Abstract
Background
Recently, in genome-wide analyses of DNA copy number abnormalities using single nucleotide polymorphism microarrays, genetic alterations targeting PAX5 were identified in over 30% of pediatric patients with acute lymphoblastic leukemia. So far the occurrence of PAX5 alterations and their clinical correlation have not been investigated in adults with BCR-ABL1-positive acute lymphoblastic leukemia.
Design and Methods
The aim of this study was to characterize the rearrangements on 9p involving PAX5 and their clinical significance in adults with BCR-ABL1-positive acute lymphoblastic leukemia. Eighty-nine adults with de novo BCR-ABL1-positive acute lymphoblastic leukemia were enrolled into institutional (n=15) or GIMEMA (Gruppo Italiano Malattie EMatologiche dell’Adulto) (n=74) clinical trials and, after obtaining informed consent, their genome was analyzed by single nucleotide polymorphism arrays (Affymetrix 250K NspI and SNP 6.0), genomic polymerase chain reaction analysis and re-sequencing.
Results
PAX5 genomic deletions were identified in 29 patients (33%) with the extent of deletions ranging from a complete loss of chromosome 9 to the loss of a subset of exons. In contrast to BCR-ABL1-negative acute lymphoblastic leukemia, no point mutations were found, suggesting that deletions are the main mechanism of inactivation of PAX5 in BCR-ABL1-positive acute lymphoblastic leukemia. The deletions were predicted to result in PAX5 haploinsufficiency or expression of PAX5 isoforms with impaired DNA-binding. Deletions of PAX5 were not significantly correlated with overall survival, disease-free survival or cumulative incidence of relapse, suggesting that PAX5 deletions are not associated with outcome.
Conclusions
PAX5 deletions are frequent in adult BCR-ABL1-positive acute lymphoblastic leukemia and are not associated with a poor outcome.
Keywords: ALL, BCR-ABL1, PAX5
Introduction
PAX5 is a member of the highly conserved paired-box (PAX) domain family of transcription factors necessary for many types of cell differentiation. PAX5 is the only PAX family member expressed within the hematopoietic system, and its expression is restricted to certain stages along the B-cell differentiation pathway.1 Its expression is initiated in pre-pro-B cells and maintained throughout subsequent stages of B-cell development before it is down-regulated in plasma cells.2,3 It may function both as a transcriptional activator and a repressor, because it positively controls target genes encoding essential components of (pre-) B-cell receptor signaling such as the signal-transducing chain Igα (also called CD79a and mb-1),4 the co-stimulatory receptors CD19 and CD21,5 the inhibitory co-receptor CD725,6 and the central adaptor protein BLNK (also called SLP65).7 It negatively controls PD-1, NOTCH1 (which is necessary for the commitment of lymphoid progenitors to the T-cell pathway), and the transcription of M-CSFR, thus rendering B-cell precursors unresponsive to myeloid cytokines such as macrophage colony-stimulating factor (M-CSF).1 The PAX5 gene is located at 9p13 and is juxtaposed to IGH@ in cases with t(9;14)(p13;q32), a rare but recurring translocation found in a subset of B-cell non-Hodgkin’s lymphomas8 and resulting in deregulated expression of the gene product because of the proximity of IgH enhancers. Recently, in genome-wide analyses of DNA copy number abnormalities and loss of heterozygosity (LOH) using single nucleotide polymorphism (SNP) arrays, genetic alterations (deletions, focal internal amplification, sequence mutations and translocations) targeting the B-lymphoid transcription factor PAX5 have been identified in over 30% of cases of childhood B-progenitor acute lymphoblastic leukemia (ALL).9,10
The structural rearrangements included fusion of PAX5 to FOXP1 (3p13),9,11 AUTS2 (7q11),11 ELN (7q11),12 ETV6 (12p13),9,11,13 PML (15q24),14 ZNF521 (18q11)9 and C20orf112 (20q11),11,15 LOC392027 (7p12.1),15 SLCO1B3 (12p12),15 ASXL1 (20q11.1),15 KIF3B (20q11.21),15 HIPK1 (1p13),16 POM121 (7q11),16 DACH1 (13q21)16 and BRD1 (22q13.33).16 In each predicted fusion protein, the DNA-binding paired domain of PAX5, and a variable amount of the C-terminal transactivating domains are fused to functional domains of the partner genes. Using quantitative polymerase chain reaction (PCR) analysis, An et al.15 demonstrated that both the deletion and gene fusion events resulted in the same under-expression of PAX5, which extended to the differential expression of the PAX5 target genes, EBF1, ALDH1A1, ATP9A and FLT3, suggesting a potential pathogenic role of PAX5 deletion in pediatric acute leukemias.9
The impact of PAX5 alterations on clinical outcome was investigated for the first time by Mullighan et al.17 in children with high-risk B-cell-progenitor ALL in whom it was found that abnormalities of PAX5 copy-number failed to act as a predictor of poor outcome. However, the occurrence and the role of PAX5 alterations have so far not been investigated in adult patients with BCR-ABL1-positive ALL, the most frequent and prognostically the most unfavorable subtype of ALL in adults.18 The outcome of patients with BCR-ABL1-positive ALL has improved with current therapies that include tyrosine kinase inhibitors such as imatinib,19 nilotinib20,21 and dasatinib.22 Complete hematologic remissions can be obtained in 98–100% of patients treated with a tyrosine kinase inhibitor alone19 or in association with conventional chemotherapy,23 but relapse is still an expected event in the majority of cases.21
Here, we used different molecular approaches, including high resolution determination of DNA copy number alterations (Affymetrix GeneChip® Human Mapping 250K NspI and Genome-Wide Human SNP 6.0 array GeneChip microarrays), fluorescence in situ hybridization (FISH) and re-sequencing to study rearrangements on 9p involving the PAX5 locus in 89 adults with BCR-ABL1-positive ALL.
