Abstract
DEAD-box RNA helicases play various, often critical, roles in all processes where RNAs are involved. Members of this family of proteins are linked to human disease, including cancer and viral infections. DEAD-box proteins contain two conserved domains that both contribute to RNA and ATP binding. Despite recent advances the molecular details of how these enzymes convert chemical energy into RNA remodeling is unknown. We present crystal structures of the isolated DEAD-domains of human DDX2A/eIF4A1, DDX2B/eIF4A2, DDX5, DDX10/DBP4, DDX18/myc-regulated DEAD-box protein, DDX20, DDX47, DDX52/ROK1, and DDX53/CAGE, and of the helicase domains of DDX25 and DDX41. Together with prior knowledge this enables a family-wide comparative structural analysis. We propose a general mechanism for opening of the RNA binding site. This analysis also provides insights into the diversity of DExD/H- proteins, with implications for understanding the functions of individual family members.
Introduction
DExD/H-box RNA helicases, from virus and bacteria to eukaryotes, play important roles in processes including ribosome biogenesis, RNA processing and folding, ribonucleoprotein (RNP) remodeling, RNA nuclear export, the regulation of RNA translation and transcription, and nonsense-mediated RNA decay. DExD/H-box RNA helicases have multiple functions in these processes: They can act as RNA chaperones, ATP-dependent RNA helicases and unwindases, as RNPases by mediating RNA-protein association and dissociation [1]–[4] or as co-activators and co-repressors of transcription ([5]–[7] and refs. therein). Cancer cell lines often feature deregulated expression or impaired functioning of RNA helicases [5], [8]. In addition, several family members are captured and regulated by viral proteins [9], are involved in viral RNA maturation [10], or mediate antiviral host defense [11], [12]. Inhibition of individual RNA helicases as a therapeutic route is currently being explored (e.g., [13]–[17]).
DExD/H-box proteins often contain accessory regulatory domains and localization modules, but their cores consist of two RecA-like domains joined by a short flexible linker. The N-terminal domain is commonly referred to as conserved domain-1, or DEAD-domain, and the C-terminal domain as conserved domain-2, or helicase domain [3], [4], [18]. Both domains contribute to the binding site for RNA substrates and both contribute to ATP hydrolysis. These activities are coupled to one another by allostery throughout the protein molecules. Consequently, a detailed understanding of how these proteins convert chemical energy into RNA remodeling requires knowledge of the structures of the two conserved domains independent of each other and interacting in the closed active state. To date, crystal structures of tandem domains are available for several DExD/H-box helicases, also in complex with RNA substrates [19]–[22]. To understand the RNA remodeling event and the underlying structural rearrangements, it is important to compare these structures with those of each domain in isolation.
We have solved crystal structures of single domains from eleven human DExD/H-box helicases of the DEAD-motif subfamily. A comparative analysis of these structures uncovered not only isoform specific features, but also nucleotide specific positioning of flexible elements that are common to several proteins. We suggest a structural mechanism for the linkage between binding of ATP and activation of the RNA binding site.
Results and Discussion
We used X-ray crystallography to determine the structures of the DEAD-domains of DDX2A, DDX2B, DDX5, DDX10, DDX18, DDX20, DDX47, DDX52, and DDX53, as well as the helicase domains of DDX25 and DDX41. While the physiological roles of these proteins are diverse (Table 1) all structures show the RecA-like fold. Superposition of the DEAD-domain structures gives root mean square deviations of Cα-atom positions between 0.6 and 1.9 Å for proteins with sequence identity between 86 and 27%. The two helicase domains have a sequence identity of 23% and their structures superimpose with an r.m.s.d. of 3 Å. Details of the synchrotron data collection, structure determination, and refinement statistics are presented in Table 2.
Table 1. Summary of previously established roles and functions for the RNA helicases covered in this study.
| Helicase | Function |
| DDX2A | DDX2A (eIF4A1) is essential for translation initiation. It is part of the eIF4F complex that consists of eIF4G, eIF4E and eIF4A [51]–[53]. Its activity is strongly enhanced by eIF4G, eIF4B and eIF4H [54]. The eIF4F complex and eIF4A are potential targets for anti cancer drugs [55]–[57]. |
| DDX2B | Also known as eIF4A2, an isoform of DDX2A. |
| DDX5 | DDX5 is a co-regulator of different transcription factors including ERα, p53, MyoD and Runx2, but ATPase/helicase activity is not required for transcriptional co-regulation. DDX5 also participates in pre-RNA processing, alternative splicing, microRNA and ribosomal RNA processing (reviewed in ref. [6]). |
| DDX10 | DDX10 is probably involved in ribosome assembly. Fusion of the nucleoporin gene NUP98 with the DDX10 gene leads to the NUP98-DDX10 gene product. This fusion protein is involved in leukemogenesis [58], [59]. |
| DDX18 | DDX18 (Myc-regulated DEAD-box protein, or MrDP; [60]) is a nucleolar protein that is specifically upregulated in highly proliferating cells [61]. |
| DDX20 | DDX20 (Gemin3) is a component of the SMN (Survival of Motor Neurons) complex that is involved in assembly and reconstruction of different RNP (ribonucleoprotein) complexes [62]. DDX20, Gemin4 and eIF2C2 form a separate complex that contains numerous miRNAs [63]. DDX20 also binds to the Epstein-Barr Virus Nuclear Proteins EBNA2 and EBNA3C. The poliovirus-encoded proteinase 2Apro cleaves DDX20 resulting in DDX20 inactivation and reduced snRNP assembly [64]. |
| DDX25 | DDX25 (GRTH) is a testis specific, gonadotropin and androgen regulated protein that is essential for completion of spermatogenesis [65]. DDX20 acts as a shuttling protein in the gene-specific nuclear export of RNA messages. Furthermore it regulates the translation of specific genes in germ cells [66]. |
| DDX41 | DDX41 (Abstrakt) post-transcriptionally regulates the expression levels of the insc protein that is essential for control of cell polarity and spindle orientation [67]. |
| DDX47 | DDX47 is involved in pre-rRNA processing. It interacts with NOP132 which recruits pre-rRNA processing proteins to the region within the nucleolus were rRNA is transcribed [68]. |
| DDX52 | DDX52 (Rok1) is required for the release of snR30 (small nucleolar RNA-30) from pre-ribosomes. snR30 is one of three snoRNAs that are critical for pre-rRNA processing in yeast. DDX52 ATPase activity is important for optimal pre-ribosomal RNA processing, but not essential for release of snR30 [69]. |
| DDX53 | DDX53 (CAGE) is expressed in testis and various tumors, but not in other tissues. Expression of the CAGE-gene is determined by its methylation status [70]. |
Table 2. Summary of crystallographic data analysis and refinement statistics*.
