Abstract
Background
Mosquitoes in the Culex pipiens complex are among the most medically important vectors for human disease worldwide and include major vectors for lymphatic filariasis and West Nile virus transmission. However, detailed genetic studies in the complex are limited by the number of genetic markers available. Here, we describe methods for the rapid and efficient identification and development of single locus, highly polymorphic microsatellite markers for Cx. pipiens complex mosquitoes via in silico screening of the Cx. quinquefasciatus genome sequence.
Methodology/Principal Findings
Six lab colonies representing four Cx. pipiens and two Cx. quinquefasciatus populations were utilized for preliminary assessment of 38 putative loci identified within 16 Cx. quinquefasciatus supercontig assemblies (CpipJ1) containing previously mapped genetic marker sequences. We identified and validated 12 new microsatellite markers distributed across all three linkage groups that amplify consistently among strains representing the complex. We also developed four groups of 3–5 microsatellite loci each for multiplex-ready PCR. Field collections from three cities in Indiana were used to assess the multiplex groups for their application to natural populations. All were highly polymorphic (Mean = 13.0 alleles) per locus and reflected high polymorphism information content (PIC) (Mean = 0.701). Pairwise FST indicated population structuring between Terre Haute and Fort Wayne and between Terre Haute and Indianapolis, but not between Fort Wayne and Indianapolis. In addition, we performed whole genome comparisons of microsatellite motifs and abundance between Cx. quinquefasciatus and the primary vectors for dengue virus and malaria parasites, Aedes aegypti and Anopheles gambiae, respectively.
Conclusions/Significance
We demonstrate a systematic approach for isolation and validation of microsatellites for the Cx. pipiens complex by direct screen of the Cx. quinquefasciatus genome supercontig assemblies. The genome density of microsatellites is greater in Cx. quinquefasciatus (0.26%) than in Ae. aegypti (0.14%), but considerably lower than in An. gambiae (0.77%).
Introduction
Mosquitoes in the Culex pipiens complex are major vectors for a number of important human pathogens including West Nile virus, St. Louis encephalitis virus, and Wuchereria bancrofti, a causative agent of lymphatic filariasis [1]–[3]. Cx. pipiens complex mosquitoes can be found on every continent except Antarctica [4], and include two widespread species: Cx. quinquefasciatus (Say 1823) and Cx. pipiens (Linnaeus 1758). Cx. quinquefasciatus inhabits tropical, subtropical, and warm temperate zones while Cx. pipiens inhabits temperate zones [4]. Sympatric populations occur where their ranges overlap [5], [6]. A recent study on the Cx. pipiens complex along a north-south transect in North America revealed hybrid populations as far north as Illinois and as far south as Alabama [6]. Distinct physiological differences between species in the Cx. pipiens complex are known and are thought to influence pathogen transmission as well as their geographic distribution [7], [8]. Identification of the genes contributing to these physiological processes could provide novel targets for genetic control methods. Though genetic marker development has facilitated construction of detailed linkage maps in dengue virus vector, Aedes aegypti [9]–[12] and the Plasmodium falciparum vector, Anopheles gambiae [13]–[16] mosquitoes, genetic studies based on linkage analyses in Cx. pipiens are limited due to the paucity of available marker loci [8], [17].
Microsatellites have been preferred as genetic markers due to their high polymorphism, co-dominance, and potential for high throughput analysis. Approximately 33 microsatellite markers have been developed for Cx. pipiens complex to date [18]–[21], yet none of these have been mapped to their respective linkage group. One disadvantage of microsatellite markers is they typically need to be developed for each species of interest [22]. Indeed, cross-species utility of individual microsatellite loci within the Cx. pipiens complex can be limited [20]. Because spatial distributions of individual Cx. pipiens complex members often overlap across freely interbreeding hybrid zones [5], [6], population studies would benefit greatly with availability of additional microsatellites loci with broad species compatibility.
Here we employ a method we recently reported for Ae. aegypti [23] toward the rapid and efficient development of microsatellite markers for Cx. pipiens complex mosquitoes by screening Cx. quinquefasciatus whole genome shotgun sequence (wgs) supercontig assemblies for microsatellite motifs [24]. We utilize 12 microsatellites to assess structuring among Cx. pipiens populations from three cities in Indiana, USA. Additionally, we perform a comparative genome analysis of microsatellite repeat motifs and abundance in Cx. pipiens, An. gambiae and Ae. aegypti.
Materials and Methods
Ethics Statement
Our protocol for maintenance and care of experimental animals was reviewed and approved by the Institutional Animal Care and Use Committee at the University of Notre Dame. Animals are maintained and cared for in the Freimann Life Science Center, an AAALAC accredited facility.
In silico identification of microsatellites
Sequences previously mapped using restriction fragment length polymorphism (RFLP) markers based on random cDNAs [8], [17] were used for BLASTn analysis against the Cx. quinquefasciatus whole genome sequence assembly (CpipJ1) at VectorBase [24]. Genome supercontigs containing the RFLP marker sequences were downloaded from VectorBase and screened with Tandem Repeats Finder (TRF) software using the default parameters [25]. TRF output data were evaluated for regions containing microsatellites with a period size of 2–4 bp and number of repeats less than 30.