Design and Methods
Patients
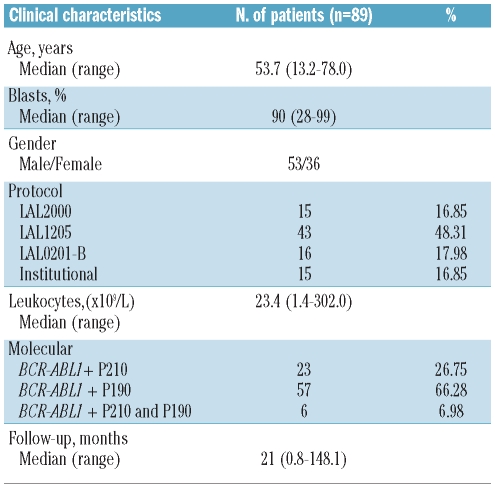
All patients gave their informed consent to blood collection and biological analyses, in agreement with the Declaration of Helsinki. The study was approved by the Seràgnoli Department of Hematology and Medical Sciences, University of Bologna, Italy. We analyzed 89 patients (53 males and 36 females; median age, 54 years; range, 13.2–78; median blast percentage, 90%; range, 28–99%) with de novo BCR-ABL1-positive ALL enrolled between 04/1996 and 08/2008 in different clinical trials of the GIMEMA AL Working party or in institutional protocols (Table 1). In detail, 74 patients (83%) were enrolled in GIMEMA clinical trials (16 patients in the GIMEMA LAL0201-B protocol, 15 in the LAL2000 protocol and 43 in the LAL1205 protocol), while 15 patients (17%) were enrolled into institutional protocols. Details of the treatments schemes have been previously reported.24 Briefly, patients enrolled for the LAL0201-B protocol were elderly (>60 years), Philadelphia chromosome (Ph)-positive ALL patients who received imatinib 800 mg daily, associated with steroids as front-line treatment.19 Patients enrolled in the LAL2000 study were adults (>18 years) with ALL, including Ph-positive cases, who received induction and consolidation chemotherapy followed by maintenance therapy with imatinib. The LAL1205 trial25,26 enrolled adult Ph-positive ALL patients who received dasatinib 70 mg bid for 84 consecutive days, as induction therapy, initially associated with steroids, without further chemotherapy as frontline treatment.
Table 1.
Demographic and clinical characteristics of patients with BCR-ABL1-positive acute lymphoblastic leukemia.
Nineteen patients underwent allogeneic stem cell transplantation in complete hematologic response as consolidation therapy (5 had an HLA-identical donor, 9 had a matched unrelated donor, 1 had an allogeneic graft and 4 an autologous transplant). At diagnosis, all patients were found to be BCR-ABL1-positive. The percentages of BCR-ABL fusion transcripts corresponding to p210 versus p190 versus p190+p210 were 27%, 66% and 7%, respectively.
Single nucleotide polymorphism microarray analysis
All 89 BCR-ABL1-positive ALL patients were analyzed by SNP array for PAX5 deletions. Genomic DNA was extracted using the DNA Blood Mini Kit (Qiagen, Valencia, CA, USA) from mononuclear cells isolated from peripheral blood or bone marrow aspirate samples by Ficoll gradient centrifugation. DNA was quantified using the Nanodrop Spectrophotometer and quality was assessed using the Nanodrop and by agarose gel electrophoresis.
Samples were genotyped with GeneChip® Human Mapping 250K NspI (69 cases, 78%) and Genome-Wide Human SNP 6.0 (20 cases, 22%) arrays (Affymetrix, Santa Clara, CA, USA) as previously described.27 Copy number aberrations were scored using the hidden Markov model and the segmentation approach available within the Partek Genomic Suite (Partek Inc, Saint Louise, MO, USA) software package as well as by visual inspection. Five consecutive markers for the Human Mapping 250K NspI and ten for the Genome-Wide Human SNP 6.0 arrays were considered as alterations. All aberrations were calculated by a comparison with a set of 270 HapMap normal individuals and a set of samples obtained from people with acute leukemia in remission (n=20) in order to reduce the noise of raw copy number data. Copy number aberrations were also analyzed and eventually confirmed using Genotyping Console 3.0 (Affymetrix, Santa Clara, CA, USA).
Fluorescence in situ hybridization and probes
FISH analysis for the PAX5 locus was performed as previously described.27 Overlapping bacterial artificial chromosome (BAC) probes specific for the PAX5 gene [RP11-465P6 (36,899,192-37,065,371), RP11-243F8 (36,834,935-37,023,204), and RP11-344B23 (36,764,163-36,939,832)], as well as a BAC for BCR [RP11-164N13 (chr22:21,897,904-22,091,572)] and a fosmid probe specific for the IKZF1 gene [G248P800745C8 (chr7:50,381,496-50,422,338)], were properly selected according to the March 2006 release of the University Santa Cruz (UCSC) Human Genome Browser (http://genome.ucsc.edu/). At least 15 metaphases were analyzed for each patient.
PAX5 genomic quantitative polymerase chain reaction
PAX5 genomic quantitative PCR (Q-PCR) was performed in order to confirm SNP results and to characterize the extension of the deletion in 20/29 (69%) patients with PAX5 deletion and in three normal germline cases. It was performed as described by Mullighan et al.18 using a 7900 Real-Time PCR system and 7900 System software (Applied Biosystems, Foster City, CA, USA).
