al. Impacts of biodiversity loss on ocean ecosystem services. Science. 2006;314:787–790. doi: 10.1126/science.1132294. [DOI] [PubMed] [Google Scholar]
- 44.Food and Agricultural Organization of the United Nations (FAO) The State of World Fisheries and Aquaculture 2008 (SOFIA) Rome: FAO Fisheries and Aquaculture Department; 2009. pp. 1–176. [Google Scholar]
- 45.Naylor RL, et al. Feeding aquaculture in an era of finite resources. Proc Natl Acad Sci USA. 2009;106:15103–15110. doi: 10.1073/pnas.0905235106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bell JG, Tocher DR. In: Oils and Fats Handbook Volume 4; Fish Oils. Rossell B, editor. Leatherhead: Leatherhead Food International; 2009. pp. 171–184. [Google Scholar]
- 47.Saitou N, Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 48.Destaillats F, Angers P. One-step methodology for the synthesis of FA picolinyl esters from intact lipids. J Am Oil Chem Soc. 2002;79:253–256. [Google Scholar]