Abstract
Celiac disease (CD) is an enteropathy triggered by the ingestion of gluten proteins from wheat and similar proteins from barley and rye. The inflammatory reaction is controlled by T cells that recognize gluten peptides in the context of human leukocyte antigen (HLA) DQ2 or HLA-DQ8 molecules. The only available treatment for the disease is a lifelong gluten-exclusion diet. We have used RNAi to down-regulate the expression of gliadins in bread wheat. A set of hairpin constructs were designed and expressed in the endosperm of bread wheat. The expression of gliadins was strongly down-regulated in the transgenic lines. Total gluten protein was extracted from transgenic lines and tested for ability to stimulate four different T-cell clones derived from the intestinal lesion of CD patients and specific for the DQ2-α-II, DQ2-γ-VII, DQ8-α-I, and DQ8-γ-I epitopes. For five of the transgenic lines, there was a 1.5–2 log reduction in the amount of the DQ2-α-II and DQ2-γ-VII epitopes and at least 1 log reduction in the amount of the DQ8-α-I and DQ8-γ-I epitopes. Furthermore, transgenic lines were also tested with two T-cell lines that are reactive with ω-gliadin epitopes. The total gluten extracts were unable to elicit T-cell responses for three of the transgenic wheat lines, and there were reduced responses for six of the transgenic lines. This work shows that the down-regulation of gliadins by RNAi can be used to obtain wheat lines with very low levels of toxicity for CD patients.
Keywords: RNAi, post-transcriptional gene silencing, gluten intolerance, celiac sprue
Celiac disease (CD) is a food-sensitive enteropathy caused by the ingestion of wheat-gluten proteins and similar proteins from barley and rye (1–3). The disease has high heritability, and the genes encoding the human leukocyte antigen (HLA) DQ2 or HLA-DQ8 molecules are a major genetic risk factor. The disease occurs almost worldwide, but the prevalence is particularly high in Western countries (1%) (4, 5). CD can be diagnosed in early childhood with symptoms like diarrhea and malabsorption, but it is often diagnosed later in life and then, the spectrum of symptoms is wider. The formation of disease lesion in the small intestine, which is characterized by villous blunting, crypt cell hyperplasi, and infiltration of leukocytes, involves the activation of gluten-reactive CD4 (cluster of differentiation 4) T cells (2). These T cells recognize particular gluten peptides presented by the predisposing HLA-DQ2 or HLA-DQ8 molecules, and most of the gluten peptides are better recognized after conversion of certain glutamine residues to glutamate by the enzyme transglutaminase 2 (TG2) expressed in the gut mucosa. The only treatment available for CD patients is a lifelong strict gluten-free diet. Adhering to a strict gluten-free diet is demanding, because gluten is a ubiquitous additive in various foods. Thus, the development of wheat varieties with reduced gluten-toxicity profiles may contribute to the improvement of the diet of CD patients and the reduction of CD incidence, because it is also observed that the initiation of CD is associated with the level and duration of exposure to gluten (6, 7).
Wheat gluten can be divided into two protein families: the glutenins and the gliadins (8). The glutenins comprise high molecular weight (HMW) and low molecular weight (LMW) fractions, whereas the gliadins can be divided into three structural types: α-, γ-, and ω-gliadins (9). Isolation and characterization of intestinal T cells from CD patients have revealed several distinct but similar DQ2 and DQ8 epitopes. These epitopes are generally very rich in proline and glutamine residues (10). Although a number of epitopes are derived from glutenins (11), the majority of the epitopes reside in the gliadin fraction (10, 12). Wheat gliadin genes occur in tightly linked clusters, termed blocks, located at complex loci on group 1 and 6 chromosomes. The ω- and γ-gliadins are coded by clusters of genes at the Gli-1 loci (Gli-A1, Gli-B1, and Gli-D1) on the short arms of the homologous group 1 chromosomes, whereas the α-gliadins are controlled by the Gli-2 loci (Gli-A2, Gli-B2, and Gli-D2) present on the short arms of the group 6 chromosomes (13). The estimated copy numbers in hexaploid wheat of genes encoding α-gliadins ranges from 25 to 150 copies (14), from 15 to 18 copies for ω-gliadins, and from 17 to 39 copies for γ-gliadins (15). The sequences of individual gene copies within the same gliadin family are very similar, and they may contain multiple and different T-cell epitopes (16, 17). This high level of complexity and the fact that gliadin genes are inherited in blocks make conventional breeding approaches to obtain wheat varieties with reduced content of T-cell stimulatory sequences very difficult.
RNAi is a posttranscriptional process triggered by dsRNA that leads to gene silencing through a two-step mechanism (18, 19). Most current RNAi technology is based on hairpin RNA (hpRNA) vectors, which are composed of a promoter and terminator regions in between an inversely repeated sequence of the target gene, with a spacer region between the repeats. In plants, this approach is being used to engineer metabolic pathways to overproduce products with health, yield, or environmental benefits (20–22). We previously have used this approach to down-regulate the expression of γ-gliadins in bread wheat (23), and this shows that RNAi technology can be used to down-regulate groups of proteins encoded by multigene families (24). Here, we have made a set of hairpin constructs containing a fragment of 361 bp highly conserved among α-, ω-, and γ-gliadins. Down-regulation of α-, γ-, and ω-gliadins in transgenic lines by use of these constructs lead to strongly reduced expression of CD-related gliadin T-cell epitopes.
