Abstract
Background
Natural killer cell-type lymphoproliferative disease of granular lymphocytes is a disorder characterized by chronic proliferation of CD3−CD16+ granular lymphocytes. By flow cytometry analysis, we previously demonstrated a dysregulation in killer immunoglobulin-like receptor (KIR) expression in natural killer cells from patients with this lymphoproliferative disease, the activating KIR receptors being mostly expressed. We also found that patients with natural killer cell-type lymphoproliferative disease of granular lymphocytes usually had KIR genotypes characterized by multiple activating KIR genes.
Design and Methods
We investigated the mRNA levels of the KIR3DL1 inhibitory and the related KIR3DS1 activating receptors in 15 patients with natural killer cell-type lymphoproliferative disease of granular lymphocytes and in ten controls. These genes are usually expressed when present in the genome of the Caucasian population.
Results
We demonstrated the complete lack of KIR3DL1 expression in most of the patients analyzed, with the receptor being expressed in 13% of patients compared to in 90% of controls (P<0.01). Interestingly, studies of the methylation patterns of KIR3DL1 promoter showed a significantly higher methylation status (0.76 ± 0.12 SD) in patients than in healthy subjects (0.49±0.10 SD, P<0.01). The levels of expression of DNA methyl transferases, which are the enzymes responsible for DNA methylation, did not differ between patients and controls.
Conclusions
In this study we showed, for the first time, a consistent down-regulation of the inhibitory KIR3DL1 signal due to marked methylation of its promoter, thus suggesting that together with the increased expression of activating receptors, the lack of the inhibitory signal could also play a role in the pathogenesis of natural killer cell-type lymphoproliferative disease of granular lymphocytes.
Keywords: natural killer cells, lymphoproliferative disease of granular lymphocytes, KIR3DL1 down-regulation
Introduction
Lymphoproliferative disease of granular lymphocytes (LDGL) is a rare disorder characterized by the chronic proliferation of granular lymphocytes.1–3 Based on the surface phenotype of the proliferating lymphocytes, LDGL can be classified into two subclasses. In the majority of cases (85%), the proliferating granular lymphocytes belong to the T-cell lineage (CD3+CD16+); however, in nearly 15% of cases, patients’ granular lymphocytes originate from the expansion of natural killer (NK) cells (CD3−CD16+). The etiology and pathogenesis of NK-LDGL proliferation is still a matter of debate; however, accumulating data suggest that exogenous agents, in particular viruses, might play a role.4
In healthy individuals, NK cells represent a subset of lymphocytes involved in immunosurveillance against virus-infected and tumor-transformed cells.5,6 The ability of NK cells to recognize and kill susceptible targets while sparing normal autologous cells is commonly determined by the concerted action of different surface receptor families, recognized as NK cell receptors, which either trigger or inhibit NK cell-mediated cytolytic activity.7–10 Among these, killer immunoglobulin-like receptor (KIR) molecules are critical for the human NK cell’s ability to discriminate aberrant cells from normal self playing a role in autoimmune diseases, transplantation, infectious diseases and cancer.11,12 Within an individual’s NK cell population the expression of KIR is very heterogeneous due to the expression of different numbers of KIR receptors and different combinations of KIR genes.13,14 Moreover, long-term NK cell clones maintain uniform KIR gene and allele-specific expression, indicating that KIR transcription patterns are stable in mature NK cells through many rounds of DNA replication.15 Recently, it has been demonstrated that DNA methylation plays an important role in the regulation of KIR gene expression.16–18 CpG dinucleotides located upstream of the translation initiation codon of each KIR gene (KIR2DL3, 3DL2, 3DL1) are either densely methylated in the case of repressed KIR genes or demethylated in expressed KIR genes. Expression of formerly silenced KIR genes can be readily induced using a demethylating agent. Reporter gene experiments using in vitro methylated 5′ untranslated region constructs indicated that KIR CpG islands largely overlap with regions having promoter activity.17
Previous flow cytometry data highlighted a dysregulation in KIR expression in NK cells from patients with NK-LDGL.19,20 Particularly, in LDGL patients we found that pathological NK cells mostly express activating KIR and are positively selected, suggesting that these receptors may be directly involved in the priming of the proliferation of granular lymphocytes.19 Thus, stimulation through an activating KIR may represent the triggering event at the onset of NK cell proliferation. We also demonstrated that NK-LDGL patients usually have KIR genotypes containing multiple activating KIR genes.21 Interestingly most NK-LDGL cells freshly isolated from patients consistently exhibited altered surface KIR3DL1/3DS1 expression, with lack of the inhibitory receptor and the cells providing cytotoxic effects in vitro.22
Building on the findings described above, we used semi-quantitative polymerase chain reaction (PCR) analysis to investigate the mRNA levels of the inhibitory KIR3DL1 and the related activating KIR3DS1. Since it has recently been demonstrated that DNA methylation is the main mechanism involved in the regulation of KIR gene expression, we also analyzed the methylation pattern of CpG islands in the promoter of these genes. Despite the high homology between KIR promoters (>91%15), which makes it difficult to choose primers specific for each KIR gene, in the case of KIR3D1 the primers used efficiently identified the specific promoter.17 Finally we evaluated the expression levels of DNA methyltransferases (DNMT), the enzymes responsible for DNA methylation.
