Abstract
Background
Ets-1 is a widely expressed transcription factor implicated in several biological processes including hematopoiesis, where it contributes to the regulation of cellular differentiation. The functions of Ets-1 are regulated by transcription factors as well as by phosphorylation events: phosphorylation of threonine 38 activates Ets-1, whereas phosphorylation of a cluster of serines within exon VII reduces DNA binding activity. This study focuses on the role of Ets-1 during granulocytic differentiation of NB4 promyelocytic and HL60 myeloblastic leukemia cell lines induced by all-trans retinoic acid.
Design and Methods
Ets-1 expression was measured by real-time reverse transcriptase polymerase chain reaction and western blotting. The role of Ets-1 during all-trans retinoic acid-induced differentiation was analyzed by using a transdominant negative molecule or small interfering RNA.
Results
NB4 and HL60 cell lines expressed high levels of p51 Ets-1, while the splice variant isoform that lacks exon VII (p42) was almost undetectable. The addition of all-trans retinoic acid reduced p51 Ets-1 levels and induced inhibitory phosphorylation of the remaining protein. Expression of Ets-1 was also reduced during dimethylsulfoxide-induced differentiation and during granulocytic differentiation of human CD34+ hematopoietic progenitor cells but not in NB4.R2 and HL60R cells resistant to all-trans retinoic acid. In line with these observations, transduction of a transdominant negative molecule of Ets-1, which inhibited DNA binding and transcriptional activity of the wild-type Ets-1, significantly increased chemical-induced differentiation. Consistently, Ets-1 knockdown by small interfering RNA increased the number of mature neutrophils upon addition of all-trans retinoic acid. Interestingly, p51 Ets-1 over-expression was frequently observed in CD34+ hematopoietic progenitor cells derived from patients with acute myeloid leukemia, as compared to its expression in normal CD34+ cells.
Conclusions
Our results indicated that a decreased expression of Ets-1 protein generalizes to granulocytic differentiation and may represent a crucial event for granulocytic maturation.
Keywords: hematopoiesis, granulocytic differentiation, transcription factors
Introduction
Ets-1 is the founding member of the ETS family of transcription factors, which is defined by a conserved DNA binding domain spanning 85 amino acids that forms a winged helix-turn-helix structural motif.1
The human ETS-1 gene produces two major protein isoforms, a 51 kDa protein (p51 Ets-1) and a 42 kDa protein (p42 Ets-1).2–3 Ets-1 is expressed in a variety of tissues and in several cases the p42 Ets-1 is co-expressed with full-length p51 Ets-1. p51 and p42 have common and diverse physical properties allowing for both functional redundancy and isoform-specific activity.2 Shared domains include the DNA binding and the pointed transactivation domain. p42 Ets-1 lacks exon VII3 which encodes an auto-inhibitory module4 that regulates Ets-1 DNA binding activity. Cooperative binding with other proteins counteracts auto-inhibition modulating Ets-1 DNA binding affinity.2,5 Ets-1 activity can also be regulated by phosphorylation. Phosphorylation of threonine 38 (T38) within the pointed domain by ERK1/2 activates Ets-1,6,7 whereas phosphorylation of serines S251, S257, S282, S285 (4S) within the N-terminal part of exon VII, by calmodulin-dependent kinase II (CaMKII) and myosin light-chain kinase (MLCK), stabilizes and reinforces the auto-inhibitory module lowering DNA affinity.8–10 The splice variant isoform lacking the inhibitory module may, therefore, function differently from the full-length protein.
Ets-1 plays an important role in cell proliferation, apoptosis, transformation, differentiation, angiogenesis and hematopoiesis. Ets-1 expression is induced in a variety of human tumors, and its level has been associated with the grade of malignancy and prognosis.11 A high level of Ets-1 was also found in leukemic T cells and seems an attractive candidate oncogene involved in the 11q23 amplifications detected in some adult patients with acute myeloid leukemia (AML) with complex karyotypes.12–14
In normal hematopoiesis Ets-1 is involved in the regulation of lymphopoiesis,15,16 in the development of natural killer cells17 and in megakaryopoiesis.18,19 Ets-1 is involved in the regulation of eosinophil-specific promoter,20 but no data are currently available on its role, if any, during granulocytic differentiation.
The human NB4 promyelocytic and HL60 myeloblastic leukemia cell lines represent in vitro models for studying the molecular events taking place during terminal differentiation of myeloid cells. NB4 and HL60 cells are morphologically similar and can undergo granulocytic differentiation when treated with all-trans retinoic acid (ATRA), or other chemical compounds.21,22
NB4 cells carry the t(15;17) translocation which results in the fusion of the promyelocytic leukemia gene (PML) and retinoic acid receptor α gene (RARA). The PML/RARα fusion protein blocks the normal maturation of granulocytes, leading to the accumulation of abnormal promyelocytes. Pharmacological doses of ATRA (10−6 M) overcome this block, inducing differentiation of the immature leukemic promyelocytes into mature granulocytes.
HL60 cells can also be induced to differentiate by ATRA, even though HL60 cells lack the t(15;17) chromosomal translocation. The underlying mechanism of this induction of HL60 cell differentiation remains unclear.
Using these model systems, we analyzed ETS-1 gene expression during ATRA-induced granulocytic differentiation.
Design and Methods
Cell cultures and infection
The NB4, HL60, NB4.R223 and HL60R24 cell lines were grown in RPMI 1640 medium supplemented with 10% fetal calf serum. Granulocytic differentiation was induced with 10−6 M ATRA or 1.4% dimethylsulfoxide (DMSO).
A Phoenix packaging cell line was cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum and transfected by calcium-phosphate/chloroquine with retroviral vectors. The retroviral vector Pinco/IRES-GFP (Pi) containing Ets-1 cDNA has been described previously.19 The Ets-1 transdominant negative mutant (TM)25 was cloned into the same Pi vector. Infectious particles were produced as previously described19 and cells were infected by the spin inoculation method.26
Purification of hematopoietic progenitor cells and granulocytic culture
Adult peripheral blood was obtained from buffy coats of blood donation units. CD34+ hematopoietic progenitor cells were isolated as previously described.27 CD34+ hematopoietic progenitor cells were cultured in serum-free medium in the presence of 1 U/mL interleukin-3, 0.1 ng/mL granulocyte-monocyte colony-stimulating factor and a saturating level of granulocyte colony-stimulating factor (500 U/mL) (PeproTech, Rocky Hill, NJ, USA).
