Abstract
The Nuclear Pore Complex (NPC) mediates all exchange between the cytoplasm and the nucleus. Small molecules can passively diffuse through the NPC, while larger cargos require transport receptors to translocate1. How the NPC facilitates the translocation of transport receptor/cargo complexes remains unclear. Here, we track single protein-functionalized Quantum Dot (QD) cargos as they translocate the NPC. Import proceeds by successive sub-steps comprising cargo capture, filtering and translocation, and release into the nucleus. The majority of QDs are rejected at one of these steps and return to the cytoplasm including very large cargos that abort at a size-selective barrier. Cargo movement in the central channel is subdiffusive and cargos that can bind more transport receptors diffuse more freely. Without Ran, cargos still explore the entire NPC, but have a markedly reduced probability of exit into the nucleus, suggesting that NPC entry and exit steps are not equivalent and that the pore is functionally asymmetric to importing cargos. The overall selectivity of the NPC appears to arise from the cumulative action of multiple reversible sub-steps and a final irreversible exit step.
Receptors of the importin–β/karyopherin family utilise a steep RanGTP gradient across the nuclear envelope to carry cargos directionally across the NPC2. Unstructured phenylalanine-glycine repeat (FG-repeat) proteins in the NPC are essential for its function but how they contribute to NPC selectivity is controversial. Contemporary NPC transport models1,3 include the ‘selective phase’4–6, ‘virtual gate’7, ‘dimensionality reduction’8 and ‘polymer brush’9–11 models. In vitro studies suggest that FG-repeat proteins may interact12 to form a ‘selective phase’ hydrogel4,5, through which cargos possessing transport receptors selectively diffuse. Alternatively, FG-proteins may act as an entropic brush9,10, forming an occlusion that can be collapsed by transport receptors9–11. Current transport models differ significantly with respect to the assumed arrangement of the FG repeats, the physical basis for selectivity, and the translocation dynamics.
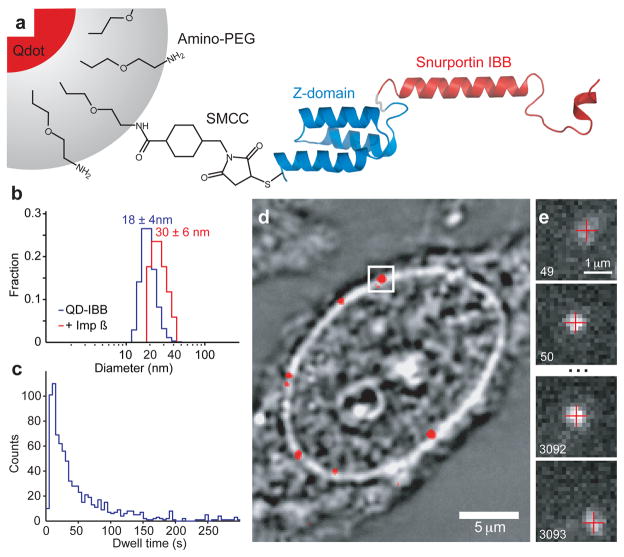
We tracked single QD cargos to discriminate among these models and observe possible transport sub-steps. Previous single molecule studies13–15 have begun to address transport mechanisms using conventional fluorophores and small, rapidly translocating cargos, yielding ~30 nm localisation precisions and trajectories composed of tens of data-points. Unlike conventional fluorophores, QDs are sufficiently bright to be localized with nanometre precision at millisecond temporal resolution16. Extensive QD functionalisation studies yielded a QD-based cargo small enough to be imported into the nucleus when covalently coupled to multiple copies of the importin-β binding (IBB) domain of Snurportin-1, the import receptor for m3G-capped snRNPs17,18 (Fig. 1a, Supplementary Materials). QD-IBBs have a mean hydrodynamic diameter of 18±4 nm and can bind ~40 importin-βs, increasing the diameter to 30±6 nm (Fig. 1b, Supplementary Materials). While larger than single proteins, the size of our QD-IBB is similar to biological substrates that translocate intact through the NPC, such as ribosomal subunits, the Parvovirus capsid, HIV reverse transcription complex, and the human Hepatitis B capsid19–22.
Figure 1. Schematic of Experiment.