PAX5 point mutation screening
Genomic re-sequencing of PAX5 coding exons was performed in 34 cases of BCR-ABL1-positive ALL. One hundred nanograms of genomic DNA was amplified with 2 U of FastStart Taq DNA Polymerase (Roche Diagnostics, Mannheim, Germany), 0.8 mM dNTP, 1 mM MgCl2, and 0.2 M forward and reverse primers in 25 μL reaction volumes (Online Supplementary Table S1). PCR cycling parameters were: one cycle of 95°C for 5 min, 35 cycles of 95°C for 30 s, 62°C for 30 s and 72°C for 1 min, followed by one cycle of 72°C for 7 min. PCR products were purified using a QIAquick PCR purification kit (Qiagen) and then directly sequenced using an ABI PRISM 3730 automated DNA sequencer (Applied Biosystems) and a Big Dye Terminator DNA sequencing kit (Applied Biosystems). In some cases the PCR products were sub-cloned into the PCR®2.1-TOPO vector using the TOPO TA Cloning Kit (Invitrogen, San Diego, CA, USA). The cloned PCR products were purified and sequenced as described above. All sequence variations were detected, using the BLAST software tool (www.ncbi.nlm.nih.gov/BLAST/), by comparison to reference genome sequence data obtained from the UCSC browser (http://genome.ucsc.edu/cgibin/hgGateway?db=hg1 8;March 2006 release).
PAX5 real-time polymerase chain reaction and gene expression analysis
Mononuclear cells from 31 samples were separated as previously described27 and total cellular RNA was extracted using the RNeasy total RNA isolation kit (Qiagen, Valencia, CA, USA). One microgram of total RNA was reverse transcribed using the High Capacity cDNA Archive Kit (Applied Biosystems). PCR using primers specific for exon 1 and exon 10 (F1 and R1, respectively, in Online Supplementary Table S1) of PAX5 and nucleotide sequencing were performed to identify the isoforms derived from focal deletions. RNA integrity was confirmed by PCR amplification of GAPDH mRNA, which is expressed ubiquitously in human hematopoietic cells. PAX5 gene expression was analyzed using the Hs00277134_m1 assay, a 7900 real-time PCR system and 7900 System software (Applied Biosystems). Results were expressed as 2−ΔΔCt. GAPDH was used as a control gene (Hs99999905_m1).
Statistical analysis
The primary end-points of the study were achievement of complete remission, duration of first complete remission (in terms of disease-free survival and cumulative incidence of relapse), and overall survival. The median follow-up time was estimated by reversing the codes for the censoring indicator in a Kaplan-Meier analysis.28 Differences in the distributions of prognostic factors in subgroups were analyzed by the χ2 or Fisher’s exact test, and by the Kruskal-Wallis test. Overall survival was defined as the time from diagnosis to date of death or date of the last follow-up. Disease-free survival and the cumulative incidence of relapse were calculated from the time of achieving complete remission to the date of first relapse, death or date of last follow-up. The probabilities of overall and disease-free survival were estimated using the Kaplan-Meier method28 and the probability of the cumulative incidence of relapse29 was estimated using the appropriate non-parametric method, considering death in complete remission as a competing risk. The log-rank test was used to compare treatment effect and risk factor categories for the Kaplan-Meier curves and Gray’s test was used for the incidence curves; 95% confidence intervals (95% CI) were estimated using the method of Simon and Lee.30 Cox proportional hazard regression models31 were performed to examine and check for treatment results and the risk factors affecting disease-free survival.
All tests were two-sided, with P values of 0.05 or less indicating a statistically significant difference. All analyses were performed using the SAS software (SAS Institute, Cary, NC, USA).
Results
Deletion of PAX5 occurs in 33% of adult cases of BCR-ABL1-positive acute lymphoblastic leukemia
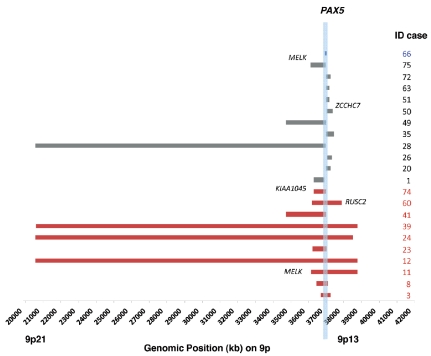
We determined the occurrence of PAX5 alterations in a large cohort of adult patients with BCR-ABL1-positive ALL (n=89). These patients were enrolled from 1996 to 2008 in different clinical trials of the GIMEMA AL Working Party or in institutional trials (Table 1). Overall, by SNP array analysis we identified a loss of PAX5 in 29 patients (33%). In all cases, the deletion was heterozygous. Four patterns of PAX5 deletion were observed: (i) focal deletion involving only the PAX5 gene in one case (3%); (ii) deletions involving only a portion of PAX5 and both telomeric and centromeric flanking genes in 11 cases (38%) with a median size of 310 kb (range, 101 kb–16,395 kb) and ranging from 9p13.3 to 9p21.3; (iii) broader deletions involving the entire PAX5 locus and a variable number of flanking genes in ten patients (34%) with a median size of 1,999 kb (range, 567 kb–18,208 kb) and ranging from 9p13.3 to 9p21.3; (iv) deletion of all chromosome 9 or 9p in seven patients (24%) (Figure 1 and Online Supplementary Table S2).
Figure 1.
Diagrammatic representation of 29 PAX5 deletions as assessed by SNP arrays. The size of the monoallelic deletion is indicated by the horizontal bars. Each row represents a type of deletion: the focal deletion involving only the PAX5 gene is in blue; deletions involving only a portion of PAX5 and flanking genes are in gray; broader deletions involving PAX5 and a variable number of flanking genes are in red. Deletions of the entire chromosome 9 are omitted. The genomic location of PAX5 is shown by the vertical sky-blue box.