Fresh leukemic blasts were isolated from either bone marrow or peripheral blood obtained from 15 AML patients, using Ficoll-Hypaque density-gradient centrifugation. Further details are provided in the Online Supplementary Design and Methods.
RNA extraction and reverse transcriptase polymerase chain reaction
RNA was extracted using Trizol reagent. Further details on the real-time reverse transcriptase polymerase chain reaction (qRT-PCR) and the semi-quantitative reverse transcriptase polymerase reaction (RT-PCR) are provided in the Online Supplementary Design and Methods.
Whole cell extracts, treatment with calf intestinal phosphatase and western blotting
Extracts were obtained as previously described.19 For treatment with calf intestinal phosphatase (CIP), extracts were prepared without protease inhibitors and chelators and incubated with CIP (Promega, Madison, WI USA). Cell extracts were loaded onto a sodium dodecyl sulfate polyacrylamide gel, transferred onto Hybond-C paper (Amersham Biosciences), incubated with anti-Ets-1 (C-20, Santa Cruz Biotechnology, Santa Cruz, CA, USA), anti-Ets-1 pS282/285 and anti-Ets-1 pT38 phospho-specific (Biosource International, Inc, Camarillo, CA, USA), and anti-actin (Oncogene Research, San Diego, CA, USA) antibodies and detected using an ECL detection kit (Pierce, Rockford, IL, USA). The levels of expression were analyzed using Scion Image Software (Scion, Frederich, MD, USA).
DNA electrophoretic mobility shift assay
The electrophoretic mobility shift assay (EMSA) is described elsewhere19 and further detailed in the Online Supplementary Design and Methods.
Flow cytometry analysis
Annexin V/propidium iodide staining
Cells were incubated with annexin V-binding buffer (2.5 mM CaCl2, 140 mM NaCl, 10 mM Hepes) and Alexa Fluor 647 annexin V conjugate (Invitrogen) for 15 min, re-suspended in the same buffer containing 5 μg/mL propidium iodide (PI) and analyzed by LSR II (Becton Dickinson).
Hoechst 33342 staining
Cells were incubated with phosphate-buffered saline (PBS)/Hoechst 33342 10 μg/mL for 1 h at 37°C then re-suspended in PBS/PI 5 μg/mL and analyzed by LSR II. The detection of cell surface markers is detailed in the Online Supplementary Design and Methods.
Small interfering RNA transfection
Cells were transfected with 160 nM small interfering RNA (siGENOME SMARTpool reagent M-003887-00-0020; Dharmacon Lafayette, CO, USA) and a fluorescein isothiocyanate (FITC)-conjugated double-stranded RNA (Dharmacon), all combined with the INTERFERin™ transfection kit (Polyplus-transfection, New York, NY, USA).
Statistical analysis
Data are presented as mean values and error bars indicate the standard deviation. The groups were compared by two-way analysis of variance (ANOVA) using Bonferroni’s test. P values less than 0.05 are considered statistically significant.
Results
Ets-1 is down-regulated in all-trans retinoic acid-induced neutrophil differentiation of NB4 and HL60 cell lines
Endogenous expression of Ets-1 alternatively spliced isoforms p51 and p42 was analyzed during ATRA-mediated NB4 and HL60 cells differentiation by qRT-PCR, semi-quantitative RT-PCR and western blotting. Cells were treated with 10−6 M ATRA and harvested every 24 h. Cellular differentiation was monitored evaluating increased expression of myeloid-specific surface antigens by flow cytometry (Online Supplementary Figure S1A).
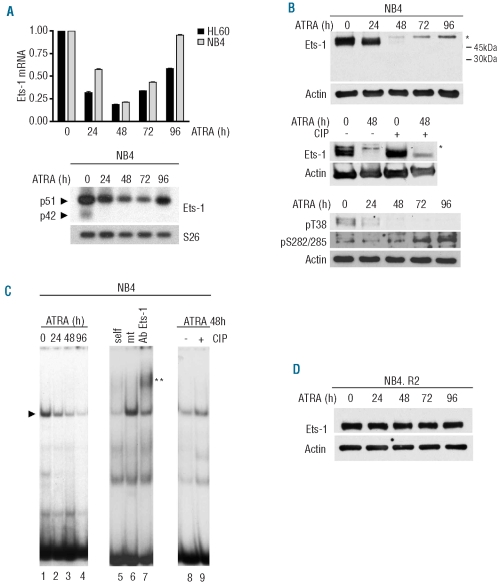
Ets-1 p51 mRNA, highly expressed in untreated cells, was down-regulated upon addition of ATRA (Figure 1A). After 3 days of treatment, Ets-1 mRNA started to increase, reaching relevant levels of expression in differentiated cells. Conversely, the p42 isoform was barely detectable only in untreated samples, as evaluated by semi-quantitative RT-PCR using specific primers recognizing both isoforms (Figure 1A, bottom panel).
Figure 1.
ATRA reduced p51 Ets-1 expression and DNA binding affinity in NB4 and HL60 cells but not in their ATRA-resistant counterpart. NB4 and HL60 cells were treated with 10−6M ATRA and collected at the indicated time points. (A) (Top) Ets1 expression by qRT-PCR. GAPDH endogenous control was used for normalization. Error bars represent standard deviation and indicate the average values from three independent experiments. (Bottom) Semi-quantitative RT-PCR of alternatively spliced isoforms of Ets-1 mRNA in NB4 cells. S26 was used for normalization. (B) (Top) Western blot of Ets-1 protein during ATRA-induced differentiation of NB4 cells and (middle) after CIP treatment. Asterisks represent the size shift of the phosphorylated protein. (Bottom) Western blot using antibodies against Ets-1 pT38 or pS282/285. (C) EMSA of untreated and ATRA-treated NB4 cells (lanes 1–4). Complex formation is indicated by arrows. Extracts were incubated with a 200-fold molar excess of wild-type (lane 5) or mutated (lane 6) cold DNA competitors and anti-Ets-1 antibody (lane 7). The double asterisk indicates the supershift of the complex. EMSA of cell extracts untreated (lane 8) and treated with CIP after 48 h of exposure to ATRA (lane 9). (D) western blot of Ets-1 in ATRA-treated NB4.R2. In all Western Blots, equal loading was confirmed by probing with anti-actin antibody.