Primer design
Sequences of 400–600 bp containing a microsatellite of interest were extracted from individual supercontigs and subjected to BLASTn analysis against the Cx. quinquefasciatus genome at VectorBase to determine the degree of repetitive sequences flanking the target microsatellite sequence. PCR primers were designed for those regions with minimal repetitive sequences using Primer3 v.4.0 [26], with a target amplicon size of 120–400 bp. The resulting primer sequences were subjected to BLASTn analysis against the Cx. quinquefasciatus genome to assess copy number and potential nontarget amplification. Primer adjustments were made with the assistance of OligoCalc [27]. In addition, BLASTn analysis was performed using PCR primer and amplicon sequences of previously described microsatellite loci [18]–[21] against the Cx. quinquefasciatus genome at VectorBase to determine if any of these loci were within supercontigs with known genetic map positions based on RFLP marker loci [8], [17].
Mosquito samples
Individuals from six laboratory colonies were utilized in the preliminary screening of all microsatellite loci. These included four colonies of Cx. pipiens (Gose, Shinkura, Shasta and South Bend strains) and two of Cx. quinquefasciatus (Boana and Johannesburg strains). Shinkura is an autogenous strain founded from samples collected in 1998 from Tokushima, Japan. The South Bend and Gose strains are described elsewhere [8]. The Johannesburg strain was the source for the Cx. quinquefasciatus genome project [24]. The Shasta and Boana strains were kindly provided by Anton Cornel, University of California at Davis.
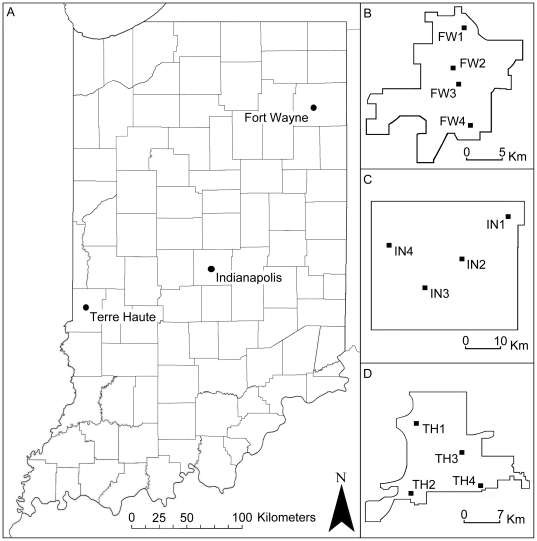
Field samples of Cx. pipiens populations were collected from three cities in Indiana (Fort Wayne, Indianapolis, and Terre Haute) during August-October, 2008 (Figure 1). Four collection sites within each city were chosen according to recommendations from local health department personnel (Figure 1, Table S1). Distances between cities ranged from ∼260 km (Ft. Wayne to Terre Haute) to ∼110 km (Indianapolis to Terre Haute). Egg rafts were collected using oviposition traps made from 17 L plastic pans (Sterilite) containing an aged (3–5 days) alfalfa-infusion [28]. Larvae (∼20 from each egg raft) were reared in 8 oz. plastic deli cups with ∼150 mL of aged tap water and fed a slurry of water and crushed Tetramin® fish food (Spectrum Brands Co.). Larvae were reared to the 3rd or 4th instar at which time they were identified to species level [29] and preserved in 95% ethanol.
Figure 1. Maps showing the spatial relationships among the three cities sampled in Indiana and locations of the collection sites within city boundaries.
A: City locations in Indiana. B: Fort Wayne. C: Indianapolis. D: Terre Haute. Coordinates listed in Table S1.
DNA extraction and PCR amplification
DNA extractions on individuals from the laboratory colonies were performed using a simple alkaline method [30]. DNA from each alkaline extraction was suspended in a final volume of 1500 µL containing 0.01 M NaOH and 0.018 M Tris-HCL, pH 8.0. Genomic DNA extractions from field samples were performed using the DNeasy® Tissue Kit (Qiagen) or a standard phenol/chloroform method [31]. Only one larva from an egg raft was extracted for PCR amplification.
PCR amplification was performed in 25 µL reactions in 96-well PCR plates (Dot Scientific). Each reaction contained 1X Taq buffer (50 mM KCl, 10 mM Tris pH 9.0, 0.1% Triton X), 1.5 mM MgCl2, 200 µM dNTPs, 5 pmoles each primer, 1 unit of Taq DNA polymerase, and 1 µL of genomic DNA (∼20 ng). Thermal cycling was performed using Mastercycler® thermocyclers (Eppendorf) under the following conditions: initial denaturation for 5 min at 94°C followed by 30 cycles of denaturation for 1 min at 94°C, annealing for 1 min at 60°C, extension for 2 min at 72°C, with a final extension for 10 min at 72°C.
