Abstract
The leucine-rich repeat (LRR) proteins LRIM1 and APL1C control the function of the complement-like protein TEP1 in Anopheles mosquitoes. The molecular structure of LRIM1 and APL1C and the basis of their interaction with TEP1 represent a new type of innate immune complex. The LRIM1/APL1C complex specifically binds and solubilizes a cleaved form of TEP1 without an intact thioester bond. The LRIM1 and APL1C LRR domains have a large radius of curvature, glycosylated concave face, and a novel C-terminal capping motif. The LRIM1/APL1C complex is a heterodimer with a single intermolecular disulfide bond. The structure of the LRIM1/APL1C heterodimer reveals an interface between the two LRR domains and an extensive C-terminal coiled-coil domain. We propose that a cleaved form of TEP1 may act as a convertase for activation of other TEP1 molecules and that the LRIM1/APL1C heterodimer regulates formation of this TEP1 convertase.
Keywords: host-pathogen interactions, innate immunity, malaria, thioester
Complement is an innate immune system in vertebrates comprising a number of serum proteins that function to detect and destroy microorganisms. Insects possess a complement-like immune response to bacteria, fungi, and protozoan parasites (1–4). Anopheles gambiae thioester-containing protein 1 (TEP1) is structurally and functionally homologous to complement factor C3 (3, 5–7), including an intramolecular thioester bond that mediates covalent labeling of pathogens. Covalent attachment of TEP1 to bacteria promotes their phagocytosis (3), whereas binding of TEP1 to Plasmodium berghei ookinetes targets them for lysis (1).
Complement activation centers upon a conformational change in C3 that is regulated by proteolysis. C3 is cleaved twice; first during intracellular processing, then a second activating cleavage that dissociates a small protein domain known as anaphylatoxin, or C3a. This dissociation triggers a large conformational change in the remaining molecule, known as C3b, exposing the thioester bond (8, 9). The proteolytic activation of C3 is regulated by the formation of a transient complex between a protease and a complement factor, known as a convertase.
The alternative pathway involves self-activation of C3. Hydrolysis of the C3 thioester bond induces a conformational change, producing C3(H2O) which has a similar conformation to C3b. C3(H2O) can recruit the protease Factor B. Cleavage of Factor B by Factor D produces the transient complex C3(H2O)Bb that is a C3 convertase (10). The alternative pathway is inhibited in the fluid phase and the presence of self surfaces by the binding of Factor H, which competes with Factor B for binding to the C345C domain of C3 (11, 12).
TEP1 lacks both the anaphylatoxin and C345C domains, and is secreted as a full-length molecule. Constitutive cleavage is observed in the hemolymph within a protease-sensitive region similar to that of C3. Proteolysis of TEP1 in the protease-sensitive region in vitro does not appear to cause a conformational change, and the thioester remains present in the resulting cleaved form of TEP1 (13), referred to here as TEP1cut, implying that additional factors regulate activation of TEP1 in vivo..
Two leucine-rich repeat (LRR) proteins were recently shown to control the function of TEP1 in A. gambiae (13, 14). Leucine-Rich Immune Molecule 1 (LRIM1) was initially identified as having an antimalarial phenotype within the P. berghei model system and also as a factor required for efficient phagocytosis of Gram-negative but not Gram-positive bacteria (15–17). Anopheles Plasmodium-responsive Leucine-rich repeat protein 1 (APL1) was identified in a genetic association study within a locus of variation in wild mosquito populations that correlated with resistance to P. falciparum (18). The APL1 locus encodes a family of three closely-related genes, designated A, B, and C; only APL1C is responsible for the immune phenotype against P. berghei (19).
LRIM1 and APL1C are members of a family of proteins secreted by mosquitoes that contain an N-terminal LRR domain followed by a cysteine-rich region and a variable C terminus (14). LRIM1 and APL1C are required for efficient binding of TEP1 to P. berghei parasites (13). RNAi knockdown of either of LRIM1 or APL1C leads to depletion of TEP1cut from the hemolymph and its deposition on self-tissues. LRIM1 and APL1C also stabilize each other within the hemolymph and form an intermolecular disulfide-bridged complex, as evidenced by migration as a high-molecular weight species on nonreducing SDS/PAGE (14). It has been proposed that the LRIM1/APL1C complex may be similar to multimeric complexes formed by variable lymphocyte receptor antibodies in jawless vertebrates (20), or other complexes known to activate complement, such as component 1 of the classical complement pathway (C1q), immunoglobulin M (IgM), and mannose-binding-lectin (MBL).
We have now structurally characterized the LRIM1/APL1C complex and the nature of its interaction with TEP1. These data show that LRIM1 and APL1C form a heterodimer via interaction of their C-terminal coiled-coil domains and a single disulfide bond. This heterodimeric complex stabilizes a particular form of TEP1cut that lacks an intact thioester. Our results suggest that the LRIM1/APL1C heterodimer either stabilizes an active form of TEP1cut or regulates the formation of a TEP1 convertase that catalyzes the activation of other TEP1 molecules in the proximity of pathogens.
Results
LRIM1 and APL1C Form a Heterodimer Mediated by a Single Disulfide Bond.