The expression levels of Ets-1 protein were then examined by western blotting. As shown in Figure 1B (upper panel), p51 Ets-1 protein was expressed in untreated cells, whereas the p42 isoform was undetectable. The level of p51 decreased after 24 h of treatment and, instead, a protein of slightly higher molecular weight appeared, which increased in a time-dependent manner. The size shift of p51 Ets-1 suggested that the protein may be phosphorylated upon ATRA treatment. In order to establish whether Ets-1 was phosphorylated, we treated the whole cell lysate with CIP, to release the phosphate groups. CIP treatment of 48 h ATRA-treated cell extracts reduced the size shift of Ets-1 protein, thus suggesting that Ets-1 is phosphorylated following ATRA treatment (Figure 1B, middle panel). ATRA could trigger both MAP kinase and calcium-dependent signaling pathways which induce activating (T38) and inhibitory (4S) phosphorylation of Ets-1, respectively. To determine which type of phosphorylation is induced by ATRA we probed the western blot membrane with either an anti-Ets-1 antibody specifically recognizing phosphorylated T38 of Ets-1 (pT38) or with an antibody specifically recognizing the phosphorylated form in serines 282 and 285 (pS282/285) (Figure 1B, bottom panel). As shown in Figure 1B pT38 was clearly detected only in untreated cells, whereas pS282/285 antibody detected a protein which increased over the 4 days of treatment. The simultaneous presence of both activating and inhibitory phosphorylation events has already been described in other cellular systems.28
To determine whether serine phosphorylation may account for decreased Ets-1 binding activity, during ATRA-induced differentiation we performed an EMSA. We examined the binding activity of Ets-1 protein to 32P-labeled synthetic double-stranded oligonucleotides containing a consensus binding motif for Ets-1. A specific Ets-1-DNA complex was clearly evidenced in untreated cells (Figure 1C, lanes 1, 6, 7), whereas ATRA treatment markedly reduced Ets-1-DNA complex formation throughout the differentiation process (Figure 1C, lanes 2–4). As expected,8 CIP treatment of 48 h-treated NB4 whole cell extracts resulted in increased Ets-1 binding activity (Figure 1C, lanes 8, 9). Similar results were obtained during ATRA-induced differentiation of HL60 cells (Online Supplementary Figure S1B and data not shown).
These results indicate that ATRA treatment reduced Ets-1 protein expression and induced inhibitory phosphorylation of the remaining protein in a time-dependent manner.
We next analyzed the expression of Ets-1 in NB4.R2 and HL60R cells, which are resistant to ATRA-induced differentiation. Treatment of NB4.R2 and HL60R cells with ATRA did not decrease Ets-1 expression or induce Ets-1 phosphorylations size shift (Figure 1D and Online Supplementary Figure S1C).
Collectively, these data indicated that Ets-1 down-modulation is part of the ATRA pathways leading to granulocytic differentiation.
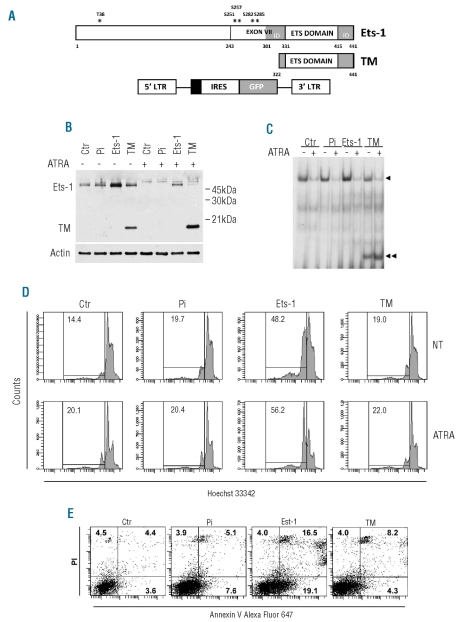
Enforced expression of a dominant negative mutant of Ets-1 in NB4/HL60 enhances all-trans retinoic acid-induced granulocytic differentiation
To further investigate the functional relevance of Ets-1 down-modulation related to ATRA-induced differentiation we infected NB4 cells with either a previously generated retroviral vector carrying Ets-1 cDNA19 or a trans-dominant negative mutant (TM) of Ets-125 (Figure 2A). This mutant contains the DNA binding domain of Ets-1, but lacks the transactivation domain and most of the auto-inhibitory module, thus competing with the wild-type protein for binding to DNA. NB4 cells were infected with Pi/Ets-1, or Pi/TM or its backbone control Pi. After transduction, the cells efficiently expressing green fluorescent protein (GFP) were selected by fluorescence-activated cell sorting (FACS) to establish stable cell lines. Ectopic expression of Ets-1 and TM was confirmed by western blotting. The results were consistent with ectopic expression of Ets-1 close to a physiological range (about 2–2.5 fold) (Online Supplementary Figure S2A). Similarly, the Ets-1 transdominant negative molecule was expressed at a level comparable to that of the endogenous Ets-1 protein (Figure 2B). Experiments showing higher Ets-1 and TM expression (>2.5–3 fold) were not considered to avoid spurious results of excessively over-expression or ‘squelching’. As expected, 48 h of ATRA treatment markedly reduced the expression of Ets-1 protein in control, NB4/TM and NB4/Pi cells; in NB4/Ets-1 cells Ets-1 was also decreased but remained at a level comparable to that in untreated control cells (Figure 2B). After 4 days of treatment, Ets-1 expression was also down-regulated in NB4/Ets-1 cells (data not shown).
Figure 2.
