Abstract
Background
Cdx4 is a homeobox gene essential for normal blood formation during embryonic development in the zebrafish, through activation of posterior Hox genes. However, its role in adult mammalian hematopoiesis has not been extensively studied and its requirement in leukemia associated with Hox gene expression alteration is unclear.
Design and Methods
We inactivated Cdx4 in mice through either a germline or conditional knockout approach and analyzed requirement for Cdx4 in both normal adult hematopoiesis and leukemogenesis initiated by the MLL-AF9 fusion oncogene.
Results
Here, we report that loss of Cdx4 had a minimal effect on adult hematopoiesis. Indeed, although an increase in white blood cell counts was observed, no significant differences in the distribution of mature blood cells, progenitors or stem cells were observed in Cdx4-deficient animals. In addition, long-term repopulating activity in competitive transplantation assays was not significantly altered. In vitro, B-cell progenitor clonogenic potential was reduced in Cdx4-deficient animals but no significant alteration of mature B cells was detected in vivo. Finally, induction of acute myeloid leukemia in mice by MLL-AF9 was significantly delayed in the absence of Cdx4 in a retroviral transduction/bone marrow transplant model.
Conclusions
These observations indicate that Cdx4 is dispensable for the establishment and maintenance of normal hematopoiesis in adult mammals. These results, therefore, outline substantial differences in the Cdx-Hox axis between mammals and zebrafish and support the hypothesis that Cdx factors are functionally redundant during mammalian hematopoietic development under homeostatic conditions. In addition, our results suggest that Cdx4 participates in MLL-AF9-mediated leukemogenesis supporting a role for Cdx factors in the pathogenesis of myeloid leukemia.
Keywords: Cdx4, homeobox, leukemia, MLL-AF9, hematopoietic stem cell, Hox
Introduction
The Cdx genes are the mammalian homolog of the Drosophila caudal gene and encode homeobox transcription factors that regulate axial elongation and anterior-posterior patterning during embryogenesis through modulation of Hox gene expression.1–5 The Cdx gene family consists of three members: Cdx1, Cdx2, and Cdx4.6–8
Several studies have indicated a role for Cdx genes in normal hematopoiesis during embryonic development. In the zebrafish embryo, cdx1 and cdx4 are important for blood formation through activation of posterior Hox genes.9,10 In addition, during in vitro hematopoietic differentiation of murine embryonic stem cells (mESC), inactivation of Cdx4 results in reduced hematopoietic colony-forming potential, which is almost completely abolished upon compound Cdx1, Cdx2, and Cdx4 inactivation.6 Conversely, over-expression of Cdx1, Cdx2, or Cdx4 in mESC was shown to facilitate early hematopoietic progenitor formation through up-regulation of Hox gene expression.6,7,9,11,12 Of note, Cdx4 over-expression in this system resulted in enhanced formation of progenitors with lymphoid repopulation capacity, suggesting a specific role for Cdx4 during lymphopoiesis.11 Interestingly, several studies have suggested a connection between Cdx4 and the mixed lineage leukemia gene, MLL, a master regulator of Hox gene expression13–15 implicated in normal and malignant hematopoiesis.16 In support of this connection, a similar pattern of Hox gene dysregulation is observed in Mll−/− and Cdx4−/− embryos.13,14 In addition, in vitro hematopoietic differentiation of Mll−/− mESC can be rescued by over-expression of Cdx4.14 Finally, Cdx4 has been found to interact with menin, a co-factor of MLL in myeloid leukemogenesis17,18 which participates in the control of Hox gene expression.19 Taken together, these observations suggest that Cdx4 plays an important role in the control of normal embryonic hematopoiesis, likely through a regulatory network involving Cdx4, Mll, and Hox genes; however, they do not directly establish the normal function of Cdx4 in adult mammalian hematopoiesis.
CDX genes have also been implicated in human hematopoietic malignancies. CDX2 was found to be fused to the ETV6 gene in the rare chromosomal translocation t(12;13)(p13;q12) associated with acute myeloid leukemia (AML), resulting in its deregulated expression.20 Subsequent studies identified aberrant CDX2 expression in the majority of adult AML patients regardless of karyotype.21,22 In addition, CDX2 expression correlates with persistence of minimal residual disease and has been proposed as a negative prognostic marker in acute lymphoblastic leukemia,23,24 suggesting that CDX2 could be involved in both myeloid and lymphoid malignancies. Similarly, aberrant expression of CDX4, located on chromosome X, was also observed in patients with AML.25 Consistent with a causal role in malignant transformation of myeloid stem and progenitor cells, over-expression of Cdx2 or Cdx4 in murine bone marrow transplant models alters Hox gene expression and results in AML.22,25,26
Despite their well-established role in leukemogenesis, the physiological function of Cdx family members during normal mammalian hematopoiesis remains incompletely understood. We, therefore, assessed the consequences of germline and conditional Cdx4 deletion in mice. Furthermore, we investigated the impact of Cdx4 loss on the initiation and maintenance of AML induced by the chimeric MLL-MLLT3 (also known and hereafter referred to as MLL-AF9) fusion gene in a murine bone marrow transplantation model.