Design and Methods
Patients and control blood donors
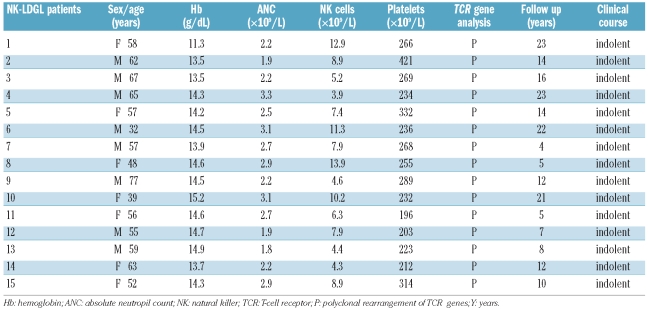
Fifteen Caucasian patients meeting the criteria for the diagnosis of NK-type LDGL3 were consecutively enrolled in the study (Table 1) and investigated for the presence and expression of KIR3DL1/DS1 (Figure 1). Informed consent was obtained from all patients according to the Declaration of Helsinki and the study was approved by the local Ethics Committee. In all cases under study there was chronic lymphocytosis, with at least 2,000 granular lymphocytes/mm3 in the peripheral blood, lasting more than 6 months. Most patients (14 of 15) were affected by LDGL characterized by a proliferation of CD3−CD16+CD56bright NK cells (range, 40–85%); only patient #1 had a proliferation of CD3−CD16+CD56− cells. At the time of the study none of the patients had received treatment. The control experiments were performed on ten blood donors, representative of the healthy population.
Table 1.
Laboratory and clinical features of the 15 LDGL patients studied.
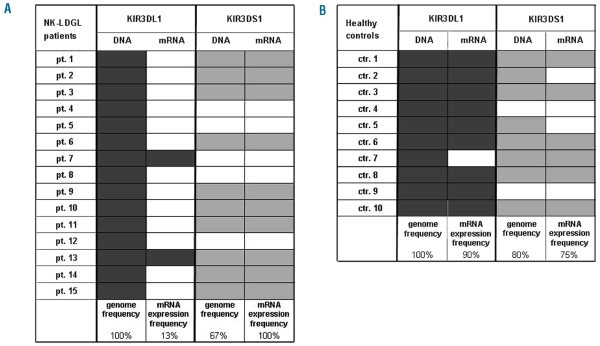
Figure 1.
Genotyping of KIR3DL1 and KIR3DS1 alleles and frequency of KIR3DL1/3DS1 mRNA expression in patients and normal controls. Genomic DNA was extracted from peripheral blood mononuclear cells of 15 patients and ten controls. The presence or absence of KIR3DL1/3DS1 gene was established by PCR-SSP using two different primer sets specific for both alleles. The figure (columns genome frequency) summarizes the KIR3DL1/3DS1 locus profiles observed in the patients (A) and controls (B). Filled boxes indicate the presence of KIR allele; open boxes indicate the absence. The difference of presence in the genome of KIR3DS1 between patients and controls was not statistically significant (P=0.28). For the investigation of KIR3DL1/DS1 expression total mRNA was isolated from sorted NK cells of patients and controls. Semi-quantitative PCR was carried out to determine the presence or absence of transcription for both KIR3DL1/3DS1 alleles (columns mRNA expression frequency). Amplification of β-actin was used as a control. The filled boxes indicate the corresponding expression of the mRNA, for both activating and inhibitory receptors in patients (A) and controls (B).
Flow cytometry analysis
The surface expression of KIR3DL1/KIR3DS1 receptors was assessed by flow cytometry analysis in freshly isolated NK cells using direct or indirect immunofluorescence assays as previously described.22,23 Briefly, cells were stained with the appropriate unlabeled or labeled monoclonal antibodies; staining with unlabeled monoclonal antibodies was followed by phycoerythrin (PE)– or fluorescein isothiocyanate (FITC)–conjugated goat anti-mouse immunoglobulins (Southern Biotechnology, Birmingham, AL, or Caltag, Burlingame, CA, USA). The analysis on fresh peripheral blood mononuclear cells was performed on the CD3−CD16+ gated cells. Samples were analyzed by multi-color flow cytometry analysis (FACSCalibur, Becton Dickinson, Mountain View, CA, USA) and data were processed using CELLQuest Software (Becton Dickinson).
Purification of peripheral blood mononuclear cells and sorting of natural killer cells from patients and control blood donors
Peripheral blood mononuclear cells were derived from healthy donors and from patients by Ficoll-Hypaque gradient. To obtain enriched NK cells, the peripheral blood mononuclear cells from patients were incubated (for 20 min at 4°C) with FITC- or PE-conjugated monoclonal antibodies specific for CD16 or CD3, respectively; peripheral blood mononuclear cells from healthy donors were, instead, incubated with FITC-, PE- or PE-Cy5-conjugated monoclonal antibodies specific for CD16, CD3, or KIR3DL1/KIR3DS1, respectively. In samples from patients with LDGL, the CD3−CD16+ NK cell fraction was sorted, whereas in healthy donors, beyond the CD3−CD16+ NK cell population, the CD3−CD16+KIR3DL1/KIR3DS1− and CD3−CD16+KIR3DL1/KIR3DS1+ fractions were further sorted. Cell sorting was performed with a FACSAria cell sorter (BD Biosciences, San Jose, CA, USA) equipped with blue, red, and violet lasers. Dead cells were excluded by propidium iodide (10 μg/mL) versus forward scatter dot plots and viable cells positive for the above mentioned antibodies were selected and sorted, using 530 nm, 575 nm and 610 band pass filters and an argon ion laser (488 nm, 100 mW) for excitation and the cell sorter’s purity option at a rate of 15,000 events per second. Sorted NK populations were reanalyzed for purity and viability, which both resulted greater than 95%.
Monoclonal antibodies
The commercially available FITC- or PE-labeled mouse antihuman Z27 (anti-KIR3DL1/KIR3DS1),19 CD3 and CD16 monoclonal antibodies used in immunofluorescence assays and sorting experiments were from Becton Dickinson (Sunnyvale, CA, USA).
