Abstract
Three-dimensional tumor models have emerged as valuable in vitro research tools, though the power of such systems as quantitative reporters of tumor growth and treatment response has not been adequately explored. We introduce an approach combining a 3-D model of disseminated ovarian cancer with high-throughput processing of image data for quantification of growth characteristics and cytotoxic response. We developed custom MATLAB routines to analyze longitudinally acquired dark-field microscopy images containing thousands of 3-D nodules. These data reveal a reproducible bimodal log-normal size distribution. Growth behavior is driven by migration and assembly, causing an exponential decay in spatial density concomitant with increasing mean size. At day 10, cultures are treated with either carboplatin or photodynamic therapy (PDT). We quantify size-dependent cytotoxic response for each treatment on a nodule by nodule basis using automated segmentation combined with ratiometric batch-processing of calcein and ethidium bromide fluorescence intensity data (indicating live and dead cells, respectively). Both treatments reduce viability, though carboplatin leaves micronodules largely structurally intact with a size distribution similar to untreated cultures. In contrast, PDT treatment disrupts micronodular structure, causing punctate regions of toxicity, shifting the distribution toward smaller sizes, and potentially increasing vulnerability to subsequent chemotherapeutic treatment.
Keywords: ovarian cancer, three-dimensional models, image processing, photodynamic therapy, carboplatin
Introduction
Three-dimensional (3-D) tumor models have emerged as valuable in vitro research platforms to evaluate tumor growth properties,1, 2, 3, 4 and therapeutic response in a more physiologically relevant manner than monolayer cultures.5, 6, 7, 8, 9 In pioneering studies, the Bissel laboratory established in vitro 3-D breast cancer models in which normal and malignant breast epithelial cells grown on a bed of growth-factor-reduced (GFR) Matrigel™ form polarized 3-D acinar structures.10 Implementing these 3-D models to understand cell signaling in relation to position within a acinus, their group and others were able to make basic tumor biology insights into breast carcinogenesis and progression, which would not be possible in monolayer cultures.1, 2, 3, 4 Treatment response studies have shown that cancer cells induced to form 3-D spheroids are vastly less sensitive to chemotherapy than monolayer cells.5 Finlay et al.,6 Bigelow et al.,7 Georgakoudi and Foster,8 and Foster et al.,9 also demonstrated the utility of tumor spheroids to study dose rate effects and other key determinants of photodynamic therapy (PDT) efficacy. While these reports demonstrate the importance of restoring key architectural cues in vitro, the full capability of 3-D tumor models as a biologically relevant platform for analysis of tumor growth and cytotoxic response has not yet been adequately explored. In vitro 3-D tumor systems, could, with the appropriate statistical characterization methods, be used as tools to provide a window into tumor growth mechanisms in vivo, while providing a level of access for imaging and manipulation of the system that is difficult to achieve in animal models. Similarly, 3-D models provide a unique but untapped opportunity to quantify cytotoxic response across large numbers of individual tumor nodules of different sizes in a system where physical variables such as penetration of drugs through multiple cell layers actually reflect the physiological scenario.
To tap into the inherent capability of in vitro 3-D systems as reporters of growth properties and cytotoxic response, optical microscopy emerges as a natural research tool. Cultures can be plated on coverslip bottom dishes or multiwell plates to allow for imaging with an inverted microscope, which requires no disruption of the sample and is highly conducive to longitudinal monitoring. In contrast to previous studies examining cellular-level signaling in high-resolution images of single acini,1, 2, 3, 4 here we focus on quantification of large data sets, by acquisition of microscope fields each containing hundreds of tumor nodules. Model systems such as those just described, in which cells are overlaid on a basement membrane layer, are particularly conducive to this type of imaging as 3-D structures are formed in a single focal plane on the surface of the gel (as opposed to being embedded inside it). These fields can then be automatically segmented and batch processed to obtain lists of sizes, eccentricities, and positions for each nodule. By acquiring sets of images across independently prepared cultures at sequences of time points, evolution of the full size distribution (rather than just representative measurements) for thousands of 3-D structures can be monitored. Similarly, by using the appropriate fluorescence agents to stain live and dead cells following cytotoxic treatments, 2-D arrays of pixel data can again be segmented to report treatment response nodule by nodule, correlating viability with structural or morphological changes in response to treatment. With appropriate ratiometric quantification, this approach provides a level of sophistication in reporting of cytotoxic response that cannot be achieved with traditional colorimetric assays in which the viability of an entire culture dish is reduced to a single number.