| Structure | DDX2A | DDX2B | DDX5 | DDX10 | DDX18 | DDX20 | DDX25 | DDX41 | DDX47 | DDX52 | DDX53 |
| Domain | DEAD | DEAD | DEAD | DEAD | DEAD | DEAD | helicase | helicase | DEAD | DEAD | DEAD |
| PDB entry | 2G9N | 3BOR | 3FE2 | 2PL3 | 3LY5 | 3B7G | 2RB4 | 2P6N | 3BER | 3DKP | 3IUY |
| Ligand | - | - | ADP | ADP | PO4 | AMPPNP | - | - | AMP | ADP | AMP |
| Beamline | ESRF ID14-2 | APS 19-ID | BESSY 14.2 | MAX II I911-2 | DIAMOND I04 | ESRF ID29 | BESSY 14.2 | ESRF ID14-4 | ESRF ID29 | ESRF ID23-1 | ESRF ID14-2 |
| Wavelength (Å) | 0.93300 | 0.97242 | 0.9184 | 1.04123 | 0.9789 | 1.00595 | 0.95373 | 1.04005 | 0.97472 | 1.00000 | 0.97930 |
| Space group | P 1 21 1 | P 21 21 2 | C 2 2 21 | P 61 2 2 | P 31 | P 31 2 1 | P 43 21 2 | P 65 2 2 | C 1 2 1 | P 1 21 1 | P 1 21 1 |
| Cell dimensions | |||||||||||
| a, b, c (Å) | a = 47.8,b = 78.25, c = 59.09 | a = 58.09, b = 80.1, c = 42.74 | a = 84.57, b = 106.87, c = 117.32 | a = 63.5, b = 63.5, c = 304.01 | a = 41.34, b = 41.34, c = 230.54 | a = 63.56, b = 63.56, c = 214.6 | a = 70.31, b = 70.31, c = 187.12 | a = 68.01, b = 68.01, c = 305.6 | a = 93.05, b = 70.37, c = 35.86 | a = 40.63, b = 38.36, c = 73.84 | a = 56.44, b = 61.25, c = 65.79 |
| α, β, γ (°) | α = 90, β = 103.43, γ = 90 | α = 90, β = 90, γ = 90 | α = 90, β = 90, γ = 90 | α = 90, β = 90, γ = 120 | α = 90, β = 90, γ = 120 | α = 90, β = 90, γ = 120 | α = 90, β = 90, γ = 90 | α = 90, β = 90, γ = 120 | α = 90, β = 90.7, γ = 90 | α = 90, β = 90.37, γ = 90 | α = 90, β = 96.36, γ = 90 |
| Highest resolution shell range(Å) | 2.37–2.25 | 1.92–1.85 | 2.67–2.60 | 2.30–2.15 | 2.85–2.7 | 2.00–1.90 | 2.90–2.80 | 2.80–2.60 | 1.50–1.40 | 2.20–2.10 | 2.53–2.40 |
| Rmeas (a) | 0.07 (0.24)(b) | 0.14 (0.67)(c) | 0.14 (0.74) | 0.05 (0.16) | 0.24 (1.29) | 0.081 (0.13) | 0.11 (0.82)(b) | 0.10 (0.45) | 0.05 (0.14) | 0.09 (0.38) | 0.20 (0.61) |
| I/σ(I) | 7.4 (5.3) | 27.6 (3.8)(d) | 11.0 (2.4) | 54.1 (24.5) | 6.2 (2.0) | 32.10 (21.3) | 15.65 (2.6) | 35.9 (12.6) | 28.7 (14.1) | 12.8 (4.6) | 13.4 (4.2) |
| Completeness (%) | 99.7 (100.0) | 100 (99.9) | 99.4 (99.8) | 99.7 (100.0) | 99.98 (100.0) | 99.7 (100.0) | 100.0 (100.0) | 99.9 (100.0) | 99.5 (100.0) | 99.5 (99.8) | 100.0 (100.0) |
| Redundancy | 7.4 (3.5) | 11.4 (10.1) | 4.0 (4.0) | 25.4 (26.5) | 5.1 (5.5) | 20.5 (18.2) | 8.8 (9.1) | 40.0 (41.8) | 7.3 (7.3) | 3.7 (3.7) | 9.3 (9.5) |
| Refinement | |||||||||||
| Resolution range (Å) | 20–2.25 | 30–1.85 | 35–2.6 | 30–2.15 | 38.4–2.8 | 38.43–1.90 | 19.44–2.80 | 29.72–2.60 | 28.24–1.40 | 19.56–2.10 | 45.13–2.40 |
| No. reflections | 19100 | 17560 | 15770 | 19842 | 10358 | 38606 | 11208 | 13220 | 43193 | 12804 | 17613 |
| Rwork/Rfree | 0.1761/0.2573 | 0.185/0.226 | 0.205/0.273 | 0.210/0.248 | 0.246/0.274 | 0.172/0.202 | 0.233/0.267 | 0.243/0.294 | 0.165/0.189 | 0.182/0.236 | 0.201/0.251 |
| B-factor (Å2) | |||||||||||
| Protein | 15 | 21 | 27 | 48 | 47 | 18 | 82 | 57 | 13 | 23 | 23 |
| Water | 17 | 17 | 20 | 45 | 29 | 29 | 53 | 28 | 17 | 25 | |
| Ligand | 27 | 43 | 36 | 26 | 14 | 12 | 17 | ||||
| R.m.s deviations | |||||||||||
| Bond lengths (Å) | 0.018 | 0.015 | 0.012 | 0.017 | 0.007 | 0.014 | 0.01 | 0.016 | 0.014 | 0.013 | 0.007 |
| Bond angles (°) | 1.732 | 1.344 | 1.354 | 1.695 | 0.952 | 1.648 | 1.244 | 1.602 | 1.638 | 1.414 | 1.157 |
| Ramachandran plot | |||||||||||
| Favored regions (%)(e) | 96.0 | 99.5 | 98.3 | 98.2 | 96.88 | 98.8 | 91.4 | 95.2 | 99.6 | 99.6 | 98.8 |