Design and Methods
Generation of Cdx4 knockout mice
To create the conditional Cdx4 mouse strain, a targeting vector containing loxP sites flanking the first exon, 5′ untranslated region and the proximal promoter region of the Cdx4 gene was generated (Figure 1B). This exon encodes the majority of the coding sequence of CDX4 (165 out of 282 amino acids including the homeodomain) and its excision would preclude expression of a stable, functional protein. This construct was transfected into mESC, and ESC clones were selected with hygromycin and screened by polymerase chain reaction (PCR) and Southern blot analyses. Correctly targeted ESC (i.e. clone 9 shown in Figure 1) were injected into Balb/C blastocysts to obtain chimeric animals that were crossed with C57/B6 wild-type mice to obtain germline transmission of the floxed Cdx4 allele (termed Cdx4F). The Cdx4F allele was back-crossed for at least six generations with C57/B6 wild-type animals (Stock # 000664, The Jackson Laboratory). Cdx4F/WT were then bred with Mx1-Cre transgenic animals and interbred to obtain Cdx4F/F Mx1-Cre (abbreviated Cdx4F/F-Cre+) animals and Cdx4F/F control (abbreviated Cdx4F/F-Cre− or F/F-Cre−) animals that were treated with polyinosinic-polycytidylic acid (pIpC; Sigma, St. Louis, MO, USA; five intraperitoneal injections of 500 μg every other day). Excision efficiency was assessed by PCR with the following primers: (i) Cdx4−/− mice: WT-F: 5′-GCA CCT GCG GTA TAA ATT CT-3′; WT-R: 5′-GCA ACT CAG AAC AGG TCC TT-3′; GFP-F: 5′-TCA TCT GCA CCA CCG GCA A-3′; GFP-R: 5′-GTT GTA GTT GTA CTC CAG CT-3′. Wild-type and mutant alleles gave 200-bp and 300-bp PCR products, respectively; (ii) Cdx4F/F mice: WT-F5: 5′-CTT TAC GGA TGG TTG TGA GC-3′; WT-R5: 5′-AGG ACA GGA ACT CAT GGA GTT T-3′; Exc-R1: 5′-GGC CGC TCT AGA ACT AGT GGA-3′. Wild-type, floxed, and excised alleles generated 200-bp, 250-bp, and 300-bp PCR products, respectively. Mice bearing a germline deletion of Cdx4 exon 1 (termed Cdx4− allele) were described previously.6 Approval for the use of animals in this study was granted by the Children’s Hospital Boston Animal Care and Use Committee.
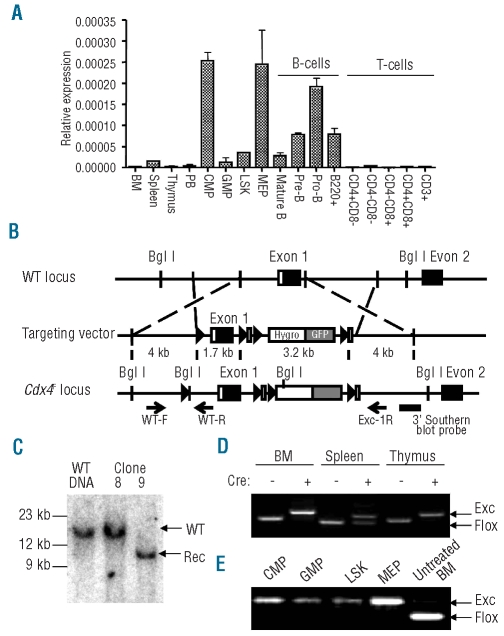
Figure 1.
Cdx4 expression and conditional inactivation strategy. (A) Quantitative real-time RT-PCR was used to measure Cdx4 mRNA expression levels relative to β-actin in normal hematopoietic tissues and flow sorted cells. BM: bone marrow; PB: peripheral blood; LSK, enriched for hematopoietic stem cells; CMP common myeloid progenitors; GMP, granulocyte-monocyte progenitors; MEP, megakaryocyte-erythroid progenitors. (B) Homologous recombination targeting strategy to obtain Cdx4F/F mice. (C) Southern blot analysis using the 3′ probe indicated in Panel B. WT: wild-type Cdx4 locus, Rec: Cdx4 locus target by the conditional knock-out construct. Clone 9 showed homologous recombination and was used to generate the conditional knock-out mouse line. Clone 8 was not correctly targeted, and demonstrates the germline configuration. (D) PCR to assess Cdx4 excision 5 weeks after pIpC treatment was performed on DNA from the indicated organs of Cdx4F/F Mx1Cre+ or Cdx4F/F Mx1Cre− animals using the three primers shown in Panel B (WT-F, WT-R, Exc-1R). Flox: unexcised allele, Exc: Excised allele. (E) LSK cells and myeloid progenitor populations from Cdx4F/F Mx1Cre+ were flow sorted 6 weeks after pIpC treatment and DNA was extracted to assess excision efficiency as described in (D). DNA from untreated Cdx4F/F Mx1Cre+ total bone marrow cells was used as a control.
Real-time quantitative reverse transcription polymerase chain reaction
Total RNA from sorted cells or hematopoietic tissues was isolated using the Trizol reagent (Invitrogen). RNA samples were reverse-transcribed with the Superscript II kit (Invitrogen). Real-time quantitative PCR assays for Cdx4 and β-actin (reference gene) were obtained from Applied Biosystems (Mm00432451_m1 and Mm01205647_g1, respectively). PCR were performed on an ABI-7000 sequence detection system. The ΔΔCt method was used to calculate expression of Cdx4 relative to β-actin. Hox genes were quantified as described previously.25 All reactions were performed on an ABI-7000 sequence detection system using SYBR Green PCR Master Mix or Taqman Universal PCR Master Mix reagents (Applied Biosystems).