Stable expression of Ets-1 protein and the Ets-1 dominant negative mutant. (A) Representation of Ets-1 protein, Ets-1 transdominant negative (TM) and retroviral vector used for cloning. The black box represents the polycloning site. (B) Western blot of Ets-1 in untreated and 48 h ATRA-treated NB4-transduced cells. Equal loading was confirmed by probing with anti-actin antibody. (C) EMSA of untreated and ATRA-treated (96 h) NB4 transduced cells. Complex formation for Ets-1 and TM is indicated by one or two arrows, respectively. (D) Hypodiploid DNA content FACS analysis in untreated and 96 h ATRA-treated NB4 transduced cells. (E) FACS analysis of apoptotic events by double-staining with annexin V Alexa Fluor 647 conjugate and propidium iodide in NB4, NB4/Pi, NB4/Ets-1 or NB4/TM cells.
Suppression of Ets-1 binding activity by the ectopically expressed Ets-1 transdominant negative mutant was then analyzed by EMSA. Ets-1 activity was detected to a similar extent in both control cells and cells infected with empty vector, enhanced in NB4/Ets-1 cells and partially reduced in TM-infected cells (Figure 2C). However, upon 96 h of ATRA treatment, which decreased both protein level and binding activity, the Ets-1 transdominant negative mutant efficiently bound to the consensus binding sequence for Ets-1.
In a cell viability assay (trypan blue staining) we observed reduced proliferation in both transdominant negative and Ets-1 transduced cells (Online Supplementary Figure S2B).Conversely, only NB4/Ets-1 cells showed a marked increase in cell mortality. As expected, during ATRA-induced differentiation, reduced proliferation was observed in all cell clones (Online Supplementary Figure S2B). In order to investigate the mechanisms responsible for the observed anti-proliferative effect, we analyzed cell cycling. No significant differences in cell cycle phases were observed among the different cell populations (data not shown). Conversely, the percentage of cells with hypodiploid DNA content, characteristic of apoptotic cells, was significantly higher in Ets-1 over-expressing cells (Figure 2D). Induction of differentiation was associated with a slightly increased apoptosis in all cell populations (Figure 2D).
To confirm the apoptotic effect of Ets-1 over-expression, cells were stained with annexin V and PI and subjected to flow cytometric quantification. The percentage of early (annexin V+/PI−) and late (annexin V+/PI+) apoptotic cells was significantly higher in Ets-1 over-expressing NB4 cells than in NB4/TM, backbone control and control cells (Figure 2E). Similar results were obtained with transduced HL60 cells (data not shown).
We then analyzed cell differentiation. May-Grunwald-Giemsa staining showed similar morphology in all three transduced cell populations and in NB4 cells. However, after induction for 4 days with 10−6 M ATRA, TM-expressing cells showed a significantly greater fraction of more mature cells corresponding to band cells and metamyelocytes (Online Supplementary Figure S2C).
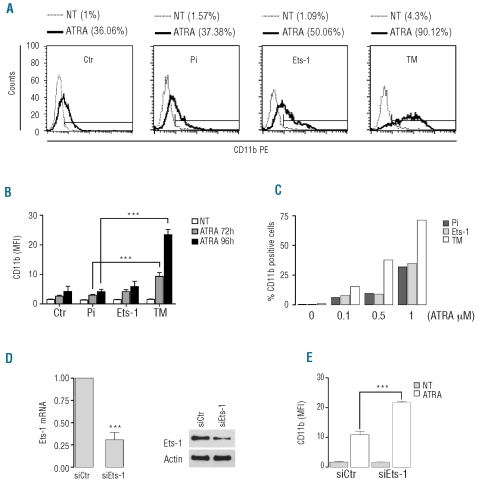
We next analyzed by flow cytometry the expression of the maturation-specific cell surface marker CD11b after 4 days of culture with ATRA. NB4/Ets-1 showed a modest increase of CD11b when compared with control cells. NB4 cells over-expressing the Ets-1 transdominant negative mutant showed a significant increase both in the frequency of CD11b positive cells (up to 90%) (Figure 3A) and in the level of CD11b protein expression as compared to control cells (Figure 3B). Moreover, when cells were treated with a suboptimal amount of ATRA (0.1, 0.5 μM), we observed a marked induction of CD11b expression only in TM-expressing cells (Figure 3C). Similar results were observed in Pi, Ets-1 and TM HL60 transduced cells (data not shown).
Figure 3.
Inhibition of Ets-1 functions increased ATRA-induced differentiation in NB4 cells. (A) FACS analysis of CD11b, in untreated and 96 h ATRA-treated NB4 transduced cells. One representative experiment out of four is presented. (B) Mean fluorescence intensity (MFI) of CD11b in NB4 transduced cells after ATRA treatment. The values reported are the ratio between MFI of the sample and the isotype control. The error bars represent mean ± SD (n=4). (C) Representative experiment of CD11b expression in NB4 transduced cells treated with different concentrations of ATRA for 72 h. (D) NB4 cells transfected with control siRNA (siCtr) or Ets-1 siRNA (siEts-1). (Left) Analysis of Ets-1 mRNA by qRT-PCR at 48 h after transfection. The error bars represent the mean ± SD (n=3). (Right) A representative western blot of Ets-1 performed on NB4 cells transfected with siCtr and siEts-1. (E) Flow cytometric analysis of CD11b MFI in NB4 cells transfected with siCtr and siEts-1 after 72 h of treatment with ATRA. Mean values ± SD from three independent experiments.
To further elucidate the role of Ets-1 during ATRA-induced granulocytic differentiation we knocked-down Ets-1 by using small interfering RNA (siRNA). NB4 cells were transiently transfected with siRNA against Ets-1 (siEts-1) or with control non-targeting siRNA (siCtr) together with a FITC-conjugated double-stranded RNA. Twenty-four hours after transfection, FITC-positive cells were sorted and then cultured. The decreases in the levels of Ets-1 mRNA and protein were confirmed by qRT-PCR and western blot analysis (Figure 3D). Despite the decrease of Ets-1 protein, no difference in the differentiation status was observed when siEts-1 cells were compared with control cells (Figure 3E). Transfected cells were then treated with ATRA and analyzed after 72 h; when compared with the control group, siEts-1 cells showed a significant increase of CD11b expression (Figure 3E).