| Allowed regions (%)(e) | 99.2 | 100 | 100 | 100 | 100 | 100 | 99.7 | 100 | 100 | 100 | 100 |
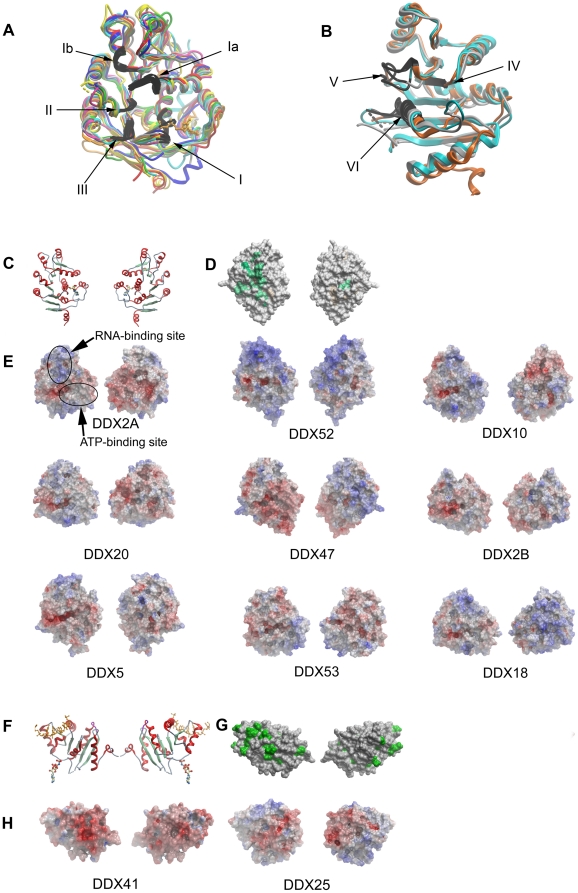
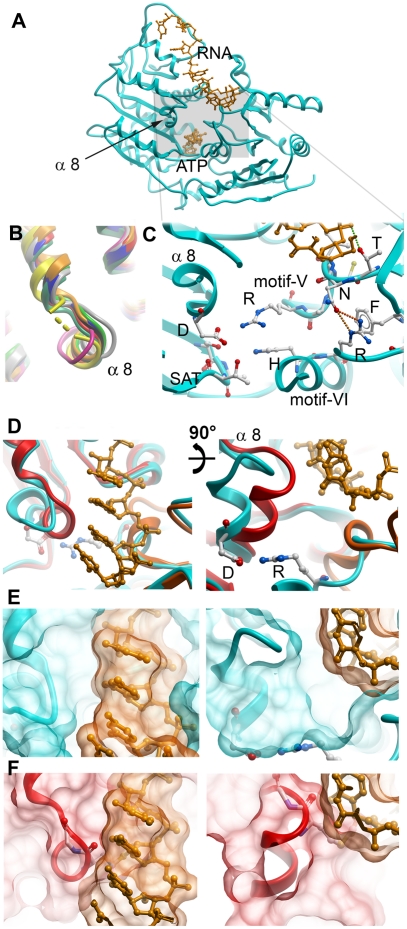
Superpostition of the different crystal structures illustrates the location of flexible regions (Figure 1A, 1B). In general, regions of high sequence conservation (the conserved motifs in particular) contribute to the binding sites for nucleotide and for RNA, and these sites coincide with the highest structural similarity (Figure 2). Conversely, unconserved regions in the DEAD-domains determined here show a higher r.m.s.d. in their Cα-atom positions. Some of the unconserved regions in the structures are flexible, as documented by high B-factors and partially missing electron density.
Figure 1. Crystal structures of DEAD-box conserved domains-1 and -2.
(A) Superposition of the DEAD-domains of DDX2A (green), DDX2B (brown), DDX5 (red), DDX10 (turquoise), DDX18 (grey), DDX47 (dark blue), DDX52 (yellow), and DDX53 (dark yellow). The positions of conserved motifs I–III (black) are indicated. (B) Superposition of the helicase domains of DDX19 (light blue), DDX25 (grey) and DDX41 (orange). The positions of conserved motifs IV–VI (black) are indicated. (C) Cartoon representations of the DDX5 helicase domain in the same orientations as in the following two panels. (D) Conserved surface patches (green), projected onto the DDX47 DEAD-domain surface. (E) Electrostatic surface representation of DEAD-domains. Negative charges are shown in red and positive charges in blue. (F) Cartoon representations of the DDX41 helicase domain in the same orientation as in the following two panels. The RNA and AMPPNP (sticks representation) of the superposed DDX19 structure mark the RNA and nucleotide binding sites. (G) Conserved surface patches (green), projected onto the DDX25 helicase-domain surface. (H) Electrostatic surface representation of helicase domains.
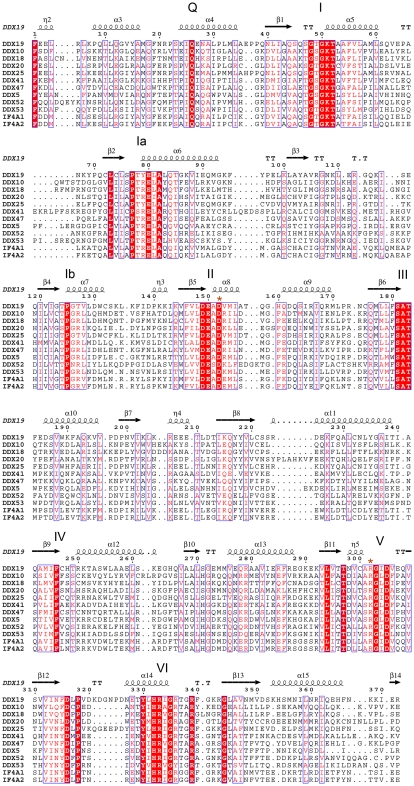
Figure 2. Sequence alignments of the two RecA-like domains of the DEAD-box proteins described in this study.
Conserved sequence motifs are indicated. Secondary structural elements are given for DDX19 (PDB entry 3G0H) above the alignment. Asterisks mark the terminal aspartate of the DEAD motif and the arginine of motif V, the interaction of which is central to positioning of α-helix 8 (see also Figure 5C, D). Sequences shown are human DDX19B (gene accession number: 13177688); DDX10 (13514831); DDX18 (38327634); DDX20 (23270929); DDX25 (29792166); DDX41 (21071032); DDX47 (45786091); DDX5 (16359122); DDX52 (27697141); DDX53 (45709415); eIF4A1/DDX2A (16307020); and eIF4A2/DDX2B (45645183).
Diverse surface properties among DEAD domains
We compared the surface charge distributions of the DEAD-domain structures (Figure 1E, 1H). All DEAD-domains feature a conserved patch that constitutes the nucleotide binding site and part of the RNA binding site. This patch forms a negatively charged channel between α-helices 8 and −10 that extends to the Mg2+-binding site. The negative charges originate from the side chains of the two helices, including the DEAD-motif on α-helix 8. As expected, the RNA binding cleft is positively charged in all DEAD domains, but the charged patches differ in size. The remainder of the DEAD-domain surfaces differs in electrostatic surface properties among the family members.