In this paper, we utilize this imaging-based approach to elucidate growth characteristics and patterns of cytotoxic response in an in vitro 3-D model of micrometastatic ovarian cancer. For the majority of patients, this disease is diagnosed at a late stage characterized by disseminated studding of the peritoneal surfaces with tumor nodules ranging in size from microscopic clusters of a few cells to large cakes of disease spanning several centimeters.11, 12, 13 The low survival rate of 31%, which has improved only marginally over the course of decades, points to the need for new physiologically relevant research platforms to meaningfully examine treatment response and devise more effective strategies. We adopt an ovarian cancer model that draws on the established in vitro models of breast cancer in which cells overlaid on a bed of GFR Matrigel basement membrane spontaneously form 3-D acini.1, 4, 10, 14, 15 In contrast to 3-D ovarian spheroids, which are grown in rotary culture to prevent adhesion to the underlying substrate,16, 17 this system recapitulates the spontaneous formation of tumor nodules from individual adherent cells. Following a characterization of the growth kinetics in this system, we examine the response to two mechanistically distinct treatments: carboplatin and PDT. Carboplatin is an established chemotherapy in clinical use for the treatment of ovarian cancer.18, 19 PDT is a light-based therapy in which wavelength-specific activation of a photosensitizer generates cytotoxicity to the illuminated region.20 PDT has demonstrated promise for treatment of ovarian cancer in clinical and preclinical studies, though additional studies are required to establish optimal treatment parameters.21, 22, 23, 24, 25, 26
Materials and Methods
Three-Dimensional Cell Culture
Epithelial ovarian cancer NIH:OVCAR-5 cells were obtained from Thomas Hamilton (Fox Chase Cancer Institute, Philadelphia, Pennsylvania). The cells were grown in RPMI-1640 (Roswell Park Memorial Institute) medium (Mediatech Inc., Herndon, Virginia, USA) supplemented with 10% heat-inactivated fetal calf serum (GIBCO Life Technologies, Grand Island, New York, USA), 100 U∕mL penicillin, and 100 μg∕mL streptomycin. For 3-D cell culture, growth factor reduced (GFR) Matrigel (BD Biosciences, San Jose, California) was used as a basement membrane, which has appropriate gel structure and established biological activity to promote growth and differentiation of a variety of cells.27 To prepare 3-D cultures 500-μL volumes of NIH:OVCAR-5 cells in single cell suspension of 15,000 cells∕mL were grown on beds of GFR Matrigel (BD Biosciences) on the glass slide inset of 35-mm MatTek culture dishes (MatTek Corporation, Ashland, Massachusetts, USA). GFR Matrigel beds were initially prepared by ejecting 150 μL of GFR Matrigel solution at ∼4 °C on the chilled MatTek plates and carefully rotating the dish at a slight angle to evenly distribute it over the 10-mm-radius glass portion of the MatTek dishes that were then incubated at 37 °C for 30 min prior to plating cells to allow gelation to occur. Following initial plating of cells, they were allowed to adhere to the Matrigel bed before addition of complete growth medium with 2% GFR Matrigel. All cultures were maintained in an incubator at 37 °C in an atmosphere of 5% CO2.
Microscopy for Growth Characterization
To characterize development of size distributions over time, 3-D cultures were routinely imaged by darkfield microscopy at 5× using a Zeiss Axiovert inverted microscope (Carl Zeiss MicroImaging, Inc., Thornwood, NY, USA) fitted with a QuantiFIRE™ cooled 12-bit monochrome CCD camera with 2048 ×2048-pixel chip (Optronics, Goleta, California). Images were acquired in a focal plane just above the surface of the gel on which 3-D structures are formed. For each plate, five dark-field images of each culture dish were acquired at each time point in the tagged image file format (TIFF) and saved for off-line processing. Time-lapse microscopy sequences were obtained using a Nikon TE2000-S inverted microscope with a 10× phase contrast objective in an enclosed weather station (Nikon Instruments, Inc, Melville, New York, USA).
Image Processing for Growth Analysis
Image data was processed using custom scripts developed using the Image Processing Toolbox in the MATLAB software package (Mathworks, Natick, Massachusetts, USA). To calculate size distributions from image data we developed a batch analysis routine in which sets of high-contrast dark-field images were thresholded (calling on a built-in automated routine based on Otsu’s method), made binary and segmented to identify in vitro nodules. The routine would count the number of pixels in each region while rejecting partial features at the edge of each field of view and calibrate sizes to square micrometers. Features of size less than 200 μm2 were rejected. Lists of statistics for each feature (size in square micrometers, equivalent diameter in micrometers, eccentricity, and the x and y positions in the image) were tabulated and grouped by time point. Size distribution histograms were then generated from lists of sizes at each time point. In general we report size in units of square micrometers as 2-D cross-sectional area A, which is directly reported from calibration of pixel counts, and strictly speaking, is the most accurate quantification of size. However, nodules are approximately spherical and it is a reasonable approximation to calculate equivalent diameter deq as a more intuitive metric of nodule size, which is obtained from area by π(deq∕2)2=A.
Image data were grouped into directories by imaging time point so that all routines were run as batch processes, enabling rapid generation of statistics on large lists of micronodules at each time point. The same automated segmentation approach was used to analyze the size distribution of residual viable disease following treatment by application of the same batch process to sets of calcein green fluorescence image data, as described in the following. To verify the accuracy and precision of high-throughput automated image segmentation approximately 1 out of every 50 images was randomly selected for manual verification by having MATLAB overlay arrows at each software-identified nodule centroid. This comparison of manual identification to software identification in the images tested revealed a 97.3% rate of agreement for positive identification of features. Additional data analysis and graphing of image data including multipeak nonlinear least-squares Gaussian curve fits was conducted using the Origin software package (OriginLab Corporation, Northampton, Massachusetts).
Carboplatin Treatments
Carboplatin treatments were performed by incubating 3-D cultures in OVCAR-5 complete culture medium with 1 mM carboplatin (Teva Pharmaceuticals, Petach Tikva, Israel) for a total of 96 h, including a carboplatin media refresh at 48 h. This dose was selected based on calculation of the concentration with respect to the area of the culture dish to provide a dosage estimated to be equivalent to the clinical dose of 400 mg∕m2, which was reported to be effective in human patients.18 Treatment response was assessed immediately after removal of carboplatin media as described in the following.
PDT Treatments
At day 10 following plating, cultures were incubated with 250 nM BPD-MA (or Verteporfin, QLT, Inc., Vancouver, British Columbia, Canada) in complete culture media for 90 min. Immediately prior to irradiation, the media were replaced with regular complete media. Laser irradiation was delivered through the bottom of each culture dish on a clear plastic tray via a vertically mounted Thorlabs FT600EMT multimode fiber (Thorlabs, Newton, New Jersey, USA), which was collimated to overfill the dish area to deliver light nearly uniformly over the central 1-cm-radius portion of a MatTek dish containing the Matrigel layer with 3-D nodules on the surface. Irradiation times were calculated to achieve total fluence of 10 J∕cm2 at a fluence rate of 50 mW∕cm2, which was measured using an Ophir Vega power meter with photodiode sensor (Ophir Optronics, Israel) prior to each experiment. After treatments, cultures were returned to the incubators for 24 h prior to assessment of treatment response.