Altogether, these results confirm that Ets-1 down-regulation is a relevant event for ATRA-induced granulocytic differentiation. However, since transcriptional suppression or siRNA knockdown are not per se sufficient to trigger the differentiation process, our data suggest that the Ets-1 decrease contributes to and/or enhances ATRA-induced differentiation.
Ets-1 down-modulation is required for granulocytic differentiation and maturation of peripheral blood CD34+ hematopoietic progenitor cells
We then investigated whether down-modulation of Ets-1 expression is specific to ATRA-induced granulocytic differentiation or also takes place when cells are treated with other granulocytic differentiation-inducing agents. DMSO-induced granulocytic differentiation of NB4 cells (Online Supplementary Figure S3A) was associated with down-regulation of Ets-1 (Online Supplementary Figure S3B); however, the rate at which Ets-1 decreases upon DMSO treatment is slower than that in ATRA-treated cells, consistent with the weaker effect of DMSO as a promoter of granulocytic differentiation. However, following 72 h of exposure to DMSO, TM-transduced cells showed a higher fraction of differentiated cells as compared to cells over-expressing Ets-1 and cells infected with an empty vector (Online Supplementary Figure S3C).
To confirm the association between the decrease of Ets-1 and granulocyte differentiation, we next analyzed Ets-1 expression in a serum-free culture system for unilineage differentiation and maturation of peripheral blood CD34+ hematopoietic progenitor cells through the granulocytic pathway. In this type of culture, cell growth is associated with a decrease of CD34+ cells and with a progressively increased expression of the granulocytic-specific surface antigen CD15. This type of culture recapitulates physiological hematopoiesis, allowing the analysis of up to 95% neutrophil cells at late days of culture29 (Online Supplementary Figure S3D).
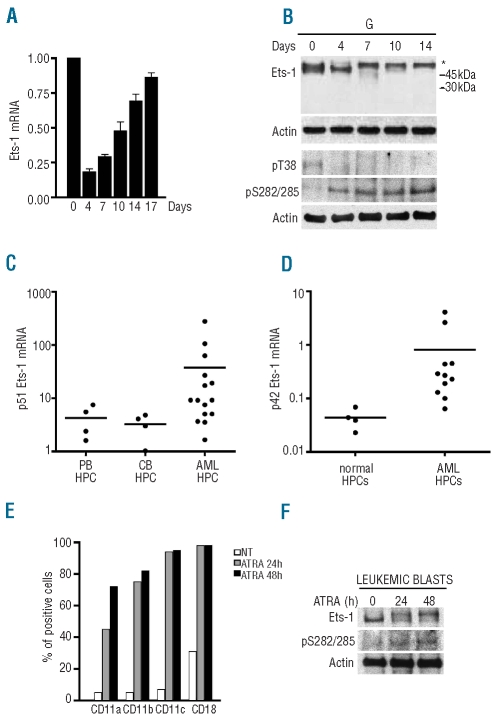
Ets-1 mRNA and protein levels were evaluated by qRT-PCR and western blot analysis; starting from quiescent CD34+ cells, total RNA and proteins were extracted from granulocytic cells at different days of culture until their complete differentiation. As shown in Figure 4A, Ets-1 mRNA was expressed in hematopoietic progenitor cells and its level strongly decreased during the initial days of culture; starting from day 7, predominantly corresponding to the myeloblastic/promyelocytic stages of differentiation, Ets-1 mRNA progressively increased until the end of the maturation process.
Figure 4.
Ets-1 expression levels in CD34+ hematopoietic progenitor cells purified from normal and leukemic samples. (A) qRT-PCR of Ets-1 mRNA was performed on the indicated days in hematopoietic progenitor cells grown in liquid phase unilineage granulocytic culture. GAPDH endogenous control was used for normalization. Error bars represent mean ± SD (n=3). (B) Western blot of Ets-1 protein in CD34+ hematopoietic stem cell granulocytic culture with anti- Ets-1, -pS282/285, -pT38 and -actin antibodies. (C) qRT-PCR of p51Ets-1 mRNA in CD34+ cells derived from peripheral blood (PB) and cord blood (CB) of healthy donors and AML patients (AML). (D) qRT-PCR of p42Ets-1 mRNA in normal CD34+ cells and AML patients. (E) FACS analysis of cell surface antigens following ATRA treatment of leukemic blasts from an t(15;17) AML patient. (F) Western blot of Ets-1 protein in ATRA-treated APL blasts.
The total amount and the phosphorylation level of Ets-1 protein were then examined by western blot (Figure 4B). During unilineage granulocytic differentiation of CD34+ hematopoietic progenitor cells, the level of p51 Ets-1 was reduced and the remaining protein, observed from day 7 of culture onward, was phosphorylated. The increase of inhibitory Ets-1 phosphorylation during normal granulocytic differentiation was confirmed by western blot analysis with either anti-Ets-1 pT38 or pS282/285 antibodies (Figure 4B).
Since Ets-1 levels seem to be elevated in normal CD34+ cells and decrease during granulocytic differentiation, we were particularly interested in exploring Ets-1 levels in CD34+ cells purified from AML samples. The CD34+ fractions, enriched for leukemia-initiating cells, were isolated from patients with various French-American-British subsets of AML, as reported in the Online Supplementary Design and Methods section. Leukemic CD34+ hematopoietic progenitor cells showed a mean 5- to 10-fold increase in Ets-1 expression compared to the levels of Ets-1 expression observed in CD34+ cells purified from either normal peripheral blood or cord blood control samples (Figure 4C). Assuming a cut-off of 10 for Ets-1 mRNA expression in normal CD34+ cells, it appeared evident that nine of 15 cases had significantly higher Ets-1 mRNA levels in their CD34+ fractions than the levels in their normal counterpart (Figure 4C). Interestingly, the p42 isoform, barely detectable in normal CD34+ cells, was clearly expressed in AML CD34+ cells as evaluated by qRT-PCR using primers specifically recognizing the p42 Ets-1 (Figure 4D).