Since PAX5 point mutations have been described to occur in B-lineage ALL,17,32 we sought to determine whether this could also be the case in adult BCR-ABL1-positive ALL. We, therefore, investigated both normal and deleted PAX5 subgroups for point mutations by PCR and subsequent deep exon-sequencing analysis but we failed to find any nucleotide substitutions, indicating that deletions were the main mechanism of PAX5 alterations in our cases (Online Supplementary Table S2).
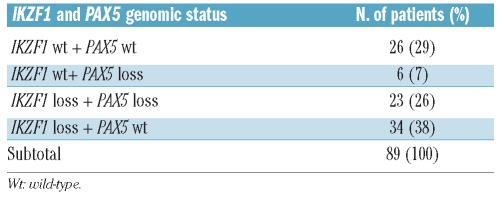
Furthermore, we investigated a molecular association between PAX5 and IKZF1 deletions. In 23/29 cases (88% of PAX5-deleted patients and 26% of the whole cohort studied) both PAX5 and IKZF1 losses were detected whereas in six patients (7%), loss of PAX5 was associated with a normal pattern of IKZF1. In 34/89 cases (38%) a deletion of IKZF1 was not associated with loss of PAX5, while the remaining 26 patients (29%) were normal for both PAX5 and IKZF1 (Table 2 and Online Supplementary Table S2).
Table 2.
Frequency of PAX5 and IKZF1 copy number losses in 89 BCR-ABL1-ALL cases studied by SNP arrays.
PAX5 deletions were confirmed by fluorescence in situ hybridization analysis and quantitative polymerase chain reaction
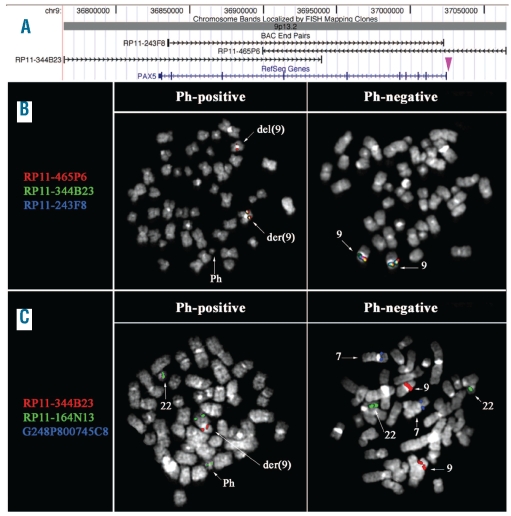
FISH analysis with three overlapping BAC probes encompassing the whole PAX5 gene (Figure 2A) was performed in two patients with BCR-ABL1-positive ALL, both harboring homozygous IKZF1 deletions and heterozygous PAX5 deletions as determined by SNP arrays. The results obtained indicate that in case #8 the breakpoint mapped within clone RP11-465P6, while in case #12 the deletion removed all the PAX5 probes (Figure 2B).
Figure 2.
FISH results. (A) Map of the clones used in FISH experiments to detect PAX5 deletions. The purple arrowhead indicates the breakpoint region mapped in case-#8 (B) and (C) FISH results obtained in case-#8 showing: (B) heterozygous deletion of clones RP11-344B23 (green) and RP11-243F8 (blue) and partial heterozygous deletion of RP11-465P6 (red) only in Ph-positive cells (middle column). Ph-negative cells showed co-localization of all three probes in both chromosome 9 homologs (right column); (C) revealing the occurrence of the PAX5 heterozygous deletion (only one signal of RP11-344B23, in red) as well as IKZF1 homozygous deletion (no signal of fosmid G248P800745C8, in blue) only in Ph-positive cells, as shown by the FISH pattern of the RP11-164N13 (BCR) probe (in green). Ph-negative cells did not show deletion of either PAX5 or of IKZF1.
Moreover, PAX5 deletions, together with IKZF1 losses, were only detected in Ph-positive cells, while no deletion was ever observed in Ph-negative cells (Figure 2C and data not shown).
Real-time genomic Q-PCR of PAX5 was performed in order to confirm the SNP results and to characterize the extension of the deletions in 20/29 (69%) patients with deletions. There was complete concordance between the extent of the deletion previously identified by SNP array and the quantitative PCR results (Online Supplementary Table S3).
Furthermore, to investigate the consequences of genomic PAX5 alterations in patients with BCR-ABL1-positive ALL, real-time quantitative PCR (RQ-PCR) was used to assess the expression of PAX5 in cases with copy number changes on 9p13.2. RQ-PCR showed a down-modulation of PAX5 transcript levels (P<0.03) in patients (n= 6) with deletion compared to those without (n=25) (Online Supplementary Figure S1). Interestingly, in one patient the deletion of PAX5 involved only a subset of exons (2–6) resulting in an alternative transcript predicted to encode a prematurely truncated protein, due to a frame-shift, lacking key PAX5 functional domains (Online Supplementary Figures S2 and S3).
PAX5 deletions and their clinical significance
PAX5 deletions were equally distributed between age, time (i.e. DNA sample stability-integrity) and between protocols (Online Supplementary Table S4). As previously reported,24 all patients (100%) enrolled into protocol LAL1205 and all patients (100%) enrolled into protocol LAL-0201-B obtained a complete hematologic response and for this reason it was not possible to correlate this with other parameters. Therefore, only patients enrolled into the LAL2000 protocol and into institutional protocols (n=30) were evaluable for a correlation between complete hematologic response and PAX5 deletion. Since one patient enrolled in LAL2000 never started therapy, 29 patients were evaluable (Online Supplementary Table S5). At the end of induction chemotherapy, 23/29 patients (79%) obtained a complete hematologic response whereas six (21%) were resistant to induction chemotherapy (ie, >25% blasts persisting in the marrow) (data not shown): 12/23 patients (52%) obtaining a complete hematologic response had wild-type PAX5, whereas 11/23 patients (58%) had a PAX5 deletion (P=0.057), suggesting that PAX5 deletion is not associated with a reduced probability of obtaining a complete hematologic response.