(a) Schematic of QD based cargo. The Snurportin-1 IBB/Z-domain fusion protein is coupled via a bifunctional SMCC crosslinker to the amino-PEG polymer coat of a fluorescent QD. The three helix Z-domain acts as a spacer to correctly present the IBB for biological function. Not to scale. (b) Dynamic Light Scattering size distributions of QD-IBB cargos in the presence and absence of importin-β. (c) Dwell time distribution of all QD interactions with the NPC. The time axis is truncated at 300 s. (d) Brightfield image of a nucleus with a quantum dot fluorescence image (with background subtraction applied) overlaid in red. A single quantum dot cargo at the nuclear envelope is highlighted. (e) Individual consecutive frames from a single-molecule experiment showing the arrival (first frame) from the cytoplasm and departure (final frame) of the cargo into the nucleus. The centroids determined from fitting of the PSF are overlaid as red crosses. Frame numbers are in the bottom left hand corner of each frame. Movies were captured at 40 Hz.
QD-IBB nuclear import was studied via an established in vitro transport assay using digitonin permeabilized HeLa cells23 in the presence of recombinant transport factors (importin-β, RanGDP (+GTP), and NTF-2, the RanGDP importer). QD import was specific to the importin-β pathway: import required importin-β and RanGDP (+GTP) and all interactions between the QD cargo and the NPC were abolished when the IBB was replaced with the inert ribonuclease S-peptide (Supplementary Materials).
Of 849 QD-IBB cargo trajectories, 177 featured successful import (as judged by detection of rapid diffusion of the cargo into the nucleus at the end of the trajectory) and 672 aborted; these cargos interacted with an NPC but ultimately returned to the cytoplasm (Fig. 1d, Supplementary Materials). Cargo dwell times varied widely (min: 2 s, median: 34 s, max: 15 min). We analyzed in detail 56 trajectories (mean length, 465 data-points/trajectory) featuring high precision and limited but non-zero blinking, indicating single QDs. Our mean localisation precision was 6 nm at 40 Hz. Trajectories were rotated to a common transport axis, X, using the path of the nuclear envelope (NE) determined by fitting the intensity profile of brightfield images of the NE. This approach was validated using a fluorescent marker of the NE (YFP-importin-β, Supplementary Materials). All 56 cargos ultimately entered a narrow channel within the NPC (Fig. 2a, green). Since all successful import trajectories transit this area we refer to it as the central channel, and use this common feature as a fiducial to align the trajectories to one another (Supplementary Materials). We define 0 on the transport axis as the cytoplasmic edge of the central channel, thus X is negative in the cytoplasm, X = 0 at the cytoplasmic edge of the central channel, and X increases towards the nucleus (Fig. 2a, left).
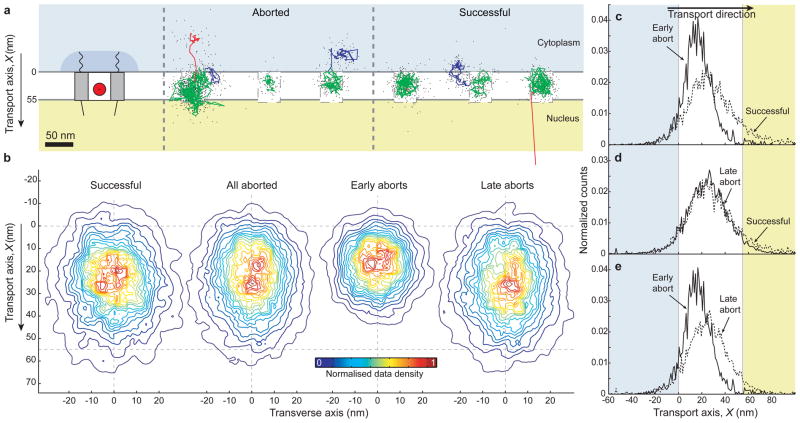
Figure 2. Examples of single-molecule trajectories and a functional map of the NPC interior.
(a) Left, Schematic of the nuclear pore complex showing approximate sizes from the literature. The QD-IBB cargo is shown as a red circle to scale. Right, Three examples each of aborted and successful import trajectories (0.1 s running mean filtered) and coloured according to sub-steps: Docking events are coloured in blue, motion in the central channel in green and undocking in red. Raw, unfiltered data of cargos showing rapid cargo exit into the nucleus are provided as Supplementary Fig. S12. (b) Contour maps of the density of found cargo positions in the central channel for all successful, all aborted, early aborts, and late aborts. The contours are coloured according to the normalised density of observations, with red representing the highest density of found cargo positions and blue the lowest (See supplementary materials for details of trajectory alignment). (c–e) Positional histograms along the transport axis, for successful, early and late aborting cargos.