Imaging-Based Assessment of Cytotoxic Response
Assessment of cytotoxic response in the 3-D culture model posed an additional challenge as the conversion products from traditional MTT and MTS assays were found to adhere to the Matrigel substrate preventing reliable reporting of viability by these methods. To overcome this we developed a quantitative fluorescence-imaging-based approach using the LIVE∕DEAD® cytotoxicity kit (Invitrogen Corporation, Carlsbad, California, USA). Media was removed from culture dishes of 3-D nodules by gentle aspiration followed by a wash with phosphate-buffered saline (PBS) and incubation at 37 °C for 40 min with 2 μM calcein AM and 4 μM ethidium homodimer-1 (Invitrogen) diluted in PBS prior to imaging. Addition of reagents to each culture dish was staggered such that the incubation period was exactly 40 min for each dish. Following incubation, three digital images were acquired by sequentially rotating filter cubes on a Zeiss Axiovert 100 TV inverted microscope (Zeiss, Germany) at each of four spatial fields imaged in each culture dish: (1) a dark-field image, (2) a calcein fluorescence emission image (494∕517 nm, excitation∕emission), and (3) an ethidium bromide fluorescence emission image (528∕617 nm excitation∕emission on intercalation into DNA). All images were acquired using a 5× objective in inverted geometry imaging through the glass window on the bottom of each MatTek culture dish and collecting light from the focal plane on which 3-D cultures were grown on the surface of the Matrigel bed. Images were captured with a CCD camera (Quantifire XI, Optronix, Israel) and saved as 12-bit TIFFs for later analysis. Again, using a batch-process routine developed in the MATLAB software package, sets of images were analyzed in high throughput to report the mean fluorescence signal from the calcein and ethidium bromide channels with the ratio of calcein fluorescence to total fluorescence (calcein plus ethidium) as a reporter of viability. The ratio was computed in this manner for all treatment groups. To make comparisons between carboplatin and PDT treatments assessed at different imaging sessions, values were normalized to the respective no-treatment control group to calculate percentage viability for each treatment following standard general practice for quantification of cytotoxic response by any method. This routine was then run inside the size distribution code already discussed to identify each nodule in a corresponding dark-field image of a given field and then calculate the viability based only on the pixels in that nodule. Iterating over all in vitro nodules, this method produces lists of hundreds of nodules for each treatment group for each experimental run, and viability for each one.
Immunofluorescence Protocol
The 3-D cultures were stained following a protocol previously described by Debnath and Brugge.1 To prevent the degradation of the Matrigel during the staining protocol and subsequent loss of 3-D nodules from the Matrigel surface, culture dishes were maintained at 37 °C between all steps. The exchange of buffers and staining solutions was accomplished using a micropipetter to minimize disruption of the Matrigel. Anti-E-cadherin antibodies (BD Transduction Laboratories, BD Bioseciences, San Jose, California, USA) were conjugated with Alexa Fluor 568 labeled goat antimouse (Invitrogen) for targeted E-cadherin molecular imaging. DAPI (4′,6-diamidino-2-phenylindole) was obtained from Sigma and diluted accordingly. Images were obtained using an Olympus FV1000 confocal microscope (Olympus of America, Inc., Center Valley, Pennsylvania, USA).
Results and Discussion
Growth Characterization
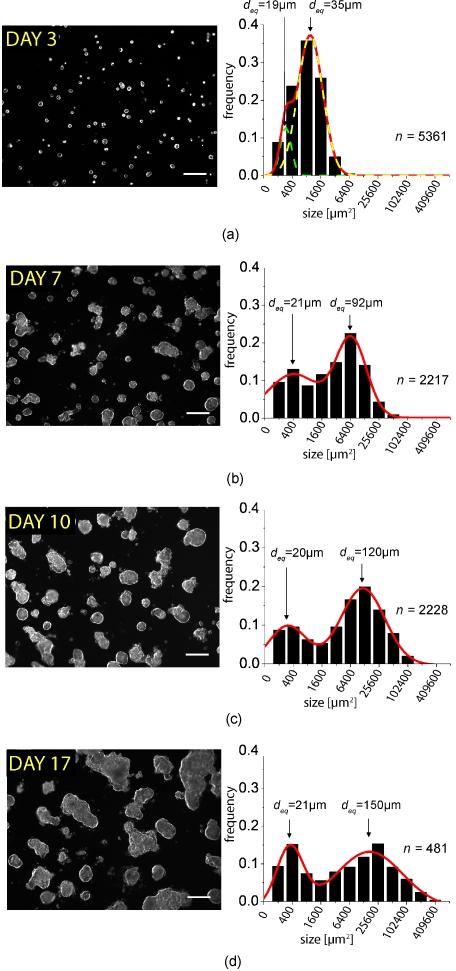
OVCAR-5 human ovarian cancer cells plated on GFR Matrigel formed solid, multicellular 3-D acini that reached sizes of the order of ∼110 μm in diameter by 10 days following plating, consistent with previously reported observations from optical coherence tomography (OCT) imaging of individual nodules plated by the same method.28 We observed that the physical characteristics of the 3-D acini (Fig. 1) was highly varied, with structures ranging from clusters of just a few cells to large acini to caked aggregates of multiple acini, qualitatively resembling the heterogeneity of micronodular disease in vivo.23 To quantify growth kinetics in this system, multiple independently plated batches were longitudinally imaged by dark-field microscopy (providing high-contrast images conducive to processing) at regular intervals for up to 24 days following initial plating. To obtain statistically significant data sets, large fields of view containing (initially) of the order 200 cells were obtained using a 5× objective. For each independently plated culture, five fields of view were obtained for each time point and saved into directories organized by time point. Using image processing routines developed in the MATLAB software package, we batch-processed these large sets of dark-field microscopy image data to produce lists of statistics for thousands of individual 3-D acini at multiple time points. We observed that the distribution of acinar sizes was initially homogeneous through days 3 to 4, but consistently broadened at late time points with a significant fraction of acini with sizes from 200 to 400 μm2 (in cross-sectional area from 2-D microscopy data) but with numerous larger acini with sizes up to 400,000 μm2. As this skewed distribution of sizes suggested a log-normal distribution we generated histograms for each time point with logarithmically (base 2) spaced bins (Fig. 1) consistent with the basic premise that growth processes are driven by multiplicative, rather than additive events.29 In these histograms, we consistently observed the emergence of two log-normal modes at later time points fitting well to a two-peak Gaussian model of the form
| (1) |
where
| (2) |
and A, w, and sc are amplitude, width, and center position of each peak, respectively. This bimodal form indicates the presence of two distinct growth behaviors. These distinct growth modes correspond to a population of cells in which Sc1 remains constant over time, corresponding to a constant equivalent diameter of ∼20 μm over the time probed. The other peak, with position Sc2, shifts to the right at subsequent imaging time points corresponds to a more rapidly assembling subdistribution of micronodules with mean equivalent diameters deq of 92, 120, and 150 μm at days 7, 10, and 17, respectively [Figs. 1b, 1c, 1d]. Extrapolating the constant position of Sc1 back to the day 3 image data and subjecting it to the same two-peak model (even though the distribution initially appears to have only one peak), we found that the nonlinear least-squares curve fit rapidly converged, indicating that this bimodal behavior also describes the early time points following plating. The other fit parameters associated with the population of the more aggressive growth mode also seem to have time dependent behavior, with w2 monotonically increasing, while A2 monotonically decreases over the time probed (but with the area under the peak remaining roughly constant). In contrast, the width and amplitude of the smaller size peak, w1 and A1, respectively, remain roughly constant.