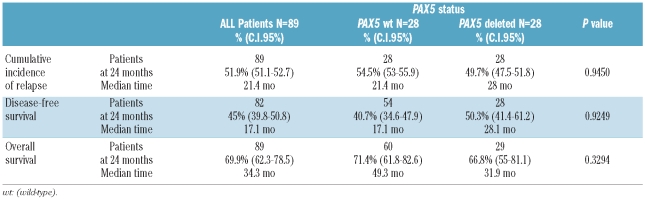
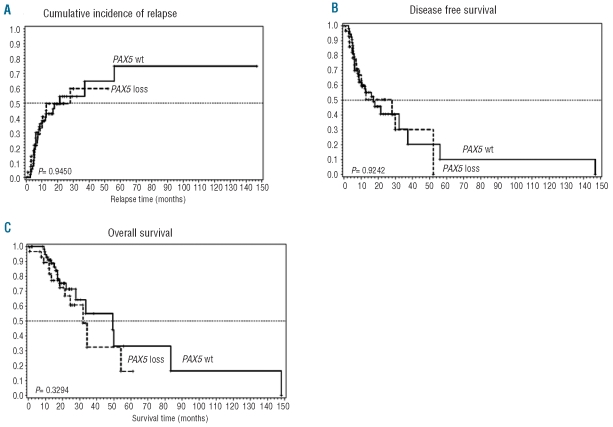
Deletions of PAX5 were also not significantly correlated with overall survival (P=0.3294), disease-free survival (P=0.9249) or cumulative incidence of relapse (P=0.945), suggesting that PAX5 deletions are not associated with outcome (Table 3 and Figure 3).
Table 3.
Clinical outcome related to PAX5 loss in univariate analysis.
Figure 3.
Cumulative incidence of (A) relapse, (B) disease-free survival and (C) overall survival of patients with de novo BCR-ABL1-ALL treated with conventional or investigational therapy including TKI (imatinib or dasatinib) regimens and with PAX5 deletion compared with patients treated with the same protocols without PAX5 deletion.
Discussion
In this study, we aimed at characterizing, by high resolution determination of genomic copy number alterations (Affymetrix GeneChip® Human Mapping 250K NspI and Genome-Wide Human SNP 6.0 microarrays), the rearrangements on 9p involving the paired box 5 gene (PAX5) and their clinical significance in adult BCR-ABL1-positive ALL. The rationale of this study was based on the observation that alterations of PAX5 occur in different B-cell malignancies, suggesting a role of PAX5 as an oncogene. In non-Hodgkin’s lymphoma PAX5 is often over-expressed or targeted by aberrant somatic hyper-mutation,33 whereas some cases of childhood ALL demonstrate monoallelic loss of PAX5, point mutations or even novel fusion genes.9,11 Here, PAX5 deletions affecting the entire gene or only some exons were identified in 33% of cases of adult BCR-ABL1-positive ALL. Deletions were predicted to result in either PAX5 haploinsufficiency or expression of PAX5 alleles with impaired DNA-binding. The latter was the case of a focal deletion affecting only a subset of exons (2–6) and resulting in an alternative isoform encoding a never previously described prematurely truncated protein lacking key PAX5 functional domains. Like other members of the PAX gene family, the PAX transcript is known to be alternatively spliced34 to produce several distinct transcripts that modify the amino acid sequence of the putative PAX5 proteins.35,36 Our results suggest that alternative splicing isoforms may derive from genomic deletion. Since, genomic deletions have been demonstrated to be important prognostic factors in ALL,17,24 we sought to determine whether PAX5 deletions affected outcome in adult patients with BCR-ABL1-positive ALL. However, when we investigated the association of PAX5 deletions with clinical outcome, no association was detected. Thereafter, the contribution of both PAX5 and IKZF1 alterations to clinical outcome was assessed showing that only the following combinations, PAX5 normal and IKZF1 loss versus PAX5 loss and IKZF1 normal and PAX5 loss and IKZF1 loss versus PAX5 loss and IKZF1 normal, were associated with significant P values (P=0.027 and P=0.031, respectively) (Online Supplementary Table S6). These data confirmed that IKZF1 status strongly affects the prognosis of adults with BCR-ABL1-positive ALL.24 From a biological point of view, this difference could be attributed to the different roles the two transcription factors have during B-cell development, as hypothesized by Georgopoulos.37 In multipotent hematopoietic stem cells, Ikaros promotes the priming of a cascade of lymphoid genes and at subsequent stages of lineage restriction and represses the expression of hematopoietic stem cell-specific genes.17 In B-cell-restricted progenitors, PAX5 is required for the transition from the pro-B cell to the pre-B cell.1,38 It is likely that the loss of PAX5 is not associated with a poor outcome because of a lack of deregulation in stem cell-associated genes.
In conclusion, PAX5 deletions are frequent in adult BCR-ABL1-positive ALL and are often associated with a loss of IKZF1. A better understanding of the role of these genes in the development of BCR-ABL1-positive ALL may lead to the identification of new targets for novel therapies.
Acknowledgments
we specially thank Charles Mullighan for help in molecular analysis and validation of PAX5 deletions by quantitative PCR. We also thank Serena Formica (University of Bologna) and Annalisa Astolfi (University of Bologna) for help in performing SNP arrays, Luciana Impera for performing FISH analysis, and Barbara Lama and Maria Chiara Abbenante (University of Bologna) for collecting clinical data. Finally, we acknowledge all the people who take care of the patients involved in the GIMEMA studies (see Appendix).