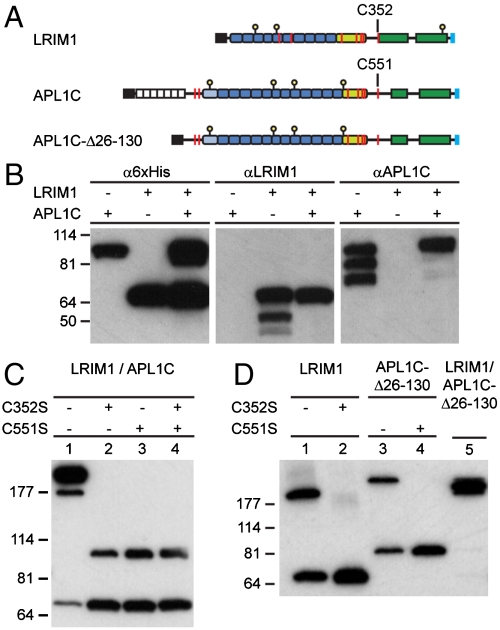
Recombinant LRIM1 and APL1C were overexpressed in insect cell cultures using the baculovirus expression system. LRIM1 and APL1C share similar domain architectures (Fig. 1A): an N-terminal LRR domain (preceded by a low-complexity region in APL1C), a cysteine-rich region of ∼40 residues, and two predicted coiled-coil domains. We first compared coexpression of C-terminal 6xHis-tagged LRIM1 and APL1C with individual expression, using the same antibodies previously used to observe their presence in vivo (Fig. 1B). Western blotting with α6xHis antibody detected both full-length proteins. Western blotting with αLRIM1 and αAPL1 antibodies, however, revealed that both LRIM1 and APL1C were subject to proteolysis when expressed individually. Proteolysis involves the C-terminal domains of each protein, as the degradation products were not detected with α6xHis antibody. Hence, interaction between the C-terminal domains of LRIM1 and APL1C protects both from proteolytic degradation, stabilizing both proteins in circulation.
Fig. 1.
Coexpression of LRIM1 and APL1C and complex formation. (A) Schematic diagram of LRIM1 and APL1C constructs. Colored boxes: black, signal peptide; white, low-complexity; blue, LRR repeat; pale-green, cysteine-rich region; green, coiled-coil domain; and pale-blue, 6xHis tag. Features: yellow stalk, N-linked glycosylation site; red line, cysteine residue. (B) Individual and coexpression of full-length APL1C and LRIM1, reducing SDS/PAGE, Western blot with α6xHis, αLRIM1, and αAPL1 antibodies. (C) Coexpression of LRIM1/APL1C (lane 1), LRIM1-C352S/APL1C (lane 2), LRIM1/APL1C-C551S (lane 3), and LRIM1-C352S/APL1C-C551S (lane 4), nonreducing SDS/PAGE, and α6xHis Western blot. (D) Expression of LRIM1 (lane 1), LRIM1-C352S (lane 2), APL1C-Δ26-130 (lane 3), APL1C-Δ26-130-C551S (lane 4), and coexpression of LRIM1/APL1C-Δ26-130 (lane 5), nonreducing SDS/PAGE, α6xHis Western blot.
Since LRIM1 and APL1C appear to interact via their C-terminal domains, and given that cysteine-rich regions flanking LRR domains are often folded motifs that serve to initiate or conclude the solenoid structure (21), intermolecular disulfide formation between LRIM1 and APL1C should involve cysteines other than those in the cysteine-rich region. Both LRIM1 and APL1C possess a single cysteine, LRIM1 Cys 352 and APL1C Cys 551, between their cysteine-rich region and coiled-coil domain. To test if these residues formed an intermolecular disulfide bond, we coexpressed C-terminal 6xHis-tagged constructs of LRIM1 and APL1C and analyzed their apparent molecular weight by SDS/PAGE under nonreducing conditions (Fig. 1C). A high-molecular weight species was observed between LRIM1 and APL1C (lane 1). This species was lost upon mutation of either LRIM1 C352S or APL1 C551S. The disulfide-bridged complex formed by LRIM1 and APL1C is therefore mediated by a single disulfide bond between LRIM1 Cys 352 and APL1C Cys 551. Thus the only disulfide-bridged complex possible is a dimer, which is observed at a high apparent molecular weight.
Under conditions of coexpression, the amount of LRIM1 was higher than that of APL1C. We observed a second high-molecular-weight species (Fig. 1C, lane 1) that was postulated to be a homodimer of LRIM1. We also hypothesized that the N-terminal low-complexity region of APL1C would be dispensable for the purposes of complex formation. These predictions were tested by comparing separate and coexpression of LRIM1 and APL1-Δ26-130 (Fig. 1D). Full-length LRIM1 (lane 1) formed both monomers and disulfide-linked homodimers that were disrupted by mutation of Cys 352 (lane 2). Likewise, APL1C-Δ26-130 (lane 3) formed both monomers and disulfide-linked homodimers that were disrupted by mutation of Cys 551 (lane 4). Coexpression of LRIM1 and APL1-Δ26-130 produced a heterodimer of intermediate apparent mass relative to the homodimers (lane 5).
Hence LRIM1 and APL1C possess a C-terminal coiled-coil region commencing with a free cysteine, that has the capacity to form homo- and heterodimeric disulfide-bridged complexes. When LRIM1 and APL1C were coexpressed and a cysteine was mutated in only one however (Fig. 1C, lanes 2–3), a homodimer of the other was not observed. Thus heterodimerization is more efficient or stable than homodimerization, as the presence of the mutant form of one protein may competitively inhibit formation of disulfide-bridged homodimers of the other.
Although the only disulfide-bridged complex formed between LRIM1 and APL1C is a heterodimer, a larger complex could be formed via noncovalent interactions. To test this hypothesis we determined the oligomeric state of the LRIM1/APL1C-Δ26-130 complex by quantifying its molecular weight in solution, using analytical ultracentrifugation (AUC) and dynamic light scattering (SI Appendix: Fig. S1). The resulting molecular weight calculated directly from the Svedberg equation is M = 131 ± 1 kDa. The combined mass of the mature peptides of LRIM1 (55 kDa) and APL1C-Δ26-130 (69 kDa) plus the mass of carbohydrate (6.5 kDa, SI Appendix: Table S1) yields a total mass of 130.5 kDa, equal within error to the experimental mass. Therefore, the LRIM1/APL1C complex is a heterodimer mediated by interaction of the respective C-terminal coiled-coil domains and a single disulfide bond between Cys 352 of LRIM1 and Cys 551 of APL1C.