Flow cytometry and cell sorting analyses
Single-cell suspensions were prepared from bone marrow cells after red blood cell lysis (Puregene). Cells were stained following standard procedures using antibodies purchased from BD-Pharmingen. Briefly, Lin−Sca1+cKit+ (LSK) cells and myeloid progenitors, including common myeloid progenitors (CMP), granulocyte-macrophage progenitors (GMP), and megakaryocyte-erythrocyte progenitors (MEP), were purified as follows. Total bone marrow cells from 6- to 8-week old wild-type C57/Bl6 animals were obtained after flushing the femora, tibiae and humeri and red blood cells were lysed (Puregene RBC lysis buffer, Qiagen). Cells were then stained with a cocktail of rat-anti-mouse antibodies against mature cells (lineage-positive cells), including Ter119, B220, CD3, CD4, CD8, IL7-R, CD19, and Gr1. After incubation with sheep anti-rat antibody coated magnetic beads (Dynabeads M-450, Dynal, Invitrogen), lineage-positive cells were physically depleted using a magnet (Invitrogen). Cells were then incubated with goat-anti rat PE-Cy5.5 conjugated antibody. After washing, cells were blocked with rat IgG prior to incubation with anti-CD34 FITC conjugated antibody, c-Kit APC conjugated antibody, Sca-1 PE-Cy7 conjugated antibody (BD Pharmingen) and Fcγ-RII/III PE conjugated antibody (Abcam). Cells were flow sorted using a double laser (488 nm/350 nm Enterprise II and 647nm Pectrum) FACS (FACSAria, BD Biosciences). For T-cell populations, thymocytes were obtained from 6- to 8-week old wild-type C57/Bl6 animals. Cells were stained with anti-CD4, anti-CD8 and anti-CD3 antibodies (BD Pharmingen) and populations were purified as indicated in Figure 1A. For B-cell populations, total bone marrow cells from 6- to 8-week old wild-type C57/Bl6 animals were stained with anti-B220, anti-CD19, anti-IgM and anti-CD43 and were purified using the gates exemplified in Figure 2B. Cells were flow sorted using a double laser (488 nm/350 nm Enterprise II and 647nm Pectrum) FACS (FACSAria, BD Biosciences) and analyses were carried out on a four-color FACSCalibur (Becton Dickinson). Raw data were analyzed using FlowJo software.
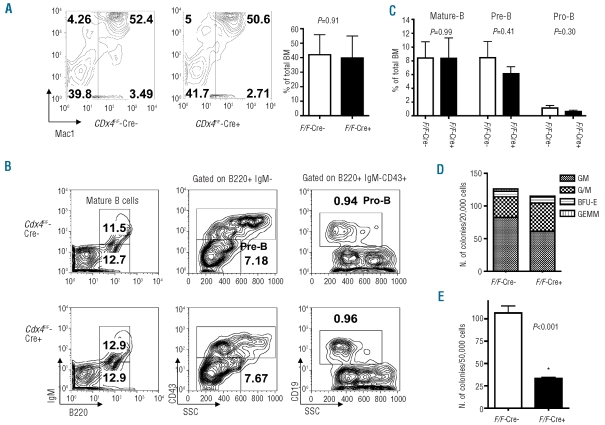
Figure 2.
Analysis of the hematopoietic compartment in Cdx4F/F-Cre+ mice. Flow cytometric analysis of the myeloid lineage (A) and B-cell lineage (B) analyzed on bone marrow cells (n=4). Except where otherwise indicated, analyses were gated on total bone marrow cells. (C) Histogram representation of results presented in Panel B (n=4). (D) Myeloid colony-forming potential of total bone marrow cells in SCF, IL3, IL6 and EPO-supplemented methylcellulose cultures (M3434, StemCells). Colonies were scored 7 days after plating and the mean ± SD is shown (n=4). GM, granulocyte macrophage; G/M, granulocyte or monocyte; GEMM, granulocyte erythroid macrophage megakaryocyte; BFU-E, burst-forming unit-erythroid. (E) B-cell colony-forming potential of total bone marrow cells in IL7-supplemented methylcellulose cultures (M3630, StemCells). Colonies were scored 10 days after plating and mean ± SD is shown (n=4).
Colony assays
Myeloid and pre-B colony-forming assays were performed by plating 20,000 and 50,000 bone marrow cells per dish in duplicate into methylcellulose medium M3434 and M3630 (Stem Cell Technologies, Vancouver, BC, Canada), respectively. Colonies were counted on day 7 and 10, respectively. Bone marrow cells from leukemic mice were plated in methylcellulose medium M3434 at 10,000 cells per dish in duplicate. Colonies were counted on day 7 and serially re-plated every 7 days at a density of 10,000 cells per dish.
Retroviral production, bone marrow transduction and transplantation assays
Retroviral supernatant production and bone marrow transplants were performed as previously described27 using MSCV-MLL-AF9-GFP and MSCV-GFP. Briefly, 8-week old Cdx4−/− or wild-type litter-mate donor mice were injected with 5-fluorouracil (5-FU: 150 mg/kg) 5 days prior to bone marrow collection. On day 0, primary bone marrow cells were obtained from femora and tibiae by flushing with PBS 1x supplemented with 2% FBS; red blood cells were lysed (Puregene, Qiagen) and cells were then cultured overnight in RPMI 1640 supplemented with 10% FBS and IL3 (10 ng/mL), IL6 (20 ng/mL), SCF (10 ng/mL). Cells were mixed with similar titer viral supernatants twice on day 1 and day 2, centrifuged for 90 min at 2500 rpm and 33°C each time (spinfection) and returned to the incubator for 2–3 hours. After the second spinfection, cells were washed and re-suspended in Hank’s balanced salt solution 1x (HBSS 1x) and 1×106 cells were injected into the tail veins of each lethally irradiated recipient. Animals were monitored daily for disease development and sacrificed according to institutional guidelines.
Non-competitive and competitive transplants were carried out with two sets of donor mice in two independent experiments, with five recipient mice per group in each experiment. CD45.2+ bone marrow cells from Cdx4F/F−Cre+ and control donor mice were injected into the lateral tail veins of lethally irradiated CD45.1+ C57/B6.SJL recipient mice, either alone for non-competitive transplants, or mixed with competitor bone marrow cells from wild-type F1 C57/B6 mice (CD45.1/2+) for competitive transplants (Figure 3c). Recipient mice were bled from the eyes every 4 weeks up to 16 weeks after transplantation for analysis of CD45 in the peripheral blood. Bone marrow cells were obtained from recipient mice 16 weeks after transplantation for further analysis of the CD45.2 donor contribution.