Preliminary screening and multiplex PCR
Preliminary assessment of microsatellites for PCR amplification and copy number was performed by size fractionation of PCR products by electrophoresis in 2% agarose gels stained with ethidium bromide and visualized using UV light. Microsatellites amplifying across at least five of the six lab strains with single copy amplicons were assessed for allelic polymorphism on 6% denaturing polyacrylamide gels using the GenePrint® STR System (Promega). Sequences of single-copy microsatellite loci were submitted to the GenBank STS database (Table S2). Multiplexes were assembled according to amplicon size and tested on two individuals from each lab strain (n = 12) and three individuals from each of the three cities sampled (n = 9) by size fractionation using the 3730 Genetic Analyzer (Applied Biosystems) [32]. Mendelian inheritance was assessed based on conformity to Hardy-Weinberg expectations in samples from the Johannesburg (n = 28) and South Bend (n = 28) colonies.
Genotyping
Fluorophore-labeled (6-FAM®, HEX®, NED®) forward primers were used in PCR amplification for fragment analysis as described above. Prior to fragment analysis, multiplex PCR products were diluted 1∶10 in sterile H2O, and 1 µL of this dilution was added to 9 µL of a mixture containing 1 mL HiDi Formamide® (Applied Biosystems) and 15 µL ROX 400HD® (Applied Biosystems) in 96-well PCR plates. The samples were denatured for 2 minutes at 94°C and immediately placed on ice. Amplification products were sized using the ABI PRISM 3730 Genetic Analyzer (Applied Biosystems) and ROX 400HD size standard. Alleles were called using GENEMAPPER® v.4.0 software (Applied Biosystems), with subsequent visual verification of each sample.
Comparative genome analysis
The whole genome sequence assemblies of Cx. quinquefasciatus (CpipJ1.2), Ae. aegypti (AaegL1.1) and An. gambiae (AgamP3.4) were scanned for microsatellite motifs using the SciRoKo3.4 software program [33]. Our search was restricted to identify perfect di-, tri-, tetra-, penta- and hexa-nucleotide repeats with no less than 7, 5, 4, 4 and 4 repeats in each, respectively. The flanking regions of microsatellites (200 bp either side) were extracted using the ‘Little Helper’ module of SciRoKo. The absolute counts and average motif lengths in each microsatellite category obtained from the program output were used to calculate the relative proportions of microsatellite sequences in each genome.
Statistical analysis
Genetic diversity among the field populations was based on the observed and expected heterozygote frequencies and the number of alleles at each locus. ARLEQUIN v3.0 [34] was used to calculate FIS, pairwise FST and AMOVA following Weir and Cockerham [35] and to perform an exact test of Hardy-Weinberg (HW) equilibrium following Guo and Thompson [36]. GENEPOP 4.0 [37], [38] was used to test for isolation by distance using a Mantel test. Polymorphism information content (PIC) was calculated using Excel Microsatellite Toolkit [39]. The BOTTLENECK software program was used to assess the microsatellite data for evidence of recent population reductions based on gene diversity and allele frequency distributions [40].
Results
Development of microsatellite loci
From our preliminary screening of 16 Cx. quinquefasciatus whole genome sequence supercontigs containing previously mapped RFLP marker loci [8], [17] we selected 38 putative microsatellite loci for further evaluation. These represented five dinucleotide, eight trinucleotide and 1 tetranucleotide motifs (Table 1). The majority of microsatellites were dinucleotide repeats, and among these, the TG/AC motif was the most common. We identified 12 loci within eleven supercontigs that amplified consistently, were single copy and polymorphic when tested in individuals from six laboratory colonies representing both Cx. pipiens and Cx. quinquefasciatus populations derived from diverse sites worldwide (Table S3). Of these 12 loci, 9 (75%) comprise dinucleotide repeats, while the remaining three loci comprise trinucleotide repeats. Another 21 microsatellites, while single copy, showed strain-specific amplification, were monomorphic or did not have alleles within HW expectations thus limiting their potential utility across the species complex. With the remaining five microsatellites, no amplification was obtained with any of the strains. Only one of the 16 supercontigs examined (3.626) did not contain microsatellite motifs. BLASTn analysis of microsatellites against Cx. quinquefasciatus transcripts (CpipJ1.2 Gene Build) indicated that C99TGT1 and C177GAA1 were within CPIJ005634 and CPIJ008257, respectfully, while no other microsatellites were within coding regions [24]. Additionally, BLASTn analysis indicated that none of the previously reported microsatellites were within these 16 supercontigs, but that two of them are located in other supercontigs with known genetic marker loci (CxpGT4, gb-AY423738, supercontig 3.5, chromosome 2-36.1; CxqTri4, gb-AY958079, supercontig 3.208, chromosome 3-26.3) [24]. To improve the throughput and minimize the cost of microsatellite genotyping, individuals from the six laboratory colonies and Cx. pipiens field collections were used to develop four PCR multiplexes consisting of 3–5 microsatellite primer sets (Table 2). These include one locus on chromosome 1, seven loci on chromosome 2, and four loci on chromosome 3. Except for C48GTT1 and C48CGA1, which are on the same supercontig, the loci on chromosome 2 are distributed across 76.4 cM out of a total of 85.9 cM, while the the loci on chromosome 3 are distributed across 47.9 cM out of a total of 79.2 cM (see Table S3). The number of repeats, number of alleles and heterozygosities observed per locus are similar to that observed for Cx. pipiens microsatellites described previously [7], [18]–[21], [41].