Crystal Structure of the LRIM1/APL1C Complex.
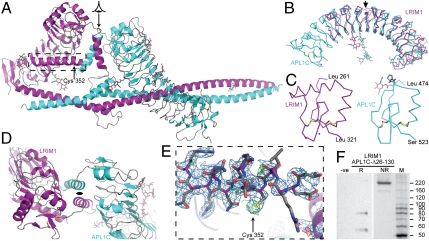
The specificity of LRIM1/APL1C heterodimerization and the fact that both are required for stabilization of TEP1 in vivo suggests that specific structural features of the LRIM1/APL1C heterodimer direct the function of these proteins. Hence we crystallized the LRIM1/APL1C-Δ26-130-6xHis heterodimer and determined its structure to 2.7 Å resolution (SI Appendix: Table S2). The final model contains one molecule of the heterodimer in the asymmetric unit, the entire LRIM1 chain except residues 342–346, the APL1C-Δ26-130 chain 138–709 except for residues 149–150 and 545–580, and all seven N-linked glycosylation sites.
The overall structure places the LRIM1 and APL1C LRR domains back-to-back with a pseudo-C2 axis of symmetry, packed sideways against the coiled-coil domain (Fig. 2A). The LRR domains of LRIM1 and APL1C (solved independently to 2.0 Å and 1.85 Å, respectively, see SI Appendix: Table S2 and Fig. S2) share a set of common features that define the LRIM1-APL1 LRR family as distinct from other LRR domains (Fig. 2B). These are: (i) an unusually large radius of curvature centered around a short LRR in the middle of the domain, (ii) N-linked glycosylation on the concave face, and (iii) a novel C-terminal capping motif (LRRCT) containing two disulfide bonds (Fig. 2C), LRIM1 Cys 273–Cys 318 and Cys 305–Cys 317, and APL1 Cys 486–Cys 520 and between Cys 508–Cys 519. The combination of shallow curvature and glycosylation suggests that LRIM1-LRR and APL1C-LRR may not engage in canonical protein-protein interactions on the concave face as observed in LRR proteins such as ribonuclease inhibitor (22, 23).
Fig. 2.
Crystal structure of LRIM1 and APL1C. (A) Structure of the LRIM1/APL1C-Δ26-130 heterodimer. Secondary structure elements colored purple (LRIM1) and cyan (APL1C). A viewing arrow indicates the pseudo-C2 axis of symmetry between the LRR domains, and a dashed box indicates the LRIM1 helix harboring C352 (arrow). (B) Aligned ribbon diagram of LRIM1 and APL1C LRR domains. An arrow indicates the unusually short repeat LRIM1-LRR V and APL1C-LRR VIII. N-linked glycans and disulfide bonds shown as sticks. (C) LRRCT of LRIM1 and APL1C. Cα ribbon with disulfide bonds and side chains and Pro499 cis-peptide shown as sticks. An asterisk indicates a site of LRIM1 polymorphism. (D) LRIM1/APL1C LRRCT interface, viewed along the pseudo-C2 axis of symmetry. (E) Electron density for the LRIM1 α-helix containing C352 but with no accompanying density for the APL1 chain and C551. Weighted 2Fo - Fc map contoured at 1σ in blue, Fo - Fc map contoured at ± 3σ in green/red. (F) Silver-stained SDS/PAGE of dissolved crystals of LRIM1/APL1C heterodimer. (left) Reducing conditions, with negative control (equivalent volume of mother liquor). (right) Nonreducing conditions. MW Markers: Benchmark™ unstained ladder (Invitrogen).
A previous study reported a polymorphism in the A. gambiae LRIM1 gene that maps to residues L310–G311 in the LRRCT (24). The resulting double mutation L310H/G311N is accommodated without steric consequence as the side chain for N311 would be solvent exposed (Fig. 2C). The corresponding residues in APL1 are Y512–Q513. A set of stable haplotypes within the APL1 genetic locus maps to the junction of the N-terminal low-complexity region with the LRRNT of APL1C (19). It is plausible that specific functions of LRIM1 and APL1 are associated not with the LRRs themselves but with their ends, the intervening repeats acting as molecular spacers. Notably, there is great diversity in the number of repeats between members of the LRIM1-APL1 family within mosquito genomes (14).
The interface between the LRIM1 and APL1C LRR domains in the structure of the LRIM1/APL1C-Δ26-130 heterodimer is a short α-helix immediately following the LRRCT motif, termed the LRRCT-helix (Fig. 2D). The LRRCT-helix of LRIM1 contacts the APL1C LRRCT-helix and the lower convex face of APL1C-LRR, and vice versa. A surface area of 1,120 Å2 is buried by this interaction. The LRIM1 LRRCT-helix ends with three disordered residues (G342–G344) followed by another α-helix packed against the bottom face of LRIM1-LRR at the start of the first predicted coiled-coil region. This helix contains LRIM1 C352 that forms a disulfide bond with APL1C C551 (Fig. 2E).
Ironically the complementary region APL1C 545–580 is disordered in the crystal. To confirm the integrity of the APL1 protein and the intermolecular bond we analyzed redissolved crystals by reducing and nonreducing SDS/PAGE (Fig. 2F). Bands corresponding to the expected molecular weights of LRIM1 and APL1C-Δ26-130 were observed under reducing conditions, and a band at ∼220 kDa as expected in the presence of an intermolecular disulfide bridge under nonreducing conditions.
The LRIM1 α-helix continues beyond C352 to Q369, where it is interrupted by an ordered loop from Y370 to D375. Another α-helix commences at Q376 and is joined by an α-helix commencing at APL1C L581 to form a true coiled-coil structure. The C-terminal coiled-coil domains adopts a helix-loop-helix (HLH) fold, ∼160 Å in length burying 3,334 Å2 surface area, the loops corresponding to a break in the coiled-coil domain predicted for each protein. HLH folds are found in numerous DNA transcription factor structures, but not previously in extracellular proteins.