Furthermore, to analyze a link between cell differentiation and Ets-1 protein expression in primary leukemic cells, freshly isolated leukemic blasts from an AML patient with t(15;17) translocation were treated with 10−6 M ATRA and harvested after 24 and 48 h. ATRA-induced differentiation was monitored by evaluating increased expression of myeloid-specific surface antigens by FACS analysis (Figure 4E). Ets-1 protein levels were then analyzed by western blotting. As shown in Figure 4F, ATRA-induced granulocytic differentiation of acute promyelocytic leukemia blasts is associated with both a decrease of Ets-1 protein expression and an increase of inhibitory phosphorylation.
Discussion
The results of the present study show that decreased expression of Ets-1 protein is associated with granulocytic differentiation. Two lines of evidence suggest that this down-regulation is a crucial event. First, in contrast to the differentiation-sensitive NB4 and HL60 cells, the ATRA-resistant NB4.R2 and HL60R cells did not show down-regulation of Ets-1 upon ATRA treatment. Second, inhibition of Ets-1 activity by enforced expression of a transdominant negative Ets-1 molecule potentiated the capability of NB4 and HL60 cells to undergo chemical-induced granulocytic differentiation. The transdominant negative molecule was successfully used to suppress Ets-1 transcriptional activity to inhibit normal and tumor angiogenesis.25,30 This molecule may potentially also interfere with the function of closely related Ets proteins, such as Ets-2.1,2 Nevertheless, specific, targeted inhibition of Ets-1 expression by siRNA resulted in a significant increase of granulocytic differentiation, thus demonstrating that a major role may be ascribed to Ets-1. Furthermore, upon ATRA treatment, the remaining p51 Ets-1 was phosphorylated within the inhibitory module in the exon VII domain. Interestingly, differentiating NB4 and HL60 cells, as well as normal CD34+ hematopoietic progenitor cells, did not express the p42 isoform which is resistant to the negative regulation and confers the ability to p42 Ets-1 to form stable complexes with DNA.
The kinases that are able to phosphorylate the cluster of serines within the inhibitory module are CaMKII8–10 and MLCK,10 both of which mediate calcium-dependent phosphorylation. In NB4 and HL60 cells, ATRA-dependent differentiation is enhanced via modulation of intracellular calcium homeostasis.31,32 Calcium-dependent inhibition of Ets-1 DNA binding could be part of a general mechanism to remove Ets-1 function from the cell during granulocytic differentiation.
Previous studies indicated that the active form of CaMKII decreases during ATRA-induced differentiation and pharmacological inhibition of CaMKII enhances ATRA-induced differentiation in these cellular systems.33 These observations indirectly indicated that CaMKII may not be involved in Ets-1 phosphorylation upon ATRA treatment. Thus MLCK or other kinases, other than CaMKII, seem to be involved in Ets-1 inactivation during granulocytic maturation. Further, it has to be considered that Ets-1 inactivation is not ATRA- dependent, but is generalized to granulocytic differentiation.
In line with the down-modulation of Ets-1 protein levels and activity, we observed a parallel decreased expression of Ets-1 mRNA following up to 48 h of ATRA treatment, while Ets-1 mRNA levels started increasing after 72 h of treatment. This up-regulation might be ascribed to the presence of a functional retinoic acid responsive element on the human Ets-1 promoter,34 but is not reflected by increased protein levels. The same trend was observed during granulocytic differentiation of CD34+ hematopoietic progenitor cells.
These data suggest that the difference between Ets-1 mRNA and protein levels in differentiated granulocytes could be ascribed to microRNA-mediated post-transcriptional regulation. The TargetScan algorithm indicated Ets-1 as a potential target of several microRNA (miR) up-regulated upon ATRA treatment of NB435 and HL6036 cells, such as miR-221, miR-222, miR-181a/b/c, miR-33, miR-124, miR-125, and miR-193. Based on these observations, we investigated the functional relevance of Ets-1 targeting by miR-221/222 as related to granulocytic differentiation. Despite miR binding to Ets-1 3’ UTR, we observed only a slight decrease of Ets-1 protein and a not significant increase of cell differentiation in miR-221/222 overex-pressing cells (V. Lulli et al., unpublished observations 2009), thus suggesting that other miR or a functional set of miR may be implicated in regulating ETS-1 gene expression during differentiation of granulocytic cells.
It is interesting to note that in AML CD34+ cells we observed a frequent over-expression of p51Ets-1 and detectable levels of p42Ets-1, as compared to in normal CD34+ cells. Although we did not explore the functional consequences of Ets-1 over-expression in AML cells, our results may support the hypothesis that elevated Ets-1 expression contributes to the defective differentiation capability of AML CD34+ cells. In line with this hypothesis, we observed that induction of granulocytic differentiation of fresh acute promyelocytic leukemia blasts was associated with decreased Ets-1 expression and increased Ets-1 inhibitory phosphorylation. Consistently, elevated levels of Ets-1 have been detected in many tumors, including breast37 and colon cancer.38
Altogether our observations indicate that: (i) during granulocytic differentiation Ets-1 oncogenic protein is both down-regulated and inactivated; (ii) Ets-1 over-expression induced apoptosis; and (iii) more than 50% of leukemic CD34+ cells analyzed expressed a higher level of Ets-1 mRNA than that in the normal counterpart, suggesting that aberrant Ets-1 expression might interfere with granulocytic maturation.
Ets-1-induced differentiation is worthy of comment. Our results showed that over-expression of the full-length Ets-1 isoform slightly enhances rather than inhibits differentiation. This apparent paradox may be explained by the changed levels of Ets-1 protein which may alter the spectrum of Ets-1 functions and modify the activity of Ets-1 containing complexes, resulting in aberrant activation of the differentiation program. Similarly, the increased apoptosis showed by Ets-1-over-expressing cells may be ascribed to unbalanced composition of Ets-1-containing complexes and/or abnormal Ets-1 activity.