Table 1. Microsatellite loci PCR screen results categorized by repeat motif.
| Strain-specific | No | |||
| Polymorphic | Monomorphic | Amplification | Amplification | |
| Repeat | (n = 15) | (n = 5) | (n = 11) | (n = 6) |
| Dinucleotide repeats | ||||
| AG/TC | 2 | 1 | ||
| AT/TA | 1 | 1 | ||
| CA/GT | 3 | 2 | 1 | |
| CT/GA | 1 | 1 | 1 | |
| TG/AC | 6 | 2 | 3 | 3 |
| Trinucleotide repeats | ||||
| ATC/TAG | 1 | |||
| CAA/GTT | 1 | |||
| CGA/GCT | 1 | |||
| CGC/GCG | 1 | |||
| CGT/GCA | 1 | |||
| GAA/CTT | 1 | |||
| GAC/CTG | 1 | |||
| TGT/ACA | 1 | |||
| Tetranucleotide repeats | ||||
| ACAT/TGTA | 1 |
Table 2. Multiplex-ready PCR groups.
| Group | Microsatellite locus | Map locationa | Predicted amplicon size (bp)b | Fluorochrome |
| CX1 | C177CA1 | 2-76.4 | 130 | HEX® |
| C68GA1 | 2-9.6 | 154 | 56-FAM® | |
| C127TC1 | 1-0.0 | 178 | NED® | |
| C99TGT1 | 3-17.9 | 214 | HEX | |
| C65AC1 | 2-15.9 | 305 | 56-FAM | |
| CX2 | C205TG1 | 3-18.5 | 150 | 56-FAM |
| C134AC1 | 2-42.3 | 195 | NED | |
| C48GTT1 | 2-29.2 | 328 | HEX | |
| CX3 | C48CGA1 | 2-29.2 | 137 | HEX |
| C68GA1 | 2-9.6 | 154 | 56-FAM | |
| C32AC1 | 2-00.0 | 184 | NED | |
| CX4 | C139TG1 | 3-26.0 | 201 | 56-FAM |
| C127TC1 | 1-0.0 | 178 | NED | |
| C446AC2 | 3-65.8 | 256 | HEX |
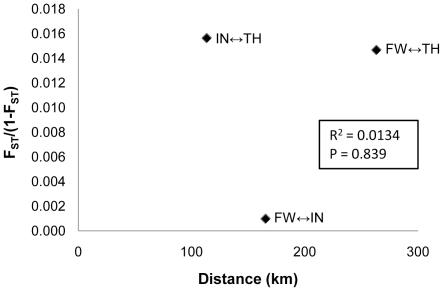
A panel of 12 microsatellites was used to assess the population structure among Cx. pipiens populations from three cities in Indiana (n = 266 individuals). All of the loci were highly polymorphic with 7 to 25 alleles (mean = 13.0) and PIC ranging from 0.509 to 0.889 (mean = 0.701) (Table 3). Allele frequencies of eight loci (C177CA1, C68GA1, C127TC1, C99TGT1, C65AC1, C48GTT1, C48CGA1, and C205TG1) were within HW expectations in all three cities, while four loci (C139TG1, C446AC2, C32AC1, and C134AC1) were within HW expectations in at least one city (Table 4). Generally, FIS was lower in Fort Wayne and Indianapolis populations than in Terre Haute (Table S4). Though pairwise FST values were relatively low among the three cities, significant structuring was evident between populations from Fort Wayne and Terre Haute and between populations from Indianapolis and Terre Haute, yet virtually no structuring was evident between populations from Fort Wayne and Indianapolis (Table 5). Tests for recent population bottlenecks based on gene diversities and allele frequency distributions were not significant. Results of AMOVA indicated that 97.02%, 1.88% and 1.10% of the estimated genetic variation was within individuals of a population, within populations, and among populations, respectively. No significant correlation between FST/(1-FST) and distance (Figure 2) was detected suggesting there was no isolation by distance (R2 = 0.0134, p = 0.839).
Table 3. Characterization of 12 microsatellite markers from Cx. pipiens collections (n = 266) from 3 cities in Indiana, USA.
| Locus | Repeat Motif | No. of Alleles | Allele Size Range | PICa |
| C177CA1 | (CA)12 | 18 | 111–152 | 0.760 |
| C68GA1 | (GA)8 | 14 | 122–155 | 0.794 |
| C127TC1 | (CT)39 | 11 | 96–129 | 0.787 |
| C99TGT1 | (TGT)6 | 10 | 184–226 | 0.759 |
| C65AC1 | (AC)13 | 13 | 266–299 | 0.684 |
| C205TG1 | (TG)12 | 14 | 114–170 | 0.697 |
| C134AC1 | (AC)7 | 7 | 185–199 | 0.554 |
| C48GTT1 | (GTT)6 | 9 | 307–337 | 0.766 |
| C48CGA1 | (CGA)9 | 7 | 117–135 | 0.570 |
| C32AC1 | (AC)11 | 13 | 207–222 | 0.798 |
| C139TG1 | (TG)10 | 25 | 198–241 | 0.889 |
| C446AC2 | (AC)7 | 15 | 217–255 | 0.579 |
PIC: allelic polymorphism information content.