Isolation of Complex Between TEP1 and LRIM1/APL1C Heterodimer.
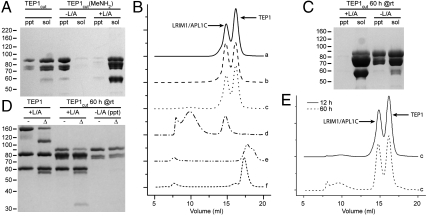
The observation that LRIM1 and APL1C stabilizes TEP1cut in the hemolymph suggests that the LRIM1/APL1C heterodimer specifically interacts with TEP1cut. We have previously used a thioester autolytic cleavage assay to examine two separate modifications to purified TEP1 (13): methylamine treatment–producing TEP1(MeNH2)–to chemically remove the thioester, and limited proteolysis with trypsin–producing TEP1cut–which leaves the thioester intact. TEP1cut (Fig. 3A, lanes 1–2) is soluble (as is TEP1(MeNH2)). When we treated TEP1cut with MeNH2 however (Fig. 3A, lanes 3–4), the protein rapidly precipitated (as did TEP1(MeNH2) upon limited proteolysis with trypsin). Yet in the presence of the LRIM1/APL1C-Δ26-130 heterodimer, TEP1cut(MeNH2) remained soluble (Fig. 3A, lanes 5–6), suggesting that the LRIM1/APL1C complex was specifically interacting with this species.
Fig. 3.
LRIM1/APL1C interaction with TEP1 analyzed by Coomassie-stained reducing SDS/PAGE and Size-exclusion chromatography (SEC). (A) Precipitate (ppt) and soluble (sol) fractions for TEP1cut after 12 h at room temperature. MeNH2 treatment causes precipitation of TEP1cut. Presence of LRIM1/APL1C-Δ26-130 (L/A) prevents precipitation. (B) SEC of mixtures containing TEP1 (T) species and LRIM1/APL1C-Δ26-130 (L/A) (arrows mark peaks): (a) TEP1 + L/A, (b) TEP1(MeNH2) + L/A, (c) TEP1cut + L/A, (d) TEP1cut(MeNH2) + L/A, (e) TEP1cut(MeNH2) + LRIM1-LRR + APL1C-LRR, (f) TEP1cut(MeNH2) + BSA. (C) ppt and sol fractions for TEP1cut after 60 h incubation in the presence and absence of LRIM1/APL1C-Δ26-130. (E) SEC of sample (c) TEP1cut + LRIM1/APL1C-Δ26-130, 60 h incubation vs. 12 h incubation.
We then analyzed the interaction between LRIM1/APL1C and all the different in vitro forms of TEP1. The specific constructs used were TEP1r-6xHis, APL1C-Δ26-130-6xHis, and LRIM1 with an internal FLAG affinity tag in place of the first turn of the LRRCT-helix. We prepared the following mixtures: (a) TEP1 and LRIM1/APL1C, (b) TEP1(MeNH2) and LRIM1/APL1C, (c) TEP1cut and LRIM1/APL1C, (d) TEP1cut(MeNH2) and LRIM1/APL1C, (e) TEP1cut(MeNH2), LRIM1-LRR and APL1C-LRR, and (f) TEP1cut(MeNH2) and BSA. After 12 h incubation at room temperature, samples (a–d) were all soluble (SI Appendix: Fig. S3A). Using size-exclusion chromatography (Fig. 3B), no evidence of direct interaction between LRIM1/APL1C and TEP1 was observed for samples (a–c), but in sample (d) TEP1cut(MeNH2) was converted to a high-molecular-weight species in complex with LRIM1/APL1C. SDS/PAGE of fractions from samples (a–d) supported a 1∶1 stoichiometry of TEP1cut(MeNH2) to LRIM1/APL1C heterodimer in the high-molecular weight species (SI Appendix: Fig. S3B). The ternary complex specifically required the LRIM1/APL1C heterodimer, as neither a twofold molar excess of LRIM1-LRR and APL1C-LRR nor an excess of BSA–samples (e–f)–prevented TEP1cut(MeNH2) precipitation (Fig. 3B).
Although initially stable in solution, after 60 h incubation we observed the slow precipitation of isolated TEP1cut (Fig. 3C). Using the autolytic cleavage assay we compared this precipitated fraction of TEP1cut to the soluble fractions of TEP1 and TEP1cut with LRIM1/APL1C (samples (a) and (c)). Precipitation corresponded to hydrolysis of the thioester (Fig. 3D). TEP1cut and LRIM1/APL1C (sample (c)) were reanalyzed by size-exclusion chromatography, and a similar high-molecular weight species as formed by TEP1cut(MeNH2) and LRIM1/APL1C was observed (Fig. 3E). Hence, slow hydrolysis of the TEP1cut thioester produces TEP1cut(H2O) that, similar to TEP1cut(MeNH2), is unstable in the absence of a specific interaction with the LRIM1/APL1C heterodimer.
The preceding experiments were performed at high protein concentrations (3 μM). Hence we performed immunoprecipitation experiments (SI Appendix: Fig. S4) for 100-fold dilutions of samples (a–d) using αFLAG antibodies to specifically immunoprecipitate LRIM1, or αTEP1 polyclonal antibodies affinity purified from rabbits immunized with purified recombinant TEP1. All three proteins were only coprecipitated in the case of TEP1cut(MeNH2) (sample (d)), indicating the formation of a strong and specific interaction between TEP1 and the LRIM1/APL1C heterodimer.
Recombinant LRIM1/APL1C Heterodimer Stabilizes Endogenous TEP1cut in A. gambiae Hemolymph.