Results
Designing of RNAi Constructs.
Two fragments of 170 bp and 191 bp, corresponding to the most conserved regions from α- and ω-gliadins (Table S1), respectively, were used for the inverted repeat (IR) fragments in plasmids pGhp-ω/α and pDhp-ω/α (Fig. S1). The IR fragment to target the γ-gliadin genes was as reported previously (23). The IR fragments showed high identity, not only with their target gliadin group but also with other gliadins (Table S2). The ω-gliadin fragment showed an average identity with α- and ω-gliadin groups of 79.2% and 75.6%, respectively, and the maximum identities were 83.8% and 99.5%, respectively. These fragments were assembled into the 361 bp full-chimeric fragment ω/α to provide two RNA silencing-triggered IR constructs: pGhp-ω/α and pDhp-ω/α. Both plasmids had the same structure but different endosperm-specific promoters: a γ-gliadin promoter (25) in the pGhp-ω/α vector and a D-hordein promoter (26) in the pDhp-ω/α vector.
Transformation with Gliadin RNAi Constructs Strongly Down-Regulates the Content of Gliadins in Bread Wheat.
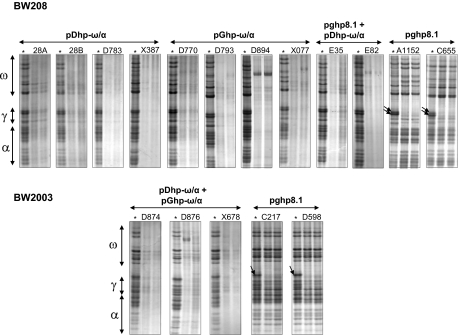
A set of 17 transgenic wheat lines was generated by particle bombardment with vectors containing the gliadin RNAi fragments (pDhp-ω/α, pGhp-ω/α, and pghpg8.1) combined with the selectable plasmid pAHC25 (27) to facilitate phosphinothricin selection. The analysis of gliadins from transgenic lines by acid-polyacrylamide gel electrophoresis (A-PAGE) showed that the ω/α RNAi chimeric fragment designed (vectors pDhp-ω/α and pGhp-ω/α) was able to effectively down-regulate gliadins from all groups, whereas in lines containing the pghp8.1 vector only, γ-gliadins were down-regulated (Fig. 1). However, there were differences among transgenic lines in the down-regulation. Thus, transgenic lines D783, X387, D793, D894, E35, E82, and X678 showed the highest levels of down-regulation compared with their wild-type control (Fig. 1). However, for lines 28A, 28B, D770, X077, D874, and D876, some bands, mainly in the region of ω-gliadins (Fig. 1), were still observed. In lines A1152, C655, C217, and D598 transformed with the pghp8.1 vector, only bands in the region of the γ-gliadins were down-regulated, with at least two major bands being clearly targeted (Fig. 1).
Fig. 1.
A-PAGE gels of gliadin extracts from wild types and transgenic wheat lines. ω, ω-gliadins fraction; γ, γ-gliadins fraction; β, β-gliadins fraction; α, α-gliadins fraction. Asterisk indicates the control line for each genotype (BW208 and BW2003). Arrows indicate γ-gliadin bands.
The total gliadin content of the transgenic lines and their controls were determined by a competitive ELISA system based on the R5 monoclonal antibody and RP-HPLC. Quantification based on the R5 monoclonal antibody assay is one of the industry reference assays for quantifying the amount of gluten in foods. The R5 assay showed a significant reduction of the gliadin content in all of the transgenic lines transformed with the pDhp-ω/α and pGhp-ω/α constructs, with the average reduction of 92.2% and a range between 89.7% and 98.1%, respectively, for lines X387 and E82 (Table 1). In contrast, transgenic lines transformed with the pghp8.1 construct did not show significant gliadin-content reduction when measured in the R5 assay. All transgenic lines, except line X387, showed a reduction in gliadin content of more than 90% by the R5 assay.
Table 1.