DNA isolation and KIR genotyping
Genomic DNA was extracted from 5–20×106 peripheral blood lymphocytes using the Genomic DNA Purification kit (Gentra Systems, Minneapolis, MN, USA) according to the manufacturer’s instructions. Sequence-specific primed PCR typing was used to detect the KIR3DL1/3DS1 genotype of LDGL patients and controls. Two different primer sets were used to establish the presence or absence of both genes: KIR3DL1a forward TACAAAGAAGACAGAATC-CACA,24 KIR3DL1a reverse.TAG GTCCCTG-CAAGGGCAA24 and KIR3DL1b forward TCCCATCTTC-CATGGCA GAT,24 KIR3DL1b reverse TAGGTCCCTG-CAAGGGCAA24 for KIR3DL1 expression, KIR3DS1a forward GGCAGAATATTCCAGGAGG,25 KIR3DS1a reverse AGGGGTCC TTAGAGATCCA25 and KIR3DS1b forward TCCCCTGGTGAAATCAGGAGAGAG and KIR3DS1b reverse AAGGGCACGCATCATGGA25,26 for KIR3DS1 expression.25 Amplifications with the primers KIR3DL1a, KIR3DL1b and KIR3DS1a were carried out using the following procedure: initial denaturation for 8 min at 95°C; then 35 cycles of 30 sec at 95°C, 45 sec at 55°C and 90 sec at 68°C, followed by 68°C for 7 min. Volume reactions of 15 μL were prepared to include 30 ng of DNA, 1.5 μL 10X PCR buffer, 1.5 mM magnesium chloride (MgCl2), 0.2 mM deoxyribonucleoside triphosphates (dNTP), 0.2U Taq gold (Applied Biosystems, Foster City, CA, USA), 0.4 μM of each primer, and 1% dimethyl sulfoxide. To amplify the KIR3DS1 gene with the primer KIR3DS1b, the following program was used: initial denaturation at 95°C for 10 min followed by 35 cycles of 30 sec at 95°C, 30 sec at 60°C, and 30 sec at 72°C, followed by 72°C for 10 min. Volume reactions of 15 μL were prepared to include 30 ng of DNA, 1.5 μL 10X PCR buffer, 3 mM MgCl2, 0.6 mM dNTP, 0.2U Taq gold (Applied Biosystems, Foster City, CA, USA), 1 μM of each primer, and 1% dimethyl sulfoxide. PCR products were analyzed for the presence or absence of an electrophoretic specific band in 1% agarose gel stained with ethidium bromide, visualized by ultraviolet light and documented with Image Master (Amersham Pharmacia Biotech, Sweden).
RNA isolation and polymerase chain reaction
A Qiagen RNeasy isolation kit (Valencia, CA, USA) was used to isolate total RNA from 4×106 cells (NK CD3−CD16+ cells) from patients and control donors. A Clontech cDNA synthesis kit (Mountain View, CA, USA) was used for cDNA synthesis. Semi-quantitative PCR was carried out to determine the presence or absence of KIR3DL1/3DS1 mRNA and transcription levels. Primers for β-actin were used as the control. Amplifications were carried out using the following procedure; initial denaturation for 8 min at 95°C; then 35 cycles of 30 sec at 95°C, 45 sec at 55°C and 90 sec at 68°C, followed by 68°C for 10 min. PCR products were analyzed for the presence or absence of an electrophoretic specific band in 1% agarose gel stained with ethidium bromide, visualized by ultraviolet light and documented with Image Master (Amersham Pharmacia Biotech, Sweden). The real-time PCR reactions to assess the levels of expression of mRNA DNMT1, DNMT3a and 3b, were performed in a volume of 15 μL containing 50 ng cDNA, 0.3 μM of each primer, deionised water, and 7.5 μL SYBR Green Master Mix (Applied Biosystems) using the following conditions: 50°C for 2 min and denaturing at 95°C for 10 min, followed by 45 cycles of 95°C for 15 sec and 60°C for 1 min. Optical data were collected during the 60°C step. The reactions were carried out in 96-well thin-wall PCR plates covered with optically clear sealing film (Applied Biosystems). Amplification, detection, and data analysis were performed using the ABI PRISM 7000 Sequence Detector system (Applied Biosystems). Each quantification target was amplified in triplicate samples. A no template control for each mix and four standard curves were generated for GAPDH, DNMT1, DNMT3a and DNMT3b, using Jurkat cell line cDNA in serial dilutions of 1, 1:5, 1:25, and 1:125. The relative amounts of mRNA were determined by comparison with standard curves. Results were normalized for GAPDH expression for each sample. A dissociation curve was generated to distinguish specific from non-specific amplifications. The primers for PCR and RT-PCR are listed in Table 2.15,25,27–29
Table 2.
Sequences of primers used in the study.
DNA methylation analysis
Bisulfite conversion of DNA leads to conversion of all unmethylated cytosines into thymidines, while methylated cytosines remain unchanged. Genomic DNA was extracted from 2–20×106 sorted cells (CD3−CD16+ NK cells from patients and CD3−CD16+KIR3D1− NK cells from control donors) using the Genomic DNA Purification kit (Gentra Systems, Minneapolis, MN, USA). The genomic DNA was modified by sodium bisulfite treatment according to the manufacturer’s instructions (EPIGENTEK, Methylamp DNA modification Kit, NY, USA).