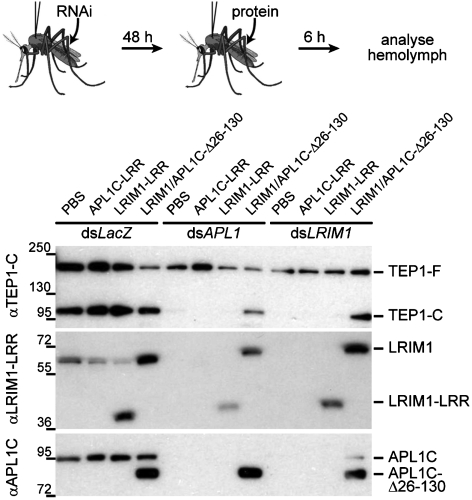
To ask whether the ternary TEPcut/LRIM1/APL1C complex isolated in vitro was relevant to the function of TEP1 in vivo, we injected purified LRIM1-LRR, APL1C-LRR and LRIM1/APL1C-Δ26-130 heterodimer into adult A. gambiae mosquitoes for which endogenous levels of LRIM1 and APL1 were depleted by dsRNA knockdown (Fig. 4). TEP1cut was lost from hemolymph extracts upon knockdown of either LRIM1 or APL1. Injection of LRIM1-LRR or APL1C-LRR had no effect, but injection of purified LRIM1/APL1C-Δ26-130 heterodimer restored the steady-state levels of TEP1cut. Thus the recombinant LRIM1/APL1C-Δ26-130 heterodimer is functional in vivo and specifically stabilizes the in vivo form of TEP1cut whose presence in the hemolymph was previously shown to correlate with TEP1-mediated killing of P. berghei (13).
Fig. 4.
Rescue of TEP1cut in vivo by injected recombinant LRIM1/APL1C. Western blotting of hemolymph for A. gambiae mosquitoes injected with dsRNA against LacZ (control) APL1, or LRIM1 and injected with PBS (control) and purified APL1C-LRR, LRIM1-LRR, or LRIM1/APL1C-Δ26-130. αTEP1-C recognizes TEP1-F and TEP1-C. αLRIM1-LRR recognizes LRIM1 and LRIM1-LRR. αAPL1C recognizes APL1C and APL1C-Δ26-130 but not APL1C-LRR. Upon dsRNA knockdown of APL1 or LRIM1, TEP1cut (TEP1-C) is lost from the hemolymph, and is rescued specifically by injection of purified LRIM1/APL1C-Δ26-130.
Discussion
Mosquitoes possess a complement-like system that plays an important role in their defense against different classes of pathogens. This system is centered on the thioester-containing protein TEP1 and includes a complex formed by two LRR proteins LRIM1 and APL1C, whose function is required for binding of TEP1 to microbes. Here we demonstrate that the LRIM1/APL1C heterodimer represents a distinct class of innate immune complex, combining the elements of an N-terminal-LRR domain with a characteristic cysteine-rich capping motif, a single intermolecular disulfide bridge and a C-terminal HLH coiled-coil domain. These facts were established by mutational analysis of LRIM1 C352 and APL1C C551, accurate molecular weight determination in solution, and x-ray crystallography. Although LRIM1 and APL1C homodimers were observed in conditions of heterologous overexpression, heterodimerization is strongly preferred and is the only form detected in vivo.
The LRIM1/APL1C heterodimer interacts with a specific form of TEP1 both in vitro and in vivo. Binding of different sets of proteins to complement factors C3, C4, and C5 in their various conformational states is well established, but no LRR proteins have presently been identified with such a function in the vertebrate complement system, illustrating both the similarities and differences between vertebrate and invertebrate complement systems. TEP1 plays a similar conceptual role in the A. gambiae innate immunity to complement factor C3 in vertebrates, but its molecular features, namely the absence of two domains important for C3 function, suggest that upstream activation and downstream effector pathways have diverged from that of vertebrates at the molecular level.
LRIM1 and APL1C cooperate in vivo to stabilize TEP1cut in the hemolymph, cleavage of TEP1 in vitro does not cause spontaneous reaction of the thioester bond, and loss of TEP1cut from the hemolymph correlates with loss of TEP1-binding to P. berghei ookinetes (13). Based on these observations, a model was previously proposed in which cleavage of TEP1 in vivo produces a mature form, TEP1cut, that retains a thioester bond. In the absence of infection, this form would be stabilized in circulation by LRIM1/APL1C. Displacement of LRIM1/APL1C in the vicinity of pathogens would then mediate activation and binding of TEP1.
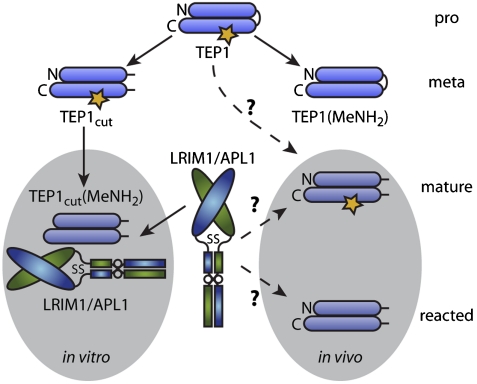
New evidence presented here however, suggests a more involved mechanism of TEP1 activation (Fig. 5). No interaction was detected between TEP1 and the LRIM1/APL1C heterodimer in vitro as predicted by the model above, but neither did the LRIM1/APL1C heterodimer interact with TEP1cut, in contrast to our prediction. A separate in vitro modification, treatment with MeNH2, abolished the thioester bond but also failed to induce any interaction between TEP1 and LRIM1/APL1C. Treatment of C3 with MeNH2 is known to produce a meta-stable species C3∗(MeNH2), that either slowly converts to a species C3(MeNH2) that has similar properties to C3b, or may revert to C3 with attendant reformation of the thioester bond (25). Although the reversibility of MeNH2 treatment was not examined here, we postulate that the species TEP1(MeNH2) and TEP1cut described may represent a similar meta-stable state as C3*, distinct from TEP1 but with local and not global conformational changes.