Analysis of the 56 high precision trajectories revealed that some cargos (9%) were confined for more than 500 ms prior to entering the central channel (Fig. 2a, blue). The location of these cytoplasmic interactions varied greatly among trajectories (up to ~50 nm into the cytoplasm or to the sides of the channel) corresponding to the position and reach of the cytoplasmic filaments24. Similar confinement was also seen at the end of some aborted trajectories, as the cargos left the inside of the NPC and returned to the cytoplasm (Fig. 2a, red). The duration and occurrence of these interactions suggests that both successful and aborted QD-IBB cargos can dock to cytoplasmic filaments but that prolonged docking is not essential for entry into the central channel.
Once within the central channel, cargos displayed one of three behaviours. Some cargos aborted early along the transport axis (X < 30 nm), some aborted late (X < 60 nm), and some entered the nucleus and then diffused away from the NPC (Supplementary Materials). The early aborts (20% of all cargos) explored an area of ~24×26 nm (Fig. 2b, Supplementary Materials). The late aborts and the successes (39% and 41%, respectively) explored a larger area of ~25×38 nm (Fig. 2b, c–e). Accounting for the cargo radius of 15 nm, the explorable area of the central channel is thus 55 nm wide and 68 nm long. The early aborts made only small displacements along the transport axis, while late aborts and successes made large displacements, sometimes moving repeatedly between the cytoplasmic and nuclear faces of the central channel (Fig. 3a–d). Some late aborts and successes were initially confined like early aborts, before escaping and exploring the remainder of the channel (Fig. 3c). We speculated that this early confinement resulted from a channel constriction, which we tested by increasing the QD’s overall diameter to 41±9 nm (Supplementary Materials). Strikingly, with these larger cargos, the ‘early abort’ behaviour dominated, occurring in 59% of trajectories (versus 20% originally). Moreover, pauses between 0 < X < 30 nm became more frequent; 88% of cargos either aborted or paused at 0 < X < 30 nm (versus 43% originally). Thus, making the cargo ~11 nm bigger reduces the overall import probability, increases ‘early aborts’, and increases the occurrence of pauses at 0 < X < 30 nm, all pointing to the presence of an internal constriction (to ~ 40 nm) within the first 30 nm of the central channel.
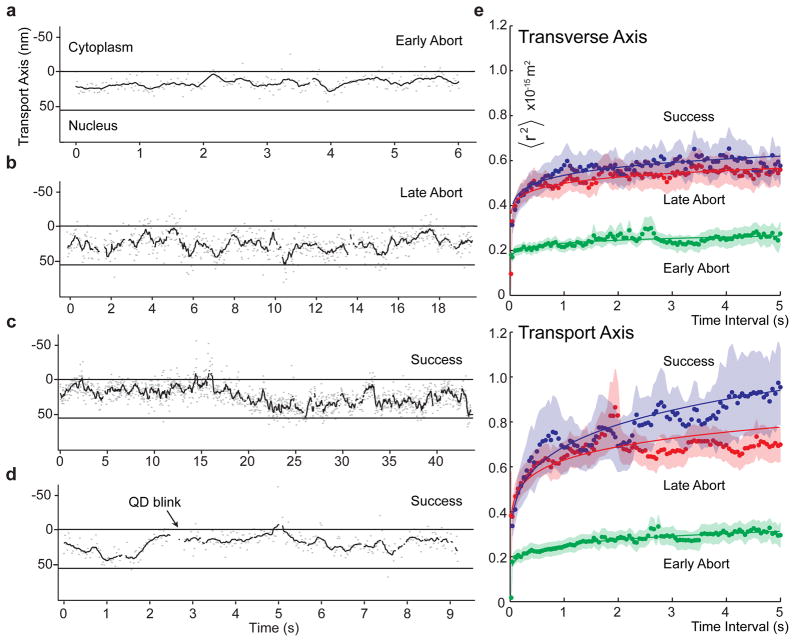
Figure 3. Cargo Motion in the NPC central channel.