However, in this study we did not define the molecular pathways and networks through which Ets-1 transcription factor acts in the various stages of granulocytic development. Considering the complexity of Ets-1 interactions, the variety of biological processes in which it takes part and the number of potential targets, it is difficult to speculate on the molecular networks/pathways in which it might be involved.
Further studies are warranted to decipher the role of Ets-1 and its implication in myeloid cell maturation, and to understand how deregulated Ets-1 expression triggers an apoptotic response while also blocking the process of differentiation.
Acknowledgments
the authors would like to thank D. Watson and Y. Sato for the kind supply of Ets1 transdominant cDNA. We also thank M. Biffoni and A. Carè for discussion and for critically reading the manuscript. We are grateful to G. Loreto for the graphics. We would also like to thank C. Nervi for the NB4, NB4R.2, HL60 and HL60R cell lines.
Footnotes
Funding: this work was supported by the Italy-USA Oncology Program, Istituto Superiore di Sanità, Rome, Italy to G.M.
The online version of this article has a Supplementary Appendix.
Authorship and Disclosures
The information provided by the authors about contributions from persons listed as authors and in acknowledgments is available with the full text of this paper at www.haematologica.org.
Financial and other disclosures provided by the authors using the ICMJE (www.icmje.org) Uniform Format for Disclosure of Competing Interests are also available at www.haematologica.org.
References
- 1.Watson DK, Seth A. Oncogene Reviews. (55) review issue 5. Vol. 19. Macmillan Publishers Ltd; 2000. Ets Gene Family. [Google Scholar]
- 2.Dittmer J. The biology of Ets-1 proto-oncogene. Mol Cancer. 2003;2:29–49. doi: 10.1186/1476-4598-2-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koizumi S, Fisher RJ, Fujiwara S, Jorcyk C, Bhat NK, Seth A, et al. Isoforms of the human ets-1 protein: generation by alternative splicing and differential phosphorylation. Oncogene. 1990;5(5):675–81. [PubMed] [Google Scholar]
- 4.Skalicky JJ, Donaldson LW, Petersen JM, Graves BJ, McIntosh LP. Structural coupling of the inhibitory regione flanking the Ets domain of murine Ets-1. Protein Sci. 1996;5(2):296–309. doi: 10.1002/pro.5560050214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garvie CW, Pufall MA, Graves BJ, Wolberger C. Structural analysis of the autoinhibition of Ets-1 and its role in protein partnerships. J Biol Chem. 2002;277(47):45529–36. doi: 10.1074/jbc.M206327200. [DOI] [PubMed] [Google Scholar]
- 6.Foulds CE, Nelson ML, Blaszczak AG, Graves BJ. Ras/mitogen-activated protein kinase signaling activates Ets-1 and Ets-2 by CBP/p300 recruitment. Mol Cell Biol. 2004;24(24):10954–64. doi: 10.1128/MCB.24.24.10954-10964.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seidel JJ, Graves BJ. An ERK2 docking site in the pointed domain distinguishes a subset of Ets transcription factors. Genes Dev. 2002;16(1):127–37. doi: 10.1101/gad.950902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cowley DO, Graves BJ. Phosphorylation represses Ets-1 DNA binding by reinforcing autoinhibition. Genes Dev. 2000;14(3):366–76. [PMC free article] [PubMed] [Google Scholar]
- 9.Pufall MA, Lee GM, Nelson ML, Kang HS, Velyvis A, Kay LE, et al. Variable control of Ets-1 DNA binding by multiple phosphates in an unstructured region. Science. 2005;309(5731):142–5. doi: 10.1126/science.1111915. [DOI] [PubMed] [Google Scholar]
- 10.Fleischman LF, Holtzclaw L, Russell JT, Mavrothalassitis G, Fisher RJ. Ets-1 in astrocytes: expression and transmitter-evoked phosphorylation. Mol Cell Biol. 1995;15(2):925–31. doi: 10.1128/mcb.15.2.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oikawa T. ETS transcription factors: possible targets for cancer therapy. Cancer Sci. 2004;95(8):626–33. doi: 10.1111/j.1349-7006.2004.tb03320.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rücker FG, Bullinger L, Schwaenen C, Lipka DB, Wessendorf S, Fröhling S, et al. Disclosure of candidate genes in acute myeloid leukemia with complex karyotypes using microarray-based molecular characterization. J Clin Oncol. 2006;24(24):3887–94. doi: 10.1200/JCO.2005.04.5450. [DOI] [PubMed] [Google Scholar]
- 13.Tyybäkinoja A, Saarinen-Pihkala U, Elonen E, Knuutila S. Amplified, lost, and fused genes in 11q23–25 amplicon in acute myeloid leukemia, an array-CGH study. Genes Chromosomes Cancer. 2006;45(3):257–64. doi: 10.1002/gcc.20288. [DOI] [PubMed] [Google Scholar]
- 14.Mrózek K. Cytogenetic, molecular genetic, and clinical characteristics of acute myeloid leukemia with a complex karyotype. Semin Oncol. 2008;35(4):365–77. doi: 10.1053/j.seminoncol.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bories JC, Willerford DM, Grevin D, Davidson L, Camus A, Martin P, et al. Increased T-cell apoptosis and terminal B-cell differentiation induced by inactivation of the Ets-1 proto-oncogene. Nature. 1995;377(6550):635–8. doi: 10.1038/377635a0. [DOI] [PubMed] [Google Scholar]
- 16.Muthusamy N, Barton K, Leiden JM. Defective activation and survival of T cells lacking the Ets-1 transcription factor. Nature. 1995;377(6550):639–42. doi: 10.1038/377639a0. [DOI] [PubMed] [Google Scholar]