Figure 1.
Representative dark-field microscopy images of in vitro 3-D acini and corresponding size distribution histograms and Gaussian fits from analysis of image data obtained at (a) day 3 following plating and (b) day 7, (c) day 10, and (d) day 17. The n values on the right of each histogram indicate the number of nodules included in each histogram. Scale bars=250 μm.
Size distributions at individual time points were also compared across individual culture dishes and dates of plating, and no statistically significant systematic variations were observed (p value>0.1 in two-tailed t test comparison of size lists between plates). The bimodal distribution was observed in all dishes with separable peaks observed in approximately the same positions in all cases with p value<0.001 in a two-tailed Student’s t test comparison of sets of sc1 and sc2 values calculated from each independently plated culture imaged at a given time point (three to nine cultures depending on the time point). Conversely, comparison of lists of sc1 values derived from different time points show no significance (p value>0.1, indicating that indeed this peak is reproducibly centered in the same position), while sc2 shifts significantly between days 7 and 17 with p value=0.01.
To examine these growth properties in real time, we conducted a series of time-lapse microscopy experiments in which 3-D cultures grown in multiwell plates were imaged at 10-min intervals over the course of several days. Individual snapshots from several timepoints of the full sequence are shown in Fig. 2. Observation of the behavior of individual cells over an extended period in time lapse sequences supports the observation of two growth behaviors; one population of cells rapidly merged together forming larger and larger multicellular aggregates [yellow arrows in Fig. 2a exemplify this behavior] while another appeared to be dominated by slow proliferation with little or no evidence of motility and assembly [the cell in the green box in Fig. 2a is consistent with this behavior]. The complete time-lapse sequence from which these frames were obtained is shown in the video. These observations give rise to the model shown schematically in Fig. 2b, indicating a rapidly comigrating group of cells (beige) juxtaposed with another group of cells (∼30% as indicated from frequency counts above) that exhibit little motility or affinity for assembly with surrounding cells, regardless of proximity. The rapid migration and assembly behavior gives rise to an overall exponential decay in the spatial density of acini [Fig. 2c] that is concomitant with increasing size [Fig. 2d] in the population of larger and more aggressively growing micronodules. It is not surprising that this behavior seems to dominate as from the frequency counts in Fig. 2 we would expect ∼70% of the cells in a given field to follow the high-motility migration and assembly kinetics. The decay is of the form n(t)=n0+A exp(−t∕τ), where τ=5.9 days−1.
Figure 2.
Characterization and quantification of growth behavior from time lapse microscopy correlated with analysis of longitudinal darkfield image data (shown in Fig. 1). (a) Individual frames from time-lapse microscopy acquired from 4 h following plating until 4 days following plating (the full time-lapse sequence is included as a video). Yellow arrows indicate clusters of cells that rapidly migrate over the Matrigel surface and assemble to form larger multicellular acini. The green box indicates a representative cell that does not participate in assembly events in the immediate vicinity, but does undergo cell division. Scale bars=100 μm. (b) Schematic representation of the bimodal growth kinetics described in Fig. 1 and (a). Approximately 30% of cells (gray), exhibit low motility and little potential to migrate toward other clusters of cells. The other 70% of cells (beige cells) exhibit a much more aggressive growth behavior, rapidly forming larger and larger multicellular aggregates. The sequential numbering 1, 2, and 3 in the diagram indicates (1) single cells following plating that (2) form small clusters and (3) subsequently form larger multicellular clusters. Inset: confocal immunofluorescence section from the center of a nodule fixed and stained for E-cadherin (red) and DAPI (blue) at day 10 of growth to reveal large multicellular 3-D structures formed from assembly and division of individual cells on the gel surface. Scale bar=20 μm. (c) Assembly events lead to an overall decay in the spatial density of 3-D acini with a decay constant of 5.9 days−1. Error bars in (c) are standard deviations of density measurements from five fields, each of three to nine independent prepared culture dishes imaged at a given time point. (d) Exponential decay in density is concomitant with an increase in mean diameter over time of rapidly growing nodules [corresponding to sc2 from Gaussian fits in Fig. 1 and Eq. 1 of the text]. Mean diameter remains constant in the remaining nodules [sc1 in Eq. 1]. Error bars in (d) are derived from the widths of Gaussian fits shown in Fig. 1. (QuickTime, 1.45 MB) .