(a,b) Examples of early and late aborting cargos. Kymographs showing motion along the transport axis, X, as a function of time (grey, raw data; red, 0.1 s running mean filtered). (c) An example of a successfully importing trajectory showing early confinement at an internal constriction. (d) A second example of a successful trajectory without visible early confinement. (e) Ensemble averaged Mean Squared Displacement (MSD) plots showing anomalous sub-diffusion in the transverse and transport axes of the central channel. Fits to a power law 〈r2〉= Γtα are shown as solid lines (Supplementary Materials). Raw frame-to-frame displacements versus channel position are provided as Supplementary Fig. S13. Errors are standard error of the mean (nsuccess = 23, nearly = 11, nlate = 22).
One key difference among transport models is the predicted motion of the cargo, ranging from directed to sub-diffusive. Frame-to-frame displacements of the QDs were largest at the entry and exit points and smallest within the central channel (Supplementary Materials). The Mean Squared Displacements (MSD) of the cargos in the channel obeyed a power law 〈r2〉 = Γtα with α < 1, indicating anomalous subdiffusion (Fig. 3e). Simple crowding reduces α equally in all directions, whereas anisotropic obstruction (such as by a channel wall) reduces α for motion normal to the wall. We find that inside the NPC, cargo motion in both directions is subdiffusive, consistent with diffusion through a crowded environment, and also anisotropic, consistent with diffusion through a channel. Cargos moved significantly more freely along the transport axis (α||=0.19±0.01, Fig. 3f) than laterally (α⊥ =0.09±0.01) (Supplementary Materials). The large back and forth movements along the transport axis (Fig. 3b,d) are compatible with anomalous subdiffusion but incompatible with models invoking directed movement25 during translocation.
A second key difference among transport models is the predicted effect of receptor density on the ease with which cargos transit. More IBBs (and hence importin-βs) per cargo could either facilitate passage due to greater solubility in the channel or hinder it due to more discrete binding interactions with the NPC. We synthesised QDs with a reduced IBB density by coating the QDs with a 1:1 mixture of IBB to inert S-peptide (QD-S50%IBB50%, Supplementary Materials) and collected 43 high precision trajectories with this cargo. Remarkably, these cargos have a significantly longer dwell time than the QD-IBB100% cargos (p<0.05, Mann-Whitney test, Fig. 4b). The increased overall dwell times arise predominantly from a slowing of the cargo within the channel (p<0.05, Mann-Whitney test, Fig. 4a,c). The inverse relation between crossing time and receptor density argues against models that balance entropic exclusions from the channel with transient, but discrete, interactions between cargo and FG-repeats. In such scenarios, cargos with more IBBs would take longer to cross between the faces of the NPC. Indeed cargos with tens of receptors on their surface, as is the case here, would be unlikely to ever transit the NPC since the diffusion constant should decrease exponentially with the number of binding sites (D ~ e−N)26. By contrast, the finding that cargos with a higher receptor density translocate faster agrees with models like the ‘selective phase’, which assert that cargos with a greater solubility within the FG mesh diffuse more freely.
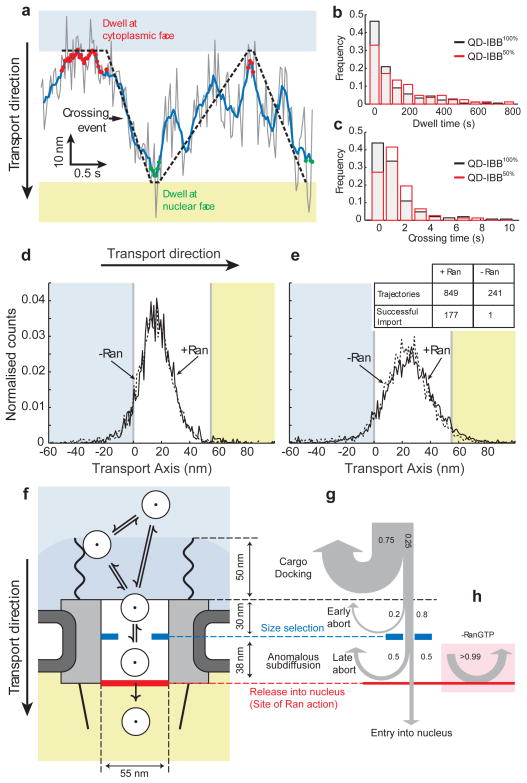
Figure 4. Effect of IBB density on cargo motion, location of Ran action, and data summary.