Appendix
The following authors contributed to this study: Leone Giuseppe - Università Cattolica del Sacro Cuore - Policlinico A. Gemelli, Rome; Torelli Giuseppe - Centro Oncologico Modenese - Dipartimento di Oncoematologia, Modena; Ferrara Felicetto -Azienda Ospedaliera di Rilievo Nazionale “A. Cardarelli”, Napoli; Majolino Ignazio - Divisione di Ematologia - Ospedale S. Camillo, Rome; Fanin Renato - Clinica Ematologica, Policlinico Universitario, Udine; Pizzolo Giovanni e Bonifacio Massimiliano - Università degli Studi di Verona - A. O. - Istituti Ospitalieri di Verona- Div. di Ematologia – Policlinico G.B. Rossi, Verona; Di Raimondo Francesco - Università di Catania -Cattedra di Ematologia - Ospedale “Ferrarotto” – Catania; Morra Enrica - Ospedale Niguarda “Cà Granda” – Milano; Mirto Salvatore and Francesco Fabbiano “Ospedali Riuniti Villa Sofia-Cervello, Palermo; Nobile Francesco - Dipartimento Emato-Oncologia A.O. “Bianchi-Melacrino-Morelli” - Reggio Calabria; Longinotti Maurizio - Serv. di Ematologia Ist. di Ematologia ed Endocrinologia – Sassari; Quarta Giovanni -Divisione di Ematologia Osp. Reg. A. Di Summa – Brindisi; Liso Vincenzo - Unità Operativa Ematologia 1 - Università degli Studi di Bari - Padiglione Chini −3° piano – Bari; Peta Antonio -Azienda Ospedaliera Pugliese Ciaccio - Presidio Ospedaliero A.Pugliese - Unità Operativa di Ematologia – Catanzaro; Rotoli Bruno - Azienda Ospedaliera Universitaria - Università degli Studi di Napoli “Federico II” Facoltà di Medicina, Napoli; Fioritoni Giuseppe - U.O. Ematologia Clinica - Azienda USL di Pescara; Olivieri Attilio - Ematologia - Ospedale San Carlo –Potenza; De Fabritiis Paolo - U.O.C. Ematologia - Ospedale S.Eugenio – Roma; Boccadoro Mario - Div. di Ematologia Ospedale “S.Giovanni Battista” – Torino; Saglio Giuseppe -Dip. di Scienze Cliniche e Biologiche - Ospedale S. Luigi Gonzaga - Orbassano (TO); Montanaro Marco - Azienda Sanitaria Locale Viterbo - Polo Ospedaliero Centrale - Ospedale Di Ronciglione - U.O. di Ematologia - Ronciglione (Viterbo); Zaccaria Alfonso - Dipartimento Oncologico - Ospedale S.Maria delle Croci – Ravenna; D’arco Alfonso Maria - U.O. Medicina Interna Ematologia ed Oncologia P.O. Umberto I -Nocera Inferiore (SA); Brugiatelli Maura - Divisione di Ematologia - Azienda Ospedaliera “Papardo” – Messina; Gaidano Gianluca - S.C.D.U. Ematologia - DIMECS e Dipartimento Oncologico - Università del Piemonte Orientale Amedeo Avogadro – Novara; Amadori Sergio - Università degli Studi - Policlinico di Tor Vergata – Roma.
Footnotes
Funding: this work was supported by: European LeukemiaNet, AIL, AIRC, Fondazione Del Monte di Bologna e Ravenna, FIRB 2006, Ateneo RFO grants, Project of integreted program (PIO), Programma di Ricerca Regione –Università 2007 – 2009. Progetti di Ricerca di Interesse Nazionale (PRIN), Fondo per gli Investimenti della Ricerca di Base (FIRB), Rome; Compagnia di San Paolo, Turin; Progetto “Oncologia”, Ministero della Salute, Rome; Fondazione per le Biotecnologie and Fondazione Internazionale di Ricerca in Medicina Sperimentale (FIRMS), Turin.
The online version of this article has a Supplementary Appendix.
Authorship and Disclosures
The information provided by the authors about contributions from persons listed as authors and in acknowledgments is available with the full text of this paper at www.haematologica.org.
Financial and other disclosures provided by the authors using the ICMJE (www.icmje.org) Uniform Format for Disclosure of Competing Interests are also available at www.haematologica.org.