ATP binding site: The flexible P-loop
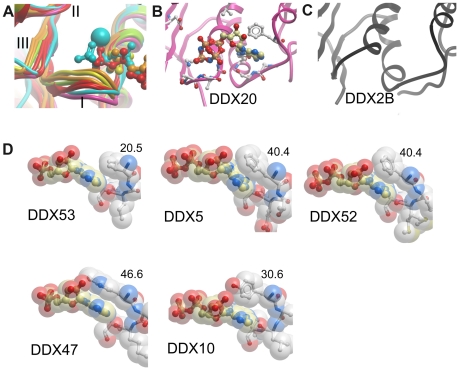
Conserved motifs I (the P-loop), Ia, II, and the Q-motif participate in nucleotide binding [3], [23]. The P-loop and motif II coordinate the nucleotide phosphates and the magnesium ion, whereas residues of the Q-motif bind and recognize the adenine moiety. The side chains that participate in nucleotide and magnesium binding are highly conserved (Figure 2). The nucleotide phosphates interact with backbone atoms, a conserved lysine, and the divalent cation. Superposition of the DEAD-domains shows that the structures of the P-loop and motif III are determined by the state of nucleotide hydrolysis. The P-loop is in a wide-open conformation when ATP is bound, as seen in DDX20 as well as in the previously published structures of DDX19 [22] and eIF4AIII [19]. In the crystal complexes with either ADP or AMP the loop closes up, resulting in a shift in Cα-atom positions by up to 3.5 Å between the ATP- and the AMP-states, or by up to 2.5 Å between the ADP- and the AMP-state (Figure 3A). Thus the conformation of the P-loop is determined by the nucleotide phosphates, and longer phosphate tails result in a more open loop. This observation agrees with previous results [24]. Motif III follows the P-loop transition and its position changes by up to 3 Å toward the P-loop. Motifs Ia, Ib and II seem unaffected by the state of ATP hydrolysis, and their conformations remain unchanged even in the crystal structures in which the nucleotide binding site is not occupied.
Figure 3. Details of the ATP binding sites.
(A) Superposition of multiple DEAD-domains to illustrate variability in P-loop (Motif I) conformations. P-loops in DEAD-domain structures with bound phosphate (yellow), with bound AMP (orange), with bound ADP (red), DDX19 P-loop with bound AMPPNP and Mg2+ (blue), DDX20 P-loop with bound AMPPNP (magenta), and P-loop in nucleotide-free eIF4A/DDX2A (green) are shown. Motifs I, II and III are indicated. (B) Two different conformations of the β- and γ-phosphates in the DDX20-AMPPNP complex. Side chains that interact with the AMPPNP are shown as balls-and-sticks. (C) DDX2B with a closed P-loop. The α-helix that follows the P-loop starts one turn earlier compared to other DEAD-domain structures shown. (D) Variability of interactions with the adenosine nucleotide. The adenosine moiety is coordinated through π-stacking interactions or hydrophobic interactions. Numbers denote the interaction surface, in Å2, between the nucleotide and the stacking side chain, as determined using the PISA server [25].
Two of the structures show unique P-loop conformations. The DDX2B structure features an α-helix 4 that is longer than in other helicases, and leads into an unusually closed P-loop conformation (Figure 3C). As a consequence, the ATP binding site is not visible on the surface of the DDX2B structure. This conformation is most likely induced by a crystal contact in this region.
The AMPPNP-bound DDX20 structure contains no metal ion (Figure 3B). Lack of γ-phosphate coordination by a metal ion leads to a shift in the position of the β- and γ-phosphates, which bind where the α- and β-phosphates are bound in other ATP complexes. Since the adenine base is coordinated in the usual fashion the α-phosphate and the sugar moiety are tilted out of the expected positions. This illustrates that DDX20 (and presumably other helicases) can bind ATP also in the absence of divalent cation. However, a divalent cation is needed to allow coordination of three phosphates in the correct geometry for catalysis.
Diversity in ATP coordination
Some of the side chains that interact with the nucleotides are not conserved, and most of these are found in the Q-motif. Three hydrogen bonds between the adenine ring and the protein ensure specific binding of adenosine nucleotides. These are formed by the conserved glutamine and the backbone carbonyl five residues upstream of the glutamine (Figure 3). The 6th residue upstream of the conserved glutamine is an aromatic residue in most DEAD-box helicases. Its side chain stacks with the nucleotide base, stabilizing it in its position. Interestingly this residue is not conserved: While phenylalanine is most common, DDX10 has a tyrosine and DDX47 has a tryptophan in the corresponding position. Moreover, an aromatic residue in this position is not obligatory: DDX53 features an isoleucine, with weaker van-der-Waals interactions with the adenosine ring than the base stacking interactions with the aromatic side chains (Figure 3D). We analyzed the protein-nucleotide binding interfaces in these crystal structures using the PISA server [25]. This analysis showed that, while the overall ligand interface areas are similar in the different nucleotide complexes, the contribution by the base stacking residues vary considerably. The variability in the stacking residue position may reflect different needs for conformational flexibility in this region of the DEAD-domains.
Helicase domain variation
The helicase domain contributes to nucleotide coordination via motifs V and VI. From the closed state DDX19 structure [22] it is apparent that four side chains are of particular importance: The aspartate of motif V coordinates the O3′ of the ribose. The second arginine side chain of motif VI (HRxGRxGR) interacts with the γ-phosphate. The third arginine, which is also the putative arginine finger during ATP hydrolysis, coordinates all three ATP phosphates. The variable residue that follows this arginine coordinates the adenosine ring by different means. In the DDX19 helicase domain a phenylalanine stacks with the adenosine rings. A superposition of DDX19 with the DDX25 and DDX41 helicase domains shows that in the latter two structures part of motif VI is not visible in the electron density, indicating its flexibility. The conserved motifs IV and VI superpose well, whereas motif V shows different conformations in all three structures (Figure 1B).
The only part of motif IV that is not flexible is the histidine-arginine pair, and it superposes in all three crystal structures. The arginine points to a negatively charged pocked formed in part by side chains from motifs IV and V in the inside of the helicase domain. The aliphatic part of the arginine side chain makes a hydrophobic contact with the phenylalanine of motif IV. In the two-domain closed state structures the histidine interacts with the SAT motif from the helicase domain. Therefore, the SAT motif is indirectly linked to the ATP binding site as well as to the RNA binding sites of both domains. This explains the central importance of this motif in the coupling of ATP hydrolysis and RNA unwinding [26]. In SAT-motif mutants of eIF4A the ATPase and helicase activities were uncoupled [27]: SAT-to-AAA mutant protein is capable of binding RNA in an ATP dependent manner, but lacks RNA unwinding activity.
Conserved and variable parts constitute the RNA binding site
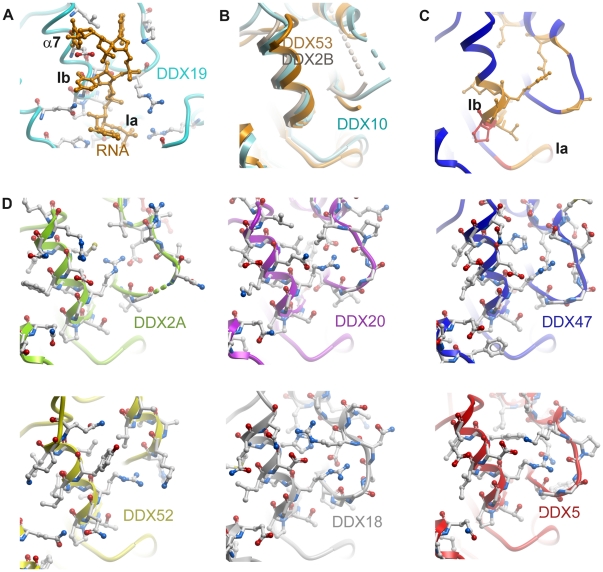
The available atomic resolution structures of DEAD-box helicases with bound RNA [19]–[22] show that the DEAD-domain contributes to RNA binding through two conserved and one variable structural element: (i) Motif Ia; (ii) α-helix 7, with its conserved motif Ib; and (iii) the variable loop connecting β-sheets 3 and 4. These interactions are illustrated for DDX19 in Figure 4A: While the variable loop clamps the RNA substrate in a specific conformation, motifs Ia and Ib each coordinate an RNA-backbone phosphate and induce a tilt of one or more RNA bases.