Converted DNA (25 ng) was used as a template in a 25μL reaction volume. The reaction contains 25 pmol of each primer F-GGAATTCCGTTTTTTATGTTAGTATAGATTTTA and R-CGGGATCCCGCCATATCTTTAC-CTCCAAATC,17 1 mM dNTP mix, 3 mM MgCl2, and 0.5 U HotStarTaq DNA polymerase (Qiagen). After initial denaturation for 10 min at 95°C, 34 cycles were performed, each consisting of 90 sec at 95°C, 55 sec at 48°C, and 1 min at 72°C, followed by 72°C for 10 min. The PCR products were analyzed for the presence of an electrophoretic specific band in 1% agarose gel stained with ethidium bromide, cloned with the TOPO-TA cloning system (Invitrogen) and sequenced. Ten clones were sequenced for each case. In all sequenced PCR clones, more than 99% of random cytosines (not part of the CpG dinucleotides) were converted to thymidines, consistent with adequate bisulfite treatment. Genomic KIR sequences were obtained from www.genecards.org (GeneID 3811).
Statistical analysis
Observed KIR gene frequencies were determined by direct counting. Differences in the gene frequencies and KIR expression frequencies between NK-LDGL patients and controls were estimated by Fisher’s exact test. The comparison of methylation frequencies of CpG dinucleotides and DNMT1 mRNA expression levels in the group of patients with respect to controls was estimated using the Student’s t-test. The data are expressed as mean ± standard deviation (SD). For all analyses, P values less than 0.05 were accepted as statistically significant.
Results
KIR3DL1/3DS1 genotyping
Since not all KIR genes are necessarily present in the genome of a person, as a first step, using sequence-specific primed PCR we analyzed the KIR3DL1/3DS1 genes in the genome of 15 NK-LDGL patients and ten healthy controls. The KIR3DL1/3DS1 gene is unique within the KIR cluster in that it is the only KIR locus coding for both activating (KIR3DS1) and inhibitory (KIR3DL1) allotypes. The KIR3DL1/3DS1 gene is also highly polymorphic with 23 sequenced alleles, 17 coding for the inhibitory receptor and 6 for the activating receptor, the different alleles sharing a sequence similarity of more than 95% in the extra-cellular domains (http://www.ebi.ac.uk/ipd/kir/align.html). To examine the different known alleles, we used two sets of primers (see the Design and Methods section), which could eventually identify the inhibitory and activating receptors. We found that a KIR3DL1 allele was present with a frequency of 100% in both patients (KIR3DL1 genome frequency column in Figure 1A) and controls (KIR3DL1 genome frequency column in Figure 1B), whereas an activating KIR3DS1 allele was present with a frequency of 67% in patients (KIR3DS1 genome frequency column in Figure 1A) and 80% in controls (KIR3DS1 genome frequency column in Figure 1B, P=0.28). These results indicate that there were not differences in gene frequency for the KIR3DL1/3DS1 between NK-LDGL patients and healthy controls.
Patients with natural killer cell lymphoproliferative disease of granular lymphocytes express the KIR3DL1 alleles differently
The mRNA expression of KIR3DL1/3DS1 was evaluated by semi-quantitative PCR in CD3−CD16+ NK cells sorted from peripheral blood mononuclear cells of the patients and controls. Interestingly we found that the inhibitory KIR3DL1 alleles were expressed in 13% of the patients (KIR3DL1 mRNA expression frequency column in Figure 1A) but in 90% of healthy controls (KIR3DL1 mRNA expression frequency column in Figure 1B; P<0.01). Conversely, we found that the activating KIR3DS1 alleles, when present in the genome, were expressed in 100% of the LDGL patients (KIR3DS1 mRNA expression frequency column in Figure 1A) and in 75% of control subjects (KIR3DS1 mRNA expression frequency column in Figure 1B; P=0.11). These results suggest that, in the LDGL patients, there is an important down-regulation of the inhibitory KIR3DL1 signal, usually associated with expression of KIR3DS1 (when the gene is present in the genome).
Analysis of the methylation pattern in the KIR3DL1/3DS1 promoter
Since KIR3DL1 was not expressed in our patients, we investigated the mechanism modulating the expression of the KIR3DL1/3DS1 alleles. To date the most important mechanism reported to be involved in the regulation of KIR gene expression is methylation of CpG dinucleotides in the promoter of each KIR gene. In fact, analyses of DNA methylation patterns of KIR genes in NK cell lines as well as in NK cells freshly isolated from peripheral blood demonstrated that a small CpG island surrounding the transcriptional start site of each KIR gene (Figure 2) is demethylated in expressed KIR and methylated in unexpressed KIR.17 Since the KIR3DL1 and KIR3DS1 alleles are reported to be under control of the same promoter30,31 the set of primers specific for the KIR3D1 promoter used in this study could not discriminate between the activating and inhibitory alleles. After DNA modification with sodium bisulfite, PCR amplification and TOPO TA cloning, we sequenced ten plasmids for each patient and each control analyzed in our study. Five patients were analyzed, including patients #1, #2, and #3, who had both genes in the genome, but expressed only the activating KIR3DS1 receptor, and patients #4 and #5, who had only the inhibitory KIR3DL1 gene which was not expressed (Figure 1A). To provide appropriate controls for methylation, NK cells that did not express either KIR3DL1 inhibitory or KIR3DS1 activating receptors (i.e. the phenotype CD3−CD16+ KIR3DL1/KIR3DS1−) at the cell surface were sorted from five healthy donors.
Figure 2.
Distribution of CpG dinucleotides in the 5′ region of the KIR3D1 gene. The transcriptional start site is indicated by a square and the CpG dinucleotides are highlighted in gray. Analysis of the methylation patterns of the CpG island surrounding the transcriptional start site of this gene revealed that the methylation status consistently correlates with the transcriptional activity in primary NK cells.17 The primers used in this work to analyze the methylation pattern of this sequence are underlined.