References
- 1.Cobaleda C, Schebesta A, Delogu A, Busslinger M. Pax5: the guardian of B cell identity and function. Nat Immunol. 2007;8(5):463–70. doi: 10.1038/ni1454. [DOI] [PubMed] [Google Scholar]
- 2.Fuxa M, Skok JA. Transcriptional regulation in early B cell development. Curr Opin Immunol. 2007;19(2):129–36. doi: 10.1016/j.coi.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 3.Nutt SL, Eberhard D, Horcher M, Rolink AG, Busslinger M. Pax5 determines the identity of B cells from the beginning to the end of B-lymphopoiesis. Int Rev Immunol. 2001;20(1):65–82. doi: 10.3109/08830180109056723. [DOI] [PubMed] [Google Scholar]
- 4.Fitzsimmons D, Hodsdon W, Wheat W, Maira SM, Wasylyk B, Hagman J. Pax-5 (BSAP) recruits Ets proto-oncogene family proteins to form functional ternary complexes on a B-cell-specific promoter. Genes Dev. 1996;10(17):2198–211. doi: 10.1101/gad.10.17.2198. [DOI] [PubMed] [Google Scholar]
- 5.Horcher M, Souabni A, Busslinger M. Pax5/BSAP maintains the identity of B cells in late B lymphopoiesis. Immunity. 2001;14(6):779–90. doi: 10.1016/s1074-7613(01)00153-4. [DOI] [PubMed] [Google Scholar]
- 6.Ying H, Healy JI, Goodnow CC, Parnes JR. Regulation of mouse CD72 gene expression during B lymphocyte development. J Immunol. 1998;161(9):4760–7. [PubMed] [Google Scholar]
- 7.Schebesta M, Pfeffer PL, Busslinger M. Control of pre-BCR signaling by Pax5-dependent activation of the BLNK gene. Immunity. 2002;17(4):473–85. doi: 10.1016/s1074-7613(02)00418-1. [DOI] [PubMed] [Google Scholar]
- 8.Morrison AM, Jager U, Chott A, Schebesta M, Haas OA, Busslinger M. Deregulated PAX-5 transcription from a translocated IgH promoter in marginal zone lymphoma. Blood. 1998;92(10):3865–78. [PubMed] [Google Scholar]
- 9.Mullighan CG, Goorha S, Radtke I, Miller CB, Coustan-Smith E, Dalton JD, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446(7137):758–64. doi: 10.1038/nature05690. [DOI] [PubMed] [Google Scholar]
- 10.Mullighan CG, Williams RT, Downing JR, Sherr CJ. Failure of CDKN2A/B (INK4A/B-ARF)-mediated tumor suppression and resistance to targeted therapy in acute lymphoblastic leukemia induced by BCR-ABL. Genes Dev. 2008;22(11):1411–5. doi: 10.1101/gad.1673908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kawamata N, Ogawa S, Zimmermann M, Niebuhr B, Stocking C, Sanada M, et al. Cloning of genes involved in chromosomal translocations by high-resolution single nucleotide polymorphism genomic microarray. Proc Natl Acad Sci USA. 2008;105(33):11921–6. doi: 10.1073/pnas.0711039105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bousquet M, Broccardo C, Quelen C, Meggetto F, Kuhlein E, Delsol G, et al. A novel PAX5-ELN fusion protein identified in B-cell acute lymphoblastic leukemia acts as a dominant negative on wild-type PAX5. Blood. 2007;109(8):3417–23. doi: 10.1182/blood-2006-05-025221. [DOI] [PubMed] [Google Scholar]
- 13.Cazzaniga G, Daniotti M, Tosi S, Giudici G, Aloisi A, Pogliani E, et al. The paired box domain gene PAX5 is fused to ETV6/TEL in an acute lymphoblastic leukemia case. Cancer Res. 2001;61(12):4666–70. [PubMed] [Google Scholar]
- 14.Nebral K, Konig M, Harder L, Siebert R, Haas OA, Strehl S. Identification of PML as novel PAX5 fusion partner in childhood acute lymphoblastic leukaemia. Br J Haematol. 2007;139(2):269–74. doi: 10.1111/j.1365-2141.2007.06731.x. [DOI] [PubMed] [Google Scholar]
- 15.An Q, Wright SL, Konn ZJ, Matheson E, Minto L, Moorman AV, et al. Variable breakpoints target PAX5 in patients with dicentric chromosomes: a model for the basis of unbalanced translocations in cancer. Proc Natl Acad Sci USA. 2008;105(44):17050–4. doi: 10.1073/pnas.0803494105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nebral K, Denk D, Attarbaschi A, König M, Mann G, Haas OA, Strehl S. Incidence and diversity of PAX5 fusion genes in childhood acute lymphoblastic leukemia. Leukemia. 2009;23(1):134–43. doi: 10.1038/leu.2008.306. [DOI] [PubMed] [Google Scholar]
- 17.Mullighan CG, Su X, Zhang J, Radtke I, Phillips LA, Miller CB, et al. Deletion of IKZF1 and prognosis in acute lymphoblastic leukemia. N Engl J Med. 2009;360(5):470–80. doi: 10.1056/NEJMoa0808253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mullighan CG, Miller CB, Radtke I, Phillips LA, Dalton J, Ma J, et al. BCR-ABL1 lymphoblastic leukaemia is characterized by the deletion of Ikaros. Nature. 2008;453(7191):110–4. doi: 10.1038/nature06866. [DOI] [PubMed] [Google Scholar]
- 19.Vignetti M, Fazi P, Cimino G, Martinelli G, Di Raimondo F, Ferrara F, et al. Imatinib plus steroids induces complete remissions and prolonged survival in elderly Philadelphia chromosome-positive patients with acute lymphoblastic leukemia without additional chemotherapy: results of the Gruppo Italiano Malattie Ematologiche dell’Adulto (GIMEMA) LAL0201-B protocol. Blood. 2007;109(9):3676–8. doi: 10.1182/blood-2006-10-052746. [DOI] [PubMed] [Google Scholar]