Gliadin and glutenin contents, total protein content, and SDS sedimentation test of transgenic and wild-type lines
| RP-HPLC (%) |
|||||||||||
| Gliadins |
Glutenins |
||||||||||
| Genotype | Line | Constructs | R5 (ppm) | ω | α | γ | Total | HMW | LMW | Protein (%) | SDS test (mL·g−1) |
| BW208 | Wild type | NA | 64,395 | 100 | 100 | 100 | 100 | 100 | 100 | 14.0 | 12.9 |
| 28A | pDhp-ω/α | 5,961* | 28.2* | 17.4* | 0.9* | 16.6* | 143 | 116 | 13.0 | 12.4 | |
| 28B | pDhp-ω/α | 5,782* | 40.7* | 26.0* | 3.0* | 24.7* | 137 | 122 | 12.8 | 12.8 | |
| D783 | pDhp-ω/α | 6,400* | 40.6* | 36.7* | 4.5* | 30.2* | 113 | 95.0 | 13.0 | 12.6 | |
| X387 | pDhp-ω/α | 6,613* | 40.4* | 34.5* | 6.2* | 29.5* | 133 | 109 | 13.1 | 12.6 | |
| D770 | pGhp-ω/α | 5,467* | 38.2* | 23.0* | 0.8* | 22.1* | 127 | 98.7 | 13.3 | 12.0 | |
| D793 | pGhp-ω/α | 2,327* | 19.0* | 13.9* | 0.2* | 12.1* | 116 | 60.5 | 13.9 | 8.9* | |
| D894 | pGhp-ω/α | 5,188* | 20.0* | 14.8* | 1.5* | 13.1* | ND | ND | 13.0 | 12.5 | |
| X077 | pGhp-ω/α | 6,188* | 42.7* | 23.5* | 2.9* | 24.1* | 151 | 120 | 13.1 | 12.0 | |
| E35 | pghpg8.1+pDhp-ω/α | 3,366* | 25.7* | 18.2* | 0.0* | 16.0* | ND | ND | 14.0 | 12.7 | |
| E82 | pghpg8.1+pDhp-ω/α | 1,227* | ND | ND | ND | ND | 75.6 | 47.2* | 13.9 | 6.2* | |
| A1152 | pghpg8.1 | 65,170 | 96.4 | 103 | 1.3* | 77.1* | 88.0 | 90.1 | 15.7* | 14.3 | |
| C655 | pghpg8.1 | 67,668 | 85.4 | 85.3 | 6.2* | 66.7* | 76.1 | 88.3 | 14.7 | 13.8 | |
| BW2003 | Wild type | NA | 114,043 | 100 | 100 | 100 | 100 | 100 | 100 | 14.6 | 12.4 |
| D874 | pDhp-ω/α+pGhp-ω/α | 10,363* | 28.7* | 11.9* | 1.7* | 15.2* | 71.6 | 18.2* | 14.4 | 9.1* | |
| D876 | pDhp-ω/α+pGhp-ω/α | 8,675* | 24.9* | 9.0* | 0.3* | 12.3* | 84.9 | 21.1* | 13.7 | 9.3* | |
| X678 | pDhp-ω/α+pGhp-ω/α | 10,613* | 31.8* | 11.2* | 0.9* | 15.7* | 102 | 25.5* | 13.5 | 9.1* | |
| C217 | pghpg8.1 | 137,194 | 95.3 | 111 | 18.4* | 85.4 | 103 | 81.0 | 14.5 | 13.9* | |
| D598 | pghpg8.1 | 130,342 | 101 | 128 | 27.8* | 97.1 | 105 | 91.0 | 14.6 | 12.9 | |
Gliadins were estimated by R5 competitive ELISA and RP-HPLC. Glutenins were determined by RP-HPLC. ppm, parts per million; ω, ω-gliadins; α, α-gliadins; γ, γ-gliadins; total, total gliadin content; HMW, high molecular weight; LMW, low molecular weight; ND, data not determined; NA, not applicable. All percent values are relative to control.
*Means are significantly different to control as determined by Dunnett's multiple comparisons at P < 0.05.
Quantification of gliadins by RP-HPLC showed clear differences in the gliadin content and pattern between the two wild types used as controls (Figs. S2 and S3). Total gliadin content as determined by RP-HPLC showed a significant correlation with data obtained using the R5 antibody (R = 0.97, P < 0.0001). The RP-HPLC analysis showed that all transgenic lines transformed with ω/α hpRNA constructs (pDhp-ω/α and pGhp-ω/α) had a significant reduction in the total content of gliadins (Table 1 and Figs. S2 and S4). The reduction of gliadins with the ω/α hpRNA constructs ranged from 69.8% in line D783 to 87.9% in line D793. Three transgenic lines, namely D793, D894, and D876, showed a reduction of total gliadin greater than 85%. A more detailed analysis of the gliadin fractions showed that, in all transgenic lines containing the ω/α hpRNA constructs, all α-, γ-, and ω-gliadin fractions were significantly lower than that of the wild types (Table 1). The down-regulation was strongest for the γ-gliadin fraction (Table 1 and Figs. S2 and S4), with reduction varying between 93.8% and 100% (lines X387 and E35), and for α-gliadins, the reduction varied between 63.3% and 91% (lines D783 and D876) of reduction (Table 1). In contrast, the ω-gliadin fraction had a lower level of down-regulation ranging between 35.6% and 81% (lines E82 and D793) of reduction (Table 1 and Fig. S2). The transgenic lines containing the pghp8.1 construct (A1152, C655, C217, and D598) had low reduction in total gliadin content, being only significant for lines A1152 and C655 (Table 1 and Fig. S3). In these two lines, the expression of the γ-gliadin fraction was reduced by 93.8% and 98.7%, respectively. Similarly, for lines C217 and D598, the γ-gliadin fraction was significantly reduced (between 72.2% and 81.6%) (Table 1 and Fig. S3).
The morphology of the seeds of transgenic plants was similar to that of wild types (Figs. S2–S4). Glutenin content (HMW and LMW) was quantified by RP-HPLC in transgenic and wild-type lines. The HMW glutenins were increased in some transgenic lines compared with wild types (Table 1). In contrast, the LMW glutenins were unaffected for most of the lines, and only four lines (E82, D874, D876, and X678) showed a significant decreased in the LMW content relative to the wild-type line. Although most of transgenic lines containing the ω/α hpRNA constructs had slightly lower protein content than that of wild types, differences in protein content were only significant for line A1152 (Table 1). The SDS sedimentation volumes were significantly lower than in the wild type for lines D793, E82, D874, and D876 (Table 1).
Ability of Transgenic Wheat to Stimulate Gliadin-Reactive T-Cell Clones from CD Patients.