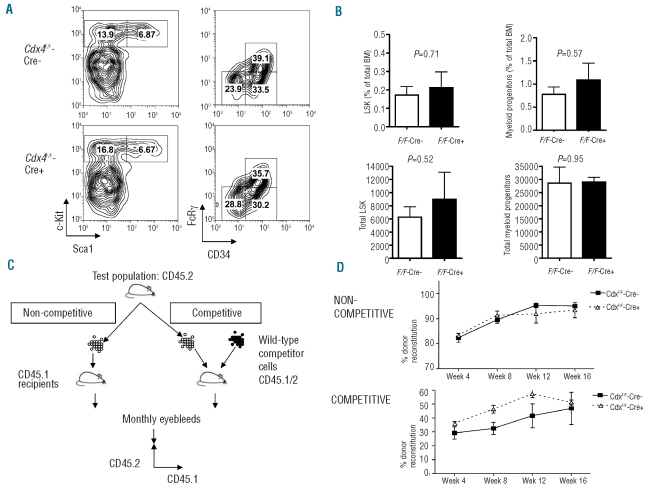
Figure 3.
Analysis of the hematopoietic stem cell compartment in Cdx4F/F-Cre+ mice. (A) Flow cytometric analysis of the LSK (Lin−c-Kit+Sca1+) and myeloid progenitor compartments 4–6 weeks after pIpC treatment. Left panels show 10000 cells gated on viable Lin− cells. Right panels show 10000 cells gated on viable Lin− c-Kit+Sca1− cells (B) Histogram representation of results presented in (A). Mean±SD are shown (n=4). (C) Diagram showing non-competitive and competitive bone marrow repopulation assays. (D) Contribution of donor cells (CD45.2+CD45.1−) to the hematopoiesis of lethally irradiated recipients in non-competitive and competitive transplants. The percentages of CD45.2+ and CD45.1+ cells in the peripheral blood of recipient mice were measured by flow cytometric analysis at 4, 8, 12 and 16 weeks after transplantations (n=5). Mean±SD of the percentage of CD45.2+ cells are shown.
Histopathology and microscopy
Peripheral blood was collected through non-lethal eye-bleeds under anesthesia with isoflurane in accordance with institutional guidelines. Complete blood counts were determined using a Hemavet 950 cell counter (Drew Scientific, Oxford, CT, USA). Paraffin-embedded tissue sections were prepared at the Dana Farber/Harvard Cancer Center Specialized Histopathology Services Core and stained with hematoxylin and eosin. Images were obtained using a Nikon Eclipse E400 microscope (Nikon, Tokyo, Japan) and a SPOT RT color digital camera (Diagnostic Instruments, Sterling Heights, MI, USA).
Statistical analysis
Statistical significance of differences between the results was assessed by a two-tailed unpaired Student’s t-test using Prism software.
Results
Cdx4 expression in normal murine hematopoeisis
We first determined the expression pattern of Cdx4 during normal hematopoietic differentiation using quantitative reverse transcription PCR on RNA from wild-type C57/B6 murine hematopoietic tissues and flow-sorted progenitors. Cdx4 mRNA was preferentially expressed in myeloid progenitor cells (Figure 1A) and during B-cell differentiation.
Generation of conditional Cdx4 knockout mice
To establish the role of Cdx4 in normal hematopoiesis, we engineered two mouse models of Cdx4 inactivation, through either straight knockout6 (named the Cdx4− allele) or conditional knockout (named the Cdx4F allele) based on inducible deletion of the entire exon 1 of the Cdx4 gene (Figure 1B). This was achieved through homologous recombination in ESC; Southern blot analysis was performed to select clones correctly targeted at the endogenous Cdx4 locus on chromosome X (i.e. clone 9 but not clone 8 in Figure 1C and Online Supplementary Figure S1). After germline transmission of the Cdx4− and Cdx4F alleles, crosses were performed to obtain homozygous animals. Both Cdx4−/− and Cdx4F/F knockout animals were born at Mendelian ratios, appeared normal, had a weight similar to their wild-type, age-matched littermates, and were fertile. Cdx4F/F mice were next crossed with Mx1-Cre transgenic animals to allow for inducible Cre recombinase expression. Cdx4 excision in Cdx4F/F-Mx1-Cre (Cdx4F/F-Cre+ or F/F-Cre+) mice was induced in 4- to 6-week old animals by pIpC treatment. Subsequent analyses were performed 4–6 weeks after pIpC treatment unless otherwise indicated. Full Cdx4 excision was observed in whole bone marrow (Figure 1D) as well as in purified hematopoietic stem and progenitor-enriched populations (Figure 1E). Full excision of Cdx4 in the bone marrow was also demonstrated at 6 and 12 months (data not shown), indicating that under homeostatic conditions, there was no selective advantage for rare “escaper” cells in which excision had not occurred.
Loss of Cdx4 results in minimal hematologic abnormalities
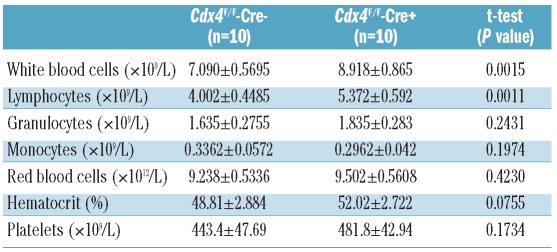
We then performed analyses of the peripheral blood and hematopoietic organs. Both Cdx4−/− and Cdx4F/F-Cre+ mice showed a significant increase in the number of lymphocytes compared to their respective wild-type controls, as assessed by an automated cell counter (Table 1, Online Supplementary Table S1). However, there was no consistent difference in myeloid, erythroid and platelets counts, absolute numbers and distribution of bone marrow cells or spleen and liver weights between Cdx4 knockout mice (Cdx4−/− or Cdx4F/F-Cre+) and their respective littermate controls (Online Supplementary Table S2). Flow cytometric analyses of bone marrow cells from Cdx4 knockout mice identified no significant differences in the major hematopoietic compartments including myeloid, erythroid, B-, and T-cell lineages (Figure 2A–C, Online Supplementary Figures S2A–C and S3). Similar results were observed in a cohort of animals analyzed 8 and 12 months after pIpC treatment (Online Supplementary Figure S4A,B and data not shown).