(a) Detail of a crossing event within the central channel (Grey, raw data; blue line, 0.1s running average). The cargo dwells at the cytoplasmic and nuclear faces of the channel for various durations. The crossing time is the time between leaving the cytoplasmic face and arriving at the nuclear face and vice versa. (b) Cargo dwell times with 50% or 100% IBB. The mean/median dwell time increases for the 50% labelled cargo. The distributions (p<0.05, Kolmogorov Smirnov test) and medians (p<0.05, two-tailed Mann-Whitney test) are significantly different (Supplementary Materials). (c) Crossing time distributions for cargos with 50% or 100% IBB. QB-IBB50% cargos take significantly longer to cross the channel (p<0.05, Mann-Whitney test). (d,e) Position histograms of early (d) and late (e) aborts with and without Ran. Inset: import statistics with and without Ran. The presence of Ran significantly increases the probability of successful import. (f) Results summary. Cargos arriving from the cytoplasm (white circles) may dock on the cytoplasmic filaments or directly enter the NPC. Once inside the central channel, cargos exhibit anomalous subdiffusion. There is a size selective constriction (blue) within the first 30nm of the channel. Efficient cargo exit into the nucleus requires Ran (red bar). (g) Probabilities of cargos being rejected versus position, highlighting the sequence of steps in cargo translocation. Many cargos interact with the cytoplasmic filaments, but most (75%) immediately rebound into the cytoplasm. The remaining 25% interact with the NPC (grey box) for longer times. Of those, 20% abort early due to a size gate and 80% reach the central channel. Once inside the channel, in the presence of Ran, 50% of cargos ultimately enter the nucleus and the remaining 50% abort. (h) In the absence of Ran, cargos do not enter the nucleus (>99% abort) and return to the cytoplasm.
Lastly, we sought to determine where the Ran gradient is coupled to cargo transport. Previous studies have shown that Ran is essential for NPC translocation and abolishing the Ran/importin-β interaction prevents cargo exiting from the nuclear side of the NPC27,28, but it is not known whether Ran affects cargo movement within the central channel. We collected 37 high precision trajectories with the 18 nm QD-IBB100% in a transport assay lacking Ran. Strikingly, the -Ran trajectories were indistinguishable from the +Ran trajectories in all aspects (including area explored, occurrence of early and late aborts, details of movement within the channel), except that the probability of cargo exit into the nucleus, dropped dramatically from 20% to <0.5% (Fig. 4d,e). The absence of an effect on cargo motion within the channel indicates that Ran acts exclusively at the nuclear face, regulating the properties of the FG network or the cargo-receptor-FG interactions to allow cargos to leave the central channel. The +/−Ran comparison also shows that the central channel is functionally asymmetric; with and without Ran, a QD-IBB within the central channel is more likely to return to the cytoplasm than enter the nucleus.
In our study we have watched single large cargos being rejected or imported by the NPC, revealing translocation sub-steps (Fig. 4f–h). (1) The cytoplasmic filaments increase the capture area for transport competent cargos, although lengthy cytoplasmic docking is not essential. (2) Inside the channel, cargos encounter a size selective barrier. (3) Transport competent cargos diffuse anomalously within the central channel, with more free diffusion of cargos with higher receptor density. (4) The central channel is functionally asymmetric. Cargos in the central channel can return to the cytoplasm with and without Ran, but Ran is needed to transit efficiently from the central channel into the nucleus. The mechanistic details of this Ran-driven cargo exit step remain unclear and await further study.
The arrangement of transport sub-steps shown in Fig. 4f-h may be the origin of the NPC’s import efficiency and specificity. The cascade of reversible steps sequentially tests potential cargos for receptor presence, cargo size, and receptor density prior to the irreversible Ran-driven exit step. Since the early steps are reversible, unsuccessful cargos can vacate the NPC and return to the cytoplasm, rather than accumulating within the central channel. This mechanism, consisting of a cascade of reversible pre-filters and a final irreversible exit step, may increase the overall selectivity of facilitated nuclear import while possibly also increasing efficiency, by rejecting most cargos early in the transport reaction.