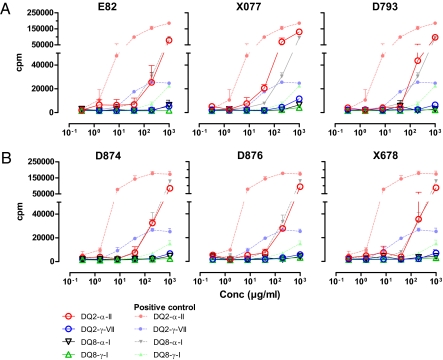
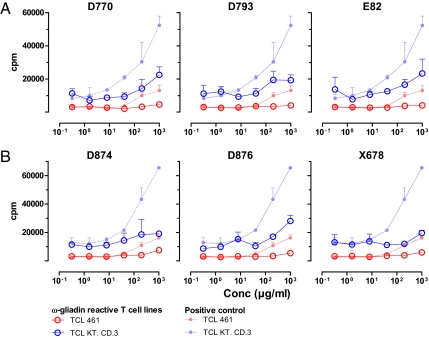
To test if the transgenic wheat had lost the ability to activate gluten-specific T cells isolated from CD patients, total gluten protein were extracted for the various lines, treated with TG2, and tested in serial dilution for stimulation of DQ2- and DQ8-restricted T-cell clones of CD patients. The T-cell clones specific for the DQ2-α-II and DQ2-γ-VII epitopes were more sensitive as based on their reactivity to the wild-type positive controls, thus providing the best estimates for the reduction in amount of epitope present. Although the extracted gluten proteins of some of the transgenic lines were highly stimulatory to the proliferation of T cells (Figs. S5 and S6), several of the transgenic lines showed markedly impaired stimulatory capacity. The transgenic lines 28B (pDhp-ω/α transgenic) (Fig. S5) and X077 (pGhp-ω/α transgenic) (Fig. 2A) required approximate 1 log higher concentration of the total gluten protein to give a similar T-cell proliferative response with T-cell clones specific for the DQ2-α-II and DQ2-γ-VII epitopes. No response was seen for these transgenic lines when tested with the T-cell clones specific for the DQ8-α-I and DQ8-γ-I epitopes. Even more pronounced reduction in proliferative responses was seen with the transgenic lines D793 (pGhp-ω/α), E82 (pDhp-ω/α+pghp8.1) (Fig. 2A), D874, D876, and X678 (pDhp-ω/α+pGhp-ω/α) (Fig. 2B). There was about a 2-log reduction in the expression of the DQ2-α-II epitope in these transgenic lines. The responses of the T-cell clones specific for the other epitopes (DQ2-γ-VII, DQ8-α-I, and DQ8-γ-I) were at or below detection level for the highest concentration of gluten protein tested, indicating that there was a 2-log or more reduction in the expression of the DQ2-γ-VII epitope, at least a 1-log reduction in the expression of the DQ8-α-I epitope, and also, a substantial reduction in the expression of the DQ8-γ-I epitope. The transgenic wheat lines were also tested for recognition by T-cell lines that are responsive to a highly stimulatory peptide from ω-gliadin (28). Optimally, this should have been tested with T-cell clones reactive with this peptide, but such clones were not available to us. The two T-cell lines gave strong proliferative responses to the positive-control antigen but a reduced response to the total gluten extract from the transgenic lines 28A (pDhp-ω/α) (Fig. S7A), E82 (pDhp-ω/α+pghp8.1), E35 (pDhp-ω/α+pghp8.1) (Fig. 3A and Fig. S6A), D770 (pGhp-ω/α), and D876 (pDhp-ω/α+pGhp-ω/α) (Fig. 3B) and no response to the transgenic lines D793 (pGhp-ω/α) (Fig. 3A), D874 (pDhp-ω/α+pGhp-ω/α), and X678 (pDhp-ω/α+pGhp-ω/α) (Fig. 3B). Overall, the transgenic lines D874 and X678, containing around 30% of ω-gliadin, 10% of α-gliadin, and only 1% of γ-gliadin compared with their wild-type control grains, were particularly inefficient to stimulate the CD lesion-derived T cells.
Fig. 2.
Proliferative responses of gliadin-specific T-cell clones stimulated with total gluten extracts from transgenic wheat lines. T-cell clones made from CD patients and specific for the epitopes DQ2-α-II, DQ2-γ-VII, DQ8-α-I, and DQ8-γ-I were stimulated with antigen-presenting cells incubated with serial dilutions of gluten antigen. Transgenic lines derived from line BW208 are depicted in A, whereas transgenic lines derived from BW2003 are depicted in B. The responses of the T-cell clones against total gluten extracts from the relevant wild-type lines are shown with corresponding symbols in the lighter colors.
Fig. 3.
Proliferative responses of ω-gliadin–reactive T-cell lines stimulated with total gluten extracts from transgenic wheat lines. Transgenic lines derived from line BW208 are depicted in A, whereas transgenic lines derived from BW2003 are depicted in B. The responses of the T-cell clones against total gluten extracts from the relevant wild-type lines are shown with corresponding symbols in the lighter colors.