Table 1.
Peripheral blood counts of Cdx4F/F-Cre+ and control animals. Blood samples were taken 6–8 weeks after pIpC treatment from Cdx4F/F-Cre+ (n=10) and Cdx4F/F-Cre+ (n=10) post-pIpC treatment and complete blood counts were obtained with a Hemavet950 cell counter.
To determine whether loss of Cdx4 affects the clonogenic potential of hematopoietic progenitor populations, we performed in vitro colony-forming unit (CFU) assays. Compared to bone marrow from wild-type littermates, Cdx4 knockout bone marrow produced similar total numbers of myeloid colonies, and no significant differences in the distribution of colony types were observed (Figure 2D, Online Supplementary Figure S2D). We did, however, observe a significant decrease in pre-B colony-forming activity in bone marrow cells from Cdx4 knockout mice compared to bone marrow from their wild-type littermate controls in both the conditional as well as the germline knockout model (Figure 2E, Online Supplementary Figure S2E). Of note, we confirmed these observations in older mice that were analyzed 8 and 12 months after pIpC treatment. (Online Supplementary Figure S4C,D and data not shown).
Loss of Cdx4 does not alter the number and repopulating activity of hematopoietic stem and progenitor cells
Since Cdx4 is expressed in the hematopoietic stem and progenitor compartment, and was reported to be essential for normal hematopoiesis in zebrafish,9 we next assessed the effect of loss of Cdx4 specifically on hematopoietic stem cell function. We first performed multiparameter flow cytometry analyses on the hematopoietic stem and progenitor compartments. No significant differences in the number of LSK, CMP, GMP, or MEP were observed between both Cdx4 knockout mouse models and their respective wild-type controls (Figure 3A,B and data not shown).
We then performed non-competitive and competitive transplantation assays to assess the repopulating ability of Cdx4-deficient bone marrow cells. CD45.1−CD45.2+ Cdx4F/F-Cre+ or control Cdx4F/F-Cre− bone marrow cells were transplanted into lethally irradiated CD45.1+CD45.2−B6/SJL recipients with or without wild-type CD45.1+CD45.2+ competitor bone marrow cells (Figure 3C). The contribution of the Cdx4-deficient and control bone marrow cells to hematopoiesis in the recipients (percentage of CD45.1−CD45.2+ cells) was assessed in the peripheral blood every 4 weeks over 16 weeks, and in the bone marrow after 16 weeks. We observed a similar contribution of donor-derived CD45.1−CD45.2+ cells from Cdx4-deficient and littermate control mice in the peripheral blood (Figure 3D) and bone marrow of recipient mice (data not shown). These results show that Cdx4-deficient bone marrow cells are not significantly altered in their normal long-term repopulating ability, indicating that Cdx4 is not essential for hematopoietic stem cell function even under the replicative stress associated with bone marrow reconstitution.
Cdx4 shortens disease latency but is not essential for MLL-AF9-induced leukemia in mice
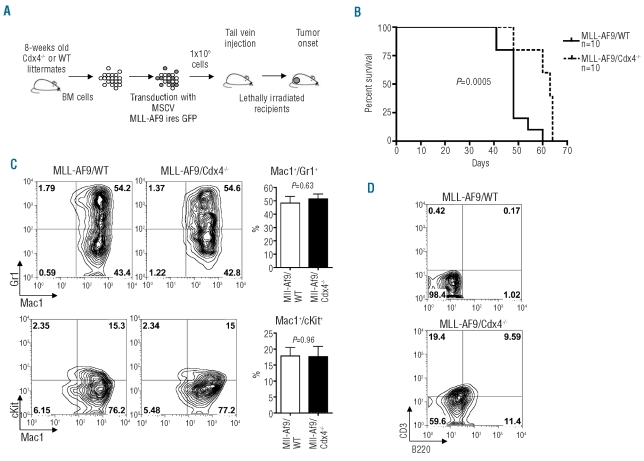
Although Cdx4 does not appear to be essential for homeostatic hematopoiesis, it was previously demonstrated that in vitro differentiation of Mll−/− ESC into blood cells could be rescued by over-expressing Cdx4, suggesting that Cdx4 might be epistatic to, or a critical downstream effector of MLL.14 Furthermore, oncogenic fusion proteins involving MLL occur frequently in patients with acute myeloid or lymphoid leukemias. We, therefore, hypothesized that Cdx4 might play a role in the context of MLL-mediated leukemogenesis. To test this idea, we used a murine model in which transplantation of bone marrow cells transduced with MLL-AF9 into lethally irradiated recipients induces a fully penetrant myeloid leukemia (Figure 4A). Recipients of wild-type bone marrow cells transduced with MLL-AF9 developed acute leukemia with a median latency of 49 days. In contrast, recipients of MLL-AF9-transduced Cdx4−/− bone marrow cells developed acute leukemia with a significantly longer latency of 63 days (P=0.0005) (Figure 4B). Histological analysis identified no morphological differences between leukemias derived from wild-type and Cdx4−/− cells (Online Supplementary Figure S5). However, flow cytometric analysis revealed that while leukemias arising in both backgrounds showed a similar expansion of Mac1+ cells in the bone marrow (Figure 4C), leukemic cells generated on the Cdx4−/− background also displayed low levels of expression of two lymphoid surface markers, CD3 and B220 (Figure 4D). Together, these studies suggest that Cdx4 is dispensable for leukemia induction by MLL-AF9, but its loss delays disease onset and alters the leukemic phenotype in a bone marrow transplantation model.
Figure 4.