Figure 4. RNA binding cleft on DEAD domains.
(A) DDX19 (light blue; PDB entry 3G0H) with bound RNA (light orange). RNA-interacting side chains are shown. (B) Flexible regions in DDX2B, DDX10 and DDX53 for which the electron density was not visible. (C) Sequence conservation in the RNA binding cleft, mapped onto the DDX47 structure (red, conserved; orange, partly conserved). (D) RNA binding sites of selected DEAD-domains to illustrate their sequence variation.
Conserved motifs Ia and Ib of DDX19 and all DEAD-domain structures described here superimpose perfectly (Figure 4). This leads us to conclude that RNA substrates are bound in a similar conformation by the conserved motifs of all these DEAD-domains. The variability in part of the RNA binding sites (Figure 4D), on the other hand, implies that different helicases could stabilize specific RNA conformations. In addition, variable side chain contribution may also reflect optimal recognition of specific nucleotide sequences.
Inspection of the RNA complexes of DDX19, vasa, and eIF4AIII [19]–[22] shows that the conserved motif that makes the most extensive contacts with the RNA-backbone phosphates is motif Ib. In two of our DEAD-domain crystal structures, anions from the crystallization buffers are bound to motif Ib (a sulfate in DDX5, and a phosphate in DDX47) highlighting the ability of this motif to bind polyanions.
Mechanism for unblocking of the RNA binding site
Our crystal structures of both DEAD-domains and helicase domains in isolation reveal that the RNA binding site on each domain is in a conformation that is incompetent to bind RNA substrate. In the free helicase domain structures motif V, an important RNA backbone interaction site [19]–[22] is in a binding incompetent conformation. In the closed state, an RNA binding competent conformation is stabilized by the interaction of the conserved arginine of motif V with the C-terminal aspartic acid of the DEAD-motif (Figure 5C). In all single DEAD-domain structures, α-helix 8 has adopted a position that would block the RNA binding site. By contrast, upon cleft closure in the two-domain ATP analog and RNA complexes, α-helix 8 has moved out of the RNA binding site (Figure 5).
Figure 5. Details of the RNA binding cleft.
(A) DDX19 closed state structure (PDB entry 3G0H). DDX19-bound RNA, Mg2+-ion and AMPPNP are in orange. (B) Superposition of several DEAD domain structures showing a conserved conformation of α-helix 8. (C) Interactions between the DEAD and helicase domains of DDX19. (D) “Top-down” view of the open and closed RNA binding cleft. DDX5 (red), the ATP-state of DDX19 (blue) and DDX41 (orange) are shown. RNA (superposed from the DDX19 complex structure) is shown in light orange. (E) Surface representation of the DDX19-RNA complex. Note that α-helix 8 does not come in contact with the RNA substrate. (F) Surface representation of DDX5 and the superposed RNA from the DDX19 complex structure. Note that α-helix 8 would clash with the RNA substrate.
Thus, superposition of single DEAD-domain structures onto the closed state structures of DDX19 and eIF4AIII suggests involvement of α-helix 8 in the formation of a competent RNA binding site. How is α-helix 8 displaced to allow access to the RNA substrate binding site? No direct interaction between α-helix 8 and the RNA have been observed; thus displacement of α-helix 8 by the RNA substrate itself seems unlikely. Also, binding of ATP itself cannot cause α-helix 8 rotation out of the RNA site: The DEAD-motif is the only link between the nucleotide and α-helix 8, but the state of nucleotide hydrolysis does not influence the conformation of the DEAD motif (motif II; Figure 1A, 3A).
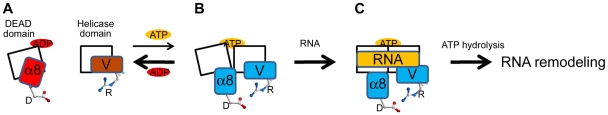
Instead, we propose direct involvement of the helicase domain in the activation of the RNA binding site on the DEAD-domain: In the complex structures, the conserved arginine of motif V in the helicase domain forms a salt bridge with the C-terminal aspartic acid of the DEAD-motif, which is also the terminal residue of α-helix 8 (Figure 5C, 5D). This interaction stabilizes a conformation where α-helix 8 is rotated out of the RNA binding site (Figure 5D). We propose that ATP binding primes the helicases for RNA substrate binding by bringing the domains together to allow motif V to push α-helix 8 out of the RNA site on the DEAD-domain. RNA binding to the DEAD-domain then completes cleft closure to allow formation of an active ATPase site (Figure 6).
Figure 6. Schematic model for the regulation of RNA binding by α-helix 8 of DEAD-box helicases.
(A) In the isolated domains, reflecting the open and substrate free states, the RNA binding sites are partially blocked by α-helix 8 in the DEAD-domain and motif V in the helicase domain. The aspartate indicated in the DEAD-domain is the second D of the DEAD sequence in motif II. The arginine indicated in the helicase domain is a conserved residue in motif V. Both residues are marked by asterisks in Fig 2. (B) Binding of ATP favors closure of the cleft, facilitating interaction of α-helix 8 with motif V across the cleft, thereby removing the blockage of the RNA binding site. (C) The closed cleft conformation is stabilized by RNA substrate to the competent site, allowing ATP hydrolysis to proceed.
This model of cleft closure and helicase activation through regulation of α-helix 8 can reconcile published data. Moreover, it can explain how substrate release in the post-hydrolysis state is achieved. DEAD-box helicases typically bind ADP with higher affinity that ATP [28]–[31], and binding of ATP and RNA are cooperative [31]–[35]. Thus, the binding energy of the RNA-protein interaction likely stabilizes a strained conformation that is competent for ATP hydrolysis. Conversely, relief of this strain upon ATP hydrolysis and phosphate release likely drives RNA substrate remodeling [36]. According to our comparative structural analysis, ATP hydrolysis and phosphate release would allow α-helix 8 to move back into its original position, releasing the RNA substrate and switching back to a binding incompetent RNA site on the DEAD domain.
DExH-box RNA helicases differ in some aspects from the DEAD-motif containing helicases. The hepatitis C virus DExH-box helicase NS3 binds RNA in the absence of ATP [37]. DExH helicase NPH-II unwinds RNA in a processive fashion [38] and thus stays bound to the RNA after each unwinding step. Our model for the role of α-helix 8 in cleft closure of DEAD-proteins is consistent also with these properties of DExH-box RNA helicases. Whereas α-helix 8 is conserved in all DEAD-box proteins, it is missing in the DExH-box proteins (refs. [37], [39]–[42] and references therein). Moreover, the DEAD-motif aspartic acid side chain that mediates opening of the RNA binding site (Figure 5) is replaced by the histidine of the DExH-motif. Thus apparently, in the absence of α-helix 8 that may block the RNA site, this terminal aspartic acid is redundant, and the histidine that substitutes it fulfills a different function [42]. We conclude that DEAD- and DExH-box helicases differ significantly in the coupling of the RNA binding event to the conformational cycle of the two RecA domains.