Discussion
RNAi technology could be useful for down-regulating gliadin genes, whose proteins are not tolerated by individuals suffering from CD. This approach has been used to down-regulate the expression of γ-gliadin (23) genes of bread wheat. In the present work, we report the use of hpRNA constructs containing IR fragment-comprising sequences from α- and ω-gliadin genes under the control of endosperm-specific promoters from γ-gliadin (pGhp-ω/α) or D-hordein (pDhp-ω/α).
Transgenic lines transformed with both ω/α hpRNA constructs had an average reduction of gliadins of 85.6%, whereas lines transformed with only the pGhp-ω/α vector (γ-gliadin promoter) had an average reduction of the gliadin content of 82.1% and lines transformed with the pDhp-ω/α vector (D-Hordein promoter) had a reduction of 74.7%. This indicates that both endosperm-specific promoters were highly effective in controlling the expression of the IR fragments and the down-regulation of the target gliadin genes. The wheat lines (D874 and X678) transformed with both the pGhp-ω/α and pDhp-ω/α vector were particularly inefficient in eliciting a T-cell response.
Gene silencing by RNA has been generally correlated with the level of homology between trigger and target sequences. Although the average percentage of identity between the α and ω IR fragments with genes from the γ-gliadin group was 69.2% and 69.3%, respectively (Table S2), the ω/α constructs showed a higher level of suppression of γ-gliadins than that exerted by pghpg8.1. Moreover, γ-gliadin RNAi fragment (pghp8.1) showed an average percentage of homology of 80.7% with the γ-gliadins group, 71.9% with α-gliadins, and 66.6% with ω-gliadins, but only the γ-gliadins genes were down-regulated with this construct. It is clear that other factors besides the homologies between trigger and target gene are also affecting the efficiency of gene silencing. One important factor is the amount of the target mRNA; hence, genes transcribed at higher levels could be silenced better than those at lower levels (29, 30). Another factor that likely affects the down-regulation efficiency is silencing spreading from the target region (31, 32). The siRNAs derived from the IR region will cleave target mRNA transcripts, leading to cleaved fragments that are converted into dsRNA, which again can produce secondary siRNAs and thereby, spread the silencing to related gliadin sequences.
The transgenic lines resulting from transformation with pghp8.1 (C217, D598, C655, and A1152) all had low expression of γ-gliadins but with retained expression of α- and ω-gliadins (Table 1). Still, the DQ2-γ-VII–specific T-cell clone gave strong response to the total gluten extract from these transgenic lines. The explanation is probably that the stimulatory material in these transgenic lines is derived from ω-gliadin, because some ω-gliadin proteins are known to harbor the native sequence of the DQ2-γ-VII epitope (9-mer core region: QQPQQPFPQ).
The transgenic wheat lines reported here will still have reasonable baking qualities, because the HMW glutenin subunits, major determinants of bread-making quality, are present in the transgenic lines. We assessed the bread-making quality of the transgenic lines using the SDS sedimentation test, because SDS volumes of this test are highly correlated with bread-making quality (33). Most of transgenic lines containing the ω/α constructs had SDS sedimentation volumes comparable with those of wild types, and five lines had SDS-volume values significantly lower than wild types. However, SDS volumes of these five lines are still comparable with those of the medium-quality bread wheat. Glutenin proteins can also elicit T-cell responses in CD patients (11, 34, 35); however, their stimulatory capacity is far less than the gliadin proteins (11). The transgenic lines produced here will not be completely devoid of sequences stimulatory to T cells of CD patients, because they contain small amounts of gliadin proteins as well as glutenin proteins. A recent meta-analysis of gluten-challenge studies concluded that a daily gluten intake between 10 and 100 mg would probably be safe for most CD patients (36), suggesting that the transgenic lines reported here could be used in foodstuff tolerated by many CD patients. Even if more optimal transgenic lines will have to be made for introduction in foodstuff, the present work shows that targeting of CD-related gene families is indeed achievable and may reduce the expression of CD-related T-cell epitopes.
Methods
RNAi Fragments and Transformation Plasmids.
The fragments and plasmids used to target the γ-gliadin genes were as reported previously (23). Plasmids pGhp-ω/α and pDhp-ω/α (Fig. S1) contain the endosperm-specific promoters from γ-gliadin and D-hordein genes, respectively (25, 26), and the nos terminator. These vectors contain the Ubi1 intron from maize as a spacer region between the ω/α RNAi IR fragments, inserted in sense and antisense orientation by recombination using the GATEWAY technology (Invitrogen). Two fragments of 170 and 191 bp, corresponding to the most conserved regions from, respectively, α- and ω-gliadins, were selected for use as IR fragments. These fragments were amplified from bread wheat, assembled to obtain the full chimeric 361-bp ω/α fragment, cloned into the recombination sites of pCR8/GW/TOPO vector (Invitrogen), and recombined to provide the final pGhp-ω/α and pDhp-ω/α vectors. Primers used are described in Table S3. All PCR primers were designed using eprimer3 (http://emboss.sourceforge.net/). All EMBOSS programs were run on the Mobyle portal (http://mobyle.pasteur.fr/cgi-bin/portal.py). Sequences of plasmids pghp8.1, pGhp-ω/α, and pDhp-ω/α are available at GenBank under accession numbers HM352557, HM352558, and HM352559, respectively.
Wheat Transformation and Identification of Transgenic Plants.