Loss of Cdx4 delays MLL-AF9-induced leukemia. (A) Schematic of the retroviral transduction/bone marrow transplant protocol. (B) Survival curves for cohorts of mice injected with wild-type (Cdx+/+) or Cdx−/− bone marrow cells transduced with MLL-AF9. Ten animals were used in each group. (C) Flow cytometric analysis of bone marrow cells from recipient animals. Histograms on the right side represent mean±SD of the percentage of the indicated population (n=3). (D) Flow cytometric analysis of expression of lymphoid markers on bone marrow cells from recipient animals.
Loss of Cdx4 does not significantly affect Hox gene expression
Cdx4 has been reported to regulate the expression of Hox genes, including Hoxa9 and Hoxb4.9,11,19 To determine the effect of Cdx4 loss on Hox gene expression in murine hematopoiesis, real-time quantitative RT-PCR was used to measure the expression of several Hox genes in bone marrow cells from wild-type mice and Cdx4−/− mice, as well as in leukemic cells generated from MLL-AF9-transduced wild-type and Cdx4−/− bone marrow cells. This analysis showed that there were no significant differences in the expression of individual Hox genes between Cdx4−/− and wild-type controls (Online Supplementary Figure S6).
Discussion
Here we report the first detailed analysis of the role of a caudal gene in adult homeostatic hematopoiesis in mammals and unexpectedly demonstrate that Cdx4 knock-out murine models do not present major hematopoietic defects. Indeed, in both germline and conditional Cdx4 knockout models, hematopoietic stem and progenitor cells as well as mature blood cells were not significantly affected by Cdx4 loss of function. Of note, our conditional Cdx4 knockout model bypasses any compensatory mechanism that might occur during embryonic development in the germline Cdx4 knockout model. These results demonstrate that Cdx4 is not essential for the establishment and maintenance of normal adult hematopoietic stem cell functions in mice.
Although these results contrast with the severe hematopoietic defects observed in cdx4 mutant zebrafish,9 it is important to note that mammals have three Cdx genes (Cdx1, Cdx2, and Cdx4), whereas only two genes have been assigned to the Cdx family in zebrafish, cdx1 and cdx4. In addition, although zebrafish cdx4 mutants have a severe hematopoietic defect,9 knockdown of cdx1 in a cdx4 mutant background results in a complete failure to specify blood,10 suggesting a minor degree of redundancy between cdx1 and cdx4 during developmental hematopoiesis. It is likely that the critical function of cdx genes in zebrafish developmental hematopoiesis can be extended to the functionally redundant homolog Cdx2. In mammals, Cdx4 deficiency was reported to cause a modest hematopoietic defect during in vitro differentiation of mESC. In addition, although yolk sac hematopoiesis was transiently altered before 9 days post-conception, no significant blood alteration was observed in Cdx4 deficient embryos in the yolk sac 9 days post-conception or in fetal liver. In contrast, Cdx2 deficiency results in a more severe defect, and a combination of Cdx1, Cdx2, and Cdx4 deficiency almost abolished blood formation from mESC.6 Furthermore, only Cdx2/4 compound mutants, but not Cdx4-deficient or Cdx1/4 double mutants, present axial elongation defects during mouse development.2 Together, these observations suggest that Cdx4 may have a non-redundant role during very early hematopoietic development but that its function in adult hematopoietic stem cells in vivo is compensated for by redundant mechanisms. This difference could be explained by a lower sensitivity to Cdx gene dosage in definitive compared to primitive hematopoiesis. An alternative possibility is that Cdx function is not essential for adult hematopoiesis. Indeed, it was previously shown that other genes essential for primitive hematopoiesis specification, such as SCL/Tal1, do not play an essential role in definitive adult stem cell function, supporting the existence of two distinct developmental pathways for the generation of embryonic hematopoietic stem cells and maintenance of adult hematopoietic stem cells.
Although Cdx4 deficiency was not associated with a gross hematopoietic phenotype, our results do suggest a previously unappreciated role for Cdx4 in lymphoid development. Indeed, we show that Cdx4 expression is up-regulated in pre-B, pro-B, and B220+ cells. We also observed a reduced capacity of bone marrow cells from Cdx4-deficient animals to form B-cell colonies in vitro. These observations are compatible with a positive role of Cdx4 in lymphopoiesis that would result in a partial block of differentiation during early B-cell development in Cdx4-deficient animals. However, we also observed that lymphocyte blood counts were consistently higher in Cdx4-deficient animals and that Cdx4-deficient MLL-AF9-transformed blasts aberrantly express some lymphoid markers, suggesting that Cdx4 deficiency accelerates and promotes lymphoid differentiation. This latter hypothesis suggests that Cdx4 restricts lymphoid identity. Although these two hypotheses may not be mutually exclusive, further studies are required to understand the precise role of Cdx factors during lymphopoiesis.
Aberrant expression of CDX2 or CDX4 genes has recently been implicated in the pathogenesis of human AML, and it has been suggested that CDX proteins may, at least in part, be responsible for the deregulated HOX gene expression observed in the majority of AML cases.8,22 In addition, Cdx4 can rescue the differentiation of Mll-deficient ESC in vitro14 and has been shown to interact with menin in the up-regulation of Hoxa cluster genes during Mll fusion-induced leukemogenesis,19 suggesting the possibility that oncogenic transformation by MLL fusions may require CDX4. In support of this hypothesis, we found that although Cdx4 is not absolutely required for leukemia induction by MLL-AF9, its absence significantly prolonged the latency of disease development. Moreover, the phenotype of the resultant disease was altered with increased expression of B- and T-lymphoid markers subsequent to Cdx4 loss. Taken together, this suggests a role for Cdx4 in MLL-induced leukemogenesis. On the other hand, the subtlety of the phenotype and the absence of a difference in Hox gene expression between MLL-AF9-transduced wild-type and Cdx4−/− bone marrow cells, also point to a degree of functional redundancy among Cdx factors in the context of leukemogenesis. In addition, it is conceivable that there are context-specific differences in the requirement for Cdx4 during AML pathogenesis, depending on the underlying mechanism that drives the leukemic phenotype. For example, mutant kinases, such as BCR-ABL1, which do not have the potential for activating self-renewal programs in hematopoietic cells, may be more reliant on Cdx4-controlled pathways than alleles such as the MLL fusion genes that can confer self-renewal properties to committed progenitors.27–29
The minimal hematopoietic impairment upon loss of Cdx4 along with its aberrant expression in acute leukemias make CDX proteins, in principle, attractive therapeutic targets. Indeed the therapeutic utility of targeting CDX2 was suggested by knockdown experiments in AML cell lines.22 Although targeting transcription factors is very challenging and has not yet been clinically realized, critical protein-protein interactions associated with transcription factor function have recently been successfully targeted with small molecule inhibitors making transcription factors potentially “druggable” targets.30 In this study, we have shown that loss of Cdx4 significantly prolongs the latency of disease onset in a mouse model of MLL-AF9-induced AML. Although further studies will be necessary to understand the full degree of redundancy between Cdx genes in both normal and malignant hematopoiesis and the precise role of these proteins in the context of other MLL- or HOX-related leukemias, specific drug targeting of CDX factors could be of value in AML.