Materials and Methods
All proteins were expressed in Escherichia coli as N-terminally hexahistidine tagged fusion proteins, and purified by nickel affinity chromatography and gel filtration. Proteins were crystallized in sitting drops at 4°C or 20°C. X-ray diffraction data were collected at the APS (Chicago, USA), the BESSY (Berlin, Germany), the Diamond (Oxfordshire, UK), the ESRF (Grenoble, France), and the MaxLab (Lund, Sweden) synchrotron radiation facilities. Data were indexed and integrated using XDS [43], MOSFLM [44], or DENZO [45], and scaled using XSCALE [43], SCALA [46] or SCALEPACK [45]. Structures were solved by molecular replacement using PHASER [47] or MOLREP [48], and refined using REFMAC [49]. Refinement rounds were complemented with manual rebuilding using COOT [50].
Detailed Materials and Methods can be found in Table S1.
Accession codes
The coordinates have been deposited in the Proteins Data Bank with accession codes 2G9N, 3BOR, 3FE2, 2PL3, 3LY5, 3B7G, 2RB4, 2P6N, 3BER, 3DKP, and 3IUY.
Supporting Information
Materials and methods detailing protein expression and purification, crystallization, X-ray data processing.
(0.15 MB PDF)
Acknowledgments
We thank the beamline staff at the APS, BESSY, Diamond, ESRF, and MaxLab synchrotron radiation facilities for excellent support. We would also like to acknowledge our colleagues at the Structural Genomics Consortium.
Footnotes
Competing Interests: Clarifying statements: 1. The Structural Genomics Consortium (SGC) is a not-for-profit organization that receives funding from a funder consortium that includes commercial sources (GlaxoSmithKline and Merck & Co., Inc.). This circumstance does not alter the authors' adherence to all the PLoS ONE policies on sharing data and materials. The SGC and its scientists are committed to making their research outputs (materials and knowledge) available without restriction on use. This means that the SGC will promptly place its results in the public domain and will not agree to file for patent protection on any of its research outputs. It will seek the same commitment from any research collaborator. 2. One of the authors (LHS) is currently employed by a commercial company. As the role of this author in the current study was terminated before her affiliation with that company, this circumstance does not alter the authors' adherence to all the PLoS ONE policies on sharing data and materials.
Funding: The Structural Genomics Consortium is a registered charity (number 1097737) that receives funds from the Canadian Institutes for Health Research, the Canada Foundation for Innovation, Genome Canada through the Ontario Genomics Institute, GlaxoSmithKline, Karolinska Institutet, the Knut and Alice Wallenberg Foundation, the Ontario Innovation Trust, the Ontario Ministry for Research and Innovation, Merck & Co., Inc., the Novartis Research Foundation, the Swedish Agency for Innovation Systems, the Swedish Foundation for Strategic Research and the Wellcome Trust. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Linder P, Tanner NK, Banroques J. From RNA helicases to RNPases. Trends Biochem Sci. 2001;26:339–341. doi: 10.1016/s0968-0004(01)01870-9. [DOI] [PubMed] [Google Scholar]
- 2.Schwer B. A new twist on RNA helicases: DExH/D box proteins as RNPases. Nat Struct Biol. 2001;8:113–116. doi: 10.1038/84091. [DOI] [PubMed] [Google Scholar]
- 3.Cordin O, Banroques J, Tanner NK, Linder P. The DEAD-box protein family of RNA helicases. Gene. 2006;367:17–37. doi: 10.1016/j.gene.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 4.Pyle AM. Translocation and unwinding mechanisms of RNA and DNA helicases. Annu Rev Biophys. 2008;37:317–336. doi: 10.1146/annurev.biophys.37.032807.125908. [DOI] [PubMed] [Google Scholar]
- 5.Clark EL, Coulson A, Dalgliesh C, Rajan P, Nicol SM, et al. The RNA helicase p68 is a novel androgen receptor coactivator involved in splicing and is overexpressed in prostate cancer. Cancer Res. 2008;68:7938–7946. doi: 10.1158/0008-5472.CAN-08-0932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fuller-Pace FV, Ali S. The DEAD box RNA helicases p68 (Ddx5) and p72 (Ddx17): novel transcriptional co-regulators. Biochem Soc Trans. 2008;36:609–612. doi: 10.1042/BST0360609. [DOI] [PubMed] [Google Scholar]
- 7.Wortham NC, Ahamed E, Nicol SM, Thomas RS, Periyasamy M, et al. The DEAD-box protein p72 regulates ERalpha-/oestrogen-dependent transcription and cell growth, and is associated with improved survival in ERalpha-positive breast cancer. Oncogene. 2009;28:4053–4064. doi: 10.1038/onc.2009.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abdelhaleem M. Do human RNA helicases have a role in cancer? Biochim Biophys Acta. 2004;1704:37–46. doi: 10.1016/j.bbcan.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 9.Schroder M. Human DEAD-box protein 3 has multiple functions in gene regulation and cell cycle control and is a prime target for viral manipulation. Biochem Pharmacol. 2010;79:297–306. doi: 10.1016/j.bcp.2009.08.032. [DOI] [PubMed] [Google Scholar]
- 10.Yount JS, Gitlin L, Moran TM, Lopez CB. MDA5 participates in the detection of paramyxovirus infection and is essential for the early activation of dendritic cells in response to Sendai Virus defective interfering particles. J Immunol. 2008;180:4910–4918. doi: 10.4049/jimmunol.180.7.4910. [DOI] [PubMed] [Google Scholar]
- 11.Takeuchi O, Akira S. MDA5/RIG-I and virus recognition. Curr Opin Immunol. 2008;20:17–22. doi: 10.1016/j.coi.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 12.Rehwinkel J, Reis e Sousa C. RIGorous detection: exposing virus through RNA sensing. Science. 2010;327:284–286. doi: 10.1126/science.1185068. [DOI] [PubMed] [Google Scholar]
- 13.Crumpacker CS, Schaffer PA. New anti-HSV therapeutics target the helicase-primase complex. Nat Med. 2002;8:327–328. doi: 10.1038/nm0402-327. [DOI] [PubMed] [Google Scholar]
- 14.Frick DN. Helicases as antiviral drug targets. Drug News Perspect. 2003;16:355–362. doi: 10.1358/dnp.2003.16.6.829307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kwong AD, Rao BG, Jeang KT. Viral and cellular RNA helicases as antiviral targets. Nat Rev Drug Discov. 2005;4:845–853. doi: 10.1038/nrd1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Manfroni G, Paeshuyse J, Massari S, Zanoli S, Gatto B, et al. Inhibition of subgenomic hepatitis C virus RNA replication by acridone derivatives: identification of an NS3 helicase inhibitor. J Med Chem. 2009;52:3354–3365. doi: 10.1021/jm801608u. [DOI] [PubMed] [Google Scholar]