Immature scutella of two lines of the bread wheat (Triticum aestivum) cv. Bobwhite (BW2003 and BW208) were isolated from primary tillers harvested 16 d after anthesis and transformed as described previously (25). Plasmids carrying the RNAi fragments were used in combination with plasmid pAHC25 containing the selectable bar gene (27). Putative transgenic plants were transferred to soil and grown to maturity in the greenhouse, and the presence of the transformation vectors was confirmed by PCR (Table S3). Transgenic plants were self-pollinated for two to three generations to obtain homozygous lines.
Gliadin and Glutenin Extraction and A-PAGE Analysis.
The gliadin fraction from 40 to 100 mg of flour was extracted stepwise three times with a 5:1 (μL/mg) ratio of 60% (vol/vol) ethanol for 45 min at room temperature with shaking every 10 min, and it was centrifuged for 20 min at 13,000 × g. Supernatants were precipitated with 1 mL of cold acetone, incubated for 30 min at 4 °C, and centrifuged for 10 min at 13,000 × g. After the supernatants were decanted and the samples were air dried, pellets were resuspended in 500 μL of 60% (vol/vol) ethanol. Gliadin fractions were separated by A-PAGE gels as described (37).
Glutenin fractions were extracted from the insoluble material of the previous step with 750 μL of 50% (vol/vol) 1-propanol, 2 M urea, 0.05 M Tris·HCl (pH 7.5), and 2% (wt/vol) DTT at 60 °C. Finally, samples were filtered through a 0.45-μm nylon filter (Teknokroma).
Analysis of Wheat Flour by R5 Competitive ELISA.
Wheat grains from transgenic lines were analyzed by the reference laboratory for gluten analysis at Centro Nacional de Biotecnología using the R5 monoclonal antibody as described elsewhere (38). The assay was performed in triplicate.
RP-HPLC Analysis.
Gliadin (20 μL) and glutenin (30 μL) extracts were applied to a 300SB-C8 reverse-phase analytical column (4.6 × 250 mm, 5-μm particle size, 300 Å pore size; Agilent Technologies) using a 1200 Series Quaternary LC System liquid chromatograph (Agilent Technologies) with a diode array ultraviolet-visible detector (DAD UV-V). The HPLC separation method was as described (39). Absorbance was monitored with the DAD UV-V module at 210 nm. The integration procedure was handled automatically by the software, with some minor manual adjustment. Three independent repetitions were carried out for each transgenic line and control.
Protein Content and SDS Sedimentation Test.
Protein content was determined using whole flour by the American Association of Cereal Chemists (AACC) Method 39–11.01 (40). The SDS sedimentation volume was determined as described (41).
Statistics.
Data were analyzed using the SPSS version 11.0 statistical software package (SPSS Inc). Data were tested for normal distribution using the Kolmogorov–Smirnov test and for homogeneity of variances with the Levene test. ANOVA and two-tailed Dunnett's test for median multiple comparisons were used to analyze the results. P values less than 0.05 were considered significant. Pearson's R was calculated to determine the correlation between gliadin contents measured by R5 competitive ELISA and RP-HPLC methods.
Extraction of Total Gluten Proteins for T-Cell Assays.
Whole-meal flour from transgenic wheat lines and wild types was used throughout as starting material. Total gluten protein was extracted from 1 g of milled seeds and treated as described (11). Total gluten protein was digested first with pepsin from porcine gastric mucosa (P7012; Sigma) and then with pancreatic protease trypsin (T9201; Sigma). Both enzymatic digestions were at a 1:100 ratio (wt/wt) at 37 °C for 4 h. The digested samples were dialyzed, lyophilized, and stored as freeze-dried proteins.
T-Cell Assays.
The pepsin- and trypsin-digested lyophilized samples were solubilized to 4.0 mg/mL in 100 mM Tris buffer containing 2 mM Ca2+ and treated with recombinant human TG2 (0.1 mg/mL final concentration) for 2 h. The TG2-treated gliadins samples were serial diluted (1,000 μg/mL to 0.19 μg/mL final concentration) and incubated with homozygous HLA-DQ2–positive (DQA1*0501/DQB1*0201; CD114) or HLA-DQ8 (DQA1*0301/DQB1*0302; #9092) B lymphoblastoid cell lines as antigen-presenting cells in 96-well U-bottom plates. The next day, T-cell clones were added to the cell cultures (∼50,000 cells/well) and incubated for 72 h, and T-cell proliferation during the last 24 h was measured by 3H thymidine incorporation. Positive control for T-cell reactivity was pepsin- and trypsin-digested and TG2-treated total gluten extracts from wild-type BW2003 and BW208 grains as well as gliadin from Sigma (G-3375). T-cell lines and clones were made from biopsies of HLA-DQ2– and HLA-DQ8–positive CD patients as described earlier (10, 42). The following T-cell clones were used: TCC364.1.0.14 DQ2-α-II–specific (PQPELPYPQ; 9-mer core region of epitope given), TCC360.11 DQ8-α-I–specific (EGSFQPSQE), TCC387.19 DQ2-γ-VII–specific (QQPEQPFPQ), and TCC544.1.1.8 DQ8-γ-I–specific (EQPQQPYPE) clones. The polyclonal T-cell lines TCL 461.0.2.1.4 and TCL KT.CD.3 reactive with the ω-gliadin–derived peptide (QPEQPFPQPEQPFPWQP) (28) were also tested.