The heterogeneity observed here qualitatively resembles the distribution of large and small nodules in peritoneal studding in vivo.23 These highly divergent behaviors, as evidenced by the differing propensities of the cells to migrate and assemble, suggest that even within a single cell line, the 3-D model reveals heterogeneity in growth properties and possibly response to treatment that are not apparent in monolayer cultures. Further studies are warranted to correlate these contrasting behaviors with relevant molecular markers and also to compare distributions of nodular size in this in vitro system with actual patient tissues. A more complete mathematical description of the combined aggregation and cell division kinetics underlying the formation of multicellular structures in this 3-D model system is part of an ongoing study.
Analysis of Carboplatin Treatment Response in 3-D Nodules
To demonstrate the value of our system as a tool to assess treatment outcome we conducted treatment response studies with carboplatin, a clinically relevant chemotherapeutic used to treat ovarian cancer. In preliminary studies, we found that viability in 3-D could not be assessed by traditional colorimetric assays as adhesion of the converted products to the Matrigel substrate obfuscated fluorescence or absorbance readings correlated with viability. For this reason the commercially available LIVE∕DEAD fluorescence imaging kit (Invitrogen L3224), which stains live cells and dead cells with a pair of fluorophores (calcein and ethidium bromide, with fluorescence emission in the green and red, respectively) was adapted into a quantitative tool to assess cell viability as described in Sec. 2. This imaging-based approach for quantitative assessment of cytotoxic response has several valuable features, as demonstrated in the following. Specifically, because we have access to a full 2-D array of pixel intensities (rather than a global average) we can combine the ratiometric quantification of viability from fluorescence intensities with the automated image segmentation (used for batch analysis of size distributions) to analyze cytotoxic response nodule by nodule. This approach provides a depth of insight into patterns of response, by size and morhphology of nodules that colorimetric assays cannot provide.
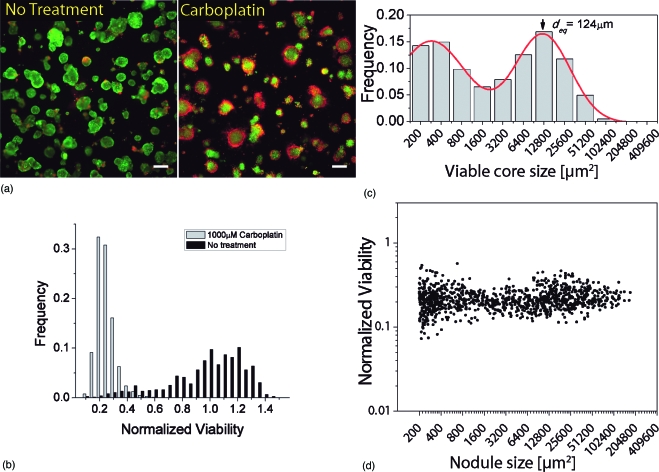
Cultures were allowed to grow to day 10 at which time treatment was initiated. Upon visual inspection of cultures stained with LIVE∕DEAD reagents [Fig. 3a] it is apparent that there is a clear cytotoxic response (larger and more intense regions of red ethidium bromide fluorescence relative to no treatment control). The mean viability of 1000 μM carboplatin-treated nodules reported as live signal divided by total fluorescence signal and normalized to mean no treatment control viability was 0.22±0.07 [minimum 0.073, maximum 0.57 Fig. 3b]. However, even with total exposure to carboplatin for an extended incubation period, cytotoxicity seems to be primarily localized to the cells on the periphery of each acinus, with large surviving (green) cores in the center [Fig. 3a]. This observation is also apparent from analyzing the size distribution of residual viable disease from segmentation of the calcein fluorescence emission images [Fig. 3c], which is essentially unchanged from the distribution of nodules prior to initiation of treatment at day 10 [Fig. 1c]. Size-dependent treatment response is summarized in Fig. 1d, in which each data point represents nodule size based on analysis of the corresponding dark-field channel (horizontal axis) and the viability based on the fluorescence intensity ratio calculated from the pixels in that nodule (vertical axis). Following carboplatin treatment the size-dependent response appears relatively flat, with all nodules uniformly reduced to a narrowly distributed range of viabilities. The characteristic bimodal growth distribution is immediately also evident here from the two dense clusters of data points to either side of a sparse region between 1600 and 3200 μm2.
Figure 3.
Carboplatin treatment response assessed by quantitative ratiometric fluorescence imaging. (a) Representative LIVE∕DEAD fluorescence image of 3-D culture subject to 1000 μM carboplatin treatment and corresponding no treatment control grown in identical conditions. The carboplatin treated plate exhibits a pattern of cytotoxic response in which cores of 3-D nodules remain viable (green) while cells at the periphery are dead. Display images adjusted with hi-lo lookup table (all quantitative analysis is based on raw unprocessed images). Scale bars=250 μm. (b) Distribution of viabilities in carboplatin treated nodules. Mean viability is 0.22 with standard deviation of 0.07 (minimum value 0.073, maximum value 0.57). (c) Size distribution of residual viable disease based on analysis of green (live) channel data of carboplatin-treated cultures by the same methods used in growth characterization as already described. The distribution resembles the untreated cultures at day 10, indicating growth arrest during treatment, with the mean diameter of larger nodules approximately the same as untreated day 10 cultures. (d) Combined plot of normalized viability versus nodule size for 1000 μM carboplatin-treated 3-D cultures plotted on the same log scale as (c). Each data point corresponds to an individual 3-D nodule and is obtained by segmentation of the dark-field image to report size and viability based on intensity data from corresponding fluorescence images of the same spatial field. The bimodal size distribution is readily evident from the two dense clusters of data points to either side of a sparse region between 1600 and 3200 μm2.