Consistent with the published data, we found that the promoters of the control individuals’ CD3−CD16+KIR3DL1/KIR3DS1− cells were methylated17 (Figure 3A). By contrast, the promoters of patients with LDGL showed a quite different status of methylation. In fact we found about 60% of sequences were consistently methylated and 40% completely unmethylated in patients #1, #2 and #3, as follows: patient #1, seven plasmids methylated and three plasmids not methylated; patient #2, five plasmids methylated and five plasmids not methylated; patient #3, six plasmids methylated and four plasmids not methylated; the results for patient #1 are illustrated in Figure 3B. Although the expected percentage of methylated and unmethylated clones in these patients should be 50%, the relatively low number of cloned plasmids could account for the slight difference. Considering the KIR3DL1/3DS1 genotype and mRNA expression, patients #1, #2, and #3 displayed each gene but expressed only the activating one (Figure 1A). For this reason it is suggested that the completely unmethylated sequences represented the expressed activating receptor KIR3DS1, whereas the methylated sequences were referred to the promoter of inhibitory KIR3DL1 receptor, which was not expressed. The results obtained from patients #4 and #5 are consistent with this interpretation. These patients presented only the inhibitory receptor, which is not expressed, in their genome and according to these feature, all the sequenced plasmids had consistently methylated CpG dinucleotides (patient #4 shown in Figure 3C). Considering only the methylated sequences, the methylation frequency of the CpG island was calculated in each clone. The methylation frequency was calculated as the number of methylated CpG dinucleotides divided by the total number of CpG dinucleotides present in the analyzed sequence. Interestingly we found that the value was higher for the patients than for the controls (0.76±0.12 SD versus 0.42±0.10 SD, respectively; P<0.01, Figure 3D). The frequency of methylation for each individual CpG dinucleotide in the analyzed region was also considered (Figure 4). The difference in frequency of each CpG between the patients and controls was also statistically significant (P<0.001). These results indicate that, in patients with LDGL, the KIR3DL1 gene is subjected to strong down-regulation by hypermethylation of its promoter.
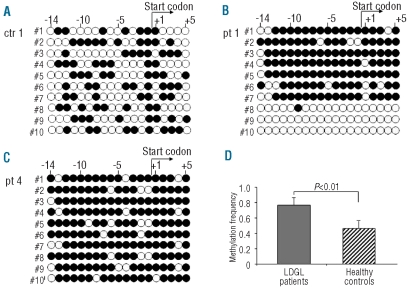
Figure 3.
Methylation of CpG dinucleotides correlates with transcriptional activity of the KIR3D1 gene in NK cells isolated from LDGL-patients. The figure reports the methylation pattern in the 5′ region of the KIR3D1 gene for one representative control (A) and two representative patients (B, C). The filled circles indicate methylated CpG dinucleotides, the empty circles unmethylated ones. The figure also shows the position of the start codon, the numbers indicate the CpG dinucleotides before and after the start codon in progressive order. The analysis of controls (5 subjects) was performed in the sorted CD3−CD16+KIR3DL1/KIR3DS1− NK cells; instead in patients (5 subjects), the analysis was performed on sorted CD3− CD16+ NK cells. The methylation frequency, for each sequenced clone, was calculated as the number of methylated CpG dinucleotides over the total CpG dinucleotides present in the analyzed sequence. As shown by the numbers of the filled circles (A, B, C), methylation was increased in the patients with respect to the controls. The difference of median methylation frequency, considering each clone for the five patients and five controls, was statistically significant 0.76±0.12 SD versus 0.49±0.10SD, respectively P<0.01 (D).
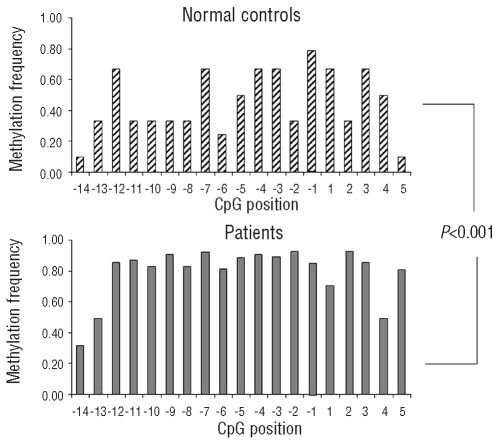
Figure 4.
The methylation frequency at individual CpG positions in normal controls and patients. The frequency for each CpG dinucleotide considered (from −14 to +5 with respect to the start codon) is reported as a median of the total sequenced clones for controls (upper panel) and patients (lower panel). The overall methylation frequency at individual positions was significantly higher in patients than in controls (P<0.001).
DNMT1, 3a and 3b reverse transcriptase polymerase chain reaction expression levels
The expression levels of DNMT1, 3a and 3b, which are mostly responsible for DNA methylation, were investigated in patients and controls. Since DNMT3a and 3b are usually expressed at very low levels in adult human tissues,32 they were not suitable for this analysis (data not shown). With regards to DNMT1, we did not found that the level of expression differed between patients and controls (5.07±2.78 versus 4.98±3.23 SD respectively; P=0.44). Accordingly, the enzymes responsible for DNA methylation do not seem to be involved in the increased methylation of KIR3DL1 promoter that we observed.
Discussion
In this study we showed that there is strong down-regulation of the KIR3DL1 inhibitory signal together with a skewed expression of the activating KIR3DS1 in NK-type LDGL patients. In fact, the KIR3DL1 inhibitory receptor was expressed in only 13% of our patients but in 90% of controls (P<0.01). By contrast, the KIR3DS1 gene, when present in the genome, was always expressed, and was found in 100% of patients and 75% of controls.