- 17.Barton K, Muthusamy N, Fischer C, Ting CN, Walunas TL, Lanier LL, et al. The Ets-1 transcription factor is required for the development of natural killer cells in mice. Immunity. 1998;9(4):555–63. doi: 10.1016/s1074-7613(00)80638-x. [DOI] [PubMed] [Google Scholar]
- 18.Jackers P, Szalai G, Moussa O, Watson DK. Ets-dependent regulation of target gene expression during megakaryopoiesis. J Biol Chem. 2004;279(50):52183–90. doi: 10.1074/jbc.M407489200. [DOI] [PubMed] [Google Scholar]
- 19.Lulli V, Romania P, Morsilli O, Gabbianelli M, Pagliuca A, Mazzeo S, et al. Ets-1 blocks erythroid and promotes megakaryocytic differentiation through modulation of GATA-2 expression. Cell Death Differ. 2006;13(7):1064–74. doi: 10.1038/sj.cdd.4401811. [DOI] [PubMed] [Google Scholar]
- 20.McNagny KM, Sieweke M, Doderlein G, Graf T, Nerlov C. Regulation of eosinophil-specific gene expression by a C/EBP–Ets complex and GATA-1. EMBO J. 1998;17(13):3669–80. doi: 10.1093/emboj/17.13.3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khanna-Gupta A, Kolibaba K, Zibello TA, Berliner N. NB4 cells show bilineage potential and an aberrant pattern of neuthrophil secondare granule protein gene expression. Blood. 1994;84(1):294–302. [PubMed] [Google Scholar]
- 22.Breitman TR, He RY. Combinations of retinoic acid with either sodium butyrate, dimethil sulfoxide, or hexamethylene bisacetamide synergistically induce differentiation of the human myeloid leucemia cell line HL60. Cancer Res. 1990;50(19):6268–73. [PubMed] [Google Scholar]
- 23.Ruchaud S, Duprez E, Gendron MC, Houge G, Genieser HG, Jastorff B, et al. Two distinctly regulated events, priming and triggering, during retinoid-induced maturation and resistance of NB4 promyelocytic leukemia cell line. Proc Natl Acad Sci USA. 1994;91(18):8428–32. doi: 10.1073/pnas.91.18.8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robertson KA, Emami B, Collins SJ. Retinoic acid-resistant HL-60R cells harbor a point mutation in the retinoic acid receptor ligand-binding domain that confers dominant negative activity. Blood. 1992;80(8):1885–9. [PubMed] [Google Scholar]
- 25.Nakano T, Abe M, Tanaka K, Shineha R, Satomi S, Sato Y. Angiogenesis Inhibition by transdominant mutant Ets-1. J Cell Physiol. 2000;184(2):255–62. doi: 10.1002/1097-4652(200008)184:2<255::AID-JCP14>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 26.Bahnson AB, Dunigan JT, Baysal BE, Mohney T, Atchison RW, Nimgaonkar MT, et al. Centrifugal enhancement of retroviral mediated gene transfer. J Virol Methods. 1995;54(2–3):131–43. doi: 10.1016/0166-0934(95)00035-s. [DOI] [PubMed] [Google Scholar]
- 27.Gabbianelli M, Testa U, Massa A, Pelosi E, Sposi NM, Riccioni R, et al. Hemoglobin switching in unicellular erythroid culture of sibling erythroid burst-forming units: kit ligand induces a dose-dependent fetal hemoglobin reactivation potentiated by sodium butyrate. Blood. 2000;95(11):3555–61. [PubMed] [Google Scholar]
- 28.Grenningloh R, Miaw SC, Moisan J, Graves BJ, Ho IC. Role of Ets-1 phosphorylation in the effector function of Th cells. Eur J Immunol. 2008;38(6):1700–05. doi: 10.1002/eji.200738112. [DOI] [PubMed] [Google Scholar]
- 29.Felli N, Pedini F, Romania P, Biffoni M, Morsilli O, Castelli G, et al. MicroRNA 223-dependent expression of LMO2 regulates normal erythropoiesis. Haematologica. 2009;94(4):479–86. doi: 10.3324/haematol.2008.002345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lefter LP, Dima S, Sunamura M, Furukawa T, Sato Y, Abe M, et al. Transcriptional silencing of ETS-1 efficiently suppresses angiogenesis of pancreatic cancer. Cancer Gene Ther. 2009;16(2):137–48. doi: 10.1038/cgt.2008.65. [DOI] [PubMed] [Google Scholar]
- 31.Launay S, Giannì M, Diomede L, Machesky LM, Enouf J, Papp B. Enhancement of ATRA-induced cell differentiation by inhibition of calcium accumulation into the endoplasmic reticulum: cross-talk between RAR alpha and calcium-dependent signaling. Blood. 2003;101(8):3220–8. doi: 10.1182/blood-2002-09-2730. [DOI] [PubMed] [Google Scholar]
- 32.Launay S, Giannì M, Kovàcs T, Bredoux R, Bruel A, Gélébart P, et al. Lineage-specific modulation of calcium pump expression during myeloid differentiation. Blood. 1999;93(12):4395–405. [PubMed] [Google Scholar]
- 33.Si J, Collins SJ. Activated Ca2+/calmodulin-dependent protein kinase II gamma is a critical regulator of myeloid leukemia cell proliferation. Cancer Res. 2008;68(10):3733–42. doi: 10.1158/0008-5472.CAN-07-2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.So EN, Crowe DL. Characterization of a retinoic acid responsive element in the human ets-1 promoter. IUBMB Life. 2000;50(6):365–70. doi: 10.1080/713803742. [DOI] [PubMed] [Google Scholar]
- 35.Fatica A, Rosa A, Ballarino M, De Marchis ML, Rasmussen KD, Bozzoni I. Role of microRNAs in myeloid differentiation. Biochem Soc Trans. 2008;36(Pt 6):1201–5. doi: 10.1042/BST0361201. [DOI] [PubMed] [Google Scholar]
- 36.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–8. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 37.Katayama S, Nakayama T, Ito M, Naito S, Sekine I. Expression of the ets-1 proto-oncogene in human breast carcinoma: differential expression with histological grading and growth pattern. Histol Histopathol. 2005;20(1):119–26. doi: 10.14670/HH-20.119. [DOI] [PubMed] [Google Scholar]
- 38.Nakayama T, Ito M, Ohtsuru A, Naito S, Sekine I. Expression of the ets-1 proto-oncogene in human colorectal carcinoma. Mod Pathol. 2001;14(5):415–22. doi: 10.1038/modpathol.3880328. [DOI] [PubMed] [Google Scholar]