- 17.Cencic R, Carrier M, Galicia-Vazquez G, Bordeleau ME, Sukarieh R, et al. Antitumor activity and mechanism of action of the cyclopenta[b]benzofuran, silvestrol. PLoS One. 2009;4:e5223. doi: 10.1371/journal.pone.0005223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Le Hir H, Andersen GR. Structural insights into the exon junction complex. Curr Opin Struct Biol. 2008;18:112–119. doi: 10.1016/j.sbi.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 19.Andersen CB, Ballut L, Johansen JS, Chamieh H, Nielsen KH, et al. Structure of the exon junction core complex with a trapped DEAD-box ATPase bound to RNA. Science. 2006;313:1968–1972. doi: 10.1126/science.1131981. [DOI] [PubMed] [Google Scholar]
- 20.Sengoku T, Nureki O, Nakamura A, Satoru KI, Yokoyama S. Structural basis for RNA unwinding by the DEAD-box protein Drosophila vasa. Cell. 2006;125:287–300. doi: 10.1016/j.cell.2006.01.054. [DOI] [PubMed] [Google Scholar]
- 21.Bono F, Ebert J, Lorentzen E, Conti E. The crystal structure of the exon junction complex reveals how it maintains a stable grip on mRNA. Cell. 2006;126:713–725. doi: 10.1016/j.cell.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 22.Collins R, Karlberg T, Lehtio L, Schütz P, van den Berg S, et al. The DEXD/H-box RNA helicase DDX19 is regulated by an {alpha}-helical switch. J Biol Chem. 2009;284:10296–10300. doi: 10.1074/jbc.C900018200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hilbert M, Karow AR, Klostermeier D. The mechanism of ATP-dependent RNA unwinding by DEAD box proteins. Biol Chem. 2009;390:1237–1250. doi: 10.1515/BC.2009.135. [DOI] [PubMed] [Google Scholar]
- 24.Högbom M, Collins R, van den Berg S, Jenvert RM, Karlberg T, et al. Crystal structure of conserved domains 1 and 2 of the human DEAD-box helicase DDX3X in complex with the mononucleotide AMP. J Mol Biol. 2007;372:150–159. doi: 10.1016/j.jmb.2007.06.050. [DOI] [PubMed] [Google Scholar]
- 25.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;3872:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 26.Banroques J, Cordin O, Doere M, Linder P, Tanner NK. A conserved phenylalanine of motif IV in superfamily 2 helicases is required for cooperative, ATP-dependent binding of RNA substrates in DEAD-box proteins. Mol Cell Biol. 2008;28:3359–3371. doi: 10.1128/MCB.01555-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pause A, Sonenberg N. Mutational analysis of a DEAD box RNA helicase: the mammalian translation initiation factor eIF-4A. EMBO J. 1992;11:2643–2654. doi: 10.1002/j.1460-2075.1992.tb05330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Talavera MA, De La Cruz EM. Equilibrium and kinetic analysis of nucleotide binding to the DEAD-box RNA helicase DbpA. Biochemistry. 2005;44:959–970. doi: 10.1021/bi048253i. [DOI] [PubMed] [Google Scholar]
- 29.Karow AR, Theissen B, Klostermeier D. Authentic interdomain communication in an RNA helicase reconstituted by expressed protein ligation of two helicase domains. FEBS J. 2007;274:463–473. doi: 10.1111/j.1742-4658.2006.05593.x. [DOI] [PubMed] [Google Scholar]
- 30.Henn A, Cao WX, Hackney DD, De La Cruz EM. The ATPase cycle mechanism of the DEAD-box rRNA helicase, DbpA. J Mol Biol. 2008;377:193–205. doi: 10.1016/j.jmb.2007.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Theissen B, Karow AR, Kohler J, Gubaev A, Klostermeier D. Cooperative binding of ATP and RNA induces a closed conformation in a DEAD box RNA helicase. Proc Natl Acad Sci U S A. 2008;105:548–553. doi: 10.1073/pnas.0705488105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Polach KJ, Uhlenbeck OC. Cooperative binding of ATP and RNA substrates to the DEAD/H protein DbpA. Biochemistry. 2002;41:3693–3702. doi: 10.1021/bi012062n. [DOI] [PubMed] [Google Scholar]
- 33.Mohr S, Stryker JM, Lambowitz AM. A DEAD-box protein functions as an ATP-dependent RNA chaperone in group I intron splicing. Cell. 2002;109:769–779. doi: 10.1016/s0092-8674(02)00771-7. [DOI] [PubMed] [Google Scholar]
- 34.Cordin O, Tanner NK, Doere M, Linder P, Banroques J. The newly discovered Q motif of DEAD-box RNA helicases regulates RNA-binding and helicase activity. EMBO J. 2004;23:2478–2487. doi: 10.1038/sj.emboj.7600272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Elles LM, Uhlenbeck OC. Mutation of the arginine finger in the active site of Escherichia coli DbpA abolishes ATPase and helicase activity and confers a dominant slow growth phenotype. Nucl Ac Res. 2008;36:41–50. doi: 10.1093/nar/gkm926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nielsen KH, Chamieh H, Andersen CBF, Fredslund F, Hamborg K, et al. Mechanism of ATP turnover inhibition in the EJC. RNA. 2009;15:67–75. doi: 10.1261/rna.1283109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gu M, Rice CM. Three conformational snapshots of the hepatitis C virus NS3 helicase reveal a ratchet translocation mechanism. Proc Natl Acad Sci U S A. 2010;107:521–528. doi: 10.1073/pnas.0913380107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jankowsky E, Gross CH, Shuman S, Pyle AM. The DExH protein NPH-II is a processive and directional motor for unwinding RNA. Nature. 2000;403:447–451. doi: 10.1038/35000239. [DOI] [PubMed] [Google Scholar]
- 39.He Y, Andersen GR, Nielsen KH. Structural basis for the function of DEAH helicases. EMBO Rep. 2010;11:180–186. doi: 10.1038/embor.2010.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Walbott H, Mouffok S, Capeyrou R, Lebaron S, Humbert O, et al. Prp43p contains a processive helicase structural architecture with a specific regulatory domain. EMBO J. 2010;29:2194–2202. doi: 10.1038/emboj.2010.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jackson RN, Klauer AA, Hintze BJ, Robinson H, van Hoof A, et al. The crystal structure of Mtr4 reveals a novel arch domain required for rRNA processing. EMBO J. 2010;29:2205–2216. doi: 10.1038/emboj.2010.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schütz P, Wahlberg E, Karlberg T, Hammarström M, Collins R, et al. Crystal structure of human RNA helicase A (DHX9): Structural basis for unselective nucleotide base binding in a DEAD-box variant protein. J Mol Biol. 2010;400:768–782. doi: 10.1016/j.jmb.2010.05.046. [DOI] [PubMed] [Google Scholar]
- 43.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Cryst. 1993;26:795–800. [Google Scholar]
- 44.Leslie AGW. Recent changes to the MOSFLM package for processing film and image plate data. 1992. No. 26.