DNA methylation of CpG dinucleotides surrounding the start codon is a mechanism for switching off the expression of genes, and has been shown to regulate the expression of KIR receptors.17 In fact, CpG islands located upstream of the translation initiation codon of each KIR gene are densely methylated in the case of repressed KIR genes and their expression can be readily induced using a demethylating agent.17 Early during embryonic development, alternating waves of methylation and demethylation regulate cell growth and differentiation and it has been suggested that the DNA methylation patterns, established during this time, remain relatively stable in normal tissue.33 However, a growing body of data has demonstrated that this mechanism of gene expression control continues life-long.33
A significant novelty found in our study is the lack of expression of the inhibitory KIR3DL1 receptor associated with significantly increased DNA methylation in the promoter in NK-type LDGL patients as compared to in controls (P<0.01). Accordingly, it can be suggested that a putative event modulating gene transcription would be crucial in the development of this disorder. In our experiments we did not find a specific alteration in the methylation status of the KIR3D1 promoter. However, our data, showing a significant increase of methylated CpG dinucleotides in the KIR3D1 promoter of patients, support the importance of the transcriptional mechanism and suggest that an exogenous event, by adding more methyls and in turn providing a strong down-regulation of gene expression, might account for this feature in LDGL patients. In other words, a crucial step in this disease would be the marked silencing of the KIR3DL1 gene, which is usually expressed in healthy individuals.
In the past few years transcriptional inactivation of tumor suppressor genes by hypermethylation of CpG dinucleotides in the promoter has been claimed as an etiological factor in hematologic malignancies.34 Epigenetic events, for example, have been demonstrated to represent a crucial step in the progression of the myelodysplastic syndromes.35–37 Hypermethylation in the promoter region of the p15INKB gene, a cyclin-dependent kinase inhibitor, occurs in approximately 50% of patients with myelodys-plastic syndrome and it has been reported to be acquired during disease progression. More recently, hypermethylation has also been reported to be involved in virus-induced disorders such as adult T-cell leukemia. In particular, it has been reported that in adult T-cell leukemia cells, methylation first occurs in the exon region and then progresses to the promoter region according to the clinical progression of the disease.38 Turning off this mechanism by a demethylating drug (e.g. 5-azacitydine) has proven a valuable strategy in the therapy of these disorders.39,40
An association between myelodysplastic syndromes and LDGL has been reported,41 indicating that granular lymphocytes can be under the same epigenetic pressure as myelodysplastic cells. Although viral agents, mostly retro-viral ones,42 have been suggested to be involved in the pathogenesis of proliferation of cytotoxic cells in LDGL patients, the precise mechanisms by which these agents act are far for being elucidated. Interestingly, it has recently been reported that Epstein-Barr virus, frequently involved in aggressive LDGL,43 is largely silenced by host-driven methylation of CpG islands during the latent stage and, in the switch to the lytic cycle, this epigenetic silencing is overturned, leading to aberrant DNA methylation and subsequently aberrant gene expression.44
The concept that, together with expression of activating KIR, significant silencing of inhibitory KIR might also favor the proliferation of pathological clones adds to the information on the mechanisms involved in the pathogenesis of NK-type LDGL. Whether the down-regulation we found is specific to the KIR3DL1 inhibitory receptor is still under investigation and further studies are in progress to evaluate the methylation pattern of others KIR promoters.
Footnotes
Funding: supported by a grant from A.I.R.C. (Milan) to GS, by a grant from Fondazione Berlucchi per la Ricerca sul Cancro on “Approccio clinico/biologico ai pazienti con leucemia linfatica cronica” and by a grant from Regione Veneto for research on chronic lymphocytic leukemia.
Authorship and Disclosures
The information provided by the authors about contributions from persons listed as authors and in acknowledgments is available with the full text of this paper at www.haematologica.org.
Financial and other disclosures provided by the authors using the ICMJE (www.icmje.org) Uniform Format for Disclosure of Competing Interests are also available at www.haematologica.org.