Acknowledgments
we are very grateful to Mahnaz Paktinat for flow cytometry analyses and Rachel Okabe and Maricel Gozo for technical assistance. This work was supported in part by NIH grants (DGG), and the Leukemia and Lymphoma Society (DGG).
Footnotes
Funding: BJH was supported by an EHA José Carreras Young Investigator Fellowship and is an MRC (UK) senior clinical fellow. SF is the recipient of an EHA José Carreras Young Investigator Fellowship. TM was supported by a Leukemia and Lymphoma Society Special Fellow Award (#3431-06), is the recipient of an EHA José Carreras Young Investigator Fellowship and is an INSERM investigator.
Authorship and Disclosures
The information provided by the authors about contributions from persons listed as authors and in acknowledgments is available with the full text of this paper at www.haematologica.org.
Financial and other disclosures provided by the authors using the ICMJE (www.icmje.org) Uniform Format for Disclosure of Competing Interests are also available at www.haematologica.org.
The online version of this article has a Supplementary Appendix.
References
- 1.van den Akker E, Forlani S, Chawengsaksophak K, de Graaff W, Beck F, Meyer BI, et al. Cdx1 and Cdx2 have overlapping functions in anteroposterior patterning and posterior axis elongation. Development. 2002;129(9):2181–93. doi: 10.1242/dev.129.9.2181. [DOI] [PubMed] [Google Scholar]
- 2.van Nes J, de Graaff W, Lebrin F, Gerhard M, Beck F, Deschamps J. The Cdx4 mutation affects axial development and reveals an essential role of Cdx genes in the ontogenesis of the placental labyrinth in mice. Development. 2006;133(3):419–28. doi: 10.1242/dev.02216. [DOI] [PubMed] [Google Scholar]
- 3.Subramanian V, Meyer BI, Gruss P. Disruption of the murine homeobox gene Cdx1 affects axial skeletal identities by altering the mesodermal expression domains of Hox genes. Cell. 1995;83(4):641–53. doi: 10.1016/0092-8674(95)90104-3. [DOI] [PubMed] [Google Scholar]
- 4.Charite J, de Graaff W, Consten D, Reijnen MJ, Korving J, Deschamps J. Transducing positional information to the Hox genes: critical interaction of cdx gene products with position-sensitive regulatory elements. Development. 1998;125(22):4349–58. doi: 10.1242/dev.125.22.4349. [DOI] [PubMed] [Google Scholar]
- 5.Chawengsaksophak K, de Graaff W, Rossant J, Deschamps J, Beck F. Cdx2 is essential for axial elongation in mouse development. Proc Natl Acad Sci USA. 2004;101(20):7641–5. doi: 10.1073/pnas.0401654101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Y, Yabuuchi A, McKinney-Freeman S, Ducharme DM, Ray MK, Chawengsaksophak K, et al. Cdx gene deficiency compromises embryonic hematopoiesis in the mouse. Proc Natl Acad Sci USA. 2008;105(22):7756–61. doi: 10.1073/pnas.0708951105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McKinney-Freeman SL, Lengerke C, Jang IH, Schmitt S, Wang Y, Philitas M, et al. Modulation of murine embryonic stem cell-derived CD41+c-kit+ hematopoietic progenitors by ectopic expression of Cdx genes. Blood. 2008;111(10):4944–53. doi: 10.1182/blood-2007-11-124644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frohling S, Scholl C, Bansal D, Huntly BJ. HOX gene regulation in acute myeloid leukemia: CDX marks the spot? Cell Cycle. 2007;6(18):2241–5. doi: 10.4161/cc.6.18.4656. [DOI] [PubMed] [Google Scholar]
- 9.Davidson AJ, Ernst P, Wang Y, Dekens MP, Kingsley PD, Palis J, et al. cdx4 mutants fail to specify blood progenitors and can be rescued by multiple hox genes. Nature. 2003;425(6955):300–6. doi: 10.1038/nature01973. [DOI] [PubMed] [Google Scholar]
- 10.Davidson AJ, Zon LI. The caudal-related homeobox genes cdx1a and cdx4 act redundantly to regulate hox gene expression and the formation of putative hematopoietic stem cells during zebrafish embryogenesis. Dev Biol. 2006;292(2):506–18. doi: 10.1016/j.ydbio.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 11.Wang Y, Yates F, Naveiras O, Ernst P, Daley GQ. Embryonic stem cell-derived hematopoietic stem cells. Proc Natl Acad Sci USA. 2005;102(52):19081–6. doi: 10.1073/pnas.0506127102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lengerke C, Schmitt S, Bowman TV, Jang IH, Maouche-Chretien L, McKinney-Freeman S, et al. BMP and Wnt specify hematopoietic fate by activation of the Cdx-Hox pathway. Cell Stem Cell. 2008;2(1):72–82. doi: 10.1016/j.stem.2007.10.022. [DOI] [PubMed] [Google Scholar]
- 13.Yu BD, Hess JL, Horning SE, Brown GA, Korsmeyer SJ. Altered Hox expression and segmental identity in Mll-mutant mice. Nature. 1995;378(6556):505–8. doi: 10.1038/378505a0. [DOI] [PubMed] [Google Scholar]