Methods summary
Amino PEG functionalised QDs, with an emission peak of 605 nm, were functionalised with sulfosuccinimidyl-4-(N-maleimidomethyl)cyclohexane-1-carboxylate, followed by reaction of the maleimide group with the free sulfhydryl group of the purified IBB/Z-domain protein. HeLa cells were permeabilised using digitonin and the QD-IBB cargos (50 pM) were introduced in a transport buffer (20 mM HEPES, 110 mM KOAc, 5 mM NaOAc, 2mM MgOAc, 2 mM DTT, pH 7.3) containing 0.5 mM importin-b, 4 mM RanGDP, 1 mM NTF2, and 1 mM GTP. Single-molecule data were acquired with 25 ms time resolution using a custom-built near-TIRF microscope, incorporating a 100× 1.65 NA objective, 100mW 532nm continuous wave laser and an Andor iXon+ camera. All measurements were taken at 20°C.
Supplementary Material
Acknowledgments
We thank Harish Agarwal for help with initial experiments. Holly Aaron (UC Berkeley Molecular Imaging Center), Ann Fischer (UC Berkeley Cell Culture Facility) and Bruce Cohen (LBNL Molecular Foundry) for use of facilities and discussions. The Bustamante, Chu and Krantz labs for use of equipment. Carlos Bustamante, Matthew Welch and David Grunwald for discussions and comments on the manuscript. This work was partially funded by the NIH (GM058065 to KW and GM77856 to JTL) and the NCI (U54CA143836 to JTL). The authors declare no competing financial interests.
Footnotes
Contributions: ARL, JJS, PK, MS, KW and JTL designed the experiments. ARL, JJS, and PK prepared materials. ARL and JJS performed the QD optimization and functionalisation, the single-molecule experiments, and the data analysis. ARL, JJS, KW and JTL wrote the manuscript.
Competing financial interests: The authors declare no competing financial interests.
Supplementary Information accompanies the paper on www.nature.com/nature.
References
- 1.Peters R. Translocation through the nuclear pore: Kaps pave the way. Bioessays. 2009;31:466–477. doi: 10.1002/bies.200800159. [DOI] [PubMed] [Google Scholar]
- 2.Stewart M. Molecular mechanism of the nuclear protein import cycle. Nat Rev Mol Cell Biol. 2007;8:195–208. doi: 10.1038/nrm2114. [DOI] [PubMed] [Google Scholar]
- 3.Weis K. The nuclear pore complex: oily spaghetti or gummy bear? Cell. 2007;130:405–407. doi: 10.1016/j.cell.2007.07.029. [DOI] [PubMed] [Google Scholar]
- 4.Frey S, Gorlich D. A saturated FG-repeat hydrogel can reproduce the permeability properties of nuclear pore complexes. Cell. 2007;130:512–523. doi: 10.1016/j.cell.2007.06.024. [DOI] [PubMed] [Google Scholar]
- 5.Frey S, Richter RP, Gorlich D. FG-rich repeats of nuclear pore proteins form a three-dimensional meshwork with hydrogel-like properties. Science. 2006;314:815–817. doi: 10.1126/science.1132516. [DOI] [PubMed] [Google Scholar]
- 6.Ribbeck K, Gorlich D. Kinetic analysis of translocation through nuclear pore complexes. EMBO J. 2001;20:1320–1330. doi: 10.1093/emboj/20.6.1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rout MP, Aitchison JD, Magnasco MO, Chait BT. Virtual gating and nuclear transport: the hole picture. Trends Cell Biol. 2003;13:622–628. doi: 10.1016/j.tcb.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 8.Peters R. Translocation through the nuclear pore complex: selectivity and speed by reduction-of-dimensionality. Traffic. 2005;6:421–427. doi: 10.1111/j.1600-0854.2005.00287.x. [DOI] [PubMed] [Google Scholar]
- 9.Lim RY, et al. Nanomechanical basis of selective gating by the nuclear pore complex. Science. 2007;318:640–643. doi: 10.1126/science.1145980. [DOI] [PubMed] [Google Scholar]
- 10.Lim RY, et al. Flexible phenylalanine-glycine nucleoporins as entropic barriers to nucleocytoplasmic transport. PNAS. 2006;103:9512–9517. doi: 10.1073/pnas.0603521103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lim RY, Koser J, Huang NP, Schwarz-Herion K, Aebi U. Nanomechanical interactions of phenylalanine-glycine nucleoporins studied by single molecule force-volume spectroscopy. J Struct Biol. 2007;159:277–289. doi: 10.1016/j.jsb.2007.01.018. [DOI] [PubMed] [Google Scholar]