- 20.le Coutre P, Ottmann OG, Giles F, Kim DW, Cortes J, Gattermann N, et al. Nilotinib (formerly AMN107), a highly selective BCR-ABL tyrosine kinase inhibitor, is active in patients with imatinib-resistant or -intolerant accelerated-phase chronic myelogenous leukemia. Blood. 2008;111(4):1834–9. doi: 10.1182/blood-2007-04-083196. [DOI] [PubMed] [Google Scholar]
- 21.Piccaluga PP, Paolini S, Martinelli G. Tyrosine kinase inhibitors for the treatment of Philadelphia chromosome-positive adult acute lymphoblastic leukemia. Cancer. 2007;110(6):1178–86. doi: 10.1002/cncr.22881. [DOI] [PubMed] [Google Scholar]
- 22.Ottmann O, Dombret H, Martinelli G, Simonsson B, Guilhot F, Larson RA, et al. Dasatinib induces rapid hematologic and cytogenetic responses in adult patients with Philadelphia chromosome positive acute lymphoblastic leukemia with resistance or intolerance to imatinib: interim results of a phase 2 study. Blood. 2007;110(7):2309–15. doi: 10.1182/blood-2007-02-073528. [DOI] [PubMed] [Google Scholar]
- 23.Ottmann OG, Wassmann B. Imatinib in the treatment of Philadelphia chromosome-positive acute lymphoblastic leukaemia: current status and evolving concepts. Best Pract Res Clin Haematol. 2002;15(4):757–69. doi: 10.1053/beha.2002.0233. [DOI] [PubMed] [Google Scholar]
- 24.Martinelli G, Iacobucci I, Storlazzi CT, Vignetti M, Paoloni F, Cilloni D, et al. IKZF1 (Ikaros) deletions in BCR-ABL1-positive acute lymphoblastic leukemia are associated with short disease-free survival and high rate of cumulative incidence of relapse: a GIMEMA AL WP report. J Clin Oncol. 2009;27(31):5202–7. doi: 10.1200/JCO.2008.21.6408. [DOI] [PubMed] [Google Scholar]
- 25.Foà R, Vitale A, Meloni G, Guarini A, De Propris S, Elia L, et al. Dasatinib as front-line monotherapy for the induction treatment of adult and elderly Ph+ acute lymphoblastic leukemia (ALL) patients: interim analysis of the GIMEMA prospective study LAL1205. Blood. 2007;110:10a. [Google Scholar]
- 26.Foà R, Fazi P, Vitale A, Guarini A, De Propris MS, Elia L, et al. Dasatinib mono -therapy as 1st line treatment of Ph acute lymphoblastic leukemia (ALL) patients: update of Gimema LAL1205 study. Haematologica. 2008;93(s1):367. Abs.0922. [Google Scholar]
- 27.Iacobucci I, Storlazzi CT, Cilloni D, Lonetti A, Ottaviani E, Soverini S, et al. Identification and molecular characterization of recurrent genomic deletions on 7p12 in the IKZF1 gene in a large cohort of BCR-ABL1-positive acute lymphoblastic leukemia patients: on behalf of Gruppo Italiano Malattie Ematologiche dell’Adulto Acute Leukemia Working Party (GIMEMA AL WP) Blood. 2009;114(10):2159–67. doi: 10.1182/blood-2008-08-173963. [DOI] [PubMed] [Google Scholar]
- 28.Kaplan E, Meier P. Non parametric estimation from incomplete observation. J Am Stat Assoc. 1958;53:457–81. [Google Scholar]
- 29.Gooley TA, Leisenring W, Crowley J, Storer B. Estimation of failure probabilities in the presence of competing risks: new representations of old estimators. Stat Med. 1999;18(6):695–706. doi: 10.1002/(sici)1097-0258(19990330)18:6<695::aid-sim60>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 30.Simon R, Lee Y. Nonparametric confidence limits survival probabilities and median survival time. Cancer Treat Res. 1982;66(1):37–42. [PubMed] [Google Scholar]
- 31.Cox D. Regression models and life-tables. J R Stat Soc. 1972;34:187–220. [Google Scholar]
- 32.Familiades J, Bousquet M, Lafage-Pochitaloff M, Béné MC, Beldjord K, De Vos J, et al. PAX5 mutations occur frequently in adult B-cell progenitor acute lymphoblastic leukemia and PAX5 haploin-sufficiency is associated with BCR-ABL1 and TCF3-PBX1 fusion genes: a GRAALL study. Leukemia. 2009;23(11):1989–98. doi: 10.1038/leu.2009.135. [DOI] [PubMed] [Google Scholar]
- 33.Busslinger M, Klix N, Pfeffer P, Graninger PG, Kozmik Z. Deregulation of PAX-5 by translocation of the Emu enhancer of the IgH locus adjacent to two alternative PAX-5 promoters in a diffuse large-cell lymphoma. Proc Natl Acad Sci USA. 1996;93(12):6129–34. doi: 10.1073/pnas.93.12.6129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zwollo P, Arrieta H, Ede K, Molinder K, Desiderio S, Pollock R. The Pax-5 gene is alternatively spliced during B-cell development. J Biol Chem. 1997;272(15):10160–8. doi: 10.1074/jbc.272.15.10160. [DOI] [PubMed] [Google Scholar]
- 35.Arseneau JR, Laflamme M, Lewis SM, Maicas E, Ouellette RJ. Multiple isoforms of PAX5 are expressed in both lymphomas and normal B-cells. Br J Haematol. 2009;147(3):328–38. doi: 10.1111/j.1365-2141.2009.07859.x. [DOI] [PubMed] [Google Scholar]
- 36.Santoro A, Bica MG, Dagnino L, Agueli C, Salemi D, Cannella S, et al. Altered mRNA expression of PAX5 is a common event in acute lymphoblastic leukaemia. Br J Haematol. 2009;146(6):686–9. doi: 10.1111/j.1365-2141.2009.07815.x. [DOI] [PubMed] [Google Scholar]
- 37.Georgopoulos K. Acute lymphoblastic leukemia–on the wings of IKAROS. N Engl J Med. 2009;360(5):524–6. doi: 10.1056/NEJMe0809819. [DOI] [PubMed] [Google Scholar]
- 38.Souabni A, Cobaleda C, Schebesta M, Busslinger M. Pax5 promotes B lymphopoiesis and blocks T cell development by repressing Notch1. Immunity. 2002;17(6):781–93. doi: 10.1016/s1074-7613(02)00472-7. [DOI] [PubMed] [Google Scholar]