Fig. 5.
Proposed model for conformational states of TEP1 and their interaction with LRIM1/APL1C. The N- and C-terminal portions of TEP1 are shown as horizontal bars linked on the right by the protease-sensitive region; the thioester bond is illustrated by a yellow star. Proteolysis within the protease-sensitive region generates TEP1cut which retains a thioester, while MeNH2 treatment removes the thioester leaving the protease-sensitive region intact; both actions are proposed to generate meta-stable species. Combined proteolysis and MeNH2 treatment in vitro generates a species TEP1cut(MeNH2) that is specifically bound by LRIM1/APL1C due to a conformational change in TEP1cut(MeNH2) that exposes a cryptic binding site. However, in vivo it remains unknown how TEP1cut is generated, whether multiple conformations of TEP1cut are present with or without an active thioester and if so which are specifically recognized by LRIM1/APL1C, and how this specifically regulates the activation of TEP1 and its immune function.
Further in vitro modification of TEP1cut by MeNH2 in the absence of LRIM1/APL1C leads to precipitation, as does limited proteolysis of TEP1(MeNH2). LRIM1/APL1C specifically binds TEP1(MeNH2) and prevents precipitation. Precipitation also accompanies slow hydrolysis of the thioester bond in TEP1cut except in the presence of LRIM1/APL1C, which binds to TEP1cut(H2O). The most plausible explanation for these observations is that the combination of limited proteolysis and reaction of the thioester causes a conformational change, revealing a cryptic binding site for LRIM1/APL1C. The mechanism(s) that may lead to the exposure of this binding site in vivo remain unknown, but we note that (i) LRIM1/APL1C specifically interacts with the form TEP1cut in A. gambiae cell culture (14), (ii) The form of TEP1cut present in conditioned medium of the A. gambiae cell line Sua5.1 does not contain a thioester bond (3), and (iii) TEP1cut precipitates on mosquito self-tissues in vivo in the absence of LRIM1/APL1C (13).
How the binding of LRIM1/APL1C to TEP1cut and its stabilization in the hemolymph relates to the TEP1-mediated immune response to gram-negative bacteria and P. berghei depends on the specific form of TEP1 stabilized by LRIM1/APL1C in vivo. One possibility is that, via a specific mechanism operating in vivo, TEP1cut undergoes a conformational change to expose the cryptic binding site for LRIM1/APL1C while retaining an intact thioester bond. The species of TEPcut stabilized would then be capable of covalent attachment to a substrate. In this case LRIM1/APL1C could play a direct activating role by maintaining this species in circulation and perhaps actively promoting recruitment to and/or activation upon nonself surfaces.
Another possibility is that the species of TEP1cut in vivo, similar to that observed for purified protein or in vitro cell culture, does not possess an intact thioester and is therefore incapable of covalent attachment to a substrate. Nevertheless this species could activate other TEP1 molecules if one postulates the formation of a TEP1 convertase, a complex that catalyzes the activation of other TEP1 molecules for covalent attachment to a substrate, analogous to the alternative complement pathway, whereby slow hydrolysis of C3 produces a low-level of C3b production (referred to as “tick-over”). In the fluid phase and on self surfaces, the catalytic production of C3b is prevented by binding of regulatory proteins such as Factor H that competitively inhibit binding of Factor B (11, 12). On nonself surfaces however, binding of Factor B predominates, leading to catalytic activation and binding of additional C3 molecules. If a “TEP1 alternative pathway” were to exist, LRIM1/APL1C could play a passive role by maintaining in circulation a TEP1 species capable of convertase formation (inhibiting its deposition on self surfaces) or, through interaction with other factors, the role of an indirect activator by recruiting this TEP1 species to nonself surfaces.
An outstanding question is how the LRIM1/APL1C heterodimer interacts with TEP1cut. LRIM1-LRR and APL1C-LRR do not bind to TEP1 in vitro or in vivo. The successful immunoprecipitation of the TEP1/LRIM1/APL1C complex using a FLAG epitope within the LRRCT-helix suggests that the pseudo-C2 interface between the LRIM1-LRR and APL1C-LRR in the heterodimer structure is absent from the ternary complex, and leads us to speculate that the LRIM1/APL1C coiled-coil domain and the HLH motif in particular may be responsible for binding TEP1cut. At 16 nm in length, the coiled-coil domain already exceeds the linear dimensions of TEP1, and there are numerous instances of loop regions within HLH domains involved in protein binding. An alternative model is one in which the LRR domains bind TEP1 cooperatively, perhaps via their convex faces in a similar manner as TLR dimers. The role of the intermolecular disulfide bridge to formation of the ternary complex is unclear as it plays no key role in the structure of the heterodimer itself. If the HLH is the site of binding to TEP1cut the function of the intermolecular disulfide bridge may be to promote formation of or to stabilize the heterodimeric HLH in order to provide the correct binding partner for TEP1cut(MeNH2) or TEP1cut(H2O). If a cooperative interface between LRIM1-LRR and APL1-LRR is responsible for binding TEP1cut then the intermolecular disulfide bridge may act to restrict the maximum separation between LRIM1-LRR and APL1C-LRR, a “local concentration” effect. Assuming that they are not involved in TEP1 binding, the LRIM1-LRR and APL1C-LRR domains may play an independent role such as recruiting other factors to the complex or targeting the complex to appropriate substrates. Notably, while the C-terminal domains of LRIM1 and APL1C are susceptible to proteolysis the LRR domains themselves, suggesting a potential function whereby the LRIM1-LRR and APL1C-LRR could be released from the TEP1-HLH complex and generate a secondary signal to the innate immune system.