Supplementary Material
Acknowledgments
We thank Ana García and Marie Johannesen for expert technical assistance. The authors thank Prof. Peter Shewry (Rothamsted Research), Prof. Paul Christou (Universitat de Lleida), and Dr. Paul Lazzeri (Agrasys SL) for critical comments on the manuscript. The authors acknowledge funding by the Spanish Comision Interministerial de Ciencia y Tecnologia (Projects AGL2007-65685-C02-01 and AGL2010-19643-C02-02), the European Regional Development Fund (ERDF), the Norwegian Foundation for Health and Rehabilitation (EXTRA fund), and the Novo Nordisk Foundation. J.G.-H. also acknowledges financial support from the I3P Program from the Consejo Superior de Investigaciones Cientificas, which is cofinanced by the European Social Fund.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. HM352557, HM352558, and HM352559).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1007773107/-/DCSupplemental.
References
- 1.Trier JS. Diagnosis of celiac sprue. Gastroenterology. 1998;115:211–216. doi: 10.1016/s0016-5085(98)70383-x. [DOI] [PubMed] [Google Scholar]
- 2.Sollid LM. Coeliac disease: Dissecting a complex inflammatory disorder. Nat Rev Immunol. 2002;2:647–655. doi: 10.1038/nri885. [DOI] [PubMed] [Google Scholar]
- 3.Kagnoff MF. Celiac disease: Pathogenesis of a model immunogenetic disease. J Clin Invest. 2007;117:41–49. doi: 10.1172/JCI30253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.West J, Logan RFA, Card TR, Smith C, Hubbard R. Fracture risk in people with celiac disease: A population-based cohort study. Gastroenterology. 2003;125:429–436. doi: 10.1016/s0016-5085(03)00891-6. [DOI] [PubMed] [Google Scholar]
- 5.Wieser H, Koehler P. The biochemical basis of celiac disease. Cereal Chem. 2008;85:1–13. [Google Scholar]
- 6.Ivarsson A, et al. Epidemic of coeliac disease in Swedish children. Acta Paediatr. 2000;89:165–171. doi: 10.1080/080352500750028771. [DOI] [PubMed] [Google Scholar]
- 7.Ventura A, Magazzù G, Greco L. Duration of exposure to gluten and risk for autoimmune disorders in patients with celiac disease. SIGEP Study Group for Autoimmune Disorders in Celiac Disease. Gastroenterology. 1999;117:297–303. doi: 10.1053/gast.1999.0029900297. [DOI] [PubMed] [Google Scholar]
- 8.Shewry PR, Miles MJ, Tatham AS. The prolamin storage proteins of wheat and related cereals. Prog Biophys Mol Biol. 1994;61:37–59. [PubMed] [Google Scholar]
- 9.Shewry PR, Halford NG. Cereal seed storage proteins: Structures, properties and role in grain utilization. J Exp Bot. 2002;53:947–958. doi: 10.1093/jexbot/53.370.947. [DOI] [PubMed] [Google Scholar]
- 10.Arentz-Hansen H, et al. Celiac lesion T cells recognize epitopes that cluster in regions of gliadins rich in proline residues. Gastroenterology. 2002;123:803–809. doi: 10.1053/gast.2002.35381. [DOI] [PubMed] [Google Scholar]
- 11.Molberg O, et al. Intestinal T-cell responses to high molecular-weight glutenins in celiac disease. Gastroenterology. 2003;125:337–344. doi: 10.1016/s0016-5085(03)00890-4. [DOI] [PubMed] [Google Scholar]
- 12.Arentz-Hansen H, et al. The intestinal T cell response to alpha-gliadin in adult celiac disease is focused on a single deamidated glutamine targeted by tissue transglutaminase. J Exp Med. 2000;191:603–612. doi: 10.1084/jem.191.4.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Payne PI. Genetics of wheat storage proteins and the effect of allelic variation on bread-making quality. Annu Rev Plant Physiol. 1987;38:141–153. [Google Scholar]
- 14.Anderson OD, Greene FC. The alpha-gliadin gene family. II. DNA and protein sequence variation, subfamily structure, and origins of pseudogenes. Theor Appl Genet. 1997;95:59–65. [Google Scholar]
- 15.Sabelli PA, Shewry PR. Characterization and organization of gene families at the Gli-1 loci of bread and durum wheats by restriction fragment analysis. Theor Appl Genet. 1991;83:209–216. doi: 10.1007/BF00226253. [DOI] [PubMed] [Google Scholar]
- 16.Spaenij-Dekking L, et al. Natural variation in toxicity of wheat: Potential for selection of nontoxic varieties for celiac disease patients. Gastroenterology. 2005;129:797–806. doi: 10.1053/j.gastro.2005.06.017. [DOI] [PubMed] [Google Scholar]
- 17.van Herpen TW, et al. Alpha-gliadin genes from the A, B, and D genomes of wheat contain different sets of celiac disease epitopes. BMC Genomics. 2006;7:1. doi: 10.1186/1471-2164-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bernstein E, Denli AM, Hannon GJ. The rest is silence. RNA. 2001;7:1509–1521. [PMC free article] [PubMed] [Google Scholar]
- 19.Hammond SM, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- 20.Ogita S, Uefuji H, Yamaguchi Y, Koizumi N, Sano H. Producing decaffeinated coffee plants. Nature. 2003;423:823. doi: 10.1038/423823a. [DOI] [PubMed] [Google Scholar]