Table 4. Summary statistics for microsatellite markers from each city representing study sites in Indiana.
| Fort Wayne (n = 86) | Indianapolis (n = 93) | Terre Haute (n = 87) | ||||
| Locus | HO | HE | HO | HE | HO | HE |
| C177CA1 | 0.732 | 0.761 | 0.739 | 0.792 | 0.693 | 0.799 |
| C127TC1 | 0.812 | 0.802 | 0.753 | 0.818 | 0.750 | 0.817 |
| C99TGT1 | 0.798 | 0.801 | 0.806 | 0.806 | 0.742 | 0.759 |
| C65AC1 | 0.639 | 0.709 | 0.736 | 0.735 | 0.747 | 0.706 |
| C205TG1 | 0.674 | 0.634 | 0.677 | 0.673 | 0.609 | 0.611 |
| C139TG1 | 0.779 | 0.864 | 0.785* | 0.881 | 0.609* | 0.861 |
| C134AC1 | 0.639 | 0.529 | 0.538* | 0.463 | 0.793* | 0.656 |
| C48GTT1 | 0.578 | 0.631 | 0.772 | 0.750 | 0.682 | 0.725 |
| C48CGA1 | 0.588 | 0.578 | 0.615 | 0.607 | 0.591 | 0.620 |
| C446AC2 | 0.553 | 0.573 | 0.618 | 0.640 | 0.477* | 0.590 |
| C32AC1 | 0.733 | 0.773 | 0.802 | 0.818 | 0.682* | 0.800 |
| C68GA1 | 0.779 | 0.800 | 0.742* | 0.825 | 0.881 | 0.822 |
HO = observed heterozygote frequency,
HE = expected heterozygote frequency under HW expectations,
* = significant deviation from HW expectations (p≤0.05).
Table 5. Pairwise FST estimates among three cities in Indiana, USA.
*Significant after permutation test [34].
Figure 2. Regression analysis of pairwise FST/(1-FST) against pairwise linear distances between three cities sampled in Indiana, USA.
Comparative genome analysis indicated that the relative abundance of microsatellite sequences varies among the three mosquitoes (Table 6). Our results show that smaller repeat motifs (di, tri-or tetra-nucleotide repeats) are more frequent in the Cx. quinquefasciatus genome (0.153%) than in the Ae. aegypti genome (0.109%), but considerably less frequent in both these genomes compared to the An. gambiae (0.75%) genome. The microsatellites with larger motifs (the penta- and hexa-nucleotide repeats) are relatively more frequent in the Cx. quinquefasciatus genome (0.109%) compared to that in the Ae. aegypti (0.035%) or the An. gambiae (0.022%) genome. However, the overall density of microsatellite sequences is relatively lower in the Cx. quinquefasciatus (2.6 kb/Mb) or the Ae. aegypti genomes (1.4 kb/Mb) compared to that in the An. gambiae genome (7.7 kb per Mb).
Table 6. Genome-wide microsatellite representation in Cx. quinquefasciatus, Ae.aegypti, and An. gambiae.
| Total Number | Mean # Repeats | Total Content (%) | |||||||
| Motif categories | Culex | Aedes | Anopheles | Culex | Aedes | Anopheles | Culex | Aedes | Anopheles |
| Dinucleotide | 15689 | 7657 | 47857 | 24.49 | 37.80 | 25.38 | 0.066 | 0.022 | 0.437 |
| Trinucleotide | 19615 | 35705 | 31639 | 17.95 | 20.03 | 22.05 | 0.061 | 0.055 | 0.251 |
| Tetranucleotide | 7119 | 18207 | 7970 | 21.10 | 22.67 | 21.68 | 0.026 | 0.032 | 0.062 |
| Pentanucleotide | 4063 | 8716 | 1093 | 56.11 | 30.55 | 27.34 | 0.039 | 0.020 | 0.011 |
| Hexanucleotide | 4673 | 3941 | 423 | 86.81 | 50.88 | 72.62 | 0.070 | 0.015 | 0.011 |
| Total | 51159 | 74226 | 88982 | 41.29 | 32.44 | 33.81 | 0.263 | 0.144 | 0.771 |
Discussion
Prior to our study there were ∼33 microsatellites available for Cx. pipiens complex mosquitoes [18]–[21]. However, not all of those amplify in both Cx. pipiens and Cx. quinquefasciatus, and even fewer amplify in Cx. pipiens pallens [20]. Furthermore, information on chromosome location is not currently available for those loci. Here we identified and validated 12 new microsatellites with broad application to Cx. pipiens complex mosquitoes, bringing the total number of microsatellites available to ∼45. All amplified in the six laboratory strains tested (Table S3), which includes colonies representing Cx. quinquefasciatus, Cx. pipiens (anautogenous), Cx. pipiens (autogenous) and Cx. pipiens from Japan (often referred to as Cx. pipiens pallens). Because we identified and scanned supercontigs containing markers previously mapped to chromosome locations, the relative locations of our microsatellites are known. An a priori assumption often made in studies of genetic variation is that the marker loci used provide reasonable coverage across the genome. Also, few problems were encountered while developing the PCR multiplexes with up to five primers sets, thus indicating the potential for the development of multiplexes other than those reported here. As expected, most primer sets worked well when genotyping our Johannesburg colony, but adjustments to primer sequences were often necessary to obtain consistent amplification (i.e., expected allele frequencies) in our South Bend strain. Once primers were adjusted for consistent amplification in both the South Bend and the Johannesburg colonies, they would usually work well for genotyping the field collections. However, several microsatellites worked well in our laboratory colonies but still reflected high null allele frequencies when they were tested on the field collections (Table S3). Nonetheless, these markers still have potential for use genetic mapping in laboratory colonies or investigating Cx. pipiens complex populations from other geographic areas.