The APL1 locus encodes three distinct genes for APL1: APL1A, APL1B, and APL1C. The experiments reported here involve APL1C that is known to be involved in the immune response of the A. gambiae G3 strain to infection with the rodent malaria parasite P. berghei (19). It has recently been reported that APL1C and LRIM1 are dispensable for efficient killing of the human malaria parasites P. falciparum (26). Instead, the APL1A gene controlled the prevalence of A. gambiae infection with P. falciparum. These results are of particular interest in light of the equally important role of TEP1 in killing both human and rodent malaria parasites (2). We propose that distinct TEP1 activation pathways operate in the presence of various pathogens. The pathway described here involving TEP1cut/LRIM1/APL1C is required for TEP1 activation in response to bacteria and rodent malaria parasites, and probably constitutes a default activation pathway, whereas during P. falciparum infections TEP1 will be activated by a separate pathway that does not require the LRIM1/APL1C complex. Further characterization of TEP1 convertase(s) shall reveal how mosquitoes, despite their lack of an adaptive immune response, are able to exhibit an effective innate immune response against a variety of invaders and may suggest novel approaches to render mosquitoes resistant to human malaria parasites.
Methods
Cloning and Expression.
LRIM1 (AGAP006348) and APL1C (AGAP007033, cDNA clone 104AF09) were produced via the Bac-to-Bac system (Invitrogen). Protein expression was performed with T. ni cells in ESF-921 medium (Expression Systems LLC). Medium was harvested at 54–60 h. Recombinant protein was purified by coaffinity, ion exchange, and gel filtration chromatography.
Coexpression of LRIM1 and APL1C.
T. ni cells were coinfected with LRIM1 and APL1C virus. 5 mL of cells were infected at an MOI of 1.0 and conditioned medium isolated at 54 h. Nonreducing/reducing SDS-PAGE was performed with 4%–12% gradient and Coomassie G-250 native PAGE with 3%–12% gradient minigels (Invitrogen). Protein was transferred to nitrocellulose (SDS/PAGE) or PVDF (native PAGE) membrane and Western blotting performed with monoclonal α6xHis/HRP antibody (Clontech).
Crystallization and Structure Determination.
LRIM1/APL1C-Δ26-130 was crystallized at 3 mg/mL in 15% PEG 1000, 0.1 M NaCl, 50 mM Na-Hepes pH 7.5, 4 °C. X-ray diffraction data was collected at beamline ID-14-4 at the European Synchrotron Radiation Facility, Grenoble. Details of the structure determination are provided in SI Appendix.
Isolation of the TEP1/LRIM1/APL1C Complex.
A FLAG tag was inserted into LRIM1 directly following the LRR domain (LRIM1-FLAG2). Purification and limited proteolysis of TEP1r-6xHis were performed as previously described (5, 13). LRIM1-FLAG2/APL1C-Δ26-130-6xHis was purified as described above. All samples were exchanged into PBS on a Sephadex200 10/30 column (GE Healthcare). Protein concentrations were adjusted to 3 μM (TEP1) and 6 μM (others) in 200 μL. Proteins were mixed at room temperature followed by addition of 20 μL 0.5 M Ches/NaOH or CHES/MeNH2 pH 9.2 (pH 9.0 final). At subsequent time points samples were centrifuged at 14,000 × g for 5 min and 200 μL analyzed on a Superose6 10/30 column (GE Healthcare).
Experiments in adult mosquitoes and antibodies against TEP1 and APL1 were as previously described (13). Protein injection was preceded by dsRNA injection by 48 h, and hemolymph extracted 6 h following injection of protein. Polyclonal rabbit antibodies for LRIM1 and APL1 were raised against the recombinant LRIM1-LRR used for crystallization.
Supplementary Material
Acknowledgments.
R.H.G.B. gratefully acknowledges Dr. C. Brautigam of the Structural Biology Laboratory for assistance in AUC data collection and analysis, Dr. D. Tomchick of the UTSW Structural Biology Laboratory for assistance with X-ray data collection, and Drs. H. Kwon, Y. Huang and other members of the Deisenhofer Laboratory, and Drs. Z. Otwinowski and D. Borek of the Otwinowski Laboratory for many fruitful discussions. Glycosylation analysis of LRIM1/APL1C was performed by Dr. M. Ishihara (18O-MS/MS site identification) and Dr. R. Sonon under direction of Dr. P. Azadi at the Complex Carbohydrate Research Center, University of Georgia. The authors acknowledge the European Synchrotron Radiation Facility for provision of synchrotron radiation facilities and would like to thank Dr. D. Flot for assistance in X-ray data collection at beamline ID-14-4. Results shown in this report are derived from work performed at Argonne National Laboratory, Structural Biology Center at the Advanced Photon Source. Argonne is operated by UChicago Argonne, LLC, for the Department of Energy, Office of Biological and Environmental Research under contract DE-AC02-06CH11357. This work was supported by grants from the Welch Foundation (to J.D. I-1185), CNRS and INSERM (to E.A.L., S.S. and G.V.), The Fondation pour la Recherche Médicale (to G.V.), the French Ministry of National Education and Research (to S.S), the European Molecular Biology Organization (EMBO) Young Investigators Program (to E.A.L.), the European Commission FP6 Network of Excellence “BioMalPar” (to E.A.L.), and FP7 Cooperation Consortium “MalVecBlok.” E.A.L. is an international Howard Hughes Medical Institute research scholar. J.D. is an investigator in the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
Data deposition: The structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes LRIM1-LRR, 3O53; APL1C-LRR, 3O6N; LRIM1/APL1C, 3OJA).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1010575107/-/DCSupplemental.