The carboplatin treatment response data indicates that although viability is reduced overall, large surviving fractions of nodules are left behind even after incubation for 4 days and in a system where the nodules are totally exposed the agent for the duration of treatment. This could point to a major concern in the clinical scenario, where even submillimeter disseminated disease (which our model system reflects) remains after chemotherapy and ultimately regrows and develops drug resistance. This scenario points toward the use of combination treatment modalities, which enhance the efficacy of chemotherapy by either improving penetration or increasing the sensitivity of cells prior to treatment.
Analysis of PDT Treatment Response in 3-D Nodules
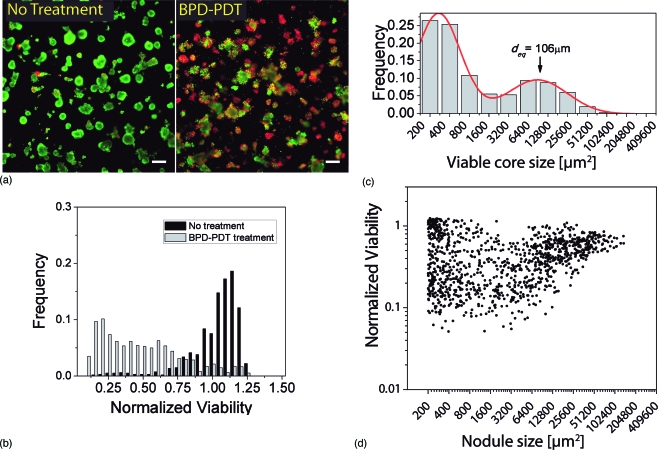
Fluorescence imaging of 3-D cultures subject to 10 J∕cm2 BPD-PDT treatment revealed a profoundly different pattern of response than the chemotherapy treatment (Fig. 4). While some nodules are mostly viable (green), others are structurally degraded with pockets of intense ethidium bromide (red) fluorescence throughout. (Color online only.) This pattern is reflected in the distribution of viabilities of treated nodules normalized to no treatment control [Fig. 4b]. Which has a mean value of 0.46 with standard deviation of 0.29 (minimum value 0.051, maximum value 1.2). Note that since nodule viabilities are normalized to the mean of the untreated control viabilities, it is possible for individual nodules to have a value higher than 1, even though the average is 46% that of the untreated. In contrast to carboplatin-treated cultures already described, in which the size distribution post treatment [Fig. 3c] is very similar to pretreatment [Fig. 1c], the size distribution of residual viable nodules (calcein green channel) following BPD-PDT treated cultures is dramatically shifted toward smaller sized nodules as a result of PDT treatment [Fig. 4c]. Integration of the first three histogram bins in Fig. 4c corresponding to sizes from 200 to 1600 μm2 (nodules up to ∼45 μm in diameter) shows that nodules in this size range account for 68% of the residual disease in the treated culture dishes as compared to 30% in untreated cultures. This is in contrast to carboplatin-treated cultures in which the same size interval accounts for 46% of residual disease, meaning that more than half of the residual nodules are larger than 45 μm equivalent diameters. To better understand how viability correlates with size for BPD-PDT-treated nodules, normalized viability is plotted against nodule size [Fig. 4d]. The vertical spread in data points corresponding to small nodules indicates that this population accounts for the overall high degree of variability. This suggests that as PDT breaks down the larger nodules, a portion of the small fractions that come off remain viable. This observation further points to the potential value of a combination treatment, using PDT to initially disrupt 3-D nodules, followed by chemotherapy to kill surviving cells. The trend evident in the scatter plot from the PDT treatment response of larger nodules (>3000 μm2) suggests that there is a size-dependent trend of increasing viability (decreasing cytotoxicity) with increasing nodule size, though additional studies are warranted to investigate this further. The ability to make such insights further points to the value of the quantitative imaging-based approach employed in this study to reveal urgently required new treatment strategies.
Figure 4.
Photodynamic therapy treatment response assessed by quantitative ratiometric fluorescence imaging. (a) Representative LIVE∕DEAD fluorescence image of 3-D culture subject to 10 J∕cm2 BPD-PDT treatment showing a profoundly different pattern of response than the chemotherapy treatment (Fig. 3). While some nodules are relatively unaffected (mostly viable, green), others are structurally degraded with pockets of deep red fluorescence throughout. Display images adjusted using the hi-lo lookup table (all quantitative analysis is based on raw unprocessed images). Scale bars=250 μm. (b) Distribution of viabilities in 10-J∕cm2 BPD-PDT treated nodules. Mean viability is 0.46 with standard deviation of 0.29 (minimum value 0.05, maximum value 1.2). (c) Size distribution of residual viable disease based on analysis of green (live) channel data of BPD-PDT treated cultures by the same methods used in growth characterization already described. Although mean viability is higher than in the case of the 1000 μM carboplatin incubation, the distribution is dramatically shifted toward smaller sized nodules. The position of the larger population peak is shifted to the left with respect to normal growth [Fig. 1c] or carboplatin incubation [Fig. 3c]. (d) Plot of normalized viability versus nodule size in 10-J∕cm2 BPD-PDT treated cultures. Each data point corresponds to an individual 3-D nodule and is obtained by segmentation of the dark-field image to report size and viability based on intensity data from corresponding fluorescence images of the same spatial field. Size-dependent response exhibits a high degree of scattering in the viabilities of small nodules likely generated by disruption of larger nodules during PDT treatment.
Conclusions
We demonstrated the utility of an imaging-based approach for analysis of tumor growth and treatment response in an in vitro 3-D model of micrometastatic ovarian carcinoma. Using this system, we introduced a model of the mechanism of growth for adherent cancer cells on Matrigel based on migration and assembly behavior that led to the development of a bimodal size distribution of 3-D nodules. The power of the approach introduced here is its ability to quantitatively evaluate and identify statistical trends in growth and cytotoxic response in distributions of micronodules that physically resemble the size and structure of tumors in physiological conditions. This system fills a unique niche in the spectrum of translational science between traditional monolayer cell culture and animal models. As demonstrated, our in vitro system is highly conducive to longitudinal imaging, requiring only the removal of culture dishes from incubation for short periods to place them on and off the microscope stage. While lacking crucial elements of in vivo tumor models such as development of vasculature, the imaging approach here also provides the ability to look at thousands of “in vivo-like” nodules simultaneously without need for anesthesia or endoscopy to obtain access. In future studies, we plan to further leverage this feature of our platform by carrying out longitudinal monitoring of the evolution of the micronodular size distribution past the treatment time point to look at longer term tumor regrowth. This will provide the ability to examine the impact of new interventions on tumor regrowth to design new strategies to most effectively target the most persistent disease.