Population structure among our study sites was consistent with findings from a similar study in which Huang et al. characterized 11 Cx. pipiens populations in the northeast United States using 12 microsatellites and detected significant structuring among several urban and rural populations with FST ranging from 0.0101 to 0.0174 in anautogenous Cx. pipiens [41]. However, Kothera et al. genotyped Cx. pipiens and Cx. quinquefasciatus populations from 14 sites in a north-south transect and reported significant FST ranging from 0.003 to 0.318 [6]. The observed structuring among populations from the three cities sampled in this study (FST ranging from 0.001 to 0.016) was due to significant differences between the Terre Haute population and both the Fort Wayne and Indianapolis populations. Because the local health departments in Indianapolis and Terre Haute were applying insecticide to potential Culex breeding sites during the period in which we collected our samples, we used the software program BOTTLENECK to determine if we could detect any recent population bottlenecks based on our microsatellite data. However, tests for recent reductions in the effective population sizes were not significant.
This study represents the first attempt to characterize microsatellite sequence representation in the Cx. quinquefasciatus assembled genome. Moreover, our comparative analysis provides a better understanding of genome-wide abundance of microsatellites among Cx. quinquefasciatus, Ae. aegypti and An. gambiae. It is well-known that microsatellite frequency is low in Ae. aegypti compared to several other insects, including An. gambiae. We note that an earlier study by Meglécz et al. [42] examined whole genome sequence (WGS) traces, but not the assembled genomes. Our results were consistent with these results after accounting for the respective differences in search strategies. That is, they also included mono-nucleotide repeats in their analysis and they used lower thresholds for di- and tri- nucleotide repeats than our thresholds for such repeats. Moreover, our estimate of microsatellites in An. gambiae closely match results of another study [43]. Microsatellites seem to be considerably more abundant (in terms of percentage of sequences per genome) in An. gambiae than in either Ae. aegypti or Cx. quinquefasciatus. On the other hand, the Cx. quinquefasciatus genome has a higher frequency of microsatellites with larger motifs (the penta- and hexa-nucleotide repeats) indicating a possible expansion of microsatellite motif length in this mosquito.
In conclusion, here we demonstrate a systematic approach for the isolation and validation of microsatellites for Cx. pipiens complex mosquitoes by screening Cx. quinquefasciatus genome supercontig assemblies for short tandem repeats. We tested and validated 12 microsatellites that amplified consistently and were polymorphic in lab colonies of Cx. quinquefasciatus and Cx. pipiens representing six populations from three continents. Additionally, microsatellite allele frequencies were within HW expectations in samples from the Johannesburg and South Bend colonies. We used four PCR multiplexes to assess the population structure of Cx. pipiens from three cities in Indiana, USA, thus demonstrating their usefulness for studies on natural populations. Lastly, we performed comparative genome analysis to characterize microsatellite type and abundance in three major disease vectors, An. gambiae, Ae. aegypti, and Cx. pipiens mosquitoes. The development of microsatellites using this approach could provide additional genetic markers to produce a linkage map with moderate resolution for Cx. pipiens mosquitoes and provide a foundation for further genetic analyses such as QTL mapping and population structure analysis. Moreover, this demonstrates the potential for the development of microsatellites from existing genome sequences for other closely related taxa.
Supporting Information
Coordinates for collection sites in Fort Wayne (FW), Indianapolis (IN), and Terre Haute (TH), Indiana, USA.
(0.04 MB DOC)
GenBank accession numbers for STS sequences of microsatellite loci.
(0.06 MB DOC)
Microsatellite variation among lab strains.
(0.11 MB DOC)
FIS estimates for each locus in each city sampled.