- 12.Patel SS, Belmont BJ, Sante JM, Rexach MF. Natively unfolded nucleoporins gate protein diffusion across the nuclear pore complex. Cell. 2007;129:83–96. doi: 10.1016/j.cell.2007.01.044. [DOI] [PubMed] [Google Scholar]
- 13.Dange T, Grunwald D, Grunwald A, Peters R, Kubitscheck U. Autonomy and robustness of translocation through the nuclear pore complex: a single-molecule study. J Cell Biol. 2008;183:77–86. doi: 10.1083/jcb.200806173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kubitscheck U, et al. Nuclear transport of single molecules: dwell times at the nuclear pore complex. J Cell Biol. 2005;168:233–243. doi: 10.1083/jcb.200411005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang W, Gelles J, Musser SM. Imaging of single-molecule translocation through nuclear pore complexes. PNAS. 2004;101:12887–12892. doi: 10.1073/pnas.0403675101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lacoste TD, et al. Ultrahigh-resolution multicolor colocalization of single fluorescent probes. PNAS. 2000;97:9461–9466. doi: 10.1073/pnas.170286097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huber J, Dickmanns A, Luhrmann R. The importin-beta binding domain of snurportin1 is responsible for the Ran- and energy-independent nuclear import of spliceosomal U snRNPs in vitro. J Cell Biol. 2002;156:467–479. doi: 10.1083/jcb.200108114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mitrousis G, Olia AS, Walker-Kopp N, Cingolani G. Molecular basis for the recognition of snurportin 1 by importin beta. J Biol Chem. 2008;283:7877–7884. doi: 10.1074/jbc.M709093200. [DOI] [PubMed] [Google Scholar]
- 19.Arhel N, et al. Quantitative four-dimensional tracking of cytoplasmic and nuclear HIV-1 complexes. Nat Meth. 2006;3:817–824. doi: 10.1038/nmeth928. [DOI] [PubMed] [Google Scholar]
- 20.Bukrinsky MI, et al. Active nuclear import of human immunodeficiency virus type 1 preintegration complexes. PNAS. 1992;89:6580–6584. doi: 10.1073/pnas.89.14.6580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pante N, Kann M. Nuclear Pore Complex Is Able to Transport Macromolecules with Diameters of ~39 nm. Mol Biol Cell. 2002;13:425–434. doi: 10.1091/mbc.01-06-0308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rabe B, Vlachou A, Pante N, Helenius A, Kann M. Nuclear import of hepatitis B virus capsids and release of the viral genome. PNAS. 2003;100:9849–9854. doi: 10.1073/pnas.1730940100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adam SA, Marr RS, Gerace L. Nuclear protein import in permeabilized mammalian cells requires soluble cytoplasmic factors. J Cell Biol. 1990;111:807–816. doi: 10.1083/jcb.111.3.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beck M, Lucic V, Forster F, Baumeister W, Medalia O. Snapshots of nuclear pore complexes in action captured by cryo-electron tomography. Nature. 2007;449:611–615. doi: 10.1038/nature06170. [DOI] [PubMed] [Google Scholar]
- 25.Pyhtila B, Rexach M. A gradient of affinity for the karyopherin Kap95p along the yeast nuclear pore complex. J Biol Chem. 2003;278:42699–42709. doi: 10.1074/jbc.M307135200. [DOI] [PubMed] [Google Scholar]
- 26.Bickel T, Bruinsma R. The nuclear pore complex mystery and anomalous diffusion in reversible gels. Biophys J. 2002;83:3079–3087. doi: 10.1016/s0006-3495(02)75312-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gorlich D, Pante N, Kutay U, Aebi U, Bischoff FR. Identification of different roles for RanGDP and RanGTP in nuclear protein import. EMBO J. 1996;15:5584–5594. [PMC free article] [PubMed] [Google Scholar]
- 28.Moore MS, Blobel G. The GTP-binding protein Ran/TC4 is required for protein import into the nucleus. Nature. 1993;365:661–663. doi: 10.1038/365661a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.