References
- 1.Blandin S, et al. Complement-like protein TEP1 is a determinant of vectorial capacity in the malaria vector Anopheles gambiae. Cell. 2004;116:661–670. doi: 10.1016/s0092-8674(04)00173-4. [DOI] [PubMed] [Google Scholar]
- 2.Dong Y, et al. Anopheles gambiae immune responses to human and rodent Plasmodium parasite species. PLoS Pathog. 2006;2:e52. doi: 10.1371/journal.ppat.0020052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Levashina EA, et al. Conserved role of a complement-like protein in phagocytosis revealed by dsRNA knockout in cultured cells of the mosquito, Anopheles gambiae. Cell. 2001;104:709–718. doi: 10.1016/s0092-8674(01)00267-7. [DOI] [PubMed] [Google Scholar]
- 4.Stroschein-Stevenson SL, Foley E, O’Farrell PH, Johnson AD. Identification of Drosophila gene products required for phagocytosis of Candida albicans. PLoS Biol. 2006;4:e4. doi: 10.1371/journal.pbio.0040004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baxter RHG, et al. Structural basis for conserved complement factor-like function in the antimalarial protein TEP1. Proc Natl Acad Sci USA. 2007;104:11615–11620. doi: 10.1073/pnas.0704967104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fredslund F, et al. The structure of bovine complement component 3 reveals the basis for thioester function. J Mol Biol. 2006;361:115–127. doi: 10.1016/j.jmb.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 7.Janssen BJ, et al. Structures of complement component C3 provide insights into the function and evolution of immunity. Nature. 2005;437:505–511. doi: 10.1038/nature04005. [DOI] [PubMed] [Google Scholar]
- 8.Janssen BJ, Christodoulidou A, McCarthy A, Lambris JD, Gros P. Structure of C3b reveals conformational changes that underlie complement activity. Nature. 2006;444:213–216. doi: 10.1038/nature05172. [DOI] [PubMed] [Google Scholar]
- 9.Wiesmann C, et al. Structure of C3b in complex with CRIg gives insights into regulation of complement activation. Nature. 2006;444:217–220. doi: 10.1038/nature05263. [DOI] [PubMed] [Google Scholar]
- 10.Volanakis JE. C3 convertases of complement. Molecular genetics, structure, and function of the catalytic domains, C2 and B. Year Immun. 1989;4:218–230. [PubMed] [Google Scholar]
- 11.Wu J, et al. Structure of complement fragment C3b-Factor H and implications for host protection by complement regulators. Nat Immunol. 2009;10:728–733. doi: 10.1038/ni.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rooijakkers SH, et al. Structural and functional implications of the alternative complement pathway C3 convertase stabilized by a staphylococcal inhibitor. Nat Immunol. 2009;10:721–727. doi: 10.1038/ni.1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fraiture M, et al. Two mosquito LRR proteins function as complement control factors in the TEP1-mediated killing of Plasmodium. Cell Host Microbe. 2009;5:273–284. doi: 10.1016/j.chom.2009.01.005. [DOI] [PubMed] [Google Scholar]
- 14.Povelones M, Waterhouse RM, Kafatos FC, Christophides GK. Leucine-rich repeat protein complex activates mosquito complement in defense against Plasmodium parasites. Science. 2009;324:258–261. doi: 10.1126/science.1171400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moita LF, et al. In vivo identification of novel regulators and conserved pathways of phagocytosis in A. gambiae. Immunity. 2005;23:65–73. doi: 10.1016/j.immuni.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 16.Osta MA, Christophides GK, Kafatos FC. Effects of mosquito genes on Plasmodium development. Science. 2004;303:2030–2032. doi: 10.1126/science.1091789. [DOI] [PubMed] [Google Scholar]
- 17.Warr E, Lambrechts L, Koella JC, Bourgouin C, Dimopoulos G. Anopheles gambiae immune responses to Sephadex beads: involvement of anti-Plasmodium factors in regulating melanization. Insect Biochem Mol Biol. 2006;36:769–778. doi: 10.1016/j.ibmb.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 18.Riehle MM, et al. Natural malaria infection in Anopheles gambiae is regulated by a single genomic control region. Science. 2006;312:577–579. [Google Scholar]
- 19.Riehle MM, et al. Anopheles gambiae APL1 is a family of variable LRR proteins required for Rel1-mediated protection from the malaria parasite, Plasmodium berghei. PLoS ONE. 2008;3:e3672. doi: 10.1371/journal.pone.0003672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herrin BR, et al. Structure and specificity of lamprey monoclonal antibodies. Proc Natl Acad Sci USA. 2008;105:2040–2045. doi: 10.1073/pnas.0711619105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bella J, Hindle KL, McEwan PA, Lovell SC. The leucine-rich repeat structure. Cell Mol Life Sci. 2008;65:2307–2333. doi: 10.1007/s00018-008-8019-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kobe B, Deisenhofer J. Crystal structure of porcine ribonuclease inhibitor, a protein with leucine-rich repeats. Nature. 1993;366:751–756. doi: 10.1038/366751a0. [DOI] [PubMed] [Google Scholar]
- 23.Kobe B, Deisenhofer J. A structural basis of the interactions between leucine-rich repeats and protein ligands. Nature. 1995;374:183–186. doi: 10.1038/374183a0. [DOI] [PubMed] [Google Scholar]
- 24.Slotman MA, et al. Patterns of selection in anti-malarial immune genes in malaria vectors: evidence for adaptive evolution in LRIM1 in Anopheles arabiensis. PLoS ONE. 2007;2:e793. doi: 10.1371/journal.pone.0000793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pangburn MK. Spontaneous reformation of the intramolecular thioester in complement protein C3 and low temperature capture of a conformational intermediate capable of reformation. J Biol Chem. 1992;267:8584–8590. [PubMed] [Google Scholar]
- 26.Mitri C, et al. Fine pathogen discrimination within the APL1 gene family protects Anopheles gambiae against human and rodent malaria species. PLoS Pathog. 2009;5:e1000576. doi: 10.1371/journal.ppat.1000576. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.