- 45.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Meth Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 46.Evans P. Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 47.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Crystallogr D Biol Crystallogr. 2005;61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 48.Vagin A, Teplyakov A. Molecular replacement with MOLREP. Acta Crystallogr D Biol Crystallogr. 2010;66:22–25. doi: 10.1107/S0907444909042589. [DOI] [PubMed] [Google Scholar]
- 49.Murshudov GN, Vagin A, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 50.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 51.Pause A, Methot N, Svitkin Y, Merrick WC, Sonenberg N. Dominant negative mutants of mammalian translation initiation factor eIF-4A define a critical role for eIF-4F in cap-dependent and cap-independent initiation of translation. EMBO J. 1994;13:1205–1215. doi: 10.1002/j.1460-2075.1994.tb06370.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gingras AC, Raught B, Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem. 1999;68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- 53.Marintchev A, Edmonds KA, Marintcheva B, Hendrickson E, Oberer M, et al. Topology and regulation of the human eIF4A/4G/4H helicase complex in translation initiation. Cell. 2009;136:447–460. doi: 10.1016/j.cell.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Grifo JA, Abramson RD, Satler CA, Merrick WC. RNA-stimulated ATPase activity of eukaryotic initiation factors. J Biol Chem. 1984;259:8648–8654. [PubMed] [Google Scholar]
- 55.Moerke NJ, Aktas H, Chen H, Cantel S, Reibarkh MY, et al. Small-molecule inhibition of the interaction between the translation initiation factors eIF4E and eIF4G. Cell. 2007;128:257–267. doi: 10.1016/j.cell.2006.11.046. [DOI] [PubMed] [Google Scholar]
- 56.Avdulov S, Li S, Michalek V, Burrichter D, Peterson M, et al. Activation of translation complex eIF4F is essential for the genesis and maintenance of the malignant phenotype in human mammary epithelial cells. Cancer Cell. 2004;5:553–563. doi: 10.1016/j.ccr.2004.05.024. [DOI] [PubMed] [Google Scholar]
- 57.Lindqvist L, Oberer M, Reibarkh M, Cencic R, Bordeleau ME, et al. Selective pharmacological targeting of a DEAD box RNA helicase. PLoS One. 2008;3:e1583. doi: 10.1371/journal.pone.0001583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Arai Y, Hosoda F, Kobayashi H, Arai K, Hayashi Y, et al. The inv(11)(p15q22) chromosome translocation of de novo and therapy-related myeloid malignancies results in fusion of the nucleoporin gene, NUP98, with the putative RNA helicase gene, DDX10. Blood. 1997;89:3936–3944. [PubMed] [Google Scholar]
- 59.Nakamura T, Yamazaki Y, Hatano Y, Miura I. NUP98 is fused to PMX1 homeobox gene in human acute myelogenous leukemia with chromosome translocation t(1;11)(q23;p15). Blood. 1999;94:741–747. [PubMed] [Google Scholar]
- 60.Grandori C, Mac J, Siebelt F, Ayer DE, Eisenman RN. Myc-Max heterodimers activate a DEAD box gene and interact with multiple E box-related sites in vivo. EMBO J. 1996;15:4344–4357. [PMC free article] [PubMed] [Google Scholar]
- 61.Dubaele S, Chene P. Cellular studies of MrDb (DDX18). Oncol Res. 2007;16:549–556. doi: 10.3727/000000007783630015. [DOI] [PubMed] [Google Scholar]
- 62.Shpargel KB, Matera AG. Gemin proteins are required for efficient assembly of Sm-class ribonucleoproteins. Proc Natl Acad Sci U S A. 2005;102:17372–17377. doi: 10.1073/pnas.0508947102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cauchi RJ, Davies KE, Liu JL. A motor function for the DEAD-box RNA helicase, Gemin3, in Drosophila. PLoS Genet. 2008;4:e1000265. doi: 10.1371/journal.pgen.1000265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Almstead LL, Sarnow P. Inhibition of U snRNP assembly by a virus-encoded proteinase. Genes Dev. 2007;21:1086–1097. doi: 10.1101/gad.1535607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tsai-Morris CH, Sheng Y, Lee E, Lei KJ, Dufau ML. Gonadotropin-regulated testicular RNA helicase (GRTH/Ddx25) is essential for spermatid development and completion of spermatogenesis. Proc Natl Acad Sci U S A. 2004;101:6373–6378. doi: 10.1073/pnas.0401855101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dufau ML, Tsai-Morris CH. Gonadotropin-regulated testicular helicase (GRTH/DDX25): an essential regulator of spermatogenesis. Trends Endocrinol Metab. 2007;18:314–320. doi: 10.1016/j.tem.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 67.Sekito A, Taira T, Niki T, Iguchi-Ariga SM, Ariga H. Stimulation of transforming activity of DJ-1 by Abstrakt, a DJ-1-binding protein. Int J Oncol. 2005;26:685–689. [PubMed] [Google Scholar]
- 68.Sekiguchi T, Hayano T, Yanagida M, Takahashi N, Nishimoto T. NOP132 is required for proper nucleolus localization of DEAD-box RNA helicase DDX47. Nucleic Acids Res. 2006;34:4593–4608. doi: 10.1093/nar/gkl603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bohnsack MT, Kos M, Tollervey D. Quantitative analysis of snoRNA association with pre-ribosomes and release of snR30 by Rok1 helicase. EMBO Rep. 2008;9:1230–1236. doi: 10.1038/embor.2008.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cho B, Lee H, Jeong S, Bang YJ, Lee HJ, et al. Promoter hypomethylation of a novel cancer/testis antigen gene CAGE is correlated with its aberrant expression and is seen in premalignant stage of gastric carcinoma. Biochem Biophys Res Commun. 2003;307:52–63. doi: 10.1016/s0006-291x(03)01121-5. [DOI] [PubMed] [Google Scholar]
- 71.Diederichs K, Karplus PA. Improved R-factors for diffraction data analysis in macromolecular crystallography. Nat Struct Biol. 1997;4:269–275. doi: 10.1038/nsb0497-269. [DOI] [PubMed] [Google Scholar]
- 72.Arndt UW, Crowther RA, Mallett JF. A computer-linked cathode-ray tube microdensitometer for x-ray crystallography. J Sci Instrum. 1968;1:510–516. doi: 10.1088/0022-3735/1/5/303. [DOI] [PubMed] [Google Scholar]
- 73.Weiss M, Hilgenfeld R. On the use of the merging R factor as a quality indicator for X-ray data. J Appl Crystallogr. 1997:203–205. [Google Scholar]
- 74.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Materials and methods detailing protein expression and purification, crystallization, X-ray data processing.
(0.15 MB PDF)