(0.04 MB DOC)
Acknowledgments
We thank the following for their assistance in field collections in Indiana: Jim Erwin, Shawn Moore, David Fiess, and Michael Grayless.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: Funding for the collection and rearing of field samples was supported by the Dunn-Foster Research Award from the Indiana Vector Control Association, Aspire Graduate Student Research Grant (Ball State University), a research grant from the Indiana Academy of Sciences, and the Russell E. Siverly Research Award from the Department of Physiology and Health Science (Ball State University). All other work was supported by grant RO1-AI079125-A1 from the National Institute of Allergy and Infectious Diseases, National Institutes of Health. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Turell MJ, O'Guinn ML, Dohm DJ, Jones JW. Vector competence or North American mosquitoes (Diptera: Culicidae) for West Nile virus. J Med Ent. 2001;38:130–134. doi: 10.1603/0022-2585-38.2.130. [DOI] [PubMed] [Google Scholar]
- 2.Day JF. Predicting St. Louis encephalitis virus epidemics: lessons from recent, and not so recent, outbreaks. Ann Rev Ent. 2001;46:111–138. doi: 10.1146/annurev.ento.46.1.111. [DOI] [PubMed] [Google Scholar]
- 3.Krida G, Bouattour A, Rodhain F, Failoux A. Variability among Tunisian populations of Culex pipiens: genetic structure and susceptibility to a filarial parasite, Brugia pahangi. Parasitol Res. 1998;84:139–142. doi: 10.1007/s004360050371. [DOI] [PubMed] [Google Scholar]
- 4.Vinogradova EB. Moscow: Pensoft; 2000. Culex pipiens pipiens Mosquitoes: taxonomy, distribution, ecology, physiology, genetics, applied importance and control.250 [Google Scholar]
- 5.Barr AR. The distribution of Culex p. pipiens and Culex p. quiquefasciatus in North America. Am J Trop Med Hyg. 1957;6:153–165. doi: 10.4269/ajtmh.1957.6.153. [DOI] [PubMed] [Google Scholar]
- 6.Kothera L, Zimmerman EM, Richards CM, Savage HM. Microsatellite characterization of subspecies and their hybrids in Culex pipiens complex (Diptera: Culicidae) mosquitoes along a north-south transect in the central United States. J Med Ent. 2009;46:236–248. doi: 10.1603/033.046.0208. [DOI] [PubMed] [Google Scholar]
- 7.Fonseca DM, Keyghobadi N, Malcolm CA, Mehmet C, Schaffner F, et al. Emerging vectors in the Culex pipiens complex. Science. 2004;303:1535–1538. doi: 10.1126/science.1094247. [DOI] [PubMed] [Google Scholar]
- 8.Mori A, Romero-Severson J, Severson DW. Genetic basis for reproductive diapause is correlated with life history traits within the Culex pipiens complex. Insect Mol Biol. 2007;16:515–524. doi: 10.1111/j.1365-2583.2007.00746.x. [DOI] [PubMed] [Google Scholar]
- 9.Severson DW, Mori A, Zhang Y, Christensen BM. Linkage map for Aedes aegypti using restriction fragment length polymorphisms. J Hered. 1993;84:241–247. doi: 10.1093/oxfordjournals.jhered.a111333. [DOI] [PubMed] [Google Scholar]
- 10.Severson DW, Meece JK, Lovin DD, Saha G, Morlais I. Linkage map organization of expressed sequence tags and sequence tagged sites in the mosquito, Aedes aegypti. Insect Mol Biol. 2002;11:371–378. doi: 10.1046/j.1365-2583.2002.00347.x. [DOI] [PubMed] [Google Scholar]
- 11.Fulton RE, Salasek ML, DuTeau NM, Black WC IV. SSCP analysis of cDNA markers provides a dense linkage map of the Aedes aegypti genome. Genetics. 2001;158:715–726. doi: 10.1093/genetics/158.2.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chambers EW, Meece JK, McGowan JA, Lovin DD, Hemme RR, et al. Microsatellite isolation and linkage group identification in the yellow fever mosquito Aedes aegypti. J Hered. 2007;98:202–210. doi: 10.1093/jhered/esm015. [DOI] [PubMed] [Google Scholar]
- 13.Dimopoulo G, Zheng L, Kumar V, della Torre A, Kafatos FC, et al. Integrated Genetic Map of Anopheles gambiae: Use of RAPD Polymorphisms for Genetic, Cytogenetic and STS Landmarks. Genetics. 1996;143:953–960. doi: 10.1093/genetics/143.2.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng L, Benedict MQ, Cornel AJ, Collins FH, Kafatos FC. Integrated Genetic Map of the African Human Malaria Vector Mosquito, Anopheles gambiae. Genetics. 1996;143:941–952. doi: 10.1093/genetics/143.2.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang R, Kafatos FC, Zheng L. Microsatellite markers and genotyping procedures for Anopheles gambiae. Parasit Today. 1999;15:33–37. doi: 10.1016/s0169-4758(98)01360-x. [DOI] [PubMed] [Google Scholar]
- 16.Sharakhov IV, Serazin AC, Grushko OG, Dana A, Lobo N, et al. Inversions and gene order shuffling in Anopheles gambiae and A. funestus. . Genetics. 2002;298:182–185. doi: 10.1126/science.1076803. [DOI] [PubMed] [Google Scholar]
- 17.Mori A, Severson DW, Christensen BM. Comparative linkage maps for the mosquitoes (Culex pipiens and Aedes aegypti) based on common RFLP loci. J Hered. 1999;90:160–164. doi: 10.1093/jhered/90.1.160. [DOI] [PubMed] [Google Scholar]
- 18.Fonseca DM, Atkinson CT, Fleischer RC. Microsatellite primers for Culex pipiens quinquefasciatus, the vector of avian malaria in Hawaii. Mol Ecol. 1998;7:1617–1619. [PubMed] [Google Scholar]
- 19.Keyghobadi N, Matrone MA, Ebel GD, Kramer LD, Fonseca DM. Microsatellite loci from the northern house mosquito (Culex pipiens), a principal vector of West Nile virus in North America. Mol Ecol Notes. 2004;4:20–22. [Google Scholar]
- 20.Smith JL, Keyghobadi N, Matrone MA, Escher RL, Fonseca DM. Cross-species comparison of microsatellite loci in the Culex pipiens complex and beyond. Mol Ecol Notes. 2005;5:697–700. [Google Scholar]
- 21.Edillo FE, Tripet F, McAbee RD, Foppa IM, Lanzaro GC, et al. A set of broadly applicable microsatellite markers for analyzing the structure of Culex pipiens (Diptera: Culicidae) populations. J Med Ent. 2007;44:145–149. doi: 10.1603/0022-2585(2007)44[145:asobam]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 22.Zane L, Bargelloni L, Patarnello T. Strategies for microsatellite isolation: a review. Mol Ecol. 2002;11:1–16. doi: 10.1046/j.0962-1083.2001.01418.x. [DOI] [PubMed] [Google Scholar]
- 23.Lovin DD, Washington KO, deBruyn B, Hemme R, Mori A, Epstein SR, et al. Genome-based polymorphic microsatellite development and validation in the mosquito Aedes aegypti and application to population genetics in Haiti. BMC Genomics. 2009;10:590. doi: 10.1186/1471-2164-10-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.VectorBase: Cx. pipiens. 2010. Available: http://cpipiens.vectorbase.org/index.php.