- 14.Ernst P, Mabon M, Davidson AJ, Zon LI, Korsmeyer SJ. An Mll-dependent Hox program drives hematopoietic progenitor expansion. Curr Biol. 2004;14(22):2063–9. doi: 10.1016/j.cub.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 15.Milne TA, Briggs SD, Brock HW, Martin ME, Gibbs D, Allis CD, et al. MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol Cell. 2002;10(5):1107–17. doi: 10.1016/s1097-2765(02)00741-4. [DOI] [PubMed] [Google Scholar]
- 16.Dorrance AM, Liu S, Yuan W, Becknell B, Arnoczky KJ, Guimond M, et al. Mll partial tandem duplication induces aberrant Hox expression in vivo via specific epigenetic alterations. J Clin Invest. 2006;116(10):2707–16. doi: 10.1172/JCI25546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yokoyama A, Somervaille TC, Smith KS, Rozenblatt-Rosen O, Meyerson M, Cleary ML. The menin tumor suppressor protein is an essential oncogenic cofactor for MLL-associated leukemogenesis. Cell. 2005;123(2):207–18. doi: 10.1016/j.cell.2005.09.025. [DOI] [PubMed] [Google Scholar]
- 18.Yokoyama A, Wang Z, Wysocka J, Sanyal M, Aufiero DJ, Kitabayashi I, et al. Leukemia proto-oncoprotein MLL forms a SET1-like histone methyltransferase complex with menin to regulate Hox gene expression. Mol Cell Biol. 2004;24(13):5639–49. doi: 10.1128/MCB.24.13.5639-5649.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yan J, Chen YX, Desmond A, Silva A, Yang Y, Wang H, et al. Cdx4 and menin co-regulate Hoxa9 expression in hematopoietic cells. PLoS ONE. 2006;1:e47. doi: 10.1371/journal.pone.0000047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chase A, Reiter A, Burci L, Cazzaniga G, Biondi A, Pickard J, et al. Fusion of ETV6 to the caudal-related homeobox gene CDX2 in acute myeloid leukemia with the t(12;13)(p13;q12) Blood. 1999;93(3):1025–31. [PubMed] [Google Scholar]
- 21.Rawat VP, Thoene S, Naidu VM, Arseni N, Heilmeier B, Metzeler K, et al. Overexpression of CDX2 perturbs HOX gene expression in murine progenitors depending on its N-terminal domain and is closely correlated with deregulated HOX gene expression in human acute myeloid leukemia. Blood. 2008;111(1):309–19. doi: 10.1182/blood-2007-04-085407. [DOI] [PubMed] [Google Scholar]
- 22.Scholl C, Bansal D, Dohner K, Eiwen K, Huntly BJ, Lee BH, et al. The homeobox gene CDX2 is aberrantly expressed in most cases of acute myeloid leukemia and promotes leukemogenesis. J Clin Invest. 2007;117(4):1037–48. doi: 10.1172/JCI30182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Riedt T, Ebinger M, Salih HR, Tomiuk J, Handgretinger R, Kanz L, et al. Aberrant expression of the homeobox gene CDX2 in pediatric acute lymphoblastic leukemia. Blood. 2009;113(17):4049–51. doi: 10.1182/blood-2008-12-196634. [DOI] [PubMed] [Google Scholar]
- 24.Thoene S, Rawat VP, Heilmeier B, Hoster E, Metzeler KH, Herold T, et al. The homeobox gene CDX2 is aberrantly expressed and associated with an inferior prognosis in patients with acute lymphoblastic leukemia. Leukemia. 2009;23(4):649–55. doi: 10.1038/leu.2008.355. [DOI] [PubMed] [Google Scholar]
- 25.Bansal D, Scholl C, Frohling S, McDowell E, Lee BH, Dohner K, et al. Cdx4 dysregulates Hox gene expression and generates acute myeloid leukemia alone and in cooperation with Meis1a in a murine model. Proc Natl Acad Sci USA. 2006;103(45):16924–9. doi: 10.1073/pnas.0604579103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rawat VP, Cusan M, Deshpande A, Hiddemann W, Quintanilla-Martinez L, Humphries RK, et al. Ectopic expression of the homeobox gene Cdx2 is the transforming event in a mouse model of t(12;13)(p13;q12) acute myeloid leukemia. Proc Natl Acad Sci USA. 2004;101(3):817–22. doi: 10.1073/pnas.0305555101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huntly BJ, Shigematsu H, Deguchi K, Lee BH, Mizuno S, Duclos N, et al. MOZ-TIF2, but not BCR-ABL, confers properties of leukemic stem cells to committed murine hematopoietic progenitors. Cancer Cell. 2004;6(6):587–96. doi: 10.1016/j.ccr.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 28.Krivtsov AV, Twomey D, Feng Z, Stubbs MC, Wang Y, Faber J, et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 2006;442(7104):818–22. doi: 10.1038/nature04980. [DOI] [PubMed] [Google Scholar]
- 29.Cozzio A, Passegue E, Ayton PM, Karsunky H, Cleary ML, Weissman IL. Similar MLL-associated leukemias arising from self-renewing stem cells and short-lived myeloid progenitors. Genes Dev. 2003;17(24):3029–35. doi: 10.1101/gad.1143403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sukhdeo K, Mani M, Zhang Y, Dutta J, Yasui H, Rooney MD, et al. Targeting the beta-catenin/TCF transcriptional complex in the treatment of multiple myeloma. Proc Natl Acad Sci USA. 2007;104(18):7516–21. doi: 10.1073/pnas.0610299104. [DOI] [PMC free article] [PubMed] [Google Scholar]