This study utilized our in vitro research platform to provide new insights into response of micronodular disease to carboplatin chemotherapy and PDT. While carboplatin reduces overall viability, even the 96-h incubation with 1 mM carboplatin leaves significant surviving fractions. Both the visual pattern of cell death and the size distribution of residual viable disease suggests that carboplatin does not penetrate into the centers of nodules, inducing cell death primarily at the periphery. In contrast, we find that BPD-PDT treatment disrupts in vitro nodules, producing a less dramatic reduction in viability of each nodule, but breaking larger nodules down into a more fragmented distribution of smaller nodules and giving rise to a significant change in the size distribution of residual nodules. These observations suggest the evaluation of a combination therapy consisting of PDT to initially disrupt nodules and enhance efficacy of a subsequent carboplatin treatment.
The imaging-based research platform introduced here provides a strong foundation to be built on in future studies. The tumor modeling and imaging tools described could be adapted to different solid tumor disease models and different treatment regimens. By incorporation of other cell types such as fibroblasts, endothelial cells, and macrophages, models could be developed that more fully recapitulate tumor stroma interactions. Such models could serve as a physiologically relevant high-throughput drug screening platform to identify the most promising agents and sequences prior to more costly in vivo testing. Furthermore, while this study focused on the evaluation of cytotoxic response to treatment, the quantitative batch processing of fluorescence image data developed here could be applied to targeted molecular imaging data. This approach could incorporate multiple channels of fluorescence input acquired in high throughput with an automated x-y positioning stage, and the analysis could be adapted for similar quantitative analysis of a broad spectrum of molecular markers in 3-D nodules. In this manner, changes in expression of relevant biomarkers could be reported in correlation with statistical changes in size distribution following treatment, providing the foundation of mechanism-based combination therapies with improved outcomes.
Acknowledgments
We thank Yupeng Tu and Drs. Bryan Spring, Daniel Neuman, and Humra Athar of the Wellman Center for Photomedicine, as well as Dr. Shyamsunder Erramilli of Boston University for helpful discussions of this work. We thank Dr. David Schoenfeld of the Department of Biostatistics at Massachusetts General Hospital and Harvard Medical School for his guidance on interpretation of statistical considerations in our results. This work was supported by National Institutes of Health, Grants R01 AR040352-09, 5R01CA119388-03, and P01 CA084203-06 (to T.H), F32 CA138153 (to C.L.E), and the Wellman Center for Photomedicine Graduate Fellowship (to I.R).
References
- Debnath J. and Brugge J. S., “Modelling glandular epithelial cancers in three-dimensional cultures,” Nat. Rev. Cancer 5(9), 675–688 (2005). 10.1038/nrc1695 [DOI] [PubMed] [Google Scholar]
- Debnath J., Mills K. R., Collins N. L., Reginato M. J., Muthuswamy S. K., and Brugge J. S., “The role of apoptosis in creating and maintaining luminal space within normal and oncogene-expressing mammary acini,” Cell 111(1), 29–40 (2002). 10.1016/S0092-8674(02)01001-2 [DOI] [PubMed] [Google Scholar]
- Muthuswamy S. K., Li D., Lelievre S., Bissell M. J., and Brugge J. S., “ErbB2, but not ErbB1, reinitiates proliferation and induces luminal repopulation in epithelial acini,” Nat. Cell Biol. 3(9), 785–792 (2001). 10.1038/ncb0901-785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissell M. J., Weaver V. M., Lelievre S. A., Wang F., Petersen O. W., and Schmeichel K. L., “Tissue structure, nuclear organization, and gene expression in normal and malignant breast,” Cancer Res. 59(7 Suppl.), 1757–1763s, discussion 1763s–1764s (1999). [PubMed] [Google Scholar]
- Ohmori T., Yang J. L., Price J. O., and Arteaga C. L., “Blockade of tumor cell transforming growth factor-betas enhances cell cycle progression and sensitizes human breast carcinoma cells to cytotoxic chemotherapy,” Exp. Cell Res. 245(2), 350–359 (1998). 10.1006/excr.1998.4261 [DOI] [PubMed] [Google Scholar]
- Finlay J. C., Mitra S., Patterson M. S., and Foster T. H., “Photobleaching kinetics of Photofrin in vivo and in multicell tumour spheroids indicate two simultaneous bleaching mechanisms,” Phys. Med. Biol. 49(21), 4837–4860 (2004). 10.1088/0031-9155/49/21/001 [DOI] [PubMed] [Google Scholar]
- Bigelow C. E., Mitra S., Knuechel R., and Foster T. H., “ALA- and ALA-hexylester-induced protoporphyrin IX fluorescence and distribution in multicell tumour spheroids,” Br. J. Cancer 85(5), 727–734 (2001). 10.1054/bjoc.2001.1977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgakoudi I. and Foster T. H., “Effects of the subcellular redistribution of two nile blue derivatives on photodynamic oxygen consumption,” Photochem. Photobiol. 68(1), 115–122 (1998). 10.1111/j.1751-1097.1998.tb03261.x [DOI] [PubMed] [Google Scholar]