References
- 1.Loughran TP., Jr Clonal diseases of large granular lymphocytes. Blood. 1993;82(1):1–14. [PubMed] [Google Scholar]
- 2.Oshimi K, Yamada O, Kaneko T, Nishinarita S, Iizuka Y, Urabe A, et al. Laboratory findings and clinical courses of 33 patients with granular lymphocyte-proliferative disorders. Leukemia. 1993;7(6):782–8. [PubMed] [Google Scholar]
- 3.Semenzato G, Zambello R, Starkebaum G, Oshimi K, Loughran TP., Jr The lymphoproliferative disease of granular lymphocytes: updated criteria for diagnosis. Blood. 1997;89(1):256–60. [PubMed] [Google Scholar]
- 4.Zambello R, Loughran TP, Jr, Trentin L, Pontisso P, Battistella L, Raimondi R, et al. Serologic and molecular evidence for a possible pathogenetic role of viral infection in CD3-negative natural killer-type lymphoproliferative disease of granular lymphocytes. Leukemia. 1995;9(7):1207–11. [PubMed] [Google Scholar]
- 5.Trinchieri G. Biology of natural killer cells. Adv Immunol. 1989;47:187–376. doi: 10.1016/S0065-2776(08)60664-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cooper MA, Fehniger TA, Caligiuri MA. The biology of human natural killer-cell subsets. Trends Immunol. 2001;22(11):633–40. doi: 10.1016/s1471-4906(01)02060-9. [DOI] [PubMed] [Google Scholar]
- 7.Moretta A, Bottino C, Vitale M, Pende D, Biassoni R, Mingari MC, Moretta L. Receptors for HLA class-I molecules in human natural killer cells. Annu Rev Immunol. 1996;14:619–48. doi: 10.1146/annurev.immunol.14.1.619. [DOI] [PubMed] [Google Scholar]
- 8.Lanier LL. NK cell receptors. Annu Rev Immunol. 1998;16:359–93. doi: 10.1146/annurev.immunol.16.1.359. [DOI] [PubMed] [Google Scholar]
- 9.Long EO. Regulation of immune responses through inhibitory receptors. Annu Rev Immunol. 1999;17:875–904. doi: 10.1146/annurev.immunol.17.1.875. [DOI] [PubMed] [Google Scholar]
- 10.Moretta A, Bottino C, Vitale M, Pende D, Cantoni C, Mingari MC, et al. Activating receptors and coreceptors involved in human natural killer cell-mediated cytolysis. Annu Rev Immunol. 2001;19:197–223. doi: 10.1146/annurev.immunol.19.1.197. [DOI] [PubMed] [Google Scholar]
- 11.Robbins SH, Brossay L. NK cell receptors: emerging roles in host defense against infectious agents. Microbes Infect. 2002;4(15):1523–30. doi: 10.1016/s1286-4579(02)00035-7. [DOI] [PubMed] [Google Scholar]
- 12.Waldhauer I, Steinle A. NK cells and cancer immunosurveillance. Oncogene. 2008;27(45):5932–43. doi: 10.1038/onc.2008.267. [DOI] [PubMed] [Google Scholar]
- 13.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16(1):6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 14.Carrington MNP. The KIR Gene Cluster. Bethesda: 2003. [Google Scholar]
- 15.Valiante NM, Uhrberg M, Shilling HG, Lienert-Weidenbach K, Arnett KL, D’Andrea A, et al. Functionally and structurally distinct NK cell receptor repertoires in the peripheral blood of two human donors. Immunity. 1997;7(6):739–51. doi: 10.1016/s1074-7613(00)80393-3. [DOI] [PubMed] [Google Scholar]
- 16.Chan HW, Kurago ZB, Stewart CA, Wilson MJ, Martin MP, Mace BE, et al. DNA methylation maintains allele-specific KIR gene expression in human natural killer cells. J Exp Med. 2003;197(2):245–55. doi: 10.1084/jem.20021127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Santourlidis S, Trompeter HI, Weinhold S, Eisermann B, Meyer KL, Wernet P, Uhrberg M. Crucial role of DNA methylation in determination of clonally distributed killer cell Ig-like receptor expression patterns in NK cells. J Immunol. 2002;169(8):4253–61. doi: 10.4049/jimmunol.169.8.4253. [DOI] [PubMed] [Google Scholar]
- 18.Gao XN, Lin J, Wang LL, Yu L. Demethylating treatment suppresses natural killer cell cytolytic activity. Mol Immunol. 2009;46(10):2064–70. doi: 10.1016/j.molimm.2009.02.033. [DOI] [PubMed] [Google Scholar]
- 19.Zambello R, Falco M, Della Chiesa M, Trentin L, Carollo D, Castriconi R, et al. Expression and function of KIR and natural cytotoxicity receptors in NK-type lymphoproliferative diseases of granular lymphocytes. Blood. 2003;102(5):1797–805. doi: 10.1182/blood-2002-12-3898. [DOI] [PubMed] [Google Scholar]
- 20.Epling-Burnette PK, Painter JS, Chaurasia P, Bai F, Wei S, Djeu JY, Loughran TP., Jr Dysregulated NK receptor expression in patients with lymphoproliferative disease of granular lymphocytes. Blood. 2004;103(9):3431–9. doi: 10.1182/blood-2003-02-0400. [DOI] [PubMed] [Google Scholar]
- 21.Scquizzato E, Teramo A, Miorin M, Facco M, Piazza F, Noventa F, et al. Genotypic evaluation of killer immunoglobulin-like receptors in NK-type lymphoproliferative disease of granular lymphocytes. Leukemia. 2007;21(5):1060–9. doi: 10.1038/sj.leu.2404634. [DOI] [PubMed] [Google Scholar]
- 22.Zambello R, Trentin L, Ciccone E, Bulian P, Agostini C, Moretta A, et al. Phenotypic diversity of natural killer (NK) populations in patients with NK-type lymphoproliferative disease of granular lymphocytes. Blood. 1993;81(9):2381–5. [PubMed] [Google Scholar]
- 23.Bottino C, Sivori S, Vitale M, Cantoni C, Falco M, Pende D, et al. A novel surface molecule homologous to the p58/p50 family of receptors is selectively expressed on a subset of human natural killer cells and induces both triggering of cell functions and proliferation. Eur J Immunol. 1996;26(8):1816–24. doi: 10.1002/eji.1830260823. [DOI] [PubMed] [Google Scholar]