- 21.Liu Q, Singh S, Green A. High-oleic and high-stearic cottonseed oils: Nutritionally improved cooking oils developed using gene silencing. J Am Coll Nutr. 2002;21(Suppl 3):205S–211S. doi: 10.1080/07315724.2002.10719267. [DOI] [PubMed] [Google Scholar]
- 22.Regina A, et al. High-amylose wheat generated by RNA interference improves indices of large-bowel health in rats. Proc Natl Acad Sci USA. 2006;103:3546–3551. doi: 10.1073/pnas.0510737103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gil-Humanes J, et al. Silencing of γ-gliadins by RNA interference (RNAi) in bread wheat. J Cereal Sci. 2008;48:565–568. [Google Scholar]
- 24.Travella S, Klimm TE, Keller B. RNA interference-based gene silencing as an efficient tool for functional genomics in hexaploid bread wheat. Plant Physiol. 2006;142:6–20. doi: 10.1104/pp.106.084517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pistón F, Marín S, Hernando A, Barro F. Analysis of the activity of a γ-gliadin promoter in transgenic wheat and characterization of gliadin synthesis in wheat by MALDI-TOF during grain development. Mol Breed. 2009;23:655–667. [Google Scholar]
- 26.Pistón F, León E, Lazzeri P, Barro F. Isolation of two storage protein promoters from Hordeum chilense and characterization of their expression patterns in transgenic wheat. Euphytica. 2008;162:371–379. [Google Scholar]
- 27.Christensen AH, Quail PH. Ubiquitin promoter-based vectors for high-level expression of selectable and/or screenable marker genes in monocotyledonous plants. Transgenic Res. 1996;5:213–218. doi: 10.1007/BF01969712. [DOI] [PubMed] [Google Scholar]
- 28.Camarca A, et al. Intestinal T cell responses to gluten peptides are largely heterogeneous: Implications for a peptide-based therapy in celiac disease. J Immunol. 2009;182:4158–4166. doi: 10.4049/jimmunol.0803181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miki D, Itoh R, Shimamoto K. RNA silencing of single and multiple members in a gene family of rice. Plant Physiol. 2005;138:1903–1913. doi: 10.1104/pp.105.063933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Piston F, et al. Down-regulation of four putative arabinoxylan feruloyl transferase genes from family PF02458 reduces ester-linked ferulate content in rice cell walls. Planta. 2010;231:677–691. doi: 10.1007/s00425-009-1077-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eamens A, Wang MB, Smith NA, Waterhouse PM. RNA silencing in plants: Yesterday, today, and tomorrow. Plant Physiol. 2008;147:456–468. doi: 10.1104/pp.108.117275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Han Y, Grierson D. The influence of inverted repeats on the production of small antisense RNAs involved in gene silencing. Mol Genet Genomics. 2002;267:629–635. doi: 10.1007/s00438-002-0696-z. [DOI] [PubMed] [Google Scholar]
- 33.Carter BP, Morris CF, Anderson JA. Optimizing the SDS sedimentation test for end-use quality selection in a soft white and club wheat breeding program. Cereal Chem. 1999;76:907–911. [Google Scholar]
- 34.van de Wal Y, et al. Glutenin is involved in the gluten-driven mucosal T cell response. Eur J Immunol. 1999;29:3133–3139. doi: 10.1002/(SICI)1521-4141(199910)29:10<3133::AID-IMMU3133>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 35.Vader W, et al. The gluten response in children with celiac disease is directed toward multiple gliadin and glutenin peptides. Gastroenterology. 2002;122:1729–1737. doi: 10.1053/gast.2002.33606. [DOI] [PubMed] [Google Scholar]
- 36.Hischenhuber C, et al. Review article: Safe amounts of gluten for patients with wheat allergy or coeliac disease. Aliment Pharmacol Ther. 2006;23:559–575. doi: 10.1111/j.1365-2036.2006.02768.x. [DOI] [PubMed] [Google Scholar]
- 37.Khan K, Hamada AS, Patek J. Polyacrylamide gel electrophoresis for wheat variety identification—effect of variables on gel properties. Cereal Chem. 1985;62:310–313. [Google Scholar]
- 38.Ferre S, García E, Méndez E. Measurement of hydrolyzed gliadins by a competitive ELISA based on monoclonal antibody R5: Analysis of syrups and beers. In: Stern M, editor. Proceedings of the 18th Meeting of the Working Group on Prolamin Analysis and Toxicity. Stockholm: Verlag Wissenschaftliche Scripten; 2003. pp. 65–69. [Google Scholar]
- 39.Wieser H, Antes S, Seilmeier W. Quantitative determination of gluten protein types in wheat flour by reversed-phase high-performance liquid chromatography. Cereal Chem. 1998;75:644–650. [Google Scholar]
- 40.AACC International . Approved Methods of Analysis. 11th Ed. St. Paul, MN: AACC International; 2000. [Google Scholar]
- 41.Williams P, Jaby el-Haramein F, Nakkoul H, Rihawi S. Technical Manual (ICARDA) Aleppo, Syria: International Center for Agricultural Research in the Dry Areas; 1986. Crop quality evaluation methods and guidelines; p. 145. [Google Scholar]
- 42.Tollefsen S, et al. HLA-DQ2 and -DQ8 signatures of gluten T cell epitopes in celiac disease. J Clin Invest. 2006;116:2226–2236. doi: 10.1172/JCI27620. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.