- Foster T. H., Hartley D. F., Nichols M. G., and Hilf R., “Fluence rate effects in photodynamic therapy of multicell tumor spheroids,” Cancer Res. 53(6), 1249–1254 (1993). [PubMed] [Google Scholar]
- Lee G. Y., Kenny P. A., Lee E. H., and Bissell M. J., “Three-dimensional culture models of normal and malignant breast epithelial cells,” Nat. Methods 4(4), 359–365 (2007). 10.1038/nmeth1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho K. R. and Shih I. M., “Ovarian cancer,” Annu. Rev. Pathol. 4, 287–313 (2009). 10.1146/annurev.pathol.4.110807.092246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jemal A., Siegel R., Ward E., Hao Y., Xu J., and Thun M. J., “Cancer statistics, 2009,” Ca-Cancer J. Clin. 59(4), 225–249 (2009). 10.3322/caac.20006 [DOI] [PubMed] [Google Scholar]
- Berek J. S. and Bast R. C. J., “Ovarian cancer,” in Cancer Medicine, Kufe D. W., Pollack R. E., Weichselbaum R. R., Bast R. C. J., Gansler T. S., Holland J. F., and Frei E. I., Eds., pp. 1831–1861, BC Decker, Hamilton, Ontario: (2003). [Google Scholar]
- Bissell M. J. and Radisky D., “Putting tumours in context,” Nat. Rev. Cancer 1(1), 46–54 (2001). 10.1038/35094059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debnath J., Muthuswamy S. K., and Brugge J. S., “Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures,” Methods 30(3), 256–268 (2003). 10.1016/S1046-2023(03)00032-X [DOI] [PubMed] [Google Scholar]
- Grun B., Benjamin E., Sinclair J., Timms J. F., Jacobs I. J., Gayther S. A., and Dafou D., “Three-dimensional in vitro cell biology models of ovarian and endometrial cancer,” Cell Prolif 42(2), 219–228 (2009). 10.1111/j.1365-2184.2008.00579.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrenson K., Benjamin E., Turmaine M., Jacobs I., Gayther S., and Dafou D., “In vitro three-dimensional modelling of human ovarian surface epithelial cells,” Cell Prolif 42(3), 385–393 (2009). 10.1111/j.1365-2184.2009.00604.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara K., “Can carboplatin replace cisplatin for intraperitoneal use?” Int. J. Gynecol. Cancer 18(Suppl. 1), 29–32 (2008). 10.1111/j.1525-1438.2007.01101.x [DOI] [PubMed] [Google Scholar]
- Markman M., “Pharmaceutical management of ovarian cancer: current status,” Drugs 68(6), 771–789 (2008). 10.2165/00003495-200868060-00004 [DOI] [PubMed] [Google Scholar]
- Hasan T., Ortel B., Solban N., and Pogue B., “Photodynamic therapy of cancer,” in Cancer Medicine, Kufe D. W., Bast R. C. J., Hait W. N., Hong W. K., Pollock R. E., Weichselbaum R. R., Holland J. F., and Frei E. I., Eds., pp. 537–548, BC Decker, Hamilton, Ontario: (2006). [Google Scholar]
- Hendren S. K., Hahn S. M., Spitz F. R., Bauer T. W., Rubin S. C., Zhu T., Glatstein E., and Fraker D. L., “Phase II trial of debulking surgery and photodynamic therapy for disseminated intraperitoneal tumors,” Ann. Surg. Oncol. 8(1), 65–71 (2001). 10.1007/s10434-001-0065-x [DOI] [PubMed] [Google Scholar]
- Hahn S. M., Fraker D. L., Mick R., Metz J., Busch T. M., Smith D., Zhu T., Rodriguez C., Dimofte A., and Spitz F., “A phase II trial of intraperitoneal photodynamic therapy for patients with peritoneal carcinomatosis and sarcomatosis,” Clin. Cancer Res. 12(8), 2517–2525 (2006). 10.1158/1078-0432.CCR-05-1625 [DOI] [PubMed] [Google Scholar]
- Zhong W., Celli J. P., Rizvi I., Mai Z., Spring B. Q., Yun S. H., and Hasan T., “In vivo high-resolution fluorescence microendoscopy for ovarian cancer detection and treatment monitoring,” Br. J. Cancer 101(12), 2015–2022 (2009). 10.1038/sj.bjc.6605436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Carmen M. G., Rizvi I., Chang Y., Moor A. C. E., Oliva E., Sherwood M., Pogue B., and Hasan T., “Synergism of epidermal growth factor receptor-targeted immunotherapy with photodynamic treatment of ovarian cancer in vivo,” J. Natl. Cancer Inst. 97(20), 1516–1524 (2005). 10.1093/jnci/dji314 [DOI] [PubMed] [Google Scholar]
- Molpus K. L., Hamblin M. R., Rizvi I., and Hasan T., “Intraperitoneal photoimmunotherapy of ovarian carcinoma xenografts in nude mice using charged photoimmunoconjugates,” Gynecol. Oncol. 76(30), 397–404 (2000). 10.1006/gyno.1999.5705 [DOI] [PubMed] [Google Scholar]
- Molpus K. L., Kato D., Hamblin M. R., Lilge L., Bamberg M., and Hasan T., “Intraperitoneal photodynamic therapy of human epithelial ovarian carcinomatosis in a xenograft murine model,” Cancer Res. 56(5), 1075–1082 (1996). [PubMed] [Google Scholar]
- Kleinman H. K., McGarvey M. L., Hassell J. R., Star V. L., Cannon F. B., Laurie G. W., and Martin G. R., “Basement membrane complexes with biological activity,” Biochemistry 25(2), 312–318 (1986). 10.1021/bi00350a005 [DOI] [PubMed] [Google Scholar]
- Evans C. L., Rizvi I., Hasan T., and de Boer J. F., “In vitro ovarian tumor growth and treatment response dynamics visualized with time-lapse OCT imaging,” Opt. Express 17(11), 8892–8906 (2009). 10.1364/OE.17.008892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limpert E., Stahel W. A., and Abbt M., “Log-normal distributions across the sciences: keys and clues,” BioScience 51(5), 341–352 (2001). 10.1641/0006-3568(2001)051[0341:LNDATS]2.0.CO;2 [DOI] [Google Scholar]