- 24.Uhrberg M, Parham P, Wernet P. Definition of gene content for nine common group B haplotypes of the Caucasoid population: KIR haplotypes contain between seven and eleven KIR genes. Immunogenetics. 2002;54(4):221–9. doi: 10.1007/s00251-002-0463-7. [DOI] [PubMed] [Google Scholar]
- 25.Halfpenny IA, Middleton D, Barnett YA, Williams F. Investigation of killer cell immunoglobulin-like receptor gene diversity: IV. KIR3DL1/S1. Hum Immunol. 2004;65(6):602–12. doi: 10.1016/j.humimm.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 26.Uhrberg M, Valiante NM, Shum BP, Shilling HG, Lienert-Weidenbach K, Corliss B, et al. Human diversity in killer cell inhibitory receptor genes. Immunity. 1997;7(6):753–63. doi: 10.1016/s1074-7613(00)80394-5. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Q, Wang HY, Marzec M, Raghunath PN, Nagasawa T, Wasik MA. STAT3- and DNA methyltransferase 1-mediated epigenetic silencing of SHP-1 tyrosine phosphatase tumor suppressor gene in malignant T lymphocytes. Proc Natl Acad Sci USA. 2005;102(19):6948–53. doi: 10.1073/pnas.0501959102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu Y, Strawn E, Basir Z, Halverson G, Guo SW. Aberrant expression of deoxyribonucleic acid methyltransferases DNMT1, DNMT3A, and DNMT3B in women with endometriosis. Fertil Steril. 2007;87(1):24–32. doi: 10.1016/j.fertnstert.2006.05.077. [DOI] [PubMed] [Google Scholar]
- 29.Thompson A, van der Slik AR, Koning F, van Bergen J. An improved RT-PCR method for the detection of killer-cell immunoglobulin-like receptor (KIR) transcripts. Immunogenetics. 2006;58(11):865–72. doi: 10.1007/s00251-006-0163-9. [DOI] [PubMed] [Google Scholar]
- 30.O’Connor GM, Guinan KJ, Cunningham RT, Middleton D, Parham P, Gardiner CM. Functional polymorphism of the KIR3DL1/S1 receptor on human NK cells. J Immunol. 2007;178(1):235–41. doi: 10.4049/jimmunol.178.1.235. [DOI] [PubMed] [Google Scholar]
- 31.Wilson MJ, Torkar M, Haude A, Milne S, Jones T, Sheer D, Beck S, Trowsdale J. Plasticity in the organization and sequences of human KIR/ILT gene families. Proc Natl Acad Sci USA. 2000;97(9):4778–83. doi: 10.1073/pnas.080588597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Okano M, Xie S, Li E. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat Genet. 1998;19(3):219–20. doi: 10.1038/890. [DOI] [PubMed] [Google Scholar]
- 33.Haaf T. Methylation dynamics in the early mammalian embryo: implications of genome reprogramming defects for development. Curr Top Microbiol Immunol. 2006;310:13–22. doi: 10.1007/3-540-31181-5_2. [DOI] [PubMed] [Google Scholar]
- 34.Esteller M. Profiling aberrant DNA methylation in hematologic neoplasms: a view from the tip of the iceberg. Clin Immunol. 2003;109(1):80–8. doi: 10.1016/s1521-6616(03)00208-0. [DOI] [PubMed] [Google Scholar]
- 35.Rowley JD, Golomb HM, Vardiman JW. Nonrandom chromosome abnormalities in acute leukemia and dysmyelopoietic syndromes in patients with previously treated malignant disease. Blood. 1981;58(4):759–67. [PubMed] [Google Scholar]
- 36.Golomb HM, Alimena G, Rowley JD, Vardiman JW, Testa JR, Sovik C. Correlation of occupation and karyotype in adults with acute nonlymphocytic leukemia. Blood. 1982;60(2):404–11. [PubMed] [Google Scholar]
- 37.Aggerholm A, Holm MS, Guldberg P, Olesen LH, Hokland P. Promoter hypermethylation of p15INK4B, HIC1, CDH1, and ER is frequent in myelodysplastic syndrome and predicts poor prognosis in early-stage patients. Eur J Haematol. 2006;76(1):23–32. doi: 10.1111/j.1600-0609.2005.00559.x. [DOI] [PubMed] [Google Scholar]
- 38.Nosaka K, Maeda M, Tamiya S, Sakai T, Mitsuya H, Matsuoka M. Increasing methylation of the CDKN2A gene is associated with the progression of adult T-cell leukemia. Cancer Res. 2000;60(4):1043–8. [PubMed] [Google Scholar]
- 39.Uchida T, Kinoshita T, Nagai H, Nakahara Y, Saito H, Hotta T, Murate T. Hypermethylation of the p15INK4B gene in myelodysplastic syndromes. Blood. 1997;90(4):1403–9. [PubMed] [Google Scholar]
- 40.Quesnel B, Guillerm G, Vereecque R, Wattel E, Preudhomme C, Bauters F, et al. Methylation of the p15(INK4b) gene in myelodysplastic syndromes is frequent and acquired during disease progression. Blood. 1998;91(8):2985–90. [PubMed] [Google Scholar]
- 41.Risitano AM, Maciejewski JP, Selleri C, Rotoli B. Function and malfunction of hematopoietic stem cells in primary bone marrow failure syndromes. Curr Stem Cell Res Ther. 2007;2(1):39–52. doi: 10.2174/157488807779316982. [DOI] [PubMed] [Google Scholar]
- 42.Sokol L, Agrawal D, Loughran TP., Jr Characterization of HTLV envelope serore-activity in large granular lymphocyte leukemia. Leuk Res. 2005;29(4):381–7. doi: 10.1016/j.leukres.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 43.Kawa-Ha K, Ishihara S, Ninomiya T, Yumura-Yagi K, Hara J, Murayama F, et al. CD3-negative lymphoproliferative disease of granular lymphocytes containing Epstein-Barr viral DNA. J Clin Invest. 1989;84(1):51–5. doi: 10.1172/JCI114168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Karlsson QH, Schelcher C, Verrall E, Petosa C, Sinclair AJ. Methylated DNA recognition during the reversal of epigenetic silencing is regulated by cysteine and serine residues in the Epstein-Barr virus lytic switch protein. PLoS Pathog. 2008;4:e1000005. doi: 10.1371/journal.ppat.1000005. [DOI] [PMC free article] [PubMed] [Google Scholar]