- 25.Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rozen S, Skaletsky HJ. Krawetz S, Misener S, editors. Primer3 on the WWW for general users and for biologist programmers. Bioinformatics methods and protocols: methods in molecular biology. Totowa: Humana. 2000. pp. 365–386. [DOI] [PubMed]
- 27.Kibbe WA. Oligocalc: an online oligonucleotide properties calculator. Nucleic Acids Res. 2007;35:W43–W46. doi: 10.1093/nar/gkm234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lewis LF, Clark TB, O'Grady JJ, Christenson DM. Collecting ovigerous Culex pipiens quinquefasciatus Say near favorable resting sites with louvered traps baited with infusions of alfalfa pellets. Mosq News. 1974;34:436–439. [Google Scholar]
- 29.Siverly RE. Indianapolis: Indiana State Board of Health; 1972. Mosquitoes of Indiana.126 [Google Scholar]
- 30.Rudbeck L, Dissing J. Rapid, simple alkaline extraction of human genomic DNA from whole blood, buccal epithelial cells, semen and forensic stains for PCR. Biotechniques. 1998;25:588–592. doi: 10.2144/98254bm09. [DOI] [PubMed] [Google Scholar]
- 31.Severson DW. RFLP analysis of insect genomes. In: Crampton JM, Beard CB, Louis C, editors. The Molecular Biology of Insect Disease Vectors: a Methods Manual. London: Chapman & Hall; 1997. pp. 309–320. [Google Scholar]
- 32.Hayden MJ, Nguyen TM, Waterman A, Chalmers KJ. Multiplex-ready PCR: a new method for multiplexed SSR and SNP genotyping. BMC Genomics. 2008;9:80. doi: 10.1186/1471-2164-9-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kofler R, Schlötterer C, Lelley T. SciRoKo: a new tool for whole genome microsatellite search and investigation. Bioinformatics. 2007;23:1683–1685. doi: 10.1093/bioinformatics/btm157. [DOI] [PubMed] [Google Scholar]
- 34.Excoffier L, Laval G, Schneider S. Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evol Bioinform Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- 35.Wier BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- 36.Guo SW, Thompson EA. Performing the exact test of Hardy-Weinberg proportion for multiple alleles. Biometrics. 1992;48:361–372. [PubMed] [Google Scholar]
- 37.Raymond M, Rousset F. GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. J Hered. 1995;86:248–249. [Google Scholar]
- 38.Rousset F. GENEPOP'007: a complete re-implementation of the GENEPOP software for Windows and Linux. Mol Ecol Res. 2008;8:103–106. doi: 10.1111/j.1471-8286.2007.01931.x. [DOI] [PubMed] [Google Scholar]
- 39.Park SDE. Trypanotolerance in West African Cattle and the Population Genetic Effects of Selection [Ph.D. thesis] University of Dublin 2001 [Google Scholar]
- 40.Cornuet JM, Luikart G. Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics. 1996;144:2001–2014. doi: 10.1093/genetics/144.4.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang S, Molaei G, Andreadis TG. Genetic insights into the population structure of Culex pipiens (Diptera: Culicidae) in the northeastern United States by using microsatellite analysis. Am J Trop Med Hyg. 2008;79:518–527. [PubMed] [Google Scholar]
- 42.Meglécz E, Anderson SJ, Bourguet D, Butcher R, Caldas A, et al. Microsatellite flanking region similarities among different loci within insect species. Insect Mol Biol. 2007;16:175–185. doi: 10.1111/j.1365-2583.2006.00713.x. [DOI] [PubMed] [Google Scholar]
- 43.Archak S, Meduri E, Kumar PS, Nagaraju J. InSatDb: a microsatellite database of fully sequenced insect genomes. Nucleic Acids Res. 2007;35:D36–D39. doi: 10.1093/nar/gkl778. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Coordinates for collection sites in Fort Wayne (FW), Indianapolis (IN), and Terre Haute (TH), Indiana, USA.
(0.04 MB DOC)
GenBank accession numbers for STS sequences of microsatellite loci.
(0.06 MB DOC)
Microsatellite variation among lab strains.
(0.11 MB DOC)
FIS estimates for each locus in each city sampled.
(0